Abstract
Even with optimal antiretroviral therapy, human immunodeficiency virus (HIV) persists in plasma, blood cells, and tissues. To develop new therapies, it is essential to know what cell types harbor residual HIV. We measured levels of HIV DNA, RNA, and RNA/DNA ratios in sorted subsets of CD4+ T cells (CCR7+, transitional memory, and effector memory) and non-CD4+ T leukocytes from blood, ileum, and rectum of 8 ART-suppressed HIV-positive subjects. Levels of HIV DNA/million cells in CCR7+ and effector memory cells were higher in the ileum than blood. When normalized by cell frequencies, most HIV DNA and RNA in the blood were found in CCR7+ cells, whereas in both gut sites, most HIV DNA and RNA were found in effector memory cells. HIV DNA and RNA were observed in non-CD4+ T leukocytes at low levels, particularly in gut tissues. Compared to the blood, the ileum had higher levels of HIV DNA and RNA in both CD4+ T cells and non-CD4+ T leukocytes, whereas the rectum had higher HIV DNA levels in both cell types but lower RNA levels in CD4+ T cells. Future studies should determine whether different mechanisms allow HIV to persist in these distinct reservoirs, and the degree to which different therapies can affect each reservoir.
Keywords: HIV, HIV-1, ART, persistence, reservoir, CD4+ T cell, gut, intestine, ileum, rectum
(See the editorial commentary by Henrich and Gandhi on pages 1189–93 and the major article by Jain et al 1202–11.)
Most patients infected with human immunodeficiency virus (HIV) achieving virologic suppression with antiretroviral therapy (ART) have low levels of HIV RNA in the plasma [1–4] and cell-associated HIV DNA and RNA in the blood [5–10], lymphoid tissues [5, 11–14], and gut [15–25]. The main obstacle to eradication of HIV is thought to be a population of latently infected resting memory CD4+ T cells [6, 7, 13, 26], though HIV has been observed in many other cell types [27] and on follicular dendritic cells [12, 13, 28]. Within CD4+ T cells, HIV DNA may be disproportionately found in certain subpopulations, including resting memory CD4+ T cells [29], activated CD4+ T cells [21], CCR6+ CD4+ T cells [30, 31], resting CD4+ TReg cells [32], and central or transitional memory CD4+ T cells [33].
In peripheral CD4+ T cells from ART-suppressed patients, central memory (TCM) and transitional memory (TTM) CD4+ T cells contribute the most to the total pool of HIV DNA in patients with high and low CD4 counts, respectively [33], whereas effector memory (TEM) cells contribute less [33]. However, little is known about the distribution of HIV DNA within CD4+ T memory subsets of the lymphoid tissues and gut, which likely represent the main anatomic reservoirs for HIV. Prior studies in ART-suppressed patients have shown that HIV DNA levels per CD4+ T cell are higher in the gut than the blood [21, 25]. Given the expected predominance of TEM cells in the gut and the observation that TEM cells in blood tend to have less HIV DNA [33], it seems paradoxical that HIV DNA would be concentrated in the gut relative to blood. We hypothesized that CD4+ T memory subsets in the gut are infected at different frequencies than the blood, or that HIV may exist in cells other than CD4+ T lymphocytes.
Another unanswered question is whether certain cell populations in either blood or gut, including TCM and TTM, are transcribing more HIV RNA. Most studies of HIV reservoirs have focused on HIV DNA, but the majority of proviral DNA is not transcribed, as revealed by RNA:DNA ratios <0.5 [25, 34]. HIV RNA-positive cells have been found in both blood and gut of ART-suppressed patients [9, 10, 15, 17, 20, 23, 25], but it is unclear whether these cells produce virus, and whether they reflect reactivation from latency, recent infection, or long-lived cells that chronically or intermittently transcribe HIV. These cells are of interest because transcription is a prerequisite for expression of HIV proteins and virions, which could be a large factor driving the persistent immune activation in patients on suppressive ART.
Given these unanswered questions, we sought to measure the levels of HIV DNA, HIV RNA, and RNA/DNA ratio (average transcription per infected cell) in subsets of CD4+ T cells and other leukocytes from blood, terminal ileum, and rectum. We hypothesized that cell frequencies, infection frequencies, and HIV transcription levels may vary by cell type and anatomic location.
METHODS
Subjects and Samples
Eight HIV-positive adults on ART were recruited from the San Francisco Veterans Affairs Medical Center (SFVAMC). Inclusion criteria included CD4+ T-cell counts ≥350 cells/µL and viral load <50 copy/mL for ≥6 months. The study was approved by the local Institutional Review Board of the University of California, San Francisco, and the SFVAMC. All participants gave informed consent. At entry, subjects underwent phlebotomy followed by colonoscopy with 15 biopsies from the terminal ileum (7 subjects) and 15 biopsies from the rectum (7 subjects). Peripheral blood mononuclear cells (PBMCs) were isolated using Ficoll [25]. Biopsies from each site were dissociated to a single cell suspension by collagenase digestion and mechanical disruption as described elsewhere [25].
Cell Sorting
Small aliquots of PBMCs, ileal cells, and rectal cells were set aside for phenotyping, and the remaining cells were sorted on a BD FACS ARIA II (BD Bioscience). Cells were stained as described elsewhere [25] with LIVE/DEAD violet stain (Invitrogen) and the fluorescently conjugated monoclonal antibodies CD45-Pacific Blue, CD3-Alexa 700, CCR7-APC, CD45RO-FITC (all BD Bioscience), CD4-ECD (Beckman Coulter), CD8-QDot605 (Invitrogen), and CD27-Alexa750 (ebioscience) to differentiate CD4+ T-cell maturation subsets (Supplementary Table 1). Cells from PBMCs, ileum, and rectum were sequentially gated to define singlets, CD45+ leukocytes, and live cells. CD4+ T cells were then defined by coexpression of CD3 and CD4 and lack of CD8. The following populations were sorted: (1) leukocytes other than CD4+ T cells (non-CD4+ T leukocytes; includes CD3+CD4−,CD3−CD4−,CD3-CD4+); (2) CCR7+ “lymphoid homing” CD4+ T cells (TR7+); (3) CCR7−CD45RO+CD27High transitional memory CD4+ T cells (TTM); and (4) CCR7−CD45RO+CD27Low effector memory CD4+ T cells (TEM). Table 1 shows the gating criteria and the expected cell types in each population. The CCR7+ population includes both naive and TCM CD4+ T cells, which were grouped together because of the extremely low frequency of CCR7+ cells in the gut. The 4 sorted populations from each site were then pelleted and flash frozen for subsequent nucleic acid extraction.
