Abstract
Pre-eclampsia is a serious complication of pregnancy that can affect both maternal and fetal outcomes. Early-onset pre-eclampsia (EOPET) is a severe form of pre-eclampsia that is associated with altered physiological characteristics and gene expression in the placenta. DNA methylation is a relatively stable epigenetic modification to DNA that can reflect gene expression, and can provide insight into the mechanisms underlying such expression changes. This case–control study focused on DNA methylation and gene expression of whole chorionic villi samples from 20 EOPET placentas and 20 gestational age-matched controls from pre-term births. DNA methylation was also assessed in placentas affected by late-onset pre-eclampsia (LOPET) and normotensive intrauterine growth restriction (nIUGR). The Illumina HumanMethylation450 BeadChip was used to assess DNA methylation at >480 000 cytosine-guanine dinucleotide (CpG) sites. The Illumina HT-12v4 Expression BeadChip was used to assess gene expression of >45 000 transcripts in a subset of cases and controls. DNA methylation analysis by pyrosequencing was used to follow-up the initial findings in four genes with a larger cohort of cases and controls, including nIUGR and LOPET placentas. Bioinformatic analysis was used to identify overrepresentation of gene ontology categories and transcription factor binding motifs. We identified 38 840 CpG sites with significant (false discovery rate <0.01) DNA methylation alterations in EOPET, of which 282 had >12.5% methylation difference compared with the controls. Significant sites were enriched at the enhancers and low CpG density regions of the associated genes and the majority (74.5%) of these sites were hypomethylated in EOPET. EOPET, but not associated clinical features, such as intrauterine growth restriction (IUGR), presented a distinct DNA methylation profile. CpG sites from four genes relevant to pre-eclampsia (INHBA, BHLHE40, SLC2A1 and ADAM12) showed different extent of changes in LOPET and nIUGR. Genome-wide expression in a subset of samples showed that some of the gene expression changes were negatively correlated with DNA methylation changes, particularly for genes that are responsible for angiogenesis (such as EPAS1 and FLT1). Results could be confounded by altered cell populations in abnormal placentas. Larger sample sizes are needed to fully address the possibility of sub-profiles of methylation within the EOPET cohort. Based on DNA methylation profiling, we conclude that there are widespread DNA methylation alterations in EOPET that may be associated with changes in placental function. This property may provide a useful tool for early screening of such placentas. This study identifies DNA methylation changes at many loci previously reported to have altered gene expression in EOPET placentas, as well as in novel biologically relevant genes we confirmed to be differentially expressed. These results may be useful for DNA- methylation-based non-invasive prenatal diagnosis of at-risk pregnancies.
Keywords: pre-eclampsia, DNA methylation, placenta, 450 K array
Introduction
Pre-eclampsia is one of the most common causes of pre-term birth and fetal growth restriction, as well as being a major cause of maternal death worldwide (Ghulmiyyah and Sibai, 2012). While the underlying causes of pre-eclampsia are heterogeneous, there is generally a failure of placental trophoblast cells to sufficiently invade and remodel maternal arteries, which may be triggered by a failed immuno-response at the maternal-fetal interface (Redman and Sargent, 2010). This causes poor perfusion leading to physiological and gene expression changes in response to hypoxia and re-oxygenation stress (Tal, 2012). Early detection and management is key to preventing maternal death and helping to maintain the pregnancy, thus postponing premature birth. While a combination of maternal serum markers (e.g. PAPP-A, Inhibin, ADAM12) and Doppler ultrasound can be useful to identify women at risk for pre-eclampsia in the first trimester (Kuc et al., 2011), the high false positive rate and lack of a consensus for preventative intervention of pre-eclampsia makes the implementation of such screening impractical at this point. Improvements in the early diagnosis of pre-eclampsia and development of new therapies depend on a better understanding of the underlying biology.
