Abstract
A diverse antibody repertoire is formed through the rearrangement of V, D, and J segments at the immunoglobulin heavy chain (Igh) loci. The C57BL/6 murine Igh locus has over 100 functional VH gene segments that can recombine to a rearranged DJH. While the non-random usage of VH genes is well documented, it is not clear what elements determine recombination frequency. To answer this question we conducted deep sequencing of 5′-RACE products of the Igh repertoire in pro-B cells, amplified in an unbiased manner. ChIP-seq results for several histone modifications and RNA polymerase II binding, RNA-seq for sense and antisense non-coding germline transcripts, and proximity to CTCF and Rad21 sites were compared to the usage of individual V genes. Computational analyses assessed the relative importance of these various accessibility elements. These elements divide the Igh locus into four epigenetically and transcriptionally distinct domains, and our computational analyses reveal different regulatory mechanisms for each region. Proximal V genes are relatively devoid of active histone marks and non-coding RNA in general, but having a CTCF site near their RSS is critical, suggesting that being positioned near the base of the chromatin loops is important for rearrangement. In contrast, distal V genes have higher levels of histone marks and non-coding RNA, which may compensate for their poorer RSSs and for being distant from CTCF sites. Thus, the Igh locus has evolved a complex system for the regulation of V(D)J rearrangement that is different for each of the four domains that comprise this locus.
Introduction
Recognizing and defending against a vast array of pathogens is an essential function of the immune system. To accomplish this, a highly diverse set of antigen receptors is created by the rearrangement of multiple variable (V), diverse (D), and joining (J) segments of the immunoglobulin and T cell receptor genes in B and T cells, respectively (1). The C57BL/6 mouse Igh locus spans a large region of ~2.8 Mb containing four JH genes, 11 D genes, and 195 VH gene segments, of which ~110 are deemed functional (2). There are 16 families of VH genes. The largest family, J558, occupies over half of the locus on the JH-distal 5′ end of the Igh locus. The second largest family, 7183, occupies the most 3′ region, proximal to the D and JH genes, and has the Q52 VH family interspersed with it. Other smaller VH families are in between (Fig. 1A). Much evidence demonstrates that individual VH genes rearrange with very different intrinsic frequencies, and that the regulation of rearrangement of the distal genes is distinct from that of proximal VH genes (3–11). However, no deep sequencing of the murine Igh repertoire has been performed to accurately enumerate the initial rearrangement frequency of each VH gene. Furthermore, it is not well understood what factors are critical to determine the frequency with which individual VH genes will rearrange other than the quality of the RSS (12).
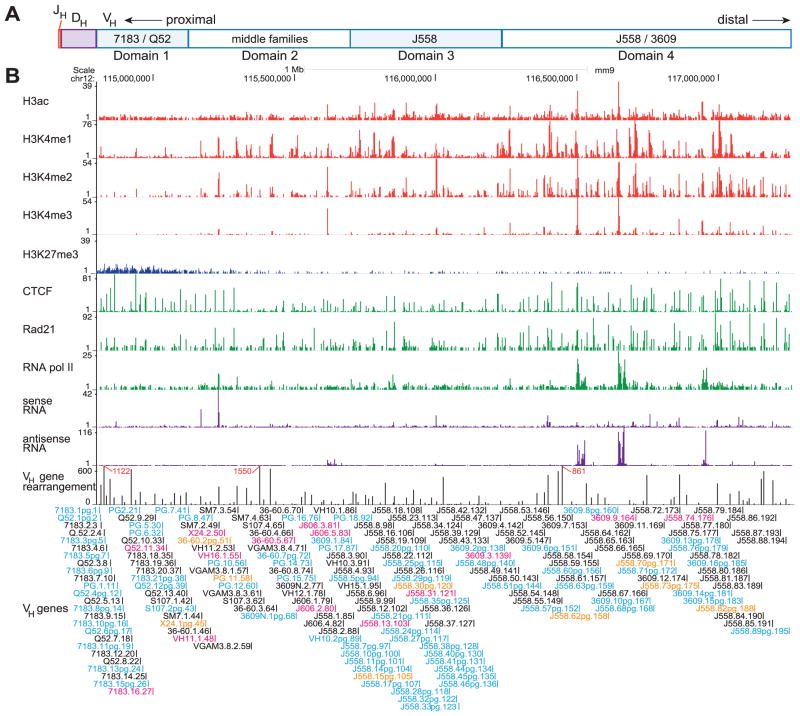
Fig. 1. Antibody repertoire and the epigenetic landscape of the mouse Igh locus.
(A) A diagram of the mouse Igh locus. The VH region was subdivided into four domains. (B) Genome browser tracks of ChIP-seq for histone modifications, CTCF, Rad21, and RNA polymerase II are shown (Red: active histone modifications, Blue: repressive modification, Green: non-histone proteins). RNA-seq results for sense and antisense transcripts are shown in purple. C57BL/6 pro-B cell VH gene rearrangement frequency is shown in black. All annotated VH genes are indicated beneath the chart (black: functional “active” gene, magenta: “inactive” gene annotated as functional, blue: “inactive” pseudogene, orange: “active” pseudogene).
Regulating the ‘openness’ of chromatin at the antigen receptor loci may be an important factor in this decision. The “accessibility hypothesis” states that gene segment rearrangement occurs only when the chromatin environment becomes permissive to bind RAG1/2 (1). This hypothesis was initially proposed when non-coding RNA (ncRNA) transcription of the unrearranged V or J/C regions, or “germline transcription”, was observed as that part of a receptor locus became poised for recombination (13, 14). Advancements in epigenetics have since been able to provide greater explanations of how accessibility may be controlled. Posttranslational modification of histone proteins is one mechanism that has been described to alter chromatin structure. Acetylation of histones is a mark of ‘open’ chromatin, whereas methylation of certain residues can indicate either accessible or repressed chromatin (15). Mono-, di-, or trimethylation of lysine 4 histone H3 (H3K4me1/2/3) is present at enhancer elements, general accessible areas, and active regions of transcription, respectively, while methylated H3K9 or H3K27 are present at repressed regions of the genome (16). Importantly, H3K4me3 has also been shown to directly recruit RAG2 through its plant homeodomain (PHD) finger (17, 18). The RAG1/2 recombinase binds to recombination signal sequences (RSS), which flank all V, D, and J gene segments. Although there is a consensus sequence for the heptamer and nonamer portions of the RSS (19, 20), individual RSSs often deviate from the consensus. Divergence from this consensus has been demonstrated to reduce recombination efficiency to varying extents (21, 22). Together these suggest that a quality RSS and a suitable chromatin environment must both be provided for efficient recruitment of the RAG complex and effective catalytic activity to occur.
Large-scale three dimensional (3D) conformational changes of chromatin structure is another proposed regulatory mechanism of V(D)J rearrangement. The Igh locus is composed of multi-looped rosettes, which have been shown by 3D-FISH to compact at the time of Igh gene rearrangement (23). This compaction of the structure of the locus will bring all V genes in much closer proximity to the DJ rearrangement to which one V gene will ultimately recombine. The insulator binding and chromatin looping protein CTCF and the cohesin complex are likely candidates for forming the rosette-like structure of the locus (24–29). When CTCF expression was knocked down, an extension in the spatial length of the Igh locus was observed (25). PAIR (Pax5-activated intergenic repeats) elements, which are bound by Pax5, E2A, and CTCF, are located in the distal intergenic regions of the VH locus (30). Extensive antisense ncRNA transcription starts from two of these PAIR elements, and these PAIR promoters directly interact with Eμ, presumably in a transcription factory (31). This movement of PAIR promoters, and possibly many other sense and antisense ncRNA promoters, to Eμ, which is within 2 kb of the DJ rearrangement, will directly result in locus compaction, and will bring many V genes in proximity to the DJ rearrangement. In addition, there may well be protein-protein interactions of various transcription factors, which could also result in looping and locus contraction. The relative position of V genes in these chromatin loops may well influence their frequency of recombination.