Table 1.
Sorting Scheme
| Sorted population | Criteria | Includes |
|---|---|---|
| Leukocytes other than CD4+ T cells (non-CD4+ T leukocytes) | CD45+ but excluding CD3+ CD4+ (ie, CD45+CD3−CD4−, CD45+CD3−CD4+, CD45+CD3+CD4−) | CD8+ T cells CD4−CD8− T cells B cells Granulocytes Monocytes Macrophages Dendritic cells NK cells Hematopoietic stem cells |
| CCR7+ “lymphoid homing” CD4+ T cells (TR7+) | CD45+CD3+CD4+CD8−CCR7+ | Naive CD4+ T cells Central memory CD4+ T cells “Other memory” CD4+ T cells |
| Transitional Memory CD4+ T cells (TTM) | CD45+CD3+CD4+CD8−CD45RO+CCR7−CD27High | Transitional memory CD4+ T cells |
| Effector memory CD4+ T cells (TEM) | CD45+CD3+CD4+CD8−CD45RO+CCR7−CD27Low | Effector memory CD4+ T cells |
HIV DNA
Total DNA and RNA were extracted from each cell population using Trireagent (Molecular Research Center), with DNA isolated using the alternative protocol with back extraction buffer. The RNA was treated with DNase and further purified on Qiagen RNEasy Mini or Micro Columns. Total DNA (up to 40% of the elution or a maximum of 100 000 cell equivalents) was tested for HIV DNA in duplicate using a modification of a published real-time polymerase chain reaction (PCR) assay that uses primers and a probe from the LTR region and can detect a single copy of HIV in brain tissue [35]. Primers were F522-43 (5′GCCTCAATAAAGCTTGCCTTGA3′; HXB2 522-543) and R626-43 (5′GGGCGCCACTGCTAGAGA3′; 626-643). The probe, 5′CCAGAGTCACACAACAGACGGGCACA3′, was dual-labeled with 6-FAM(5′) and Black Hole Quencher. Reaction volume was 50 µL with 25 µL of 2× Gene Expression Master Mix (Applied Biosystems), 10 pmol of each primer and probe. Cycling conditions were 50C for 2 minutes, 95C for 10 minutes, then cycles of 95C for 15 seconds and 59C for 1 minute. External standards were prepared from DNA extracted from known numbers of 8E5 cells (NIH AIDS Reagent Program), each of which contains one integrated HIV genome per cell.
HIV RNA
RNA (up to 36% or 100 000 cell equivalents) was assayed for total processive HIV RNA transcripts in duplicate using primers and probe from the LTR region (as above). Reaction volume was 50 µL with 25 µL of 2× One Step RNA-to-Ct mix (Applied Biosystems), 10 pmol of each primer and probe, and 1.25 µL of 40× RT (Applied Biosystems). Cycling conditions were 48C for 20 minutes, 95C for 5 minutes, then 60 cycles of 95C for 15 seconds and 59C for 1 minute. External standards of genomic HIV RNA were prepared by extracting the RNA from lab stocks of NL4-3 virions and then quantifying the RNA via replicate measurements using the Abbot Real Time assay.
Normalization of HIV Copies to Cell Numbers
HIV levels were normalized to cell equivalents using 2 methods: first, using the cell counts from the sorts and the known elution and PCR input volumes; and second, by assaying the extracted RNA and DNA for the housekeeping genes Glyceraldehyde Phosphate Dehydrogenase (GAPDH) and Telomerase Reverse Transcriptase (TERT), respectively. In total, 2 µL of sample nucleic acid was tested in duplicate for GAPDH or TERT in a reaction with 2.5 µL of 20× primer/probe mix (Applied Biosystems) using the PCR conditions described above and external standards of RNA or DNA extracted from known numbers of peripheral CD4+ T cells. Results were expressed as HIV copies/106 cells of each sorted cell type. To calculate an HIV burden that accounts for the frequency of each cell type as well as the HIV levels within each cell type, for each site, we calculate the fractional contribution of each cell type to the total measured HIV DNA or RNA in 106 leukocytes.
Statistics
Cell frequencies and HIV levels in different cell populations and anatomic sites were compared across patients using the Wilcoxon signed rank test with 2-tailed P values as calculated on GraphPad Prism 5.0.
RESULTS
Clinical Characteristics
The 8 study subjects (Table 2) were all men, with median age of 57. Other median data include HIV duration 22 years, ART duration 12.5 years, CD4 nadir 224, CD4 count 592, and CD4% of 29.5%.
Table 2.
Clinical Characteristics
| Subject | Age | Sex | Years HIV | CD4 Nadir | Time on ART, yr | Last suppression, yr | CD4/CD4% | Viral load | ART regimen |
|---|---|---|---|---|---|---|---|---|---|
| A648 | 64 | M | 26 | 0 | 12 | 3.6 | 492/27% | <20 detected | 3TC/AZT/TDF/r/LPV |
| A649 | 50 | M | 12 | 218 | 12 | 4.7 | 762/35.5% | <20 ND | 3TC/TDF/EFV |
| A650 | 56 | M | 22 | 155 | 16 | 5.0 | 548/28% | <20 detected | 3TC/ABC/r/LPV |
| A652 | 58 | M | 26 | 1 | 14 | 4.5 | 699/31.5% | <20 ND | FTC/TDF/EFV |
| A653 | 51 | M | 23 | 230 | 14 | 2.0 | 1032/29% | 41 | FTC/TDF/r/ATV |
| A654 | 57 | M | 14 | 430 | 13 | 4.0 | 636/39% | <20 detected | FTC/TDF/r/ATV |
| A727 | 44 | M | 7–16 | 303 | 1.33 | 1.1 | 547/29% | <20 ND | FTC/TDF/EFV |
| A728 | 57 | M | 22 | 264 | 3 | 2.9 | 434/30% | <20 ND | FTC/TDF/EFV |
| Median | 56.5 | 22 | 224 | 12.5 | 3.8 | 592/29.5% | <20 |
Abbreviations: ABC, abacavir; ART, antiretroviral therapy; ATV, atazanavir; AZT, azidothymidine; EFV, efavirenz; FTC, emtricitabine; HIV, human immunodeficiency virus; LPV, lopinavir; M, male; ND, not detected; r, ritonavir (boosting); TDF, tenofovir; 3TC, lamivudine.
