SUMMARY
Early embryonic patterning events are strikingly precise. The underlying molecular details are elusive and appear incompatible with stochastic gene expression observed across phyla. Using single molecule mRNA quantification in Drosophila embryos, we determine the magnitude of fluctuations in the expression of four critical patterning genes. The accumulation of mRNAs is identical across genes and fluctuates by ~8% between neighboring nuclei to generate precise protein distributions. In contrast, transcribing loci exhibit an intrinsic noise of ~45% independent of specific promoter-enhancer architecture or fluctuating inputs. The embryo recovers precise transcript distribution through straightforward spatiotemporal averaging without regulatory feedback. The common expression characteristics shared between genes suggest that the noise of fundamental physical constraints dominates the fluctuations of immediate transcriptional readout.
INTRODUCTION
A fundamental question in biology concerns the degree of precision that cellular systems exhibit in their responses to a given set of environmental conditions, extracellular signals, or other input stimuli (Lagha et al., 2012; Lander 2013, Little and Wieschaus, 2011). Production of and interactions between molecules are intrinsically stochastic, limiting the ability of cells to control gene expression and biochemical activities (Raser and O'Shea, 2005), but the propensity of cellular systems to achieve appropriate phenotypic behavior constrains the tolerable magnitude of molecular fluctuations (Rao et al., 2002). In most contexts, it is unknown how closely cellular activity and phenotypic behavior rely on precise control of gene expression.
Many features of Drosophila embryogenesis suggest that strict control of gene expression determines reproducible and precise cell fate establishment. In Drosophila embryos, patterned gene expression in the early syncytium of ~6000 nuclei is triggered by asymmetrically distributed, maternally supplied cues (Sauer et al., 1996). Among these is the transcription factor Bicoid (Bcd), the anterior-posterior (AP) concentration gradient of which shows remarkably reproducible distributions between embryos (Gregor et al., 2007). Moreover, within an embryo, the nuclei at similar AP coordinates differ in Bcd concentration by less than 10% (standard deviation (SD) over mean), a degree of precision sufficiently high for each row of cells along the AP axis to discern its position from its immediate neighbors (Gregor et al., 2007). Bcd precision correlates with highly precise protein distribution of zygotically expressed target genes (Dubuis et al., 2013; Gregor et al., 2007) that confer cells with distinct gene expression programs within under three hours following fertilization (Gergen et al., 1986; Kornberg and Tabata, 1993).
These observations suggest a model in which tightly regulated transcriptional inputs give rise to rapidly established, highly precise outputs. However, the degree of precision in developmental transcription is largely unexplored. In all contexts assayed from prokaryotes to mammalian cells, absolute levels of a given transcript differ by at least ~50% between genotypically identical cells, and for a majority of genes this variability is even higher (Cohen et al., 2009; Gandhi et al., 2011; Golding et al., 2005; Pare et al., 2009; Taniguchi et al., 2010; Raj et al., 2006; Raj et al., 2010; Reiter et al., 2011; Sigal et al., 2006; Zenklusen et al., 2008). Quantitative observations support the idea that the process of transcription is intrinsically stochastic (Kaern et al., 2005; Li and Xie, 2011). In developmental contexts, it is unknown if relatively small input transcription factor fluctuations impact the transcriptional output, and whether embryogenesis requires the activity of specialized filtering and/or feedback mechanisms to ensure fidelity in the rapid establishment of gene expression programs.
Here, we address these questions with an enhanced method of fluorescence in situ hybridization (FISH) and accompanying image analysis (Little et al., 2011) to label and detect individual zygotically expressed mRNA molecules. We measure in absolute molecular counts the magnitude and fluctuations in the earliest gene expression events of the Drosophila embryo. To separate input fluctuations from variability intrinsic to transcription, we focus on those spatial domains in which gene expression is maximally unconstrained. Here, patterning inputs do not determine expression output levels, and thus input fluctuations cannot impact output variability. These regions thereby reveal the greatest degree of precision achievable by the system. We show that in these regions the earliest expressed genes share common expression characteristics: despite their expression in spatially distinct territories, their rates of production are identical, and all display intrinsically stochastic transcriptional activity. These similarities suggest that expression rate and variability result from fundamental, global features of transcriptional regulation that limit the attainable degree of precision. Nevertheless, the stochastic expression results in precise and nearly uniform transcript accumulation, achieved by straightforward spatiotemporal averaging.
RESULTS
Measuring absolute numbers of mRNA transcripts in early Drosophila embryos
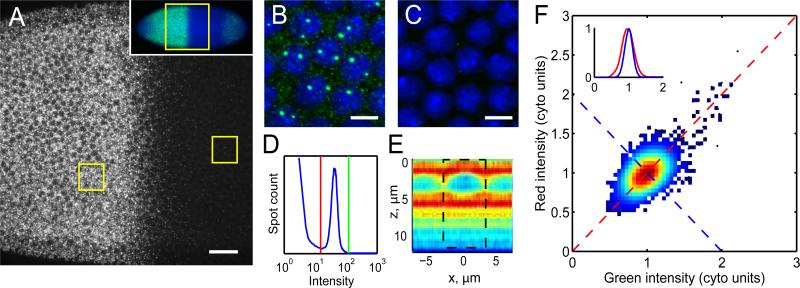
Previous work in Drosophila embryos has documented that nuclei at similar AP coordinates express nearly equivalent protein amounts of the gap gene Hunchback (Hb) with fluctuations of <10% (Gregor et al., 2007). The transcriptional activator of Hb, Bcd, displays variability on the same order as Hb (Gregor et al., 2007). A precise transcriptional response of the hb locus presents the most straightforward though as yet untested explanation of minimal Hb variation. To quantitatively evaluate transcription of hb, we adapted a FISH method developed previously (Little et al., 2011) to label hb mRNAs using multiple fluorescently labeled antisense DNA oligonucleotides (Figure 1A). By scanning confocal microscopy, we detect two broad classes of objects: sparse, bright spots representing sites of nascent transcription (e.g., Wilkie et al., 1999), and numerous diffraction limited spots, ≈90% of which are located in the internuclear space which we refer to as cytoplasmic particles (Figure 1A-C). These particles exhibit sufficiently high contrast to be readily distinguished from background imaging noise using automated image processing (Figure 1D). Each particle is found on >3 adjacent 250 nm confocal imaging sections with 3-dimensional structure identical to the measured point spread function (PSF; Figure S1A-D). To test detection efficiency, we applied probes with alternating fluorophore colors. A minimum of 85% of cytoplasmic particles detected in one channel are found in the other, indicating >94% mRNA are detected in at least one channel (Figure S1E-G).
Figure 1. Counting of absolute transcript number in Drosophila embryos.
A: Confocal section through the nuclear layer of a WT embryo during interphase 13 labeled with 114 fluorescent oligonucleotide probes against hunchback, oriented anterior to the left. Scale bar: 25 μm. Inset: Low magnification image identifying the region shown in A. B,C: Magnified views of anterior (B) and posterior (C) boxed regions in (A). Scale bars: 5 μm. D: Particle intensity histogram showing thresholds separating transcripts from noise (red line) and from the long tail of bright transcription sites (green line). E: hb transcript distribution in axial cross-section through a nucleus centered at x=0. z=0 represents apical surface. Color indicates mean particle density in relative units (red=high, blue=low). Dashed box: cylindrical summation volume. F: Intensity scatter plot in two channels using probes of alternating colors. Data point density given by color. Inset: Cross-sections of scatter plot in (F) along the correlated (red) and anti-correlated direction (blue) shows Gaussian distributions with σ=20% (red) and σ=12% (blue) after normalization to mean cytoplasmic particle intensity (1 “cyto unit”). See also Figures S1 and S2.