Until the advent of high-throughput deep sequencing, it has been impossible to accurately assess the relative usage of all of the individual VH genes during V(D)J recombination of the Igh locus. To date, no deep sequencing of the murine Igh locus has been performed. In this study, we took an unbiased approach by sequencing 5′-RACE (Rapid Amplification of cDNA Ends) products and quantitatively determining how frequently each VH gene was utilized. This data was compared to our ChIP-seq results for histone modifications, RNA polymerase II, chromatin looping factors CTCF and cohesin, and to the level of both sense and antisense ncRNA in order to obtain insight into the factors which influence VH gene rearrangement frequency. The quality of RSS sequences for the VH genes were also evaluated. Based on the epigenetic and transcriptional profile in the immediate vicinity of each VH gene, we divided the IgV locus into 4 domains, each containing different sets of VH gene families and different epigenetic characteristics. Computational analyses of all of the above factors in relationship to recombination frequency demonstrated that each domain had different factors that influenced rearrangement frequency.
Materials & Methods
Mice and cell preparation
C57BL/6 and RAG1−/− mice were maintained in our breeding colony in accordance with protocols approved by the TSRI Institutional Animal Care and Use Committee. Bone marrow cells were collected from 5–7 week old mice as described previously (32). CD19+ cells were isolated using anti-CD19 coated MACS beads (Miltenyi, Auburn, CA). Pro-B cells were sorted as B220moderate CD43+ IgM− CD2− on a BD FACS Aria-II.
RNA, cDNA preparation for 5′-RACE and RNA sequencing
RNA was extracted from pro-B cells using Trizol® (Life Technologies Corp., Carlsbad CA). Adapter ligation to RNA was performed with the 5′-RACE kit (Ambion; Life Technologies Corp.) and first strand cDNA was prepared using the Transcriptor High Fidelity cDNA Synthesis Kit (Roche Diagnostics, Indianapolis, IN). A barcoded primer against Cμ and a primer that binds to the 5′ adapter were used together for amplification of the 5′-RACE cDNA product using Phusion® High-Fidelity DNA Polymerase (New England Biolabs Inc, Ipswich, MA). 10–12 individual PCR reactions were pooled and gel purified with the QIAquick Gel Extraction Kit (QIAGEN Inc., Valencia, CA). The samples were then sequenced on a Roche 454 GS Junior at the TSRI Next Generation Sequencing (NGS) Core Facility. For RNA-seq, RNA was prepared as described above and genomic DNA was eliminated using the genomic DNA wipeout buffer in the QuantiTect Reverse transcription kit (QIAGEN). A final purification of the RNA was performed with the RNeasy kit from QIAGEN. RNA samples were submitted to the NGS Core, where they were processed with the Ovation® RNA-Seq System (NuGEN, San Carlos, CA), and sequenced on the Illumina HiSeq (San Diego, CA). RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/, accession number GSE47766).
V, D and J gene analysis
V gene identity was assigned by matching each sequence to a database of all known V gene sequences via MEGABLAST. At least a 100 bp alignment span was required for V gene assignment through a hierarchy of BLAST result parameters where the next parameter in the hierarchy was considered only if a read matched multiple genes under the previous parameter. The hierarchy, ordered to be more permissive to gaps in the BLAST alignments because of known homopolymer sequencing errors using 454 technology, was as follows: highest bit-score, highest percent-identity, longest alignment length, least number of mismatches, and finally least number of gaps. Most reads were assigned by pure bitscore. For the small number of reads for which the score matched multiple V genes exactly, the reads were given multiple assignments weighted according to the % of uniquely mapped reads for each gene in the data overall. Once the proper V gene was identified, rigorous alignments of reads to the known V gene sequence were performed using a Smith-Waterman alignment to extract the portion of the sequence downstream of the conserved cysteine at the start of the CDR3 region. This downstream region was used to identify J genes in similar fashion as V genes, filtering based on the same hierarchy but with a 20 bp alignment span requirement. Similarly, the downstream boundary of the CDR3 was extracted by identifying the conserved “WG” amino acids through a rigorous Smith-Waterman based alignment. Finally, a D gene was assigned to the extracted CDR3 sequence again via BLAST without an alignment length threshold but following the same filtering hierarchy.
Chromatin immunoprecipitation and ChIP-seq
Samples for ChIP-seq were prepared as previously described (25). ChIP-Seq was performed on the Illumina HiSeq at the NGS core. Antibodies for CTCF, H3K4me2, H3ac, and H3K27me3 were purchased from EMD Millipore (Billerica, MA), H3K4me3 from Active Motif (Carlsbad, CA), and Rad21 from Abcam (Cambridge, England). All ChIP-seq data have been deposited in the GEO database (http://www.ncbi.nlm.nih.gov/geo/, accession number GSE47766).
Computational analysis of RSSs, accessibility elements, and recombination frequency
To determine the RSS and RIC of each VH gene, we used a publically available web based program, DNAGrab (http://www.itb.cnr.it/rss (33)). For each of the chromatin and RNA-seq features analyzed, the signal intensity of each region was determined from the corresponding UCSC wig file by summing up normalized read counts. Signal intensities were then converted to log2 values, and a pseudo-count of one was added to avoid log2(0) values. Similarly, rearrangement intensities were transformed into their log2 values with an added pseudo-count. VH genes with rearrangement >4 were labeled as “active”, and Random Forest classification ((34); R package RandomForest) was used to assess variable importance; i.e. which of the features distinguish best between active and inactive genes. Cross-validated prediction performance of models with sequentially reduced number of predictors (ranked by variable importance) was used to suggest significant predictors. Regression analyses were performed on the 105 rearranging VH genes, on the full locus, and on the four domains separately. Conditional Inference trees (CI trees) and Random Forests with CI trees as base learners ((35); R package party) were used to generate the regression models. Pearson’s correlation coefficients were used in all cases to estimate which features correlate best with rearrangement intensity. A significance test of no association was used to generate p-values for the correlations.
Results
Igh repertoire deep sequencing and the epigenetic landscape of the VH locus
We analyzed the Igh sequences of cell sorter purified C57BL/6 pro-B cells to determine the initially generated repertoire before any selection could skew the primary rearrangements. To assess the repertoire in a completely unbiased manner, we performed 5′-RACE PCR. This involves ligation of a universal adapter to the 5′ end of cDNA followed by amplification with primers that anneal to the adapter and to the constant region, Cμ. In contrast, if one were to amplify genomic DNA, a multiplex PCR with a large pool of VH and JH primers is required, which can introduce significant bias caused by differential primer efficiencies as well as primer cross-reactions. To obtain accurate sampling and deep coverage of the VH locus, we sequenced 8 independent barcoded libraries of amplified 5′-RACE PCR. 10–12 PCR reactions were pooled for each library to minimize the sequencing of PCR duplicates. Duplicate sequences within each barcode were eliminated, and the VH, D and JH genes were identified. We obtained the sequences of 17,431 unique rearrangements.
We have also performed ChIP-seq for CTCF, Rad21 (a cohesin subunit), RNA polymerase II, and the histone modifications H3ac, H3K4me2, H3K4me3 and H3K27me3 (Fig. 1B). RAG1−/− pro-B cells on the C57BL/6 background were used for these ChIP-seq analyses so the entire Igh locus would be intact. The H3K4me1 data is from published work (36). The rearrangement frequency for each VH gene is displayed on the bottom line of Fig. 1B, with the RNA-seq of sense and antisense ncRNA in RAG−/− pro-B cells above it.
The 195 V genes within the Igh gene locus have been annotated previously (2). Genes were classified as potentially functional if they had no obvious defect, and as pseudogenes if there was a major defect in the promoter, RSS, splice sites, or a stop codon in the coding region. Our experimental data agreed well with the predicted functionality. There were only 14 V genes that had been classified as potentially functional that did not rearrange in our dataset (Fig. 1B, magenta). Conversely, we found a small number of rearrangements among the genes previously classified as pseudogenes (Fig. 1B, orange), but pseudogenes can rearrange if the RSS is intact as was the case for these genes.