CD4+ T Cell Maturation Subset Frequencies
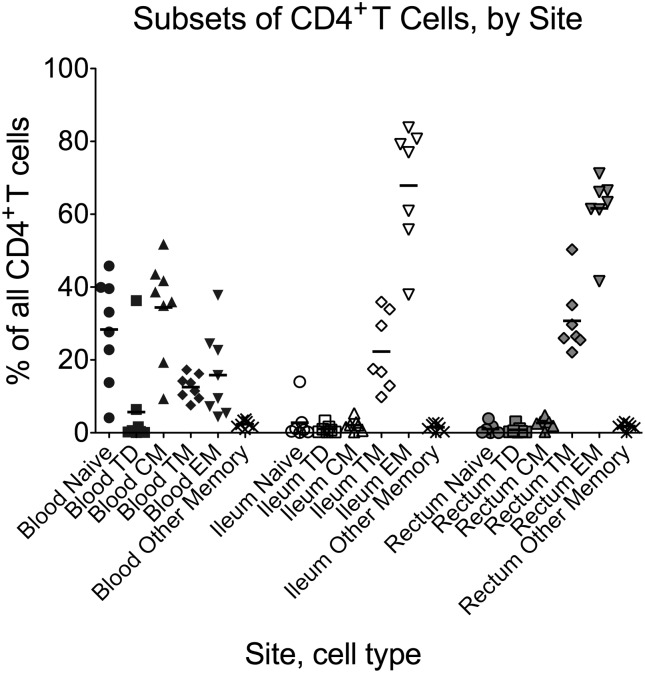
In the blood of HIV-positive subjects, the largest populations of CD4+ T cells (Figure 1) were TCM (P = .016, P = .023 for comparison to TTM, terminally differentiated [TTD]) and naive (P = .023 for comparison to TTM). In contrast, in both gut sites, most CD4+ T cells were TEM (P = .016 for comparison to all other populations, except P = .031 for rectum TEM vs TTM) and TTM (P = .016 for comparison to naive, TCM, TTD). The percentage of TCM and naive cells was greater in blood compared to either gut site, whereas the percentage of TEM cells was greater in both gut sites compared to blood, and the percentage of TTM cells was higher in rectum than blood (P = .016 for all comparisons).
Figure 1.
Flow cytometry was used to determine the proportion of all CD4+ T cells from each site (blood, ileum, rectum) that were naive, terminally differentiated effector (Td), central memory (CM), transitional memory (TM), effector memory (EM), or “other memory” cells based on expression of CD3, CD4, CD45RO, CCR7, and CD27 (Table 1). Circles represent naive T cells; squares, terminally differentiated T cells; triangles, central memory T cells; diamonds, transitional memory T cells; inverted triangles, effector memory T cells; X's, all other memory T cells. Black, blood; white, terminal ileum; grey, rectum. Bars represent the means.
Normalization Using Cell Counts and Housekeeping Genes
There was a strong linear correlation between the HIV levels as measured by cell counts and by housekeeping genes (Supplementary Figure 1A and 1B), with a slope of close to unity.
Cell-Associated HIV DNA and RNA in Each Cell Type
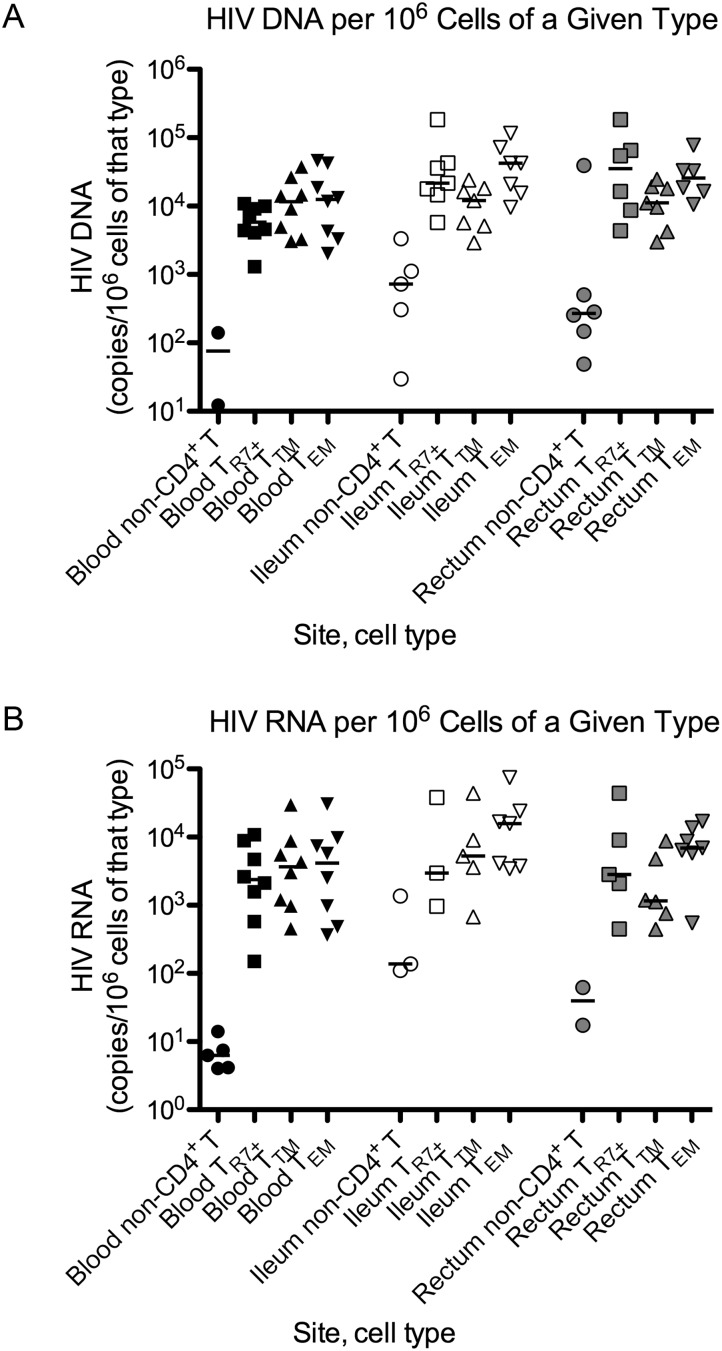
Despite low yields of sorted TR7+ and TTM CD4+ T cells from the gut, HIV DNA was detectable in all TEM, TTM, and TR7+ samples except for one rectal TR7+ sample with the lowest cell count (Supplementary Table 2). In contrast, despite generally high cell counts and TERT levels from non-CD4+ T leukocytes, HIV DNA was detected in the peripheral non-CD4+ T leukocytes in only 2 of 8 subjects, compared to 5 of 7 for ileal non-CD4+ T leukocytes and 6 of 7 for rectal non-CD4+ T leukocytes.