Tight unimodal clustering around mean intensity suggests that the cytoplasmic particles are similar in mRNA content (Figure 1D). Deviation from mean intensity results from at least two phenomena: particles can be bound by different probe numbers, and multiple particles can overlap and be detected as single spots. To determine the relative contributions of each, we examined correlation of intensities in 2-color detection. Correlation is weak (Figure 1F), implying that the fractional SD of mRNA content in detected particles is at most 16% (see Experimental Procedures). To determine the number of mRNAs per particle, we compared counts of maternally deposited hb mRNA particles in entire imaged embryos to those from quantitative RT-PCR (Figure S1H). We found an average of 1.2±0.5 mRNAs per imaged particle. The low mean indicates that the probability of detecting more than 2 mRNA per particle is essentially zero. This observation, coupled with the SD in mRNA content from imaging, determines a probability of 97% of finding 1 mRNA per particle (see Experimental Procedures). Moreover, comparing counts of wild-type (WT) and hb hemizygous embryos yields a 2-fold concentration difference (Figure S1I), lending further support to the validity of our approach.
As the density of zygotically produced hb mRNA increases above about 1 molecule per μm3, PSFs of individual molecules begin to overlap. We extend particle counting to arbitrarily high density using the naturally large dynamic expression range. We determine counts in dense regions by measuring total fluorescence collected from all mRNA per volume, and calibrating to low expressing regions where individual mRNA are counted directly (Figure S2A). We thereby measure absolute concentration and local fluctuation with accuracies of 12% and 5% respectively (Figure S2B-C). The two methods have overlapping domains of applicability: direct counts are accurate for transcript concentrations ≤0.5 molecules/μm3; and total fluorescence for concentrations ≥0.35 molecules/μm3 (Fig. S2C). Thus our FISH method is suitable for high-precision measurements of absolute mRNA counts at any density.
Cytoplasmic hb mRNA and protein distributions display similar levels of precision
To assess fluctuations in transcript number between nuclei, we measure mRNA concentration in cylinders separated by one internuclear distance to a depth of 12 μm beneath the plasma membrane, encompassing the majority of zygotically expressed transcripts (Figure 1E and S3A). As expected from prior observations (e.g. Tautz et al., 1987), hb transcripts accumulate dramatically in the embryo anterior during early blastoderm (Figure 2A). Transcription is terminated early in nc14 except near the embryo midpoint, and maternally supplied transcripts are continuously lost from the posterior (Figure 2A and S3B-C). hb mRNA expression profiles correlate well with observed protein levels (Figure S3D).
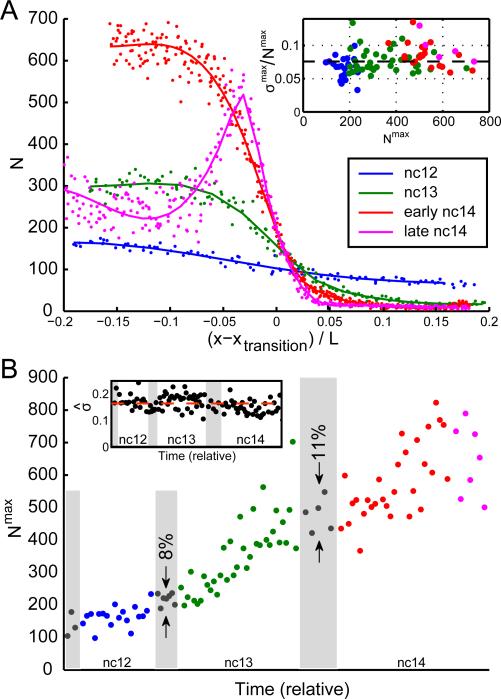
Figure 2. Precision and reproducibility of cytoplasmic hb profiles.
A: Absolute cytoplasmic hb mRNA counts per standardized volume as a function of AP position. Data for four embryos at nuclear cycle 12 (blue), 13 (green), early 14 (red), and late 14 (magenta). Position is shown as distance from inflection point xtransition (see also Figure S3C). Inset: fractional SD σmax/Nmax in the spatial domain of highest mRNA accumulation as a function of the mean count (Nmax) for 101 embryos. Dashed line at 8%. B: Cytoplasmic hb mRNA counts (Nmax) as a function of time. Ages estimated by visual inspection of DAPI staining; relative width of mitoses (gray shading) and interphases according to Alberts and Foe (1983). Reproducibility of counts in 12th and 13th mitoses is 8% and 11%, respectively. Inset: estimated reproducibility as a function of time. Data points: running averages of root-mean-square displacement from smoothed timeline over 15 consecutive data points normalized to mean. Dashed line: average (17%). See also Figure S3
As an initial quantification of precision, we ascertain the degree of variability independent of putative regulatory inputs by examining the spatial domain of maximal transcript accumulation, i.e. nuclei found in regions of highest observed gene product levels. Here expression noise (the fractional SD of hb concentration) is 8±2% as early as nc12; thus hb mRNA levels exhibit equal or better precision than Hb protein (Gregor et al., 2007). Age-ordered embryos show a monotonically increasing counts through mid-nc14 (Figure 2B), with an approximately constant fractional SD across embryos (17±3%, Figure 2B inset). Ambiguous age determination in fixed samples results in large fluctuations across embryos of approximately the same age. This effect is minimized in embryos undergoing mitosis, during which transcription ceases (Shermoen and O'Farrell, 1991) allowing unambiguously temporal ordering. Counts differed by less than 11% in mitotic embryos, similar to the degree of reproducibility in Hb protein profiles (Gregor et al., 2007). The actual precision and reproducibility are likely to be higher, since our measurements contain systematic errors arising from the FISH procedure such as physical distortion (5% measurement error) and error in counts (2-3% measurement error; see Experimental Procedures and Figure S2C). Importantly, variation of cytoplasmic profiles is nearly at the level of Poisson counting noise, i.e. at the lowest bound that can be attained by a stochastic process. For N=500 molecules per volume, as observed in late nc13 or early nc14, counting noise amounts to 5%, matching the lower bound of our measurements (Fig. 2A, inset). Large mRNA counts provide a natural buffer against potential fluctuations in translation that have been observed in other systems (Bar-Even et al., 2006; Newman et al., 2006; Taniguchi et al., 2010), yielding precise Hb expression. By comparison, in genome wide studies, the most highly (and therefore most precisely) expressed genes in yeast and E. coli exhibit cell-to-cell fluctuations exceeding 50% in mRNA count (Gandhi et al., 2011; Taniguchi et al., 2010). Thus, early embryos exhibit an extraordinary degree of precision, rarely observed in other contexts.
Our timeline suggests that hb transcript lifetime is large, as we see no decrease in counts during the 12th and 13th mitoses (Figure 2B). We verified this by measuring hb mRNA lifetime directly, disrupting transcription with α-amanitin injection and subsequently monitoring loss of zygotic hb (Figure S3E-F). We find a lifetime of ~60 minutes, consistent with an estimate from imaging (transcript loss of <11% (Figure 2B) in 5 minutes of mitosis (Foe and Alberts, 1983) corresponds to a lifetime of >45min). These results show that the accumulation of transcripts is only mildly impacted by degradation.
Determining instantaneous transcriptional activity by measuring total nuclear nascent mRNA content
The low noise of hb cytoplasmic mRNA counts suggests that nuclei in the fully active region produce transcripts at nearly equivalent rates. However, all systems studied to date, including E. coli, yeast, cultured cells, and late Drosophila embryos (Golding et al., 2005; Larson et al., 2011; Pare et al., 2009; Raj et al., 2006; Zenklusen et al., 2008), produce transcripts through brief intervals of dense output interspersed with long quiescent periods of stochastic duration (Li and Xie, 2011). This seems incompatible with near uniformity of cytoplasmic mRNA content. To determine the extent of variability in transcriptional activity, we developed a novel measure of transcription using the fluorescence intensities of nascent transcription sites.