Usage of V, D, J gene segments
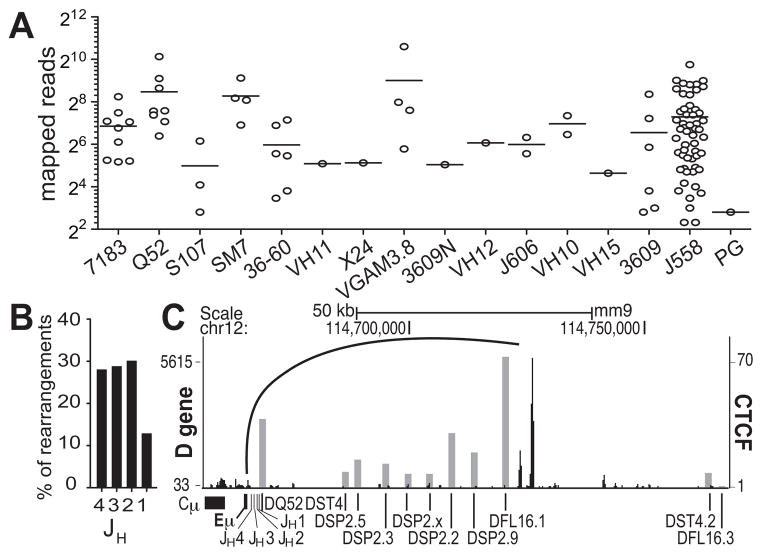
In Fig. 2A, we displayed the number of unique rearrangements sequenced for each individual VH gene. The data are grouped by VH families, and each circle represents a VH gene. It can be easily observed that there is a wide range of usage of individual VH genes in the primary repertoire in all VH gene families. Fig. 2B shows JH gene utilization and Fig. 2C shows D gene usage. We have previously demonstrated that a loop is made between Eμ and the set of CTCF/Rad21 sites 3.2 and 5.6 kb upstream of DFL16.1, and thus DFL16.1 and DQ52 are the closest D genes to the JH genes in 3D space (25). It has also been shown that more repressive histone modifications are present at the DSP2 genes (37). Together these appear to give DFL16.1 and DQ52 a distinct advantage since they were the most frequently rearranged D genes.
Fig. 2. V, D, and J gene rearrangement frequencies in pro-B cells.
(A) The rearrangement frequencies of VH genes are plotted by gene families in a log2 scale. Each circle represents an individual VH gene. Horizontal bars show the mean value for each family. (B) Percentage of JH gene rearrangements. (C) D gene rearrangement frequency in grey columns is superimposed onto the genome browser track for CTCF ChIP-seq (black).
Individual VH genes can be divided into 4 domains by their epigenetic characteristics
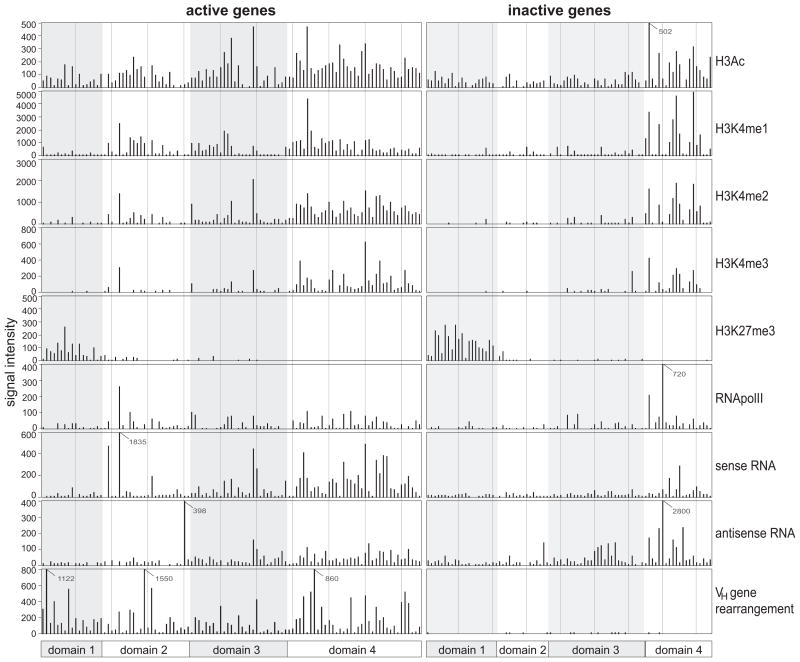
To quantitate the local level of histone modifications and protein binding at each VH gene from all of these datasets, we calculated the signal intensity for the region from 1 kb upstream to 1 kb downstream of each VH gene. Since the leader exons are not all annotated, we estimated the leader and intron to be 200 bp 5′ of the ~300 bp coding region. It is evident in Fig. 3 that, for the active (rearranged) genes, the levels of histone modifications as well as ncRNA are not evenly distributed. The distal J558/3609 part of the locus and the proximal 7183/Q52 regions showed the greatest contrast, where the distal VH genes had much higher levels of active histone marks and ncRNA.
Fig. 3. Accessibility elements at individual VH genes divide the Igh locus into 4 domains.
For each VH gene, all mapped reads were summed to calculate the signal intensity for each element indicated on the right starting from 1 kb upstream of the leader to 1 kb downstream of the RSS. The leader was estimated as 200 bp from the start of the coding region. Charts in the left column are values for the active (rearranging) genes and charts in the right column are those of the inactive (not rearranging) genes. Domains are represented in a bar beneath each column, and shaded and clear areas mark the four domains of the VH gene locus. Signal intensities beyond the y-axis range are indicated with the value.
Based on the distribution of histone modifications and transcriptional activity we divided the Igh locus into 4 domains, also considering the location of VH gene families. Domain 1 contains all 7183 and Q52 genes (genes 7183.1pg.1 through PG.7.41). Ten small VH families make up domain 2 (genes S107.1.42 through J606.5.83). The large J558 region is broken into two domains. Other than one 3609 VH gene and 2 small VH gene families, domain 3 contains 47 J558 genes. Domain 4 (genes 3609.2pg.138 to J558.89pg.195) contains the 3609 family interspersed with the remaining 42 J558 genes (Fig. 1A). Domain 1 is unique in that all but one of the active VH genes (7183.9.15) had a CTCF site in very close proximity to the RSS (9 –68 bp). This domain also differed from other domains by being generally poor in active epigenetic marks other than low levels of H3ac and H3K4me1, low in ncRNA, and it uniquely contained H3K27me3. Domains 2 and 3 displayed more genes with higher H3ac and H3K4me1/2. In domain 4, almost all functional VH genes displayed reasonably high levels of H3ac and H3K4me1/2 and this domain had the highest frequency of VH genes associated with H3K4me3. For each domain, there were many fewer genes with active histone marks and ncRNA among the inactive genes.
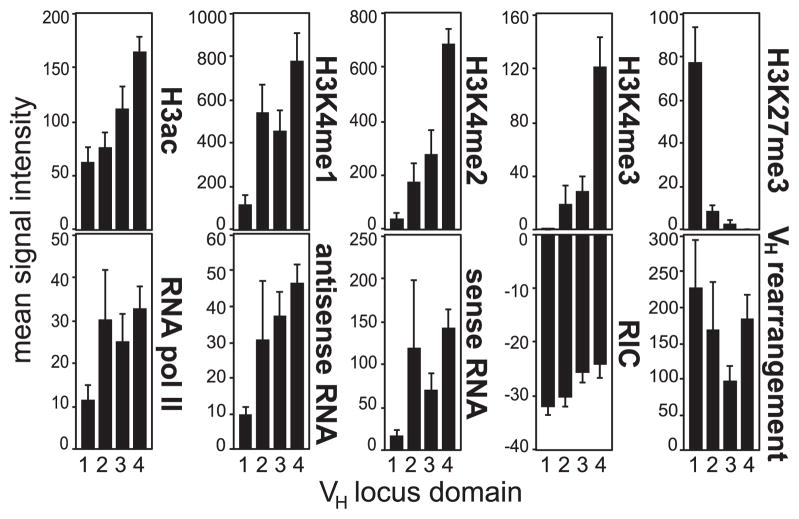
The overall differences between the four domains can be appreciated in Fig 4 that shows the mean signal intensity for each accessibility element for the active genes. In general, there were more active histone modifications and transcription at the distal domains, while domain 1 was noticeably lower in many parameters. Thus, the four domains of the Igh locus have very distinct epigenetic characteristics, suggesting that different factors are important for accessibility and rearrangement in the different parts of the Igh locus.
Fig. 4. The four domains have distinct epigenetic profiles.
The mean signal intensities for the active genes in each domain were calculated for the accessibility elements, RIC scores, and VH gene rearrangement frequencies. Error bars show the standard error of the mean.