In the blood, TTM CD4+ T cells had more HIV DNA/million cells than TR7+ cells (P = .023; Figure 2A). In the ileum, HIV DNA levels were higher in TEM cells than in TTM cells (P = .016). HIV DNA levels in ileal TR7+ and TEM cells were higher than in the corresponding cell populations in the blood (P = .016, P = .016) and tended to be higher in rectal TR7+ cells compared to TR7+ cells in the blood (P = .063).
Figure 2.
A and B, HIV DNA (A) and HIV RNA (B) in sorted cells. Cells from blood (black), ileum (white), and rectum (gray) were sorted into leukocytes other than CD4+ T cells (non-CD4+ T; circles) and 3 populations of CD4+ T cells: (1) CCR7+ (TR7+ ; squares), which includes naive, central memory, and “other memory”; (2) transitional memory (TTM; triangles); and (3) effector memory (TEM ; inverted triangles). HIV levels are expressed as copies/106 cells and graphed on a log scale. Only samples with detectable HIV levels are shown. Bars represent the medians.
HIV RNA was detected in non-CD4+ T leukocytes from the blood of 5 of 8 subjects, compared to 3 of 7 for the ileum and 2 of 7 for the rectum. In CD4+ T cells, HIV RNA was always detected in blood and in gut TEM cells but was sometimes not detected in gut TR7+ or TTM samples with lower cell counts. Differences in levels of HIV RNA/million cells tended to mirror those found for HIV DNA but did not reach statistical significance (Figure 2B).
Contribution to the Total Pool of HIV DNA and RNA in Leukocytes (WBC)
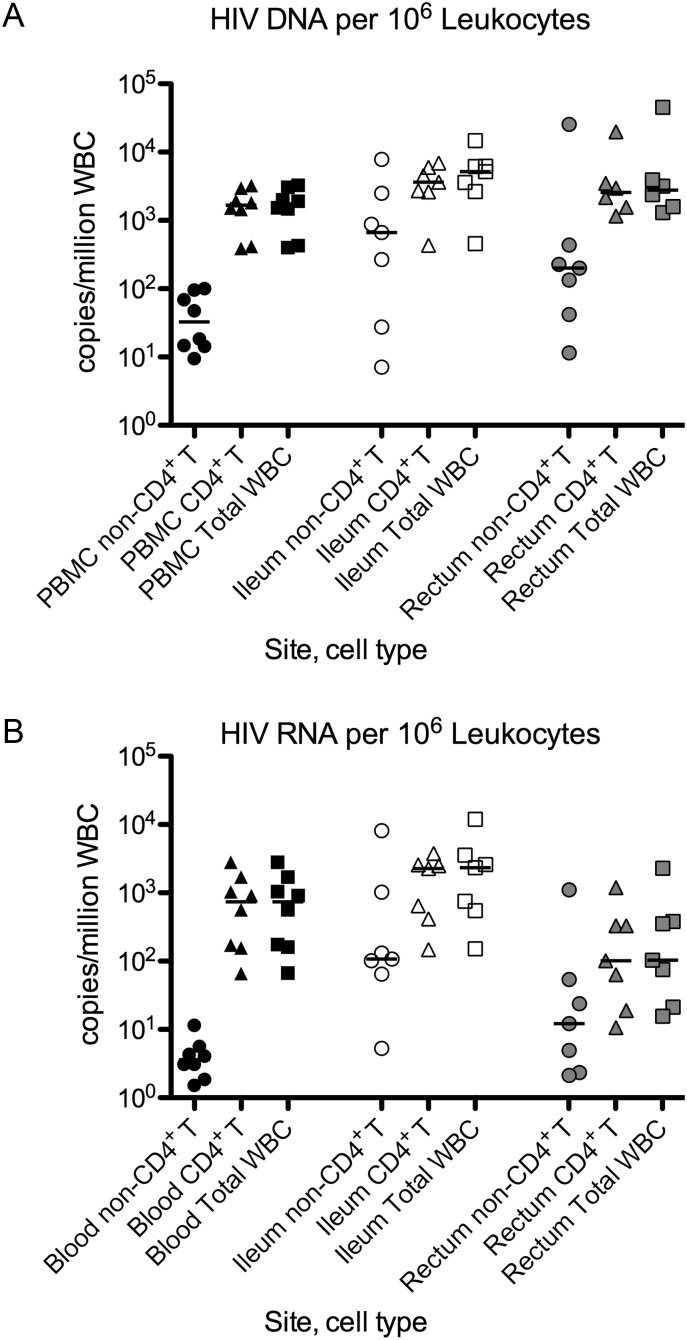
Using the HIV levels within each cell type and the frequency of each cell type (percent of live singlet CD45+ cells), we calculated the HIV content in each cell type in 106 white blood cells (WBC; Figure 3). Samples with undetectable HIV levels were assigned a value equal to the detection limit of the assay (assigning them a value of zero did not change the conclusions). The HIV levels in TR7+, TTM, and TEM CD4+ T cells were added to yield an estimate of the total HIV level in CD4+ T cells, which was added to the level in non-CD4+ T leukocytes to give an estimate of the total HIV level in WBC.
Figure 3.
A and B, HIV DNA (A) and RNA (B) with normalization to cell frequency (in 106 WBCs). For each site (blood, ileum, rectum), we used the HIV levels within each cell type (Figure 2) and the frequency of each cell type (% of live singlet CD45+ cells, as determined by flow cytometry) to calculated the HIV content within each cell type in 106 WBCs. Samples that had undetectable HIV levels were assigned a value equal to the detection limit of the assay. The HIV levels in TR7+, TTM, and TEM CD4+ T cells were added to yield an estimate of the total HIV level in CD4+ T cells (triangles), which was added to the level in non-CD4+ T leukocytes (circles) to give an estimate of the total HIV level in WBCs (squares). Data are shown on a log scale. Bars represent the medians. Abbreviations: HIV, human immunodeficiency virus; WBC, white blood cell.
In both blood and ileum, the CD4+ T-cell fraction had significantly more HIV DNA than the non-CD4+ T leukocyte fraction (P = .0078, P = .047), whereas no significant difference was observed in the rectum (P = .44; Figure 3A). Compared to the blood, both the ileum and rectum had higher HIV DNA levels per 106 WBC in the non-CD4+ T leukocyte fraction (0.031, 0.016) and the total CD4+ T-cell fraction (P = .016, .031), resulting in higher total levels in WBC (P = .016, .031).