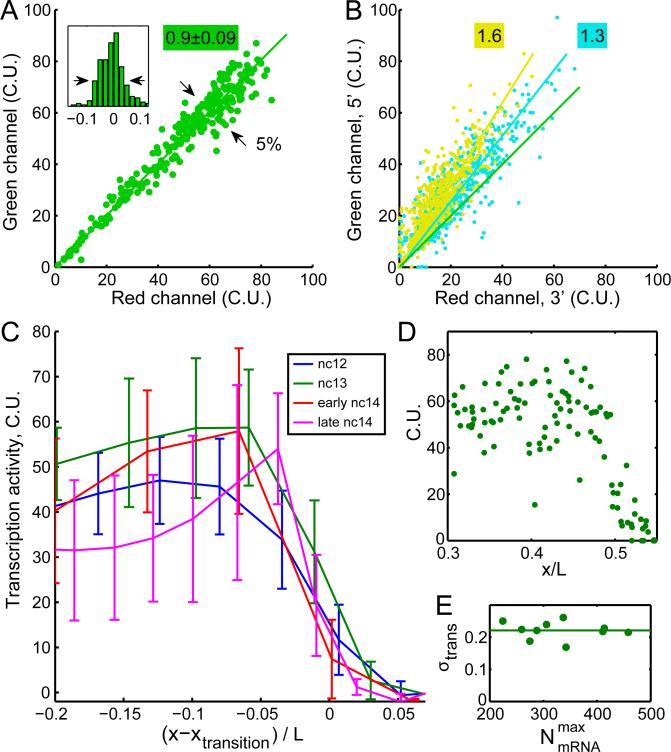
Consistent with previous results (Wilkie et al., 1999), we observe that the maximum number of detectable nascent sites per nucleus increases from DNA replication during interphase from 2 sites early to 4 at mid to late interphase (Figure S4A). Because sister chromatid loci remain in close physical proximity until mitosis, and because transcription sites occasionally occupy overlapping focal volumes, the number of active loci is challenging to discern. Instead we used the total fluorescence of all transcription sites in a nucleus as a measure of instantaneous transcriptional activity. Assuming that nascent and mature mRNAs are equally accessible to probes, nascent site intensities can be represented as an equivalent number of mature cytoplasmic mRNAs by normalizing to the mean or “unit” intensity of completed transcripts, yielding transcriptional activity in absolute units of total mRNA content. To determine the extent of measurement error arising from differences in probe binding affinity and/or the subsequent normalization procedure, we used probes of alternating fluorophore colors. Ideally, for a given nascent site, the number of cytoplasmic units (C.U.) will be identical in both colors. Plotting nascent mRNA content of one color as a function of the other yields points on a line with a slope close to unity (between 5 embryos the mean slope (± SD) is 0.90±0.09), with a scatter of 5% (Figure 3A). We thus measure transcriptional activity with an error of 5% and relate it to absolute mRNA content with an uncertainty under 20% (the largest deviation of 0.90±0.09 from 1).
Figure 3. Variability of transcriptional activity at nascent transcription sites.
A: Scatter plot of total nascent hb mRNA per nucleus using probes of alternating colors for an embryo in nuclear cycle 13 after normalization to the mean cytoplasmic particle intensity (C.U.). Intensities follow a direct proportionality relation with slope 0.90±0.09 (n=5 embryos). Inset: root mean square normalized deviation from linear fit; scatter = 5% (arrows). B: Two-color scatter plot of nascent mRNA content in which probes bearing the same fluorophore are clustered on the 5’ (green channel) and 3’ (red channel) portions of the transcript. Cyan: measurements using 57 green and 57 red-labeled probes; observed slope: 1.3. Yellow: results with 78 green and 36 red-labeled probes; observed slope: 1.6. Green line in A is plotted for comparison. C: Transcriptional activity per nucleus as a function of position along the AP axis for four embryos in nuclear cycle 12 (blue), 13 (green), 14 early (red), and 14 late (magenta) in binned averages of 10, 20, 40 and 40 nuclei, respectively. Error bars: SDs within bins. Position is shown as distance from inflection point xtransition. D: Transcriptional activity per nucleus as a function of absolute AP position for the embryo in interphase 13 in C. E: Transcription noise for 10 embryos in nuclear cycle 13 plotted as fractional SD across nuclei as a function of cytoplasmic hb counts within the spatial domain of highest accumulation. Transcription activity noise remains constant throughout interphase at 22±3%. See also Figure S4.
Three lines of evidence support the idea that nascent mRNA content reflects instantaneous transcriptional activity. First, the appearance of loci is coupled to the nuclear mitotic cycle: they are observed during interphase and absent during mitosis. Second, transcription in anterior nuclei initiates slightly earlier than in those closer to the center of the embryo, both because anterior nuclei inhabit a region of higher concentration of Bcd and because of metasynchronous nuclear divisions propagating as a wave towards the embryo center (Foe and Alberts, 1983). Consistent with expectation, during the first minute of the 13th interphase we observe a gradient of nascent mRNA content along the AP axis (Figure S4B). Third, we designed probe sets to label the 5’ and 3’ portions of the completed transcript with fluorophores of green and red colors, respectively. If nascent sites are composed of incomplete transcripts, then 5’ sequences must be more numerous than 3’ sequences (Figure S4C), resulting in an increase of green signal at the expense of red. In agreement, our measurements reveal the enrichment of green signal after normalization (Figure 3B). Importantly, greater 5’ enrichment is observed as the fraction of transcript labeled with green fluorophore increases (increasing slopes of fit lines in Figure 3B). Thus, nascent hb loci are largely composed of unfinished transcripts and serve as a measure of transcriptional activity.
Variation in nascent transcription site activity is 6-fold higher than variation in cytoplasmic output
Given the low noise in cytoplasmic counts, we expected that the nascent mRNA content at all genomic loci would rise simultaneously until saturated with RNA polymerase II (RNAP), in principle reaching and sustaining some maximum nascent mRNA content. However, our measurements of nascent mRNA content show otherwise (Figure 3C-E). The nuclear nascent mRNA content varies by 22±3%, 3-fold higher than that observed in cytoplasmic counts. To be certain that this variability does not result from the delay in attaining steady-state maximum activity after mitosis (Figure S4D), we confined our analysis to mid and late interphase 13 embryos. We observe this degree of variation even when loci are allowed the full temporal extent of interphase 13 to reach a putative maximum (Figure 3E). These results indicate that hb loci fail to sustain any amount of uniform maximum content.
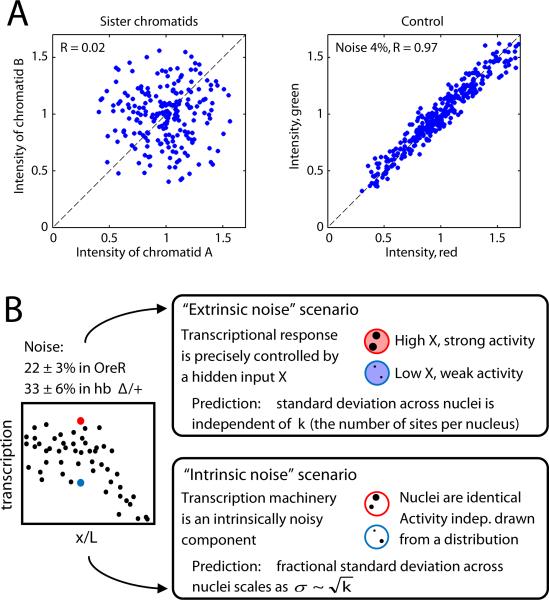
The 22% variation we observe reflects fluctuations across a maximum of 4 active genomic loci in each nucleus of WT embryos. If transcription from each locus acts independently, then the variability between nuclei must decrease in proportion to the root of the number of loci; thus the expected variability between individual loci is . To test whether loci are in fact independent, we examined embryos heterozygous for a hb deficiency in which each nucleus possesses a maximum of only 2 loci. We observed that total nascent mRNA content per nucleus varies by 33±6%, which corresponds to a transcriptional activity of in individual loci. This number is nearly identical to WT and hence consistent with independence. The variability between individual loci of ~45% represents a 6-fold increase over fluctuations in cytoplasmic counts. Analyzing closely apposed alleles on sister chromatids with sufficient separation to reliably gauge intensities reveals no correlation in their activities (Figure 4A), indicating independent activity even for recently duplicated loci. If the observed fluctuations result from variability in any input factor controlling hb expression (i.e., “extrinsic” noise), then the variation in total nuclear activity would show no dependence on the number of loci in the nucleus. However, as the noise scales with the number of loci (Figure 4B), the fluctuations we observe in the maximally expressed domain are intrinsic to the process(es) of transcription and not determined by variability in the controlling inputs.