VH gene proximity to CTCF and Rad21 sites
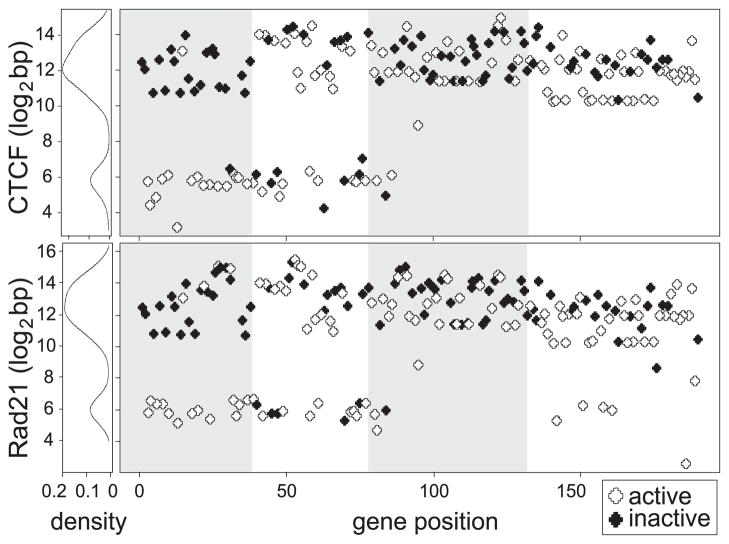
We previously observed that CTCF sites were close to the RSS of functional VH genes in the proximal part of the locus, but were intergenic in the large J558/3609 region (24). We also noted that most CTCF sites throughout the Igh locus co-localized with cohesin binding sites (24, 25). If VH genes were near the base of the rosettes that make up the 3D structure of the Igh locus, then proximity to CTCF sites may impact the efficiency of recombination. We therefore expanded our previous studies by adding more sequencing depth to the CTCF and Rad21 ChIP-seqs and determining the distance of the nearest CTCF and Rad21 binding sites from the coding end of every VH gene. These measurements were plotted against the genomic location of each gene (Fig. 5). The statistically significant CTCF and Rad21 binding peaks were determined by MACS (FDR≤1%) (38), and distance was computed from the end of each VH gene to the peak summit. All but one of the rearranging VH genes in the 7183 and Q52 families (domain 1) had a CTCF site within 68 bp of the RSS, whereas most of the non-rearranging genes had CTCF and Rad21 sites >1 kb distant from them. All of the 7183 genes and most of the Q52 genes also had Rad21 bound to the CTCF sites. It is notable that the only active 7183 gene located far from a CTCF and Rad21 site, 7183.9.15, had a low rearrangement frequency and two apparently functional but non-rearranging 7183 genes also did not have CTCF sites nearby. Domain 2 containing the 10 small VH families had a VH family-specific distribution of distance from CTCF and Rad21; the functional S107, X24, VH11, VH12 and J606 genes all had nearby CTCF and Rad21 sites (< 100 bp), while the other families did not. In domains 3 and 4 that contain the large J558 family, all genes were ~1–32 kb from a CTCF site, with the exception of the three VH10 genes that had a CTCF site within 100 bp of the RSS, and only 6 of the 89 J558 genes had nearby Rad21 binding. Thus, proximity to CTCF and Rad21 are of critical importance for domain 1 genes to rearrange, but proximity is not essential for most other VH genes.
Fig. 5. Distance to closest CTCF and Rad21 binding site has a bimodal distribution.
Distance to the closest CTCF and Rad21 sites (log2 bp) were plotted against the genomic order of genes. Active genes are depicted in white and inactive genes in black. A histogram of the density of genes at each distance is shown vertically on the left.
Evaluation of RSS quality
For a gene segment to recombine, it is essential that the RAG complex can bind to its RSS sequence. The RSS consists of a consensus heptamer and nonamer that are separated by a spacer of 12 or 23 bp (39). Despite the high degree of homology, very few RSSs have both the consensus heptamer and nonamer. Recombination substrate experiments have shown that heptamers and nonamers closer to consensus provide higher recombination efficiency (5, 40). However, every variation from consensus has not been tested by substrate competition assays, and the sequence of the spacer can also affect recombination (41, 42). Hence, one cannot a priori estimate the rearrangement efficacy of each RSS.
To determine the RSS sequence for each VH gene, we used a publically available program that predicts RSS sequences (33). This program also provides an ‘RSS information content (RIC)’ score, which is an assessment of the quality of the predicted RSS sequence (pass: RIC ≥ −58.45) (43, 44). It predicted a 23 bp spacer RSS sequence for 163 VH genes and provided each with an RIC score (Supplemental Table S1). Of the 145 genes that were assigned a passing score, 41 were inactive in our repertoire and had a lower average RIC score (−36.58 compared to −27.34 for active genes). Of the 105 active genes, 104 were assigned a passing score and a single gene, albeit with low recombination frequency (3609.5.147), was deemed to have a failed RSS. Overall 76.7% of predictions on whether a gene was able to rearrange were in concordance with the deep sequencing results.
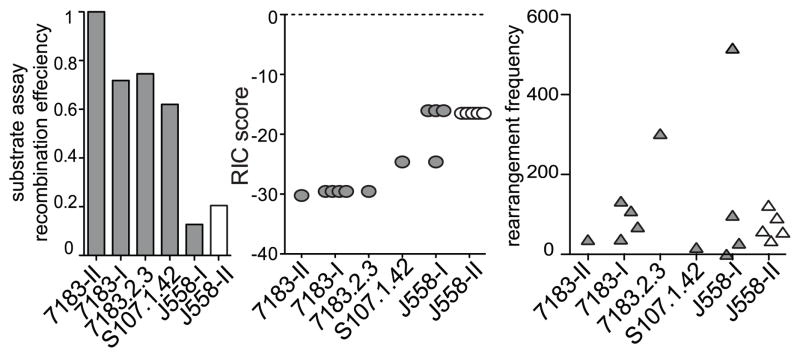
We previously examined the relative efficiencies of murine VH gene RSSs by transiently transfecting competition recombination substrates into cell lines (5, 41). Two major groups of 7183 RSSs in an Igha strain were tested in these experiments. The heptamer of Group-I (7183-I) had one base change from consensus while the heptamer of Group-II (7183-II) genes were identical to the consensus, and both groups had the same single change from consensus in the nonamer. The unique RSS for 7183.2.3 (81X) was examined due to its high level of rearrangement. The RSS for S107.1.42 and a common J558 RSS sequence (J558-I) were also included in the assay. A similar competition assay was performed by Connor et al. comparing the consensus RSS to a closely related common J558 RSS (J558-II) (45). The relative recombination efficiency for each substrate was calculated (Fig. 6, left panel). It can easily be seen that the J558 RSSs supported the lowest frequency of rearrangement. Thus, the proximal 7183 genes have the rearrangement advantage of having much better RSSs than the large distal J558 family.
Fig. 6. RSS competition substrate assays and RIC scores do not agree.
The left panel is the relative recombination efficiency assessed by plasmid-based recombination substrate assays performed previously (45, 47). 7183-II RSS is an exact match to the consensus RSS heptamer, 7183-I is a common variant RSS in the 7183 family. 7183.2.3 and S107.1.42 are individual V genes. J558-I and II are two common variants of J558 RSSs. Substrate competition assay for J558-II was performed by Connor et al. (45) and values are in white. Middle panel shows the RIC scores for all the VH genes that have the identical RSS to the recombination substrates. Right panel shows the deep sequencing recombination frequency for the same individual VH genes.
In C57BL/6 mice, which have the Ighb haplotype, there is only one 7183 V gene (7183.9.15) with an identical RSS to the 7183-II sequence and four with an RSS identical to 7183-I. For the tested J558 RSS sequences, there are four genes that have the exact J558-I RSS and five have the J558-II RSS. When the recombination efficiencies determined in substrates for these RSSs were compared to the RIC scores predicted for them in silico, it could readily be seen that there is little agreement between the two data sets (Fig. 6, left and middle panels). The 7183-II RSS was assigned the lowest RIC score despite being closest to the consensus RSS and with the highest level of recombination in the recombination plasmid substrates. Since recombination substrate assays are the gold standard for assessing the quality of an RSS, this demonstrates that the RIC score is not accurate for predicting the relative effectiveness of RSS. When the RIC and recombination substrate assay results were compared to our deep sequencing results (Fig. 6, right panel), the poor recombination of J558 genes was much better predicted by the recombination substrate results with the exception of one highly rearranged J558 gene. Also in Fig. 4, the panel showing average RIC scores for the four domains illustrates that overall the RIC scores do not agree with recombination frequency. Thus, RIC scores do not accurately predict the quality of an RSS, and recombination substrate assays are far more accurate for determining relative efficacy of an RSS. However, it can be seen in Fig. 6 that genes with identical RSS sequences rearranged at different frequencies in vivo, as we had previously observed with smaller VH gene dataset (5). Thus, although the RSS probably plays an important role in individual VH gene rearrangement, clearly factors other than the RSS must also have significant influence on rearrangement frequency in vivo.