Similar trends were observed for HIV RNA levels. The CD4+ T-cell fraction had significantly more HIV RNA than the non-CD4+ T leukocyte fraction in both blood and ileum (P = .0078, P = .047) but not rectum (Figure 3B). Compared to blood, the ileum had higher HIV RNA levels in the non-CD4+ T leukocyte fraction, total CD4+ T-cell fraction, and total WBC (P = .031, .016, .016), whereas the rectum had higher HIV RNA levels in the non-CD4+ T leukocyte fraction but lower HIV RNA levels in CD4+ T cells and lower total HIV RNA (P = .016, .031, .031).
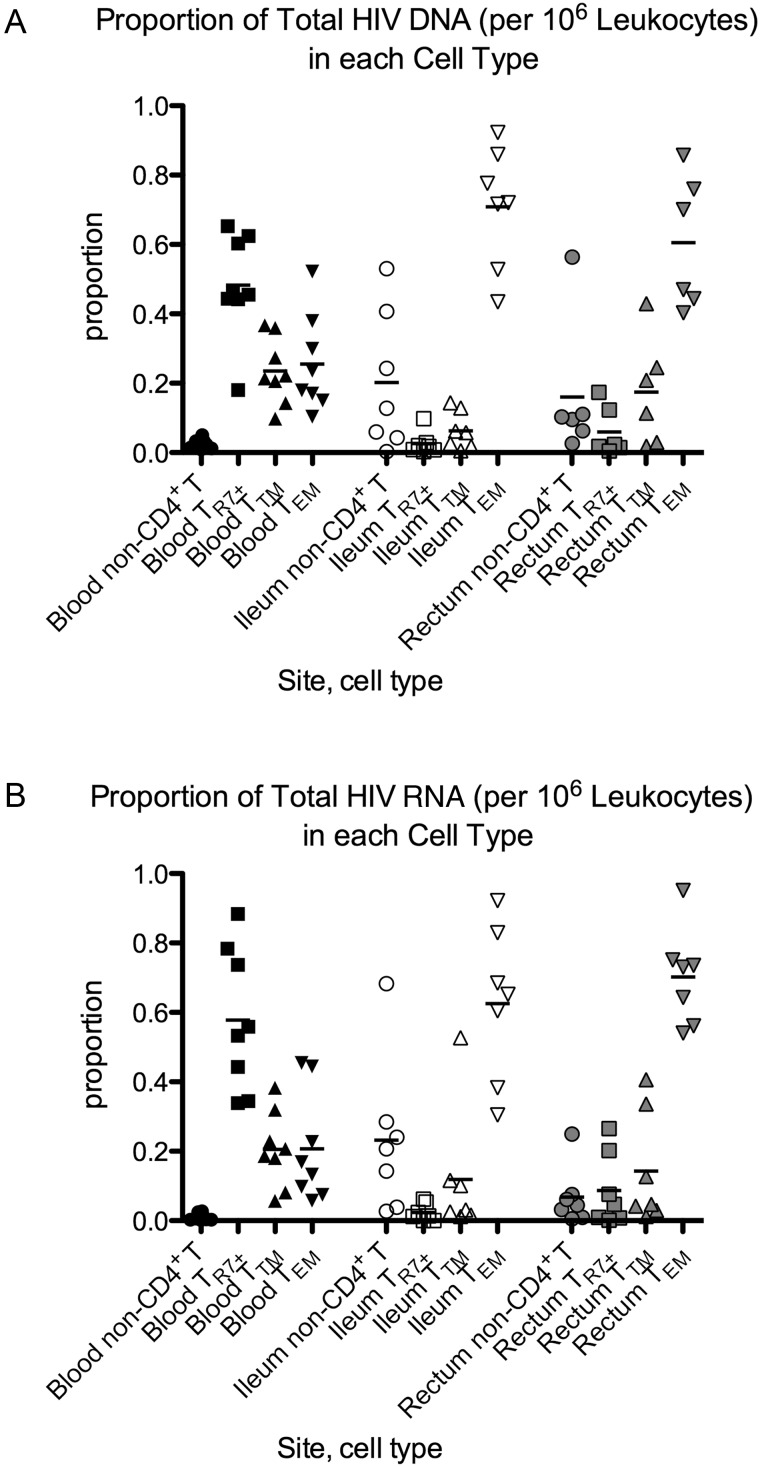
Proportion of HIV in Each Cell Type
At each site, HIV levels in each cell type per 106 WBC were divided by the total HIV level in 106 WBC to yield the proportion of HIV in that cell type (Figure 4). Samples that had undetectable HIV levels were assigned a value equal to the detection limit of the assay (assigning them a value of zero did not change the conclusions). In the blood, infected TR7+ cells comprised a larger proportion of total HIV DNA than did TTM cells (P = .039) despite TTM cells having higher HIV DNA/million sorted cells (Figure 4A). In the ileum and rectum, TEM cells comprised the largest proportion of HIV DNA (P = .016 for comparison of ileal TEM to non-CD4+ T leukocytes, TTM, or TR7+; P = .031 for comparison of rectal TEM to TTM or TR7+). The proportion of HIV DNA in TR7+ cells was larger in blood than in either gut site (P = .016, P = .031), whereas the proportion of HIV DNA in TEM cells was larger in both gut sites compared to blood (P = .016, P = .031). Rectal and ileal non-CD4+ T leukocytes tended to comprise a larger proportion of HIV DNA than corresponding cells in the blood (P = .031, P = .078).
Figure 4.
A and B, Proportion of HIV DNA (A) and RNA (B) contributed by each cell type. For each cell type, the HIV levels in 106 WBCs were divided by the total HIV level in 106 WBCs (the sum of the HIV levels in the 4 cell types at that site) to yield the proportion of HIV in that cell type. Samples that had undetectable HIV levels were assigned a maximum value equal to the detection limit of the assay. Symbols are used as in Figure 2. Bars represent the means. Abbreviations: HIV, human immunodeficiency virus; WBC, white blood cell.
Similar patterns were observed for the distribution of HIV RNA (Figure 4B). In blood, TR7+ cells comprised a larger proportion of HIV RNA than TTM (P = .0078) or TEM cells (P = .039) and non-CD4+ T leukocytes comprised the smallest proportion of HIV RNA (P = .0078 for all comparisons). In the ileum and rectum, the proportion of HIV RNA was highest in the TEM cells (P = .016, .031, .016 for comparison of ileal TEM to TR7+, TTM, and non-CD4+ T; P = .016 for comparison or rectal TEM to TR7+, TTM, or non-CD4+ T leukocytes). The proportion of HIV RNA in TR7+ CD4+ T cells was higher in blood than in either gut site (P = .016, .016), whereas the proportion of HIV RNA in TEM was higher in both gut sites compared to blood (P = .016, .016). Ileal and rectal non-CD4+ T leukocytes tended to comprise a higher proportion of HIV RNA than corresponding cells in the blood (P = .016, P = .078).