Figure 4. Fluctuations in hb transcription are dominated by intrinsic noise.
A: Transcriptional activity of loci on optically resolved sister chromatids is uncorrelated (Pearson correlation coefficient R=0.02), compared to the tight correlation (R=0.97) in a control experiment using probes of alternating colors (with 4% imaging noise). B: Transcriptional variability arises from fluctuations in inputs (extrinsic noise) and from the process of transcription itself (intrinsic noise). Two extreme scenarios are presented in cartoon form. Upper panel: a fluctuating extrinsic input leads to correlated activities of transcription sites within a given nucleus; its contribution to the fractional SD is independent of the number of transcribing loci k. Lower panel: intrinsic mechanistic noise affects all transcription sites independently; the fractional SD scales as inverse square root of available transcription sites. Left: the measured transcription noise in WT and hbΔ/+ embryos (22±3% and 33±6%, respectively) shows scaling behavior characteristic of intrinsic noise with magnitude ~45% .
Transcriptional activity will necessarily exhibit some degree of noise arising from stochastic single-molecule events, but the fluctuations we observe exceed the Poisson expectation considerably. From our observations, we can estimate the number of RNAP engaged in transcription per nucleus and thereby determine the expected degree of fluctuations; we find the predicted noise magnitude of at most 11% (see Experimental Procedures). The observed fluctuations of 22±3% are at least 2-fold greater than this prediction, ruling out a model in which transcriptional fluctuations in the region of maximum expression are determined by a single rate limiting step of RNAP loading (Figure S4C).
From these observations, we conclude that, first, even in the domain of maximal expression, hb is not saturated with the maximum possible density of RNAP; and, second, despite the near uniformity of cytoplasmic transcript concentration, instantaneous activity of individual hb loci is intrinsically stochastic. The estimated variation in transcriptional activity at an individual locus is very similar to the minimum value of ~50% observed for differences in mRNA numbers for the most highly expressed genes in yeast and E. coli (Gandhi et al., 2011; Taniguchi et al., 2010). In all contexts, variation between cells is significantly higher than that predicted for a process with a single rate-limiting step (Chubb et al., 2006; Golding et al., 2005; Le et al., 2005; Raj et al., 2006). These similarities across such diverse contexts suggest that the observed fluctuations are globally inherent features of the activity of otherwise “fully activated” genes. In the context of a rapidly developing embryo, the highest attainable expression rate would serve to minimize cell-to-cell fluctuations to the fullest possible extent and thereby promote precision. The observed tolerance of fluctuations, linked with the apparent inability to sustain saturating RNAP density, suggests that this degree of imprecision cannot be circumvented even in this highly precise developmental context.
The magnitude of expression noise is independent of autoregulation and transcriptional modulation
Our results suggest that in addition to fluctuations from controlling inputs, hb activity possesses a large inherently stochastic component. We examined whether the inherent noise could be attributed to features of hb regulation. First, the hb locus contains several binding elements for Hb protein itself, and genetic evidence indicates Hb is required for its own expression (Holloway et al., 2011; Margolis et al., 1995; Treisman and Desplan, 1989). Positive feedback will amplify fluctuations if locally produced mRNA and protein dominate autoregulation (i.e. if diffusion is limited). Second, noise necessarily decreases as RNAP approach their maximum loading density along the gene, but transcriptional repressors might disallow high densities, resulting in greater variability.
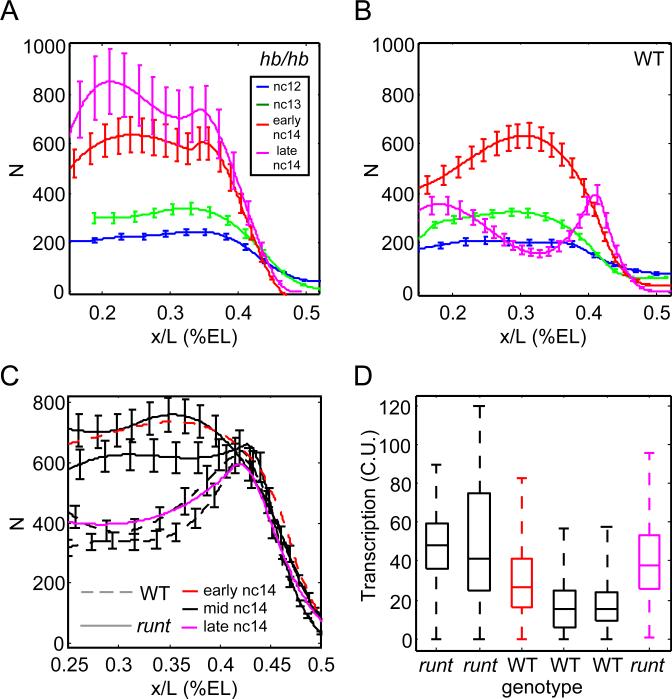
To determine the effect of positive feedback, we examined mRNA production in embryos homozygous for an early hb stop codon. Mutants and WT siblings display similar expression patterns until mid-cycle14 when WT embryos show reduced anterior expression and the loss of accumulated transcripts (Figure 5A,B). In mutants hb is maintained, resulting in continued high transcript density at late times (Figure 5A; Margolis et al., 1995). However, the absence of auto-inhibition did not alter the magnitude of variation in transcriptional activity compared to WT siblings, supporting the idea of intrinsice fluctuations and suggesting that zygotic Hb inhibits its own expression (Li et al., 2008; Perry et al., 2012; Treisman and Desplan, 1989).
Figure 5. Mutations in hb and runt impair timely repression of hb expression.
A,B: Cytoplasmic hb mRNA counts per standardized volume as a function of AP position of similarly staged zygotic hb mutants (A) and WT siblings (B). Smooth profiles and error bars obtained as in Figure 2A. C,D: runt mutants show delayed repression of hb transcription in nc14. C: Solid lines: runt mutants; dashed lines: WT siblings. Black: embryos of similar age (mid nc14) as judged by DAPI. Profile of an early nc14 WT embryo (red line) resembles mid-stage runt mutants, whereas a very late runt mutant (magenta line) is similar to earlier WT siblings. D: Transcriptional activity in anterior nuclei of the embryos shown in C. Boxplot depicts median, quartiles, and range of nascent transcription site activity for each embryo. runt mutants display consistently higher hb activity compared to WT siblings, leading to the inappropriately high hb transcript counts at late times as shown in C. See also Figure S5.
We also examined whether the removal of transcriptional inhibitors would allow the accumulation of larger numbers of RNAP. First, we examined hb expression in runt homozygous mutants, as Runt is implicated in gap gene regulation (Chen et al., 2012; Li et al., 2008;Tsai and Gergen, 1994). hb expression differs from WT starting at mid-nc14 when runt embryos maintain high levels of hb transcript in the anterior 20-40% (Figure 5C). hb profiles in runt mutants tend to resemble younger WT siblings, reaching WT levels near the end of nc14 (Tsai and Gergen, 1994), suggesting a delay in the reduction of anterior expression. In support, we find that runt mutants express slightly greater transcriptional activity than WT embryos with similar expression profiles (Figure 5D). However, at no time does expression noise differ noticeably from WT. Therefore, runt activity is required for the correct timing of hb downregulation, but plays no discernable role in determining fluctuations in transcriptional activity.
Finally, we tested the effect of manipulating the activity of maternally provided transcriptional modulators. We impaired the activity of the repressors Capicua and the co-repressor Groucho by mutation and observed that although hb expression boundaries were altered consistent with previous observations (e.g., Margolis et al., 1995), hb expression noise in the anterior was not affected. We also altered the genetic dosage of bcd to provide between 50% and 280% of WT activity, which shifted hb expression along the AP axis as expected (Liu et al., 2013) but had no effect on expression variability (Figure S5A-B). Thus our data are consistent with the view that the fluctuations in hb transcriptional activity arise from intrinsically stochastic processes, independent of variability in transcriptional modulators.