Epigenetic and accessibility elements influence recombination of V genes
Histone modifications, both sense and antisense ncRNA, as well as factors that regulate the 3D structure of chromatin such as CTCF and Rad21, all have the potential to influence the level of gene usage. While it is known at individual genes that these factors are influential, the relative weight of importance or the collective effect of all these elements are largely unknown. We therefore addressed these questions by computational classification and regression analyses.
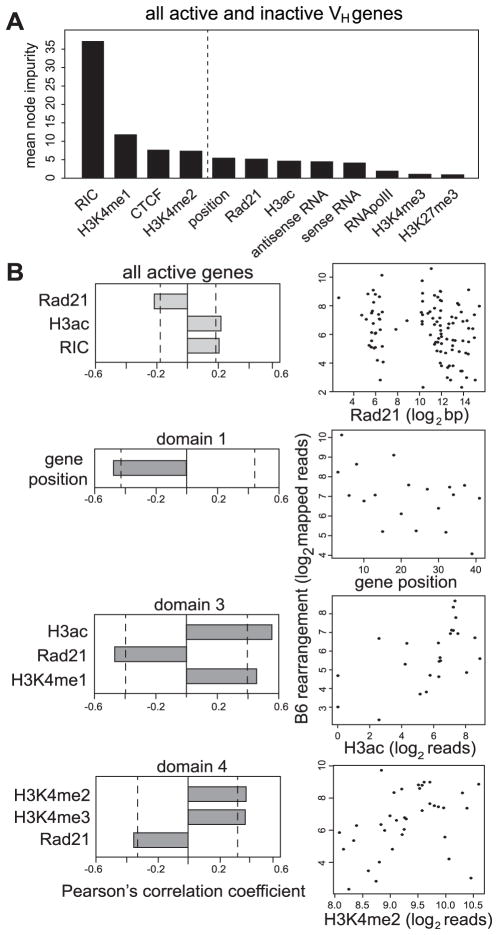
Signal intensities for all histone modifications and sense/antisense ncRNAs for each gene (Fig. 3), distance to nearest CTCF and Rad21 peaks (Fig. 5), genomic position along the locus (in order of DJH proximal to distal), and RIC scores (Table S1) were used to train a Random Forest classifier against the recombination frequency of the C57BL/6 VH genes. Although RIC scores are not nearly as accurate as recombination substrate assays, one can calculate an RIC for each V gene whereas the large number of VH genes precludes making recombination substrates for all V genes. In brief, the classifier builds a model that evaluates the most informative parameters in predicting whether a gene would be active or not based on all provided information. This model correctly predicted 68 of 84 inactive genes (classification error = 0.19) and 98 of 105 active genes (classification error = 0.067), with an overall out-of-bag estimated error of 12.17% (Fig. 7A, S1A). When using only the top four parameters that had a significant contribution for model prediction, the error rates are similar (error rate = 11.11%), suggesting that the other parameters have only minor influence on classification prediction. Having a functional RSS was determined the most informative parameter, but the accuracy of prediction was better when H3K4me1/2 and distance to the closest CTCF site were included in the analysis. Therefore, while not reliable at all in reporting the relative level of rearrangement, RIC score (in the absence of a more reliable recombination substrate score) was the best predictive criterion in determining if a VH gene was active or not.
Fig. 7. Computation of accessibility parameters and their relation to recombination.
(A) Random forest classification of all parameters for all active and inactive VH genes. Dotted line indicates the threshold of statistical significance in the contribution of each parameter to model prediction. (B) Pearson’s correlation coefficient values for each parameter for all active genes only (n=105) or for the active genes in individual domains. Dotted line indicates significance threshold (p = 0.05). Scatter plots are shown for the best correlating parameter in relation to recombination frequency; all log2 transformed.
Advancing beyond the ‘yes’ or ‘no’ question to determine whether different parameters could cooperatively predict the relative frequency of recombination of active genes, we performed linear and tree-based random forest regression analyses on the full locus and individual domains on the active VH genes only. However, model qualities (estimated with root mean square error and % variance explained after a 10-fold cross validation procedure) were generally low except for domain 3, in which the levels of H3 acetylation and H3K4me1 were predictive (Fig. S1C, D). We then performed Pearson’s correlation analyses on the active VH genes for all 12 parameters vs. the recombination frequency of each VH gene. There were three parameters that had statistically significant correlations locus wide: distance to the closest Rad21, H3 acetylation, and RIC score (Fig. 7B). As the data in Fig. 3 suggested that the locus might be governed by different mechanisms in the four domains, we also examined each domain separately. Since all genes that rearranged in domain 1, with one exception, were <100 bp from a CTCF site, relative proximity to CTCF was not a distinguishing characteristic. Only the relative position within the locus had a statistically significant correlation in domain 1, and none correlated for domain 2 (Fig. 7B). In contrast, three parameters significantly correlated with recombination frequency in domain 3 of which H3 acetylation and distance to the nearest Rad21 site were the strongest parameters. In domain 4, H3K4me2/3 had the greatest correlation followed by distance to Rad21 (Fig. S1B). Thus, each domain had distinct parameters influencing the extent of gene rearrangement.
Discussion
In this report, we determined the mouse immunoglobulin heavy chain pre-selection repertoire of C57BL/6 pro-B cells. To our knowledge, this is the first detailed description of the murine Igh repertoire by next generation sequencing. Due to the size and complexity of the Igh locus, we took an approach that amplified the rearranged sequences in a completely unbiased manner using 5′-RACE to amplify the cDNA. This allowed us to bypass potentially serious concerns of unequal amplification efficiency if we were to amplify each individual VH gene from genomic DNA by multiplex PCR. The main concern with performing deep sequencing on RNA rather than genomic DNA is that cells may vary significantly in the amount of Ig mRNA. However, although this is formally a possibility, it is unlikely that this would be much of an issue in pro-B cells.
Although it is well established that RSSs closer to the consensus support more rearrangement in recombination substrates (the gold standard for RSS quality), and also for some V genes in vivo (41, 46), we have also clearly shown previously that genes with identical RSSs, such as 7183 genes, can rearrange with different frequencies in pro-B cells (47). Here, we also demonstrated that the efficiency measured by substrate competition assays did not always correlate with the actual recombination frequency in vivo for a wider variety of VH genes. Thus, factors in addition to RSS quality, presumably epigenetic, influence rearrangement frequency in vivo. In this study, we returned to address this long-standing question with our new data from deep sequencing the Igh repertoire, plus our ChIP-seq and RNA-seq data. We determined the local level of each parameter for each VH gene by calculating the level of histone posttranslational modification or the amount of ncRNA within a 2.5 kb region containing each VH gene coding region plus 1 kb of upstream and downstream flanking DNA, and then we used computational analyses to assess the influence of each of these parameters, individually or in concert, on rearrangement frequency.
Although histone acetylation is far higher on J genes than on V genes (48), nonetheless V genes in loci that are poised for, or are undergoing, rearrangement showed higher acetylation than in loci that are not undergoing rearrangement (48–51). Furthermore, when comparing closely related VH genes, acetylation has been shown to be higher on the VH gene segments that rearrange more frequently (51, 52). Here we demonstrate that essentially every rearranging gene is associated with some level of histone acetylation, and thus histone acetylation may be considered to be strongly associated with accessibility.