RNA/DNA Ratios in Each Cell Type
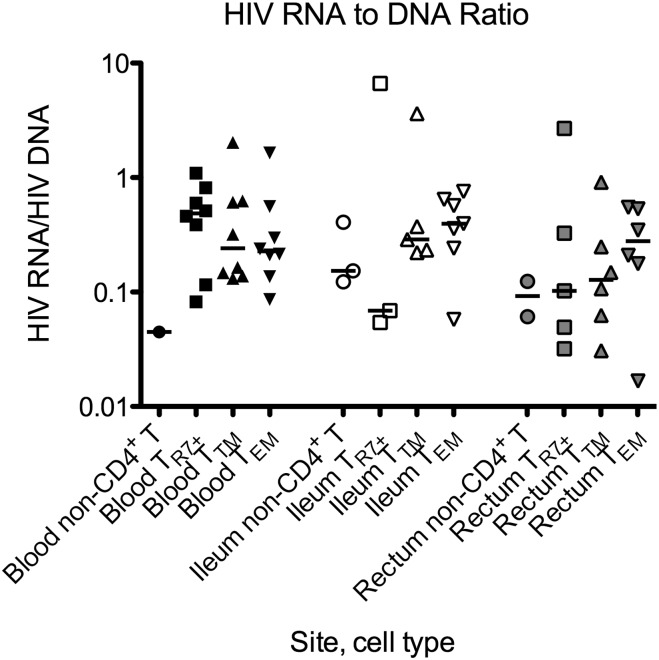
The RNA/DNA ratio (average transcription per infected cell) was calculated for all cell samples in which both HIV RNA and DNA were detectable. In the blood, the median RNA/DNA ratio was highest in TR7+ cells, whereas in both gut sites, the median ratio was highest in TEM cells (Figure 5). However, owing in part to the small numbers of calculable ratios, these differences did not reach statistical significance.
Figure 5.
Average transcription per infected cell. For all cell samples in which both HIV RNA and DNA were detectable, the HIV RNA per 106 cells (Fig. 2B) was divided by the HIV DNA per 106 cells (Fig. 2A) to determine the RNA/DNA ratio (average transcription per infected cell). Results are graphed on a log scale. Symbols are used as in Figure 2. Bars represent the medians. Abbreviation: HIV, human immunodeficiency virus.
DISCUSSION
In this study of ART-treated HIV-positive patients, HIV infection frequencies and HIV transcription levels differed by cell type and anatomic location. Most CD4+ T cells in the blood were central memory and naive CD4+ T cells, in agreement with prior reports [33]. In contrast, in the ileum and rectum, the predominant CD4+ T-cell types were TEM and, to a lesser degree, TTM. Levels of HIV DNA/million cells were higher in TR7+ and TEM cells in the ileum than in the blood, suggesting some compartmentalization between these 2 sites. When normalized by cell frequencies, most HIV DNA and RNA in the blood were found in TR7+ cells, whereas in both gut sites, most HIV DNA and RNA were found in TEM cells. These data indicate that insights provided by blood-based studies may not be indicative of what is happening in the tissues, and that the local host environment in the tissues may have dramatic effects in shaping the size and distribution of the reservoir during ART.
The observation that most HIV in the gut was in TEM cells was surprising and suggests that the factors contributing to HIV persistence in tissues differ from those that contribute to the distribution of HIV DNA in circulating cells in blood. One possibility is that tissue-resident TEM cells persist for long periods. In HIV-uninfected subjects, the half-life of circulating CD4+ TEM cells is shorter than that of TCM cells [36], but it is unclear whether these half-lives account for migration of TEM cells to the tissues, and whether TEM cells survive for longer in the tissues. CD4+ and CD8+ TEM cells persist for years after antigen exposure [37–39], suggesting a longer half-life or continuous renewal, either by homeostatic proliferation of TEM cells or by maturation from another cell type, such as TCM cells [39–41]. The enrichment of HIV in gut TEM cells could be due to maturation and proliferative expansion, or it could reflect an increased susceptibility to de novo infection.
In addition to the HIV in CD4+ T cells, we detected HIV DNA and some RNA in non-CD4+ T leukocytes, which may constitute a sizeable portion of the HIV in the gut but much less in blood. It is possible that the sorted non-CD4+ T leukocyte fraction was contaminated with low levels of CD4+ T cells, including CD4Low T cells. However, the average fold difference in per cell levels of HIV DNA in TEM cells and CD45+ non-CD4+ T leukocytes was 20-fold in the ileum and 4.66-fold in the rectum, which would require that the non-CD4+ T leukocyte fraction were contaminated with >5% TEM cells in the ileum and >20% in the rectum, whereas the predicted contamination with CD4+ T cells (based on similar sorts in other subjects) was <1%. The non-CD4+ T leukocyte population could include CD8+ T cells [29, 42], macrophages, dendritic cells, mast cells and progenitors, and natural killer (NK) cells, all of which have been shown to be reservoirs for HIV [27]. However, the PCR signals could also come from virions trapped on the cell surface or phagocytosis of virions, infected cells, or cellular debris. Future studies will be necessary to confirm whether HIV can infect these cell populations in the gut. The existence of HIV reservoirs in non-CD4+ T leukocytes would require additional consideration in developing viral eradication strategies.
In agreement with prior studies, HIV DNA levels were higher in the rectum and ileum compared to blood [25]. This difference reflects higher HIV DNA levels in both the CD4+ T-cell fraction and the non-CD4+ T leukocyte fraction. The greater HIV DNA in the CD4+ T-cell fraction likely reflects differences in the types of CD4+ T cells in the gut (more TEM cells), as well as differences in the frequency of infection of each cell type. The disproportionate frequency of infected ileal TR7+ and TEM could reflect preferential homing of infected cells from the blood, higher rates of de novo infection in the ileum, or increased survival or proliferation of infected cells in the ileum. The ileum also had higher HIV RNA levels than the blood, reflecting higher levels in both non-CD4+ T leukocytes and CD4+ T cells, whereas the rectum had lower total HIV RNA levels than the blood, reflecting lower HIV levels in CD4+ T cells. Compared to the rectum, the ileum is likely to have higher concentrations of inductive sites, with a higher density of CD4+ T cells. The ileal environment may be more likely to cause T-cell activation (resulting in more HIV RNA production) and more likely to support active replication (given that cells are in closer proximity and more likely to be activated).