Gap genes share expression characteristics and are produced at equal rates
hb transcription fluctuates around a mean polymerase density that is about half the level that is physically obtainable. What sets the magnitude of the mean activity? Specific features of the hb promoter may limit activity. Alternatively, the maximum rate may not be specific to hb but shared among early expressed genes. If so, this would suggest that the maximum obtainable output, and its related noise level, are not set by any specific promoter-enhancer arrangement or any patterning cue, and instead are determined by general physical considerations.
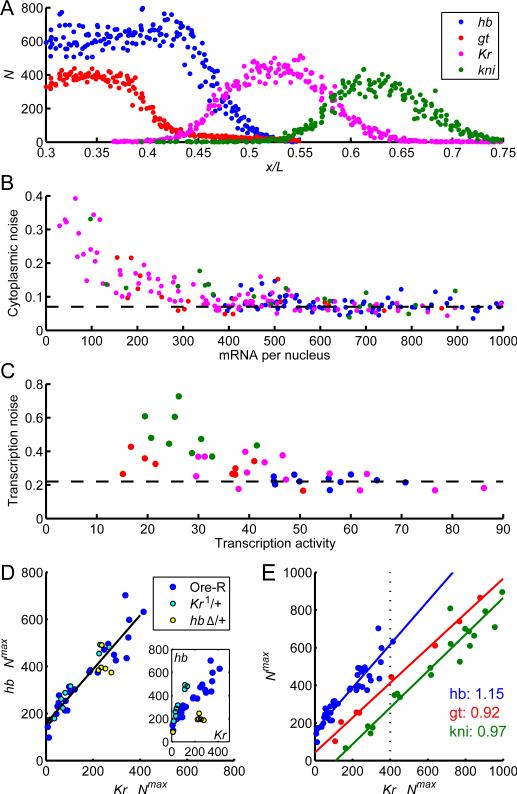
To discern between these possibilities, we measured the accumulation of transcripts of four gap genes primarily responsible for trunk patterning, hb, Krüppel (Kr), knirps (kni), and giant (gt) (Figure 6A). We found that all four genes display nearly uniform accumulation of cytoplasmic mRNAs accompanied by over 3-fold higher fluctuations in instantaneous transcriptional activity, essentially identical to the characteristics of hb (Figure 6B-C). These results strongly suggest that all early transcriptional events are subject to common constraints.
Figure 6. Universal properties of gap gene transcription.
A: Cytoplasmic profiles of four gap genes (mRNA concentration per standard volume AP position) measured in two embryos of the same age (second half of nuclear cycle 13; indicated by dotted line in panel E) processed with hb (blue) & gt (red) and with Kr (magenta) & kni (green) probes. B-E: Gap gene expression characteristics within each gene's region of maximum expression. B: Noise in cytoplasmic counts as a function of counts per nucleus (dashed line: 8%). C: Noise in transcriptional activity as a function of activity level (dashed line: 23%). D: mRNA expression (mean mRNA count per standard volume) in embryos from cycle 12 to early 14 co-stained with FISH probes against hb and Kr mRNA. Data from WT embryos (blue) coincides with those from embryos deficient for one copy of hb (hbΔ/+; yellow) or Kr (Kr1/+; cyan) when the concentration of the respective mRNA is rescaled by a factor of 2. Inset: Raw data (not rescaled). E: Levels of hb (blue), kni (green) and gt (red) versus Kr. Data from WT, hbΔ/+ and Kr1/+ embryos is combined by rescaling as in D (also see Figure S5C-D). hb data as in A; kni and gt were assessed in cycles 13 and 14. Slopes of fit lines indicate ratio of absolute production rates; all are within 15% of unity.
To closely compare transcript accumulation between genes, we took advantage of the observation that for Kr and kni cytoplasmic mRNA density increases monotonically between early nc12 and well into interphase 14 (Figure S5C-D), in contrast to hb, which ceases accumulating broadly in early nc14 (Figure 2A). We used counts of Kr or kni as a proxy for time, reducing staging uncertainty when comparing different genes. We performed dual color labeling with probes against pairs of gap genes and report cytoplasmic counts of hb, gt and kni mRNA as a function of Kr (Figure 6).
Figure 6C displays the expression of hb and Kr in WT (blue), hb hemizygous (green), and Kr1 heterozygous (red) embryos during nc12 and nc13. Counts in deficiency or mutant heterozygous embryos deviate considerably from WT (Figure 6C inset), but after multiplying the counts of the deficient gene by 2, all points collapse onto the same line (Figure 6C), showing the absence of compensatory mechanisms. We observed the same behavior for gt-Kr and kni-Kr expression pairs. Unexpectedly, for the three sets of gene pairs, linear fitting yields lines with slopes between 0.9 and 1.15 (Figure 6E); that is, in their regions of maximal expression, the four genes are produced at nearly identical rates. The differences between absolute levels within these regions (Figure 6E) reflect differences in the timing of when kni, Kr, and gt transcripts begin to accumulate, and for hb the perdurance of maternal mRNA. These maximal production rates are independent of Bcd activator concentration: although genetically altering the dosage of bcd between 50% and 280% of WT shifts the expression domains along the AP axis (Liu et al., 2013), this manipulation does not alter either accumulation rate or precision (Figure S5A-B).
These results are consistent with the idea that these transcripts are produced at the same rate. This strong similarity occurs despite the fact that these genes are expressed maximally in non-overlapping spatial domains. The transcriptional activity in the maximally expressed domain and the magnitude of transcriptional noise are therefore set independently of the inputs that determine spatially patterned expression, which are specific to each gene. By focusing on the regions of maximal expression, we could isolate the features of transcription that appear to be universal across gap genes and, furthermore, match the noise characteristics previously observed in bacteria and cell cultures. This suggests that the failure to sustain a maximal loading of RNAPs on the gene and the intrinsic noise of 45% are a common feature of transcriptional activation across diverse biological contexts.
DISCUSSION
The fundamental question of how embryos achieve precise control over the earliest transcriptional events is largely unanswered. General models of embryogenesis posit that early developmental events are dominated by molecular noise and imprecision in the control of gene expression (Arias and Hayward, 2006; Manu et al., 2009; Rao et al., 2002), a view consistent with observations of wide fluctuations in transcriptional activity in the majority of systems assayed quantitatively (Li and Xie, 2011). Indeed, the finding in fly embryos that instantaneous transcriptional activity varies between loci by nearly 50% suggests that early transcription in fly embryos obeys rules of stochastic activity observed in other systems where output can vary by a similar degree, and is often much higher (Munsky et al., 2012). Stochastic variation appears to be a universal feature of transcription from single cell organisms grown in culture (Raj et al., 2006; So et al., 2011; Stewart-Ornstein et al., 2012) and for cells in certain developmental settings (Pare et al., 2009; Raj et al., 2010; Saffer et al., 2011). Drosophila embryos display an extraordinary degree of precision in the rapid establishment of distinct gene expression programs; nevertheless, even this system cannot circumvent stochastic transcriptional activity. This finding supports the idea that control systems that might overcome stochastic molecular activity are difficult to design, costly to implement, and rarely if ever found (Lestas et al., 2010).
It is possible that cultured yeast and bacteria exist at sufficiently high densities such that precision is not required to ensure survival of a large fraction of the population; indeed, in several cases stochastic expression serves to maximize survival options (Balaban et al., 2004; Maamar et al., 2007; Mirouze et al., 2011; Nachman et al., 2007). In addition, for prokaryotes and haploid yeast and unlike early embryos, the presence of a single genomic locus precludes the possibility of noise filtering by averaging over independent loci. Alternatively, precision might be required to ensure the survival of single cell organisms when grown in their endogenous conditions, which may be difficult to study in a laboratory setting. However, in stark contrast to single cell organisms, many developing embryos possess large fields of cells that must coordinately undertake rapidly determined fate decisions, thus mandating high precision and low expression noise such that the appropriate gene expression programs are induced at the correct time and place. If in Drosophila patterning mRNAs accumulate in a precise manner minimizing expression noise, as we have shown here, how then can the embryo achieve this near uniformity?