H3K4me3 was of particular interest because the PHD finger of RAG2 specifically binds to this modification, and thus it can act to directly recruit the RAG complex (17, 18, 53). H3K4me3 is associated with actively transcribed genes, and thus is likely to be a direct functional consequence of the widespread occurrence of ncRNA at receptor loci as they become poised for rearrangement. High levels of germline transcription and H3K4me3 are only present on J genes (48, 53), however there is a low level of ncRNA throughout the large VH gene portion of receptor loci in pro-B cells (54). ncRNA is easily detected at J558 genes by PCR, while ncRNA at proximal genes is much more difficult to amplify, suggesting much lower levels (7, 54). In accord with this older data, our RNA-seq data shows much higher levels of sense and antisense RNA in the 2.5 kb environs of J558 genes in domains 3 and 4 as compared to the low levels seen in domains 1 and 2 (55). Also, our ChIP-seq demonstrates much higher levels of H3K4me3 on the VH genes in domain 4 than on other domains, and H3K4me3 is essentially absent in domain 1 (Fig. 3, 4). Since H3K4me3 recruits RAG, one would predict that the level of H3K4me3 might show a positive correlation with the rearrangement frequency of individual VH genes, but this correlation was statistically significant only in domain 4. We have previously shown that there are high levels of intergenic antisense transcription at 3 locations in domain 4 (PAIR4, 6 and 11) and, as expected, those sites also have high level of H3K4me3 and H3ac (55). Thus, it could be that high levels of transcription and/or H3K4me3 at intergenic sites are influencing the rearrangement of neighboring VH genes. However, there are no active VH genes within ~20 kb of any of those PAIR elements. It should be noted that the most frequently rearranging gene in domain 4 is 55 kb downstream of PAIR4 and that epigenetic parameters outside of the local 2.5 kb environment of each VH gene were not factored into our computational analyses. Excluding these few regions with high levels of antisense transcription, we observed that the H3K4me3 mark was predominantly located over the genes and not in intergenic regions.
Interestingly, the deposition of all of the activating histone modifications (H3K4me1/2/3 and H3ac) were skewed such that higher levels were seen towards the distal end of the locus, with this being most pronounced for H3K4me3, and least pronounced for H3ac (Fig. 3, 4). However, H3K4me1 and H3K4me2 were only positively correlated with rearrangement frequency in domains 3 and 4, respectively. Domain 1 had essentially no H3K4me2 or me3, and the level of H3K4me1 was low. In contrast to the distal regions, the 7183/Q52 region was the only domain that bore the repressive H3K27me3 modification as we previously reported using ChIP-chip (32), and this was shared with both active and inactive genes. In general, the non-rearranging pseudogenes had lower levels, or no, active H3K4 methyl marks in domains 1–3, but many nonfunctional genes in domain 4 had some level of the H3K4 methylations (Fig. 3). Sense transcription was also much lower on the non-rearranging genes. This may suggest that some level of ncRNA is required for a gene to be functional, and that selective pressure to maintain ncRNA is gone when a gene becomes a pseudogene.
Changes in higher order chromatin structure plays a critical role in V(D)J recombination. It is known that receptor loci undergo compaction at the time of rearrangement (56–58). Jhunjhunwala et al. proposed that the locus was organized into 3 rosette-like structures in pre-pro-B cells that compacted into a smaller 3D space in pro-B cells such that the distal and proximal regions were equidistant from the JH genes (23). The CTCF and cohesin proteins are likely candidates for bringing the Igh locus into this rosette structure (24). We demonstrated several years ago that CTCF and Rad21 bound at many sites throughout the Igh locus, and that the sites were close to the RSS for the proximal VH genes, but were intergenic in the distal J558-containing domains 3 and 4 (24). In the current study, we quantitated the distance from the RSS to the nearest CTCF and Rad21 binding site for all VH genes, and the bimodal distribution of distance to CTCF and Rad21 is striking. VH genes either have CTCF and Rad21 sites within 100 bp of the RSS (domain 1 and half of the small VH families in domain 2), or the sites are from 1–32 kb distant from the VH genes. We have previously demonstrated by 3C that the CTCF sites just upstream of the most V-proximal DH gene interacts with Eμ, which is located only 1–2.5 kb from the JH genes that have high levels of RAG1/2 binding (53). Thus, for the proximal part of the locus, the relative proximity of the RSS to CTCF sites may create a close cluster of RSSs in 3D space as previously suggested (25, 34). Therefore, the proximal VH genes may not need any histone modifications or extensive ncRNA to facilitate long-range movement or to recruit RAG2, and it may be that the proximity to a CTCF site is sufficient. It is striking that within domain 1, almost all of the non- rearranging VH genes do not have CTCF sites near their RSS, whereas all rearranging VH genes in the 7183 and Q52 families, with the exception of one low frequency rearranging VH gene, have nearby CTCF binding sites (25). Many of the non-rearranging domain 1 genes do not have good quality RSSs, and thus this is likely to preclude rearrangement. However, some of the VH genes without nearby CTCF binding do have fairly good RSSs, and in particular, 7183.16.27 and Q52.11.34 have good RSSs, yet no rearrangements were found in our dataset. Thus, it is likely that proximity to CTCF is a requirement for domain 1 VH genes to contribute to the repertoire.
Even though most all of the VH genes in domains 3 and 4 have CTCF sites >1 kb up to ~ 32 kb away from the RSS (with the exception of VH10 genes and one J558 gene), the distance to the nearest Rad21 site varies over a range of ~ 10 bp to ~ 45 kb. Our computational results show a negative correlation between distance to Rad21 and rearrangement frequency in those 2 domains (Fig. 7B) as can also be appreciated in the scatter plot (Fig. S1B). Thus, even for the distal VH genes, the relative position within the CTCF/Rad21-generated chromatin loops appears to be important. These results support the idea that conformational changes regulated by CTCF and Rad21 together are an important factor in determining VH gene usage throughout the locus.
To take a more integrated and comprehensive approach, we performed computational analyses that assessed the relative importance of all the elements that could regulate accessibility of a given gene and recombination. In determining whether a gene rearranges or not, the significance of a functional RSS sequence was clear (Fig. 7A). In this regard, it should be noted that the 14 genes that were deemed potentially functional but which did not rearrange in our data had RIC scores that were lower on average than the active genes overall (−40.56 vs. −27.34). The importance of accessibility was apparent in these results as well, since H3K4me1/2 at VH genes were shown to be significant decision criteria. Distance to a CTCF and/or Rad21 sites was another important factor reflecting the role of 3D structure of the locus. The 4 domains as we divided them are marked by the boundaries of gene families. From the distribution of histone modifications and RNA expression there also seem to be boundaries in the chromatin environment that are likely dependent upon the underlying cis elements that arose through duplications of VH genes. Our division into domains was very similar to that seen when the Igh locus was juxtaposed with a repeat masked alignment of itself reported by Johnston et al. (2), further suggesting a function of distinct domains within the large Igh locus. Importantly, the 4C studies of Guo et al. (59) showed that a site at the border of domains 2 and 3, and another site near the border of domains 1 and 2, interact with Eμ. Thus, these domain borders are likely to be boundaries in the three dimensional structure of the locus as well.
We performed computational analyses of the individual domains to determine whether the four domains had different rules that governed rearrangement. Domain 1 separated itself by having proximity to DJH as a unique significant criterion, as we previously demonstrated for the 7183 gene family in an Igha strain (5). Domain 2 displayed no clear correlative factors, however this may be explained by the heterogeneity of domain 2, as there are many different small VH families interspersed in this domain. In domains 3 and 4, active histone modifications and being closer to a Rad21 binding site generally correlated with higher levels of VH gene usage. We were unable to observe a strong correlation with sense or antisense transcripts and recombination in any of the domains, which we would have expected at least for domain 4. However, Pax5- and YY1-dependent antisense RNA in the intergenic region of domain 4 was previously shown to be important for bringing the distal genes into close proximity to Eμ and its adjacent DJH rearrangement, possibly in a common transcription factory (55). Because we limited our analysis to a 2.5 kb region surrounding each VH gene, the large scale structural changes in the 3D structure of the Igh locus that occur at least partially as a result of antisense transcription are not factored into these computational analyses.