Clinically, pre-eclampsia is characterized by hypertension (>140/90 mmHg) and proteinuria (>0.3 g/day). While the level of protein in the urine is often used to identify cases of ‘severe’ pre-eclampsia (at greatest risk of adverse outcomes), this measurement is not always reliable (Cote et al., 2008). A more valuable subclassification may be that of early-onset pre-eclampsia (EOPET) (diagnosis prior to 34 weeks) and late-onset pre-eclampsia (LOPET) (diagnosis after 34 weeks gestation) (von Dadelszen et al., 2003). There is accumulating evidence that these are distinct entities (Rolfo et al., 2010; Bahado-Singh et al., 2012; Escudero et al., 2013) with placental pathology playing a greater role in EOPET (Cross, 2003). Gene expression changes have also been used to sub-classify pre-eclampsia into three major molecular profiles (Cox et al., 2011) although their relationship to clinical classifications (EOPET versus LOPET, severe versus not) was not provided. Pre-eclampsia is a common cause of intrauterine growth restriction (IUGR) and it has been suggested that normotensive IUGR (nIUGR) may be associated with similar placental findings (Huppertz, 2011), but it is not clear if there is truly a shared etiology or just overlapping features.
As altered gene expression is observed in pre-eclampsia-associated placentas (Louwen et al., 2012), we expect altered DNA methylation to also be prevalent, based on the well-established link between gene expression and DNA methylation. Studying DNA methylation changes in these placentas provides a useful adjunct to characterize the pathological changes and to identify clinical subgroups of patients with differing underlying etiology. DNA methylation may also be useful in the development of new early screening approaches to identify at-risk pregnancies (Yuen et al., 2011). Lack of DNA methylation at gene promoters is generally associated with the potential for gene transcription (Weber et al., 2007). However, gene expression levels may be further modulated by DNA methylation at upstream enhancer sites (Horiuchi et al., 2012), which can affect the binding of transcription factors, and/or at the shores of cytosine-guanine dinucleotide (CpG) islands (Irizarry et al., 2009). We and others previously showed that EOPET-associated placentas exhibit hypomethylation at a number of gene promoter regions compared with controls in placenta (Yuen et al., 2010; Jia et al., 2012); Pre-eclamptic pregnancies were also associated with hypomethylation in maternal blood vessels (Mousa et al., 2012). However, our previous study was based on a small number of placentas (n = 4 EOPET placentas) and relatively small number of CpG sites tested genome-wide (n = 1506) (Yuen et al., 2010). Another study also showed altered methylation in pre-eclampsia placentas (n = 9) using a genome-wide methylated DNA immunoprecipitation approach, but did not distinguish clinical subtypes of pre-eclampsia (Jia et al., 2012). Furthermore, these studies did not compare these findings to gene expression of the complete candidate gene set.
In the present study, we sought to follow-up our initial findings (Yuen et al., 2010) with a larger set of EOPET and control placentas (n = 20 in each group) and use of a more comprehensive microarray interrogating 485 512 sites covering 99% of known genes. Importantly, this newer array includes sites in enhancers and promoter ‘shore’ regions, which were less prevalent in previous arrays and may be preferentially indicative of changes in gene expression. We used these data to (i) evaluate the ability of DNA methylation to distinguish clinical subgroups of pre-eclampsia and (ii) characterize the underlying biological pathways involved in these molecular changes and their potential relationship to changes observed in association with hypoxia exposure.
Methods
Ethical approval
This study was performed with the ethics approval of the University of British Columbia/Children's & Women’s Health Centre of British Columbia Research Ethics Board.
Sample collection
Whole chorionic villi were sampled from placentas delivered at Women's Hospital in Vancouver, Canada. Clinical criteria for EOPET were defined using the Canadian guidelines (Magee et al., 2008). For DNA methylation analysis, samples were further sub-classified by the presence/absence of severe proteinuria (>3 g/day), coincident IUGR, gestational diabetes and HELLP syndrome, defined by hemolysis, elevated liver enzymes and low platelet count. IUGR was defined as (i) birthweight below the third percentile for gender and gestational age using Canadian population parameters or (ii) birthweight below the 10th percentile with persistent uterine artery notching at 22–25 weeks, absent or reversed end diastolic velocity on umbilical artery Doppler, or oligohydramnios. As gestational age is a large determinant of DNA methylation in the placenta (Novakovic et al., 2011), it is critical that placentas from pre-eclamptic pregnancies are compared with gestational age-matched controls (Yuen et al., 2010). We therefore compared with placentas obtained from chromosomally normal losses or births due to a mix of etiologies (premature rupture of membranes, loss of amniotic fluid and cervical incompetence) with no evidence of placental abnormality based on a pathological exam (Table I; Supplementary data, Table SI). The use of such pre-term samples is validated by our previous observation that the DNA methylation profiles of these premature-controls were more similar to term-controls than to the gestational age (GA) age-matched EOPET placentas (Yuen et al., 2010). Chorionic villi from at least two sites (center and perimeter) on the fetal side of the placenta were sampled, rinsed of maternal blood. DNA was extracted from each and combined in equal quantities for better representation of placenta-wide changes. It should be noted that four of the samples in this study have been previously assessed for methylation changes (using less-comprehensive microarrays) (Yuen et al., 2009, 2010).