Spatiotemporal averaging reconciles highly variant transcription with precise accumulation and recovers the input-output relationship
Large differences in nascent transcript content sustained over sufficiently long periods would inevitably result in unequal transcript production, inconsistent with nearly homogeneous cytoplasmic transcript concentration. As noted above, the long lifetime of hb transcripts allows substantial accumulation during the course of the syncytial blastoderm stage. If instantaneous nascent mRNA content is not maintained continuously during interphase but instead fluctuates about the mean as a result of varying RNAP number, then cytoplasmic accumulation serves as a natural time-averaging filter. The impact of time averaging can be estimated in two independent ways. First, accumulation reflects temporal integration of a signal fluctuating with a characteristic time t0 , the time it takes a polymerase to traverse the 3.2 kbp of hb gene. RNAP processivity is estamtated at 1.1-1.4 kbp/min (Irvine et al., 1991; O'Brian and Lis, 1993; Shermoen and O'Farrell, 1991; Thummel et al., 1990), providing a rough estimate consistent with the observed noise filtering (see Supplementary Material). Alternatively, a more careful estimate (Figure S6B) yields a theoretical bound on the maximum efficiency of temporal averaging based on directly measured quantities, most crucially, the absolute number of engaged polymerases per nucleus. In the case of Kr mRNA profile, by the time the mean expression level reaches 800 molecules per nucleus, pure temporal averaging can at most reduce the expression noise to 8%. For these late embryos, however, our measurements show a consistently lower noise level of 6 ± 2%, suggesting an additional noise filtering mechanism.
Additional filtering can be readily provided by a small degree of spatial averaging by the exchange of mRNA between neighboring cytoplasmic volumes before the partitioning of the syncytial blastoderm. mRNA possesses some mobility: both hb and Kr transcript numbers increase at >10 μm from their sites of production in nuclei (Figures S3A, S6A). We note that cylindrical summation volumes with a diameter of one internuclear distance contact each other, so that the mRNA traveling distance required to observe spatial averaging is very small. A straightforward estimate (see Supplementary Material) shows that attributing the excess noise filtering to spatial averaging requires only 4% of produced transcripts to be exchanged between neighboring volumes. Thus, even a limited degree of spatial averaging is completely sufficient to account for the appearance of low variation in cytoplasmic accumulation from stochastic transcription.
These results have several implications. First, we note that the observed variation in cytoplasmic concentration is likely to contain error introduced by our measurement, and the variation we observe is nearly at the level of counting noise. This might indicate that spatial averaging predominates the filtering of transcription noise; however, the degree to which RNAP numbers fluctuate, and therefore the extent of purely temporal averaging, can only be determined with measurements in living embryos. Second, both spatial and temporal averaging mechanisms effectively relax a requirement for rapid, tightly controlled transcriptional responses to modulating inputs, thereby minimizing the need for additional layers of feedback or other control systems. In turn, the fluctuations of putative inputs must approach the same degree of noise as the intrinsic variability of the transcriptional process itself before any effect on gene expression is realized.
It is well established that the position of the Hb expression boundary depends on Bcd genetic dosage (Driever and Nusslein-Volhard, 1988), and that the concentration of Hb protein along the AP axis depends upon and is at least as precise as Bcd concentration (Gregor et al., 2007). Superficially, a highly stochastic transcriptional response would appear to render irrelevant any link between Bcd precision and Hb output: the 10% fluctuations observed for Bcd (Gregor et al., 2007) cannot directly impact a transcriptional process whose noise is >40%. However, each nucleus employs averaging mechanisms reducing the effect of intrinsic noise. Because of the central role played by time averaging, the relative importance of various noise sources depends on the time scale of observation. The immediate readout (on a scale of minutes) is dominated by intrinsic transcriptional noise which renders the precision of the input irrelevant. Averaging over active loci, over time, and between neighboring nuclei, the contribution of intrinsic noise becomes comparable with the input (or extrinsic) fluctuations. Thus on a long time scale, such as 3 hours of development, the precision of patterning decisions becomes limited by the extrinsic variability. A precise response to Bcd will be recovered as long as the mean activity of hb transcription is correlated with Bcd concentration, as proposed previously (Erdmann et al., 2009). This reconciles the apparently stochastic behavior of hb transcriptional activity with the precisely positioned boundary of expression (Porcher et al., 2010). In this manner, Hb activity and fluctuations in the boundary domain retain the previously observed dependence upon levels of and fluctuations in Bcd concentration.
Limitations to precise control of gene expression
We have shown that in the context of the gap genes, transcript output in the maximally expressing region does not equate with the actual maximum attainable density of RNAP loading. This maximum is attained by only a small fraction of nuclei at any given moment. Thus, it is currently unclear what determines the mean density of RNAP loading common to these four genes and what prohibits all nuclei from continuously activating the achievable maximum density. It is possible that the output rate is determined by a common, maternally supplied and spatially ubiquitous factor, for example Zelda or BSF (De Renzis et al., 2007; Liang et al., 2008), which calibrates the RNAP density of these four genes to give rise to the observed transcript output rate. Conversely, from the perspective of noise minimization, it would seem advantageous to design these genes’ promoter-enhancer architecture such as to obtain the actual maximum possible density, since higher output achieves greater noise reduction. However, a biological system likely cannot be readily engineered to produce transcripts at an arbitrarily rapid rate. Hence, it seems likely that mean RNAP loading, and hence transcript output, is strongly influenced by physical considerations, such as transcription factor binding, promoter melting, enhancer looping, and/or chromatin accessibility, that might be difficult to overcome by any simple means. Future work will determine the extent to which the mean polymerase density we observe for these four genes is a shared feature of early expression and the extent to which this rate can be manipulated according to cellular context. Moreover, further studies will be required to determine the timescale of fluctuations of an active locus during interphase: that is, whether variations arise largely from “bursts” of dense RNAP loading followed by quiescent periods, or conversely if the variations result from RNAP loading rates that are maintained continuously during interphase but differ dramatically between loci.
The formation of cellular membranes during the 14th interphase prohibits spatial exchange. It is thus improbable that spatial averaging mechanisms can play a role in ensuring precise responses at this time. Moreover, the transcripts of the pair-rule genes are directed to the apical surface where they accumulate (Davis and Ish-Horowicz, 1991). Differential cellular behavior, presaging the formation of morphological structures, emerges in the latter part of the 14th interphase. Thus it is likely that shortly after the onset of the 14th interphase, individual cells begin accumulating gene products required for their specific behaviors, thus rendering spatial averaging a hindrance to differentiation. It is therefore likely that temporal averaging and/or other mechanisms such as regulatory feedback ensure the precise distribution of patterning factors at this time. The degree of precision of transcriptional events over the course of the 14th interphase will be the subject of future investigations.
In summary this work demonstrates the potential of the Drosophila embryo as a system for quantitative evaluation of transcriptional regulation. Early fly embryos possess a number of advantages enabling such studies, including modulatory patterning inputs spanning large dynamic ranges, a complete list of essential gene network components, and an abundance of modern analysis tools. This presents the unique opportunity to uncover the biological and physical design features of a system evolutionarily constructed to achieve rapid and precise establishment of cell fates in an intact, physiologically meaningful context.
EXPERIMENTAL PROCEDURES
Fly strains and embryo manipulation
Oregon-R (Ore-R) embryos were used as WT. α-amanitin injection was performed as described (Edgar et al., 1986). RT-qPCR method is described in Supplemental Experimental Procedures. Embryos heterozygous for a deficiency spanning hb, or for the mRNA-null Kr1 mutation were collected from crosses of heterozygous adults (w1118; Df(3R)BSC477/TM6C and Kr1/SM6) and distinguished from homozygous mutant and homozygous balancer siblings by visual inspection of nascent mRNA sites. Homozygous hb12 mutants were obtained from crosses of heterozygous adults carrying a TM3 balancer marked with hb-lacZ reporter and identified by the absence of lacZ expression. runt mutant embryos were collected from crosses of a deficiency-bearing stock (Df(1)BSC645, w1118/Binsinscy) and mutants distinguished by the absence of runt expression using FISH probes. groMB36 germline clones were generated using the FLP-FRT recombinase system (Ajuria et al., 2011; Xu and Rubin, 1993). cic1 homozygous females were crossed to WT males to assay the effect of disabling capicua activity.