While accessibility of chromatin as characterized by active histone modifications and robust transcriptional activity may provide a favorable environment that recruits RAG1/2 and facilitates recombination, this does not completely explain the non-random VH gene usage in the mouse Igh repertoire. The active VH genes in domain 1, which have adjacent CTCF/Rad21 binding sites, have low levels of ncRNA and are fairly devoid of histone marks other than H3ac. Thus, the position of the V genes near the bases of the CTCF-generated rosettes, coupled with good RSSs in general and moderate levels of histone acetylation, appears to be sufficient to support good rearrangement. At the other extreme, the J558 genes in domain 4 have very poor RSSs in general as determined in recombination substrate analyses, and are the most distant from the JH genes in linear distance. However, these VH genes have the highest level of H3K4me3, which will recruit RAG2. Additionally, the three highest sites of antisense transcription are in domain 4, and this transcription moves domain 4 into proximity with the JH genes as determined by 3C (55). Thus, a strong RSS such as that of a 7183 gene may compensate for an overall lower level of active histone marks and low transcriptional activity, while the poorer RSSs for J558 genes were compensated by higher histone marks and higher ncRNA. This complex system creates an effective strategy for pro-B cells to utilize all the genomic regions to attain high diversity of the Igh repertoire. Thus, the Igh locus has evolved a complex system for the regulation of V(D)J rearrangement that is different for each of the four domains that comprise this locus.
Supplementary Material
Acknowledgments
This work was supported by NIH grants AI082918 and AI092845 to A.J.F. N.M.C. is supported by T32 AI007244. A.T. is supported by NIH-NCATS Clinical and Translational Science Award (CTSA; UL1 RR025774). E.M.O is supported by NIH grant AI081224.
We thank Amy Baumgart for her excellent technical assistance, and Drs. David Nemazee and Takayuki Ota for helpful discussions.
Abbreviations
- ncRNA
non-coding RNA
- CTCF
CCCTC-binding factor
- RSS
recombination signal sequence
- RACE
Rapid Amplification of cDNA Ends
- RIC
RSS information content
- MACS
Model-based Analysis of ChIP-Seq
References
- 1.Yancopoulos GD, Alt FW. Regulation of the assembly and expression of variable-region genes. Ann Rev Immunol. 1986;4:339–368. doi: 10.1146/annurev.iy.04.040186.002011. [DOI] [PubMed] [Google Scholar]
- 2.Johnston CM, Wood AL, Bolland DJ, Corcoran AE. Complete sequence assembly and characterization of the C57BL/6 mouse Ig heavy chain V region. J Immunol. 2006;176:4221–4234. doi: 10.4049/jimmunol.176.7.4221. [DOI] [PubMed] [Google Scholar]
- 3.Perlmutter RM, Kearney JF, Chang SP, Hood LE. Developmentally controlled expression of immunoglobulin VH genes. Science. 1985;227:1597–1600. doi: 10.1126/science.3975629. [DOI] [PubMed] [Google Scholar]
- 4.Yancopoulos GD, Desiderio SV, Paskind M, Kearney JF, Baltimore D, Alt FW. Preferential utilization of the most JH-proximal VH gene segments in pre-B-cell lines. Nature. 1984;311:727–733. doi: 10.1038/311727a0. [DOI] [PubMed] [Google Scholar]
- 5.Williams GW, Martinez A, Montalbano A, Tang A, Mauhar A, Ogwaro KM, Merz D, Chevillard C, Riblet R, Feeney AJ. Unequal VH gene rearrangement frequency within the large VH7183 gene family is not due to RSS variation, and mapping of the genes shows a bias of rearrangement based on chromosomal location. J Immunol. 2001;167:257–263. doi: 10.4049/jimmunol.167.1.257. [DOI] [PubMed] [Google Scholar]
- 6.Schelonka RL, Ivanov, Jung DH, Ippolito GC, Nitschke L, Zhuang Y, Gartland GL, Pelkonen J, Alt FW, Rajewsky K, Schroeder HW., Jr A single DH gene segment creates its own unique CDR-H3 repertoire and is sufficient for B cell development and immune function. J Immunol. 2005;175:6624–6632. doi: 10.4049/jimmunol.175.10.6624. [DOI] [PubMed] [Google Scholar]
- 7.Love VA, Lugo G, Merz D, Feeney AJ. Individual promoters vary in strength, but the frequency of rearrangement of those VH genes does not correlate with promoter strength nor enhancer independence. Mol Immunol. 2000;37:29–39. doi: 10.1016/s0161-5890(00)00023-7. [DOI] [PubMed] [Google Scholar]
- 8.Liu H, Schmidt-Supprian M, Shi Y, Hobeika E, Barteneva N, Jumaa H, Pelanda R, Reth M, Skok J, Rajewsky K, Shi Y. Yin Yang 1 is a critical regulator of B-cell development. Genes Dev. 2007;21:1179–1189. doi: 10.1101/gad.1529307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su IH, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT, Tarakhovsky A. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat Immunol. 2003;4:124–131. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- 10.Hesslein DG, Pflugh DL, Chowdhury D, Bothwell AL, Sen R, Schatz DG. Pax5 is required for recombination of transcribed, acetylated, 5′ IgH V gene segments. Genes Dev. 2003;17:37–42. doi: 10.1101/gad.1031403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malynn BA, Yancopoulos GD, Barth JE, Bona CA, Alt FW. Biased expression of JH-proximal VH genes occurs in the newly generated repertoire of neonatal and adult mice. J Exp Med. 1990;171:843–859. doi: 10.1084/jem.171.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feeney AJ. Genetic and epigenetic control of V gene rearrangement frequency. Adv Exp Med Biol. 2009;650:73–81. doi: 10.1007/978-1-4419-0296-2_6. [DOI] [PubMed] [Google Scholar]
- 13.Yancopoulos GD, Alt FW. Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell. 1985;40:271–281. doi: 10.1016/0092-8674(85)90141-2. [DOI] [PubMed] [Google Scholar]
- 14.Lennon GG, Perry RP. The temporal order of appearance of transcripts from unrearranged and rearranged Ig genes in murine fetal liver. J Immunol. 1990;144:1983–1987. [PubMed] [Google Scholar]
- 15.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 16.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature Genetics. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor- gene rearrangement. Immunity. 2007;27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, Walter KL, Utz PJ, Shi Y, Kutateladze TG, Yang W, Gozani O, Oettinger MA. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakano H, Huppi K, Heinrich G, Tonegawa S. Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature. 1979;280:288–294. doi: 10.1038/280288a0. [DOI] [PubMed] [Google Scholar]
- 20.Max EE, Seidman JG, Leder P. Sequences of five potential recombination sites encoded close to an immunoglobulin kappa constant region gene. Proc Natl Acad Sci USA. 1979;76:3450–3454. doi: 10.1073/pnas.76.7.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hesse JE, Lieber MR, Mizuuchi K, Gellert M. V(D)J recombination: a functional definition of the joining signals. Genes Dev. 1989;3:1053–1061. doi: 10.1101/gad.3.7.1053. [DOI] [PubMed] [Google Scholar]
- 22.Akira S, Okazaki K, Sakano H. Two pairs of recombination signals are sufficient to cause immunoglobulin V-(D)-J joining. Science. 1987;238:1134–1138. doi: 10.1126/science.3120312. [DOI] [PubMed] [Google Scholar]
- 23.Jhunjhunwala S, van Zelm MC, Peak MM, Cutchin S, Riblet R, van Dongen JJ, Grosveld FG, Knoch TA, Murre C. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell. 2008;133:265–279. doi: 10.1016/j.cell.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Degner SC, Wong TP, Jankevicius G, Feeney AJ. Cutting Edge: Developmental stage-specific recruitment of cohesin to CTCF sites throughout immunoglobulin loci during B lymphocyte development. J Immunol. 2009;182:44–48. doi: 10.4049/jimmunol.182.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Degner SC, Verma-Gaur J, Wong TP, Bossen C, Iverson GM, Torkamani A, Vettermann C, Lin YC, Ju Z, Schulz D, Murre CS, Birshtein BK, Schork NJ, Schlissel MS, Riblet R, Murre C, Feeney AJ. CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells. Proc Natl Acad Sci U S A. 2011;108:9566–9571. doi: 10.1073/pnas.1019391108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo C, Yoon HS, Franklin A, Jain S, Ebert A, Cheng HL, Hansen E, Despo O, Bossen C, Vettermann C, Bates JG, Richards N, Myers D, Patel H, Gallagher M, Schlissel MS, Murre C, Busslinger M, Giallourakis CC, Alt FW. CTCF-binding elements mediate control of V(D)J recombination. Nature. 2011;477:424–430. doi: 10.1038/nature10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seitan VC, Hao B, Tachibana-Konwalski K, Lavagnolli T, Mira-Bontenbal H, Brown KE, Teng G, Carroll T, Terry A, Horan K, Marks H, Adams DJ, Schatz DG, Aragon L, Fisher AG, Krangel MS, Nasmyth K, Merkenschlager M. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature. 2011;476:467–471. doi: 10.1038/nature10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shih HY, Verma-Gaur J, Torkamani A, Feeney AJ, Galjart N, Krangel MS. Tcra gene recombination is supported by a Tcra enhancer- and CTCF-dependent chromatin hub. Proc Natl Acad Sci U S A. 2012;109:E3493–3502. doi: 10.1073/pnas.1214131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bossen C, Mansson R, Murre C. Chromatin topology and the regulation of antigen receptor assembly. Ann Rev of Immunol. 2012;30:337–356. doi: 10.1146/annurev-immunol-020711-075003. [DOI] [PubMed] [Google Scholar]