Table I.
Clinical information of samples used in Illumina methylation array.
| EOPET (n = 20) mean (range) | CONTROL (n = 20) mean (range) | P-value | |
|---|---|---|---|
| Gestational age (weeks) | 31.8 (24.9–37.3) | 31.8 (25.0–37.3) | 0.931 |
| Maternal age (years) | 33.5 (19.7–42.9) | 31.5 (22.2–38.7) | 0.289 |
| Birthweight (g) | 1451 (440–3685) | 1940 (758–3470) | 0.052 |
Illumina HumanMethylation450 BeadChip
Twenty EOPET samples and 20 controls in two batches (2 batches of 10 EOPET versus 10 controls) were used for the Illumina HumanMethylation450 BeadChip (Bibikova et al., 2011) (Illumina, Inc., San Diego, CA, USA) (hereafter referred to as the ‘450 K array’). Of note, 750 ng of DNA was bisulfite converted using the EZ DNA Methylation Kit (Zymo Research, Irvine, CA, USA). Hybridization of samples was performed following manufacturer's protocol. Chips were scanned by a HiScan 2000 or iScan (Illumina). Raw and processed data have been deposited into the publically accessible database, GEO (GSE44667; http://www.ncbi.nlm.nih.gov/geo/). Raw data were read into GenomeStudio 2011 (Illumina) where background subtraction and initial processing was performed. Probes targeting a CpG with a documented single nucleotide polymorphism in the C or G were removed from analysis (Price et al., 2013). Probes targeting the sex chromosomes were also removed. Any probe that had a missing β-value or >0.01 detection P-value (probes whose signal intensity was lower than background levels) in one sample was also removed from analysis (n = 25 804). This left 430 685 probes for analysis. Signal intensities were read into R v2.14 (R Development Core Team, 2011) using methylumi (Davis et al., 2011) to create M-values (log transformations proportional to the level of methylation). Values from remaining probes were color channel corrected (Du et al., 2010), separated into Types I and II probes and normalized by subset-quantile within array normalization (Maksimovic et al., 2012). M-values were transformed back to β-values (a value between 0 and 1 representing the fraction of methylated DNA) for genome-wide analysis. Finally, probes that map away from their intended target were removed from the final candidate dataset (Price et al., 2013).
Gene expression microarray
Gene expression data were obtained using a subset of EOPET samples and controls (n = 8 each) that were also run on the 450 K array. RNA was extracted from placental villi stored in RNAlater at −80°C using an RNeasy kit (Qiagen, Heiden, Germany). RNA was assessed for quality on a Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA), reverse-transcribed to cDNA and hybridized to HT-12v4 Expression BeadChip (Illumina) following the manufacturer's protocol. This chip interrogates over 47 000 transcripts genome-wide. Raw data were quantile normalized in GenomeStudio 2011. Probes with bad detection P-values (>0.01) in all samples were removed from analysis (n = 13 125). Data points from valid probes with a bad detection P-value were removed and replaced with a value of one to represent a lack of expression. Expression values from the gene expression microarray were log2-transformed before statistical analysis. Raw and processed data have been deposited into the publically accessible database, GEO (GSE44711).
Pyrosequencing
A subset of sites with highly significant differences from the array and/or relevance for prenatal screening or hypoxia was followed up by bisulfite pyrosequencing of EOPET samples (n = 20), other disease states (LOPET (n = 21) and nIUGR (n = 17)) and a larger set of controls covering a broad range of gestational ages (n = 93). Assays (Supplementary data, Table SII) were designed using PSQ Assay design (Biotage, Upsalsa, Sweden) and run on a Qiagen Pyromark Q96 MD (Qiagen).
Statistical analysis
All statistical analysis was performed using R v2.14 (R Development Core Team, 2011). Statistically significant probes from the 450 K array were determined using the false discovery rate (FDR), which is an approach that is used to correct for multiple testing. For this, significance analysis of microarrays (SAM) was performed on M-values using the siggenes package (Schwender, 2011). To enrich for biological relevance, highly significant probes were determined by Δβ (difference in DNA methylation between cases and controls) in conjunction with an FDR cutoff. Principal component analysis (PCA) from the methylumi package (Davis et al., 2011) was used on subsets of the 450 K array data to determine stratification of the samples using several different parameters.