FISH and imaging
Embryos were fixed in 5% formaldehyde, 1x PBS for 20 min and devitelinated as described (Lecuyer et al., 2008), rinsed three times in 1x PBS, washed for 10 min in smFISH wash buffer (4x SSC, 35% formamide, 0.1% Tween-20), and hybridized for 16-24 hrs to probes conjugated to Atto 565 (Sigma 72464) or Atto 633 (Sigma 01464) and complementary to the reading frame of hb, Kr, kni, and gt and diluted to about 1 nM per probe in hybridization buffer (4x SSC, 35% formamide, 10% dextran sulfate, 2 ug/ml BSA (NEB B9001), 0.1 mg/ml salmon sperm DNA (Invitrogen 15632-011), 2 mM ribonucleoside vanadyl complex (NEB S1402S), 0.1% Tween-20). After 2 washes of 1 hr in wash buffer, embryos were rinsed twice briefly in 1x PBS, stained with DAPI and mounted in Vectashield (Vector Labs H-1000). For combined FISH and immunofluorescence, incubation in hb-Atto-565 probes was reduced to 2 hrs. Hb antibody staining was performed as described (Dubuis et al., 2013) with rat anti-Hb and goat anti-rat Alexa 647. Imaging was performed by laser scanning confocal microscopy on a Leica SP5 inverted microscope as described (Little et al., 2011) except that we used a 63x HCX PL APO CS 1.4 NA oil immersion objective with pixels of 76 × 76 nm and z-spacing of 250 nm. We typically obtained stacks representing 20 μm in total axial thickness starting at the embryo surface. Image analysis was performed as described (Little et al., 2011) with enhancements described in extended experimental procedures.
Supplementary Material
Article Highlights.
Absolute quantification of nascent and mature mRNA in Drosophila embryos
Nascent transcription is noisy whereas cytoplasmic mRNA levels are precise
All early expressed genes exhibit the same degree of transcriptional variability
Spatiotemporal averaging across multiple genomic loci generates precision
ETOC Paragraph.
Filtering Fluctuations for Fidelity of Form
Specification of gene expression programs during axis patterning in Drosophila embryos occurs in a strictly controlled manner. Using single molecule quantification, Little et al. show that despite the operation of inherently stochastic transcription machinery, the resulting output of completed transcripts for early patterning genes is highly precise. Precision arises from simple physical averaging mechanisms that minimize fluctuations of universally stochastic gene expression.
ACKNOWLEDGMENTS
We gratefully acknowledge our indebtedness to E. Wieschaus for insightful discussions and continuous support. We thank W. Bialek, S. Blythe, B. Chun, G. Deshpande, S. Di Talia, H. Garcia, S. Kyin, M. Petkova, R. Samanta, G. Schupbach, G. Tkacik, and the Bloomington Drosophila Stock Center. This work was supported by NIH Grants P50 GM071508 and R01 GM097275, and by Searle Scholar Award 10-SSP-274 to TG.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ajuria L, Nieva C, Winkler C, Kuo D, Samper N, Andreu MJ, Helman A, Gonzalez-Crespo S, Paroush Z, Courey AJ, Jimenez G. Capicua DNA-binding sites are general response elements for RTK signaling in Drosophila. Development. 2011;138:915–924. doi: 10.1242/dev.057729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias AM, Hayward P. Filtering transcriptional noise during development: concepts and mechanisms. Nat Rev Genet. 2006;7:34–44. doi: 10.1038/nrg1750. [DOI] [PubMed] [Google Scholar]
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- Bar-Even A, Paulsson J, Maheshri N, Carmi M, O'Shea E, Pilpel Y, Barkai N. Noise in protein expression scales with natural protein abundance. Nat Genet. 2006;38:636–643. doi: 10.1038/ng1807. [DOI] [PubMed] [Google Scholar]
- Boettiger AN, Levine M. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science. 2009;325:471–473. doi: 10.1126/science.1173976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Xu Z, Mei C, Yu D, Small S. A system of repressor gradients spatially organizes the boundaries of Bicoid-dependent target genes. Cell. 2012;149:618–629. doi: 10.1016/j.cell.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubb JR, Trcek T, Shenoy SM, Singer RH. Transcriptional pulsing of a developmental gene. Curr Biol. 2006;16:1018–1025. doi: 10.1016/j.cub.2006.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AA, Kalisky T, Mayo A, Geva-Zatorsky N, Danon T, Issaeva I, Kopito RB, Perzov N, Milo R, Sigal A, Alon U. Protein dynamics in individual human cells: experiment and theory. PLoS One. 2009;4:e4901. doi: 10.1371/journal.pone.0004901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis I, Ish-Horowicz D. Apical localization of pair-rule transcripts requires 3' sequences and limits protein diffusion in the Drosophila blastoderm embryo. Cell. 1991;67:927–940. doi: 10.1016/0092-8674(91)90366-7. [DOI] [PubMed] [Google Scholar]
- De Renzis S, Elemento O, Tavazoie S, Wieschaus EF. Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLoS Biol. 2007;5:e117. doi: 10.1371/journal.pbio.0050117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driever W, Nusslein-Volhard C. The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell. 1988;54:95–104. doi: 10.1016/0092-8674(88)90183-3. [DOI] [PubMed] [Google Scholar]
- Dubuis JO, Samanta R, Gregor T. Accurate measurements of dynamics and reproducibility in small genetic networks. Mol. Sys. Biol. 2013;9 doi: 10.1038/msb.2012.72. doi:10.1038/msb.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Weir MP, Schubiger G, Kornberg T. Repression and turnover pattern fushi tarazu RNA in the early Drosophila embryo. Cell. 47:747–754. doi: 10.1016/0092-8674(86)90517-9. [DOI] [PubMed] [Google Scholar]
- Erdmann T, Howard M, ten Wolde PR. Role of spatial averaging in the precision of gene expression patterns. Phys Rev Lett. 2009;103:258101. doi: 10.1103/PhysRevLett.103.258101. [DOI] [PubMed] [Google Scholar]
- Foe VE, Alberts BM. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J Cell Sci. 1983;61:31–70. doi: 10.1242/jcs.61.1.31. [DOI] [PubMed] [Google Scholar]
- Gandhi SJ, Zenklusen D, Lionnet T, Singer RH. Transcription of functionally related constitutive genes is not coordinated. Nat Struct Mol Biol. 2011;18:27–34. doi: 10.1038/nsmb.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergen JP, Coulter D, Wieschaus E. Segmental pattern and blastoderm cell identities. In: Gall JG, editor. Gametogenesis and the Early Embryo. Liss Inc; New York: 1986. pp. 195–220. [Google Scholar]
- Golding I, Paulsson J, Zawilski SM, Cox EC. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123:1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- Gregor T, Tank DW, Wieschaus EF, Bialek W. Probing the limits to positional information. Cell. 2007;130:153–164. doi: 10.1016/j.cell.2007.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway DM, Lopes FJ, da Fontoura Costa L, Travencolo BA, Golyandina N, Usevich K, Spirov AV. Gene expression noise in spatial patterning: hunchback promoter structure affects noise amplitude and distribution in Drosophila Segmentation. PLoS Comput Biol. 2011;7:e1001069. doi: 10.1371/journal.pcbi.1001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine KD, Helfand SL, Hogness DS. The large upstream control region of the Drosophila homeotic gene Ultrabithorax. Development. 1991;111:407–424. doi: 10.1242/dev.111.2.407. [DOI] [PubMed] [Google Scholar]
- Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet. 2005;6:451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- Kornberg TB, Tabata T. Segmentation of the Drosophila embryo. Curr Opin Genet Dev. 1993;3:585–594. doi: 10.1016/0959-437x(93)90094-6. [DOI] [PubMed] [Google Scholar]