In both blood and gut, HIV RNA/DNA ratios were typically <0.5, in agreement with prior reports [25, 34]. Assuming that most of the HIV DNA is integrated, and that there is roughly one copy per infected cell, at least half of the infected cells are not transcribing any HIV RNA. Most CD4+ T cells in the gut display markers associated with T-cell activation [25], which may argue against a lack of host transcription factors associated with the resting state, and may suggest another mechanism of transcriptional control. At the same time, at least a fraction of the HIV DNA-positive cells in both blood and gut are spontaneously transcribing HIV RNA, and HIV RNA levels are higher in the ileum than blood. It is possible that this HIV RNA is translated into protein with the capacity to induce immune activation, whereas some HIV RNA could be packaged as virions, which could also contribute to limited, localized replication in the ileum [43] or to residual viremia.
Limitations of our study include the small number of subjects and the low yield of sorted gut TR7+ and TTM cells. Both factors may reduce the power to detect differences between cell types or anatomic sites, particularly for RNA and RNA/DNA ratios. As noted previously, CCR7 expression was rare in the gut, making it difficult to sort out pure TCM cells, and CD27 expression in the gut was less discrete than in the blood, blurring the distinction between gut TEM and TTM cells. Finally, normalization of HIV levels to cell numbers can be challenging, particularly for RNA from the gut, although we found a fairly good correlation between GAPDH and the cell counts from the sorts.
Despite these limitations, we found significant differences in the distribution of HIV-infected cell types in the blood, ileum, and rectum. Further study is needed to determine whether different mechanisms allow HIV to persist in the different cell types and sites, and whether levels of replication-competent virus and latently infected cells differ between gut and blood. Given the magnitude of the GI reservoir, new therapies must consider these fundamental differences between cellular reservoirs in the gut and blood.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. The authors thank the following people and institutions: (1) the study participants, (2) the staff at the GI Procedures Unit of the San Francisco VA, (3) the staff at the UCSF Core Immunology Lab, and (4) the PLUS study staff.
Financial support. This work was supported by the US Department of Veterans Affairs (1 IK2 CX000520-01 [to S. Y.], I01 BX000192 [to J. W.]); the UCSF-Gladstone Center for AIDS Research (CFAR) (P30-AI027763 [to S. Y.]); and the National Institute of Allergy and Infectious Diseases at the National Institutes of Health (R56AI091573 [J. W., S. Y., A. S., D. H.], K24 AI069994 [to S. D.], U19AI096109 [to S. D., J. W.]).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Schockmel GA, Yerly S, Perrin L. Detection of low HIV-1 RNA levels in plasma. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;14:179–83. doi: 10.1097/00042560-199702010-00013. [DOI] [PubMed] [Google Scholar]
- 2.Dornadula G, Zhang H, VanUitert B, et al. Residual HIV-1 RNA in blood plasma of patients taking suppressive highly active antiretroviral therapy. JAMA. 1999;282:1627–32. doi: 10.1001/jama.282.17.1627. [DOI] [PubMed] [Google Scholar]
- 3.Havlir DV, Bassett R, Levitan D, et al. Prevalence and predictive value of intermittent viremia with combination HIV therapy. JAMA. 2001;286:171–9. doi: 10.1001/jama.286.2.171. [DOI] [PubMed] [Google Scholar]
- 4.Maldarelli F, Palmer S, King MS, et al. ART suppresses plasma HIV-1 RNA to a stable set point predicted by pretherapy viremia. PLoS Pathog. 2007;3:e46. doi: 10.1371/journal.ppat.0030046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chun TW, Carruth L, Finzi D, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–8. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 6.Chun TW, Stuyver L, Mizell SB, et al. Presence of an inducible HIV latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A. 1997;94:13193–7. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finzi D, Hermankova M, Pierson T, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 8.Wong JK, Hezareh M, Gunthard HF, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–5. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 9.Fischer M, Huber W, Kallivroussis A, et al. Highly sensitive methods for quantitation of human immunodeficiency virus type 1 RNA from plasma, cells, and tissues. J Clin Microbiol. 1999;37:1260–4. doi: 10.1128/jcm.37.5.1260-1264.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer M, Wong JK, Russenberger D, et al. Residual cell-associated unspliced HIV-1 RNA in peripheral blood of patients on potent antiretroviral therapy represents intracellular transcripts. Antivir Ther. 2002;7:91–103. [PubMed] [Google Scholar]
- 11.Lafeuillade A, Poggi C, Profizi N, Tamalet C, Costes O. Human immunodeficiency virus type 1 kinetics in lymph nodes compared with plasma. J Infect Dis. 1996;174:404–7. doi: 10.1093/infdis/174.2.404. [DOI] [PubMed] [Google Scholar]
- 12.Cavert W, Notermans DW, Staskus K, et al. Kinetics of response in lymphoid tissues to antiretroviral therapy of HIV-1 infection. Science. 1997;276:960–4. doi: 10.1126/science.276.5314.960. [DOI] [PubMed] [Google Scholar]
- 13.Wong JK, Gunthard HF, Havlir DV, et al. Reduction of HIV-1 in blood and lymph nodes following potent antiretroviral therapy and the virologic correlates of treatment failure. Proc Natl Acad Sci U S A. 1997;94:12574–9. doi: 10.1073/pnas.94.23.12574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuster H, Opravil M, Ott P, et al. Treatment-induced decline of human immunodeficiency virus-1 p24 and HIV-1 RNA in lymphoid tissue of patients with early human immunodeficiency virus-1 infection. Am J Pathol. 2000;156:1973–86. doi: 10.1016/S0002-9440(10)65070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Talal AH, Monard S, Vesanen M, et al. Virologic and immunologic effect of antiretroviral therapy on HIV-1 in gut-associated lymphoid tissue. J Acquir Immune Defic Syndr. 2001;26:1–7. doi: 10.1097/00126334-200101010-00001. [DOI] [PubMed] [Google Scholar]