- 30.Ebert A, McManus S, Tagoh H, Medvedovic J, Salvagiotto G, Novatchkova M, Tamir I, Sommer A, Jaritz M, Busslinger M. The distal VH gene cluster of the Igh locus contains distinct regulatory elements with Pax5 transcription factor-dependent activity in pro-B cells. Immunity. 2011;34:175–187. doi: 10.1016/j.immuni.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 31.Verma-Gaur J, Torkamani A, Schaffer L, Head SR, Schork NJ, Feeney AJ. Noncoding transcription within the Igh distal VH region at PAIR elements affects the 3D structure of the Igh locus in pro-B cells. Proc Natl Acad Sci USA. 2012;109:17004–17009. doi: 10.1073/pnas.1208398109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu CR, Schaffer L, Head SR, Feeney AJ. Reciprocal patterns of methylation of H3K36 and H3K27 on proximal vs. distal IgVH genes are modulated by IL-7 and Pax5. Proc Natl Acad Sci U S A. 2008;105:8685–8690. doi: 10.1073/pnas.0711758105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merelli I, Guffanti A, Fabbri M, Cocito A, Furia L, Grazini U, Bonnal RJ, Milanesi L, McBlane F. RSSsite: a reference database and prediction tool for the identification of cryptic Recombination Signal Sequences in human and murine genomes. Nucl Acids Re. 2010;38:W262–267. doi: 10.1093/nar/gkq391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lucas JS, Bossen C, Murre C. Transcription and recombination factories: common features? Current Opin in Cell Biol. 2011;23:318–324. doi: 10.1016/j.ceb.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakamoto S, Wakae K, Anzai Y, Murai K, Tamaki N, Miyazaki M, Miyazaki K, Romanow WJ, Ikawa T, Kitamura D, Yanagihara I, Minato N, Murre C, Agata Y. E2A and CBP/p300 act in synergy to promote chromatin accessibility of the immunoglobulin kappa locus. J Immunol. 2012;188:5547–5560. doi: 10.4049/jimmunol.1002346. [DOI] [PubMed] [Google Scholar]
- 36.Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, Ideker T, Glass CK, Murre C. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chakraborty T, Chowdhury D, Keyes A, Jani A, Subrahmanyam R, Ivanova I, Sen R. Repeat organization and epigenetic regulation of the DH-Cμ domain of the immunoglobulin heavy-chain gene locus. Mol Cell. 2007;27:842–850. doi: 10.1016/j.molcel.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakano H, Hüppi K, Heinrich G, Tonegawa S. Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature. 1979;280:288–294. doi: 10.1038/280288a0. [DOI] [PubMed] [Google Scholar]
- 40.Hesse JE, Lieber MR, Mizuuchi K, Gellert M. V(D)J recombination: A functional definition of the joining signals. Genes Dev. 1989;3:1053–1061. doi: 10.1101/gad.3.7.1053. [DOI] [PubMed] [Google Scholar]
- 41.Montalbano A, Ogwaro KM, Tang A, Matthews AG, Larijani M, Oettinger MA, Feeney AJ. V(D)J recombination frequencies can be profoundly affected by changes in the spacer sequence. J Immunol. 2003;171:5296–5304. doi: 10.4049/jimmunol.171.10.5296. [DOI] [PubMed] [Google Scholar]
- 42.Ramsden DA, Baetz K, Wu GE. Conservation of sequence in recombination signal sequence spacers. Nucl Acids Res. 1994;22:1785–1796. doi: 10.1093/nar/22.10.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cowell LG, Davila M, Kepler TB, Kelsoe G. Identification and utilization of arbitrary correlations in models of recombination signal sequences. Genome Biol. 2002;3:1–20. doi: 10.1186/gb-2002-3-12-research0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cowell LG, Davila M, Yang K, Kepler TB, Kelsoe G. Prospective estimation of recombination signal efficiency and identification of functional cryptic signals in the genome by statistical modeling. J Exp Med. 2003;197:207–220. doi: 10.1084/jem.20020250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Connor AM, Fanning LJ, Celler JW, Hicks LK, Ramsden DA, Wu GE. Mouse VH7183 recombination signal sequences mediate recombination more frequently than those of VHJ558. J Immunol. 1995;155:5268–5272. [PubMed] [Google Scholar]
- 46.Feeney AJ, Atkinson MJ, Cowan MJ, Escuro G, Lugo G. A defective VkA2 allele in Navajos which may play a role in increased susceptibility to Haemophilus influenzae type b disease. J Clin Invest. 1996;97:2277–2282. doi: 10.1172/JCI118669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams GS, Martinez A, Montalbano A, Tang A, Mauhar A, Ogwaro KM, Merz D, Chevillard C, Riblet R, Feeney AJ. Unequal VH gene rearrangement frequency within the large VH7183 gene family is not due to recombination signal sequence variation, and mapping of the genes shows a bias of rearrangement based on chromosomal location. J Immunol. 2001;167:257–263. doi: 10.4049/jimmunol.167.1.257. [DOI] [PubMed] [Google Scholar]
- 48.Xu CR, Feeney AJ. The epigenetic profile of Ig genes is dynamically regulated during B cell differentiation and is modulated by pre-B cell receptor signaling. J Immunol. 2009;182:1362–1369. doi: 10.4049/jimmunol.182.3.1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chowdhury D, Sen R. Stepwise activation of the immunoglobulin mu heavy chain gene locus. Embo J. 2001;20:6394–6403. doi: 10.1093/emboj/20.22.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McMurry MT, Krangel MS. A role for histone acetylation in the developmental regulation of VDJ recombination. Science. 2000;287:495–498. doi: 10.1126/science.287.5452.495. [DOI] [PubMed] [Google Scholar]
- 51.Johnson K, Angelin-Duclos C, Park S, Calame KL. Changes in histone acetylation are associated with differences in accessibility of VH gene segments to V-DJ recombination during B-cell ontogeny and development. Mol Cell Biol. 2003;23:2438–2450. doi: 10.1128/MCB.23.7.2438-2450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Espinoza CR, Feeney AJ. The extent of histone acetylation correlates with the differential rearrangement frequency of individual VH genes in pro-B cells. J Immunol. 2005;175:6668–6675. doi: 10.4049/jimmunol.175.10.6668. [DOI] [PubMed] [Google Scholar]
- 53.Ji Y, Resch W, Corbett E, Yamane A, Casellas R, Schatz DG. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 2010;141:419–431. doi: 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yancopoulos GD, Alt FW. Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell. 1985;40:271–281. doi: 10.1016/0092-8674(85)90141-2. [DOI] [PubMed] [Google Scholar]
- 55.Verma-Gaur J, Torkamani A, Schaffer L, Head SR, Schork NJ, Feeney AJ. Noncoding transcription within the Igh distal VH region at PAIR elements affects the 3D structure of the Igh locus in pro-B cells. Proc Natl Acad Sci U S A. 2012;109:17004–17009. doi: 10.1073/pnas.1208398109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fuxa M, Skok J, Souabni A, Salvagiotto G, Roldan E, Busslinger M. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 2004;18:411–422. doi: 10.1101/gad.291504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 58.Sayegh C, Jhunjhunwala S, Riblet R, Murre C. Visualization of looping involving the immunoglobulin heavy-chain locus in developing B cells. Genes Dev. 2005;19:322–327. doi: 10.1101/gad.1254305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo C, Gerasimova T, Hao H, Ivanova I, Chakraborty T, Selimyan R, Oltz EM, Sen R. Two forms of loops generate the chromatin conformation of the immunoglobulin heavy-chain gene locus. Cell. 2011;147:332–343. doi: 10.1016/j.cell.2011.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.