- Lagha M, Bothma JP, Levine M. Mechanisms of transcriptional precision in animal development. Trends in genetics. 2012;28:409–416. doi: 10.1016/j.tig.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander AD. How cells know where they are. Science. 2013;339:923–927. doi: 10.1126/science.1224186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DR, Zenklusen D, Wu B, Chao JA, Singer RH. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science. 2011;332:475–478. doi: 10.1126/science.1202142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TT, Harlepp S, Guet CC, Dittmar K, Emonet T, Pan T, Cluzel P. Real-time RNA profiling within a single bacterium. Proc Natl Acad Sci U S A. 2005;102:9160–9164. doi: 10.1073/pnas.0503311102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuyer E, Parthasarathy N, Krause HM. Fluorescent in situ hybridization protocols in Drosophila embryos and tissues. Methods Mol Biol. 2008;420:289–302. doi: 10.1007/978-1-59745-583-1_18. [DOI] [PubMed] [Google Scholar]
- Lestas I, Vinnicombe G, Paulsson J. Fundamental limits on the suppression of molecular fluctuations. Nature. 2010;467:174–178. doi: 10.1038/nature09333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GW, Xie XS. Central dogma at the single-molecule level in living cells. Nature. 2011;475:308–315. doi: 10.1038/nature10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, MacArthur S, Bourgon R, Nix D, Pollard DA, Iyer VN, Hechmer A, Simirenko L, Stapleton M, Luengo Hendriks CL, et al. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 2008;6:e27. doi: 10.1371/journal.pbio.0060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456:400–403. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little SC, Tkacik G, Kneeland TB, Wieschaus EF, Gregor T. The formation of the Bicoid morphogen gradient requires protein movement from anteriorly localized mRNA. PLoS Biol. 2011;9:e1000596. doi: 10.1371/journal.pbio.1000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little SC, Wieschaus EF. Shifting patterns: merging molecules, morphogens, motility, and methodology. Dev Cell. 2011;21:2–4. doi: 10.1016/j.devcel.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Liu F, Morrison AH, Gregor T. Dynamic interpretation of maternal inputs by the Drosophila segmentation gene network. Proc Natl Acad Sci U S A. 2013;110:6724–6729. doi: 10.1073/pnas.1220912110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maamar H, Raj A, Dubnau D. Noise in gene expression determines cell fate in Bacillus subtilis. Science. 2007;317:526–529. doi: 10.1126/science.1140818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manu, Surkova S, Spirov AV, Gursky VV, Janssens H, Kim AR, Radulescu O, Vanario-Alonso CE, Sharp DH, Samsonova M, Reinitz J. Canalization of gene expression in the Drosophila blastoderm by gap gene cross regulation. PLoS Biol. 2009;7:e1000049. doi: 10.1371/journal.pbio.1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis JS, Borowsky ML, Steingrimsson E, Shim CW, Lengyel JA, Posakony JW. Posterior stripe expression of hunchback is driven from two promoters by a common enhancer element. Development. 1995;121:3067–3077. doi: 10.1242/dev.121.9.3067. [DOI] [PubMed] [Google Scholar]
- Mirouze N, Prepiak P, Dubnau D. Fluctuations in spo0A transcription control rare developmental transitions in Bacillus subtilis. PLoS Genet. 2011;7:e1002048. doi: 10.1371/journal.pgen.1002048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munsky B, Neuert G, van Oudenaarden A. Using gene expression noise to understand gene regulation. Science. 2012;336:183–187. doi: 10.1126/science.1216379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman I, Regev A, Ramanathan S. Dissecting timing variability in yeast meiosis. Cell. 2007;131:544–556. doi: 10.1016/j.cell.2007.09.044. [DOI] [PubMed] [Google Scholar]
- Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- O'Brien T, Lis JT. Rapid changes in Drosophila transcription after an instantaneous heat shock. Mol Cell Biol. 1993;13(6):3456–3463. doi: 10.1128/mcb.13.6.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pare A, Lemons D, Kosman D, Beaver W, Freund Y, McGinnis W. Visualization of Individual Scr mRNAs during Drosophila Embryogenesis Yields Evidence for Transcriptional Bursting. Curr Biol. 2009;19:2037–2042. doi: 10.1016/j.cub.2009.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry MW, Bothma JP, Luu RD, Levine M. Precision of hunchback expression in the Drosophila embryo. Curr Biol. 2012;22:2247–2252. doi: 10.1016/j.cub.2012.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcher A, Abu-Arish A, Huart S, Roelens B, Fradin C, Dostatni N. The time to measure positional information: maternal hunchback is required for the synchrony of the Bicoid transcriptional response at the onset of zygotic transcription. Development. 2010;137:2795–2804. doi: 10.1242/dev.051300. [DOI] [PubMed] [Google Scholar]
- Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Rifkin SA, Andersen E, van Oudenaarden A. Variability in gene expression underlies incomplete penetrance. Nature. 2010;463:913–918. doi: 10.1038/nature08781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao CV, Wolf DM, Arkin AP. Control, exploitation and tolerance of intracellular noise. Nature. 2002;420:231–237. doi: 10.1038/nature01258. [DOI] [PubMed] [Google Scholar]
- Raser JM, O'Shea EK. Noise in gene expression: origins, consequences, and control. Science. 2005;309:2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter M, Kirchner B, Muller H, Holzhauer C, Mann W, Pfaffl MW. Quantification noise in single cell experiments. Nucleic Acids Res. 2011;39:e124. doi: 10.1093/nar/gkr505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffer AM, Kim DH, van Oudenaarden A, Horvitz HR. The Caenorhabditis elegans synthetic multivulva genes prevent ras pathway activation by tightly repressing global ectopic expression of lin-3 EGF. PLoS Genet. 2011;7:e1002418. doi: 10.1371/journal.pgen.1002418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer F, Rivera-Pomar R, Hoch M, Jackle H. Gene regulation in the Drosophila embryo. Philos Trans R Soc Lond B Biol Sci. 1996;351:579–587. doi: 10.1098/rstb.1996.0057. [DOI] [PubMed] [Google Scholar]
- Shermoen AW, O'Farrell PH. Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell. 1991;67:303–310. doi: 10.1016/0092-8674(91)90182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal A, Milo R, Cohen A, Geva-Zatorsky N, Klein Y, Liron Y, Rosenfeld N, Danon T, Perzov N, Alon U. Variability and memory of protein levels in human cells. Nature. 2006;444:643–646. doi: 10.1038/nature05316. [DOI] [PubMed] [Google Scholar]
- So LH, Ghosh A, Zong C, Sepulveda LA, Segev R, Golding I. General properties of transcriptional time series in Escherichia coli. Nat Genet. 2011;43:554–560. doi: 10.1038/ng.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Ornstein J, Weissman JS, El-Samad H. Cellular noise regulons underlie fluctuations in Saccharomyces cerevisiae. Mol Cell. 2012;45:483–493. doi: 10.1016/j.molcel.2011.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Lehmann R, Schnurch H, Schuh R, Seifert E, Kienlin A, Jones K, Jackle H. Finger protein of novel structure encoded by hunchback, a second member of the gap class of Drosophila segmentation genes. Nature. 1987;327:383–389. [Google Scholar]
- Thummel CS, Burtis KC, Hogness DS. Spatial and temporal patterns of E74 transcription during Drosophila development. Cell. 1990;61:101–111. doi: 10.1016/0092-8674(90)90218-4. [DOI] [PubMed] [Google Scholar]
- Treisman J, Desplan C. The products of the Drosophila gap genes hunchback and Kruppel bind to the hunchback promoters. Nature. 1989;341:335–337. doi: 10.1038/341335a0. [DOI] [PubMed] [Google Scholar]
- Tsai C, Gergen JP. Gap gene properties of the pair-rule gene runt during Drosophila segmentation. Development. 1994;120:1671–1683. doi: 10.1242/dev.120.6.1671. [DOI] [PubMed] [Google Scholar]
- Wilkie GS, Shermoen AW, O'Farrell PH, Davis I. Transcribed genes are localized according to chromosomal position within polarized Drosophila embryonic nuclei. Curr Biol. 1999;9:1263–1266. doi: 10.1016/s0960-9822(99)80509-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Zenklusen D, Larson DR, Singer RH. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol. 2008;15:1263–1271. doi: 10.1038/nsmb.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.