- 16.Guadalupe M, Reay E, Sankaran S, et al. Severe CD4+ T-cell depletion in gut lymphoid tissue during primary human immunodeficiency virus type 1 infection and substantial delay in restoration following highly active antiretroviral therapy. J Virol. 2003;77:11708–17. doi: 10.1128/JVI.77.21.11708-11717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anton PA, Mitsuyasu RT, Deeks SG, et al. Multiple measures of HIV burden in blood and tissue are correlated with each other but not with clinical parameters in aviremic subjects. AIDS. 2003;17:53–63. doi: 10.1097/00002030-200301030-00008. [DOI] [PubMed] [Google Scholar]
- 18.Brenchley JM, Schacker TW, Ruff LE, et al. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med. 2004;200:749–59. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poles MA, Boscardin WJ, Elliott J, et al. Lack of decay of HIV-1 in gut-associated lymphoid tissue reservoirs in maximally suppressed individuals. J Acquir Immune Defic Syndr. 2006;43:65–8. doi: 10.1097/01.qai.0000230524.71717.14. [DOI] [PubMed] [Google Scholar]
- 20.Belmonte L, Olmos M, Fanin A, et al. The intestinal mucosa as a reservoir of HIV-1 infection after successful HAART. AIDS. 2007;21:2106–8. doi: 10.1097/QAD.0b013e3282efb74b. [DOI] [PubMed] [Google Scholar]
- 21.Chun TW, Nickle DC, Justement JS, et al. Persistence of HIV in gut-associated lymphoid tissue despite long-term antiretroviral therapy. J Infect Dis. 2008;197:714–20. doi: 10.1086/527324. [DOI] [PubMed] [Google Scholar]
- 22.Avettand-Fenoel V, Prazuck T, Hocqueloux L, et al. HIV-DNA in rectal cells is well correlated with HIV-DNA in blood in different groups of patients, including long-term non-progressors. AIDS. 2008;22:1880–2. doi: 10.1097/QAD.0b013e32830fbdbc. [DOI] [PubMed] [Google Scholar]
- 23.Lafeuillade A, Cheret A, Hittinger G, et al. Rectal cell-associated HIV-1 RNA: a new marker ready for the clinic. HIV Clin Trials. 2009;10:324–7. doi: 10.1310/hct1005-324. [DOI] [PubMed] [Google Scholar]
- 24.Tincati C, Biasin M, Bandera A, et al. Early initiation of highly active antiretroviral therapy fails to reverse immunovirological abnormalities in gut-associated lymphoid tissue induced by acute HIV infection. Antiviral Ther. 2009;14:321–30. [PubMed] [Google Scholar]
- 25.Yukl SA, Gianella S, Sinclair E, et al. Differences in HIV burden and immune activation within the gut of HIV-positive patients receiving suppressive antiretroviral therapy. J Infect Dis. 2010;202:1553–61. doi: 10.1086/656722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chun TW, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF. In vivo fate of HIV-1 infected T cells: Quantitative analysis of the transition to stable latency. Nature Medicine. 1995;1:1284–90. doi: 10.1038/nm1295-1284. [DOI] [PubMed] [Google Scholar]
- 27.Alexaki A, Liu Y, Wigdahl B. Cellular reservoirs of HIV-1 and their role in viral persistence. Curr HIV Res. 2008;6:388–400. doi: 10.2174/157016208785861195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haase AT, Henry K, Zupancic M, et al. Quantitative image analysis of HIV-1 infection in lymphoid tissue. Science. 1996;274:985–9. doi: 10.1126/science.274.5289.985. [DOI] [PubMed] [Google Scholar]
- 29.Brenchley JM, Hill BJ, Ambrozak DR, et al. T-cell subsets that harbor human immunodeficiency virus (HIV) in vivo: implications for HIV pathogenesis. J Virol. 2004;78:1160–8. doi: 10.1128/JVI.78.3.1160-1168.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gosselin A, Monteiro P, Chomont N, et al. Peripheral blood CCR4+CCR6+ and CXCR3+CCR6+CD4+ T cells are highly permissive to HIV-1 infection. J Immun. 2010;184:1604–16. doi: 10.4049/jimmunol.0903058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monteiro P, Gosselin A, Wacleche VS, et al. Memory CCR6+CD4+ T cells are preferential targets for productive HIV type 1 infection regardless of their expression of integrin beta7. J Immun. 2011;186:4618–30. doi: 10.4049/jimmunol.1004151. [DOI] [PubMed] [Google Scholar]
- 32.Tran TA, de Goer de Herve MG, Hendel-Chavez H, et al. Resting regulatory CD4 T cells: A site of HIV persistence in patients on long-term effective antiretroviral therapy. PLoS ONE. 2008;3:e3305. doi: 10.1371/journal.pone.0003305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chomont N, El-Far M, Ancuta P, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. 2009;15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fischer M, Joos B, Niederost B, et al. Biphasic decay kinetics suggest progressive slowing in turnover of latently HIV-1 infected cells during antiretroviral therapy. Retrovirology. 2008;5:107. doi: 10.1186/1742-4690-5-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar AM, Borodowsky I, Fernandez B, Gonzalez L, Kumar M. Human immunodeficiency virus type 1 RNA levels in different regions of human brain: quantification using real-time reverse transcriptase-polymerase chain reaction. J Neurovirol. 2007;13:210–24. doi: 10.1080/13550280701327038. [DOI] [PubMed] [Google Scholar]
- 36.Macallan DC, Wallace D, Zhang Y, et al. Rapid turnover of effector-memory CD4+ T cells in healthy humans. J Exp Med. 2004;200:255–60. doi: 10.1084/jem.20040341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–12. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 38.Rivino L, Messi M, Jarrossay D, Lanzavecchia A, Sallusto F, Geginat J. Chemokine receptor expression identifies pre-T helper (Th)1, Pre-Th2, and nonpolarized cells among human CD4+ central memory T cells. J Exp Med. 2004;200:725–35. doi: 10.1084/jem.20040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baron V, Bouneaud C, Cumano A, et al. The repertoires of circulating human CD8(+) central and effector memory T cell subsets are largely distinct. Immunity. 2003;18:193–204. doi: 10.1016/s1074-7613(03)00020-7. [DOI] [PubMed] [Google Scholar]
- 40.Geginat J, Sallusto F, Lanzavecchia A. Cytokine-driven proliferation and differentiation of human naive, central memory, and effector memory CD4+ T cells. J Exp Med. 2001;194:1711–9. doi: 10.1084/jem.194.12.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lanzavecchia A, Sallusto F. Progressive differentiation and selection of the fittest in the immune response. Nat Rev Immunol. 2002;2:982–7. doi: 10.1038/nri959. [DOI] [PubMed] [Google Scholar]
- 42.Gulzar N, Diker B, Mihowich J, et al. Proportion of HIV-1 infected CD8+CD4- T lymphocytes in vivo. Curr HIV Res. 2008;6:585–96. doi: 10.2174/157016208786501544. [DOI] [PubMed] [Google Scholar]
- 43.Yukl SA, Shergill AK, McQuaid K, et al. Effect of raltegravir-containing intensification on HIV burden and T-cell activation in multiple gut sites of HIV-positive adults on suppressive antiretroviral therapy. AIDS. 2010;24:2451–60. doi: 10.1097/QAD.0b013e32833ef7bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.