Significant probes from the gene expression array were identified by SAM using an FDR cutoff of 0.05 combined with a fold change of ≥1.2. Correlation of gene expression to DNA methylation was determined using Pearson's correlation on the subset of samples used in both arrays.
DNA methylation detected by pyrosequencing was confirmed as concordant to that of the 450 K array using Pearson's correlation (average r = 0.80). Pearson's correlation was then used on the pyrosequencing data to assess if DNA methylation was affected by gestational age. If so, statistical significance was assessed by analysis of covariance (ANCOVA) with gestational age as the covariate. Bonferroni corrected pair-wise post hoc analysis determined statistical significance in DNA methylation between individual sample groups.
A χ2 test was used to determine whether probes in specific genomic elements were overrepresented in our dataset when compared with the array as a whole.
Bioinformatics
The database for annotation, visualization and integrated discovery was used to conduct gene ontology analysis (Huang da et al., 2009). Significant biological processes were elucidated by over-representation of genes in gene ontology categories. Next, genomic sequences spanning 50 bp up- and downstream of significant CpG sites were extracted using Galaxy (https://main.g2.bx.psu.edu). These sequences were submitted for recurrent motif discovery using multiple em for motif elicitation (MEME) suite (Bailey et al., 2009). Recurrent motifs were then queried for known transcription factor binding motifs using the TOMTOM tool in MEME suite.
Results
Extensive DNA methylation profile changes in early-onset pre-eclampsia
Principle component analysis was performed using all 430 685 probes (Supplementary data, Fig. S1) to determine the differences in the overall DNA methylation profile between cases and controls. Samples clustered roughly according to the presence or absence of pre-eclampsia rather than by gestational age, surprisingly because gestational age has been shown to drive dramatic differences in DNA methylation (Novakovic et al., 2011). This implies that DNA methylation changes in EOPET are very dramatic and widespread throughout the genome. However, it should be noted that the majority (65%) of cases were within a narrow GA range of 31–34 weeks.
Overall, an extremely large number of CpG sites (over 100 000) showed statistically significant differences between groups at an FDR of <0.05, suggesting dramatic epigenetic effects of EOPET on the placenta. Most array studies use a combination of significance (e.g. FDR of <0.01 or 0.05) in combination with a cutoff for mean difference (Δβ) to detect the most biologically meaningful changes. As there is no standard set of criteria, we considered various combinations of Δβ values and FDRs to establish values that would allow us to focus on the most significant probes for the follow-up (Table II). Changing the requirement for Δβ has a dramatic effect on the number of significant candidates, with 1312 candidates using Δβ of 0.1 and only 87 candidates with Δβ of 0.15. Based on this analysis we opted to focus our analysis using a relatively stringent cutoff of 12.5% Δβ and 1% FDR, providing a list of 286 CpGs that were differentially methylated in placental samples between EOPET and controls. After elimination of four probes that map to multiple locations in the genome, 282 candidate CpGs located in or near 248 genes remained.
Table II.
Number of candidate sites, without removing cross-mapping probes, obtained using different cutoff parameters for Δβ and FDR.
| FDR | Δβ cutoff |
|||
|---|---|---|---|---|
| Cutoff | 0% | 10% | 12.5% | 15% |
| 1 | – | 1333 | 313 | 88 |
| 0.1 | 154 604 | 1328 | 312 | 87 |
| 0.05 | 102 343 | 1312 | 311 | 87 |
| 0.01 | 38 840 | 1175 | 286 | 82 |
Of the candidate CpGs, 74.5% were hypomethylated and 25.5% were hypermethylated in EOPET as compared with controls (Supplementary data, Table SIII). Among the genes associated with hypomethylated CpGs were several coding for proteins suggested to be useful in maternal serum screening for pre-eclampsia including INHBA (2 CpGs), ADAM12, PAPPA2 (3 CpGs) and FLT1, as well as several genes we previously reported as exhibiting hypomethylation in pre-eclampsia (e.g. TIMP3, MEST, CXCL9) (Yuen et al., 2010). However, many of the altered CpGs were associated with genes of unknown placental function.
EOPET, but not associated clinical features, presents a distinct DNA methylation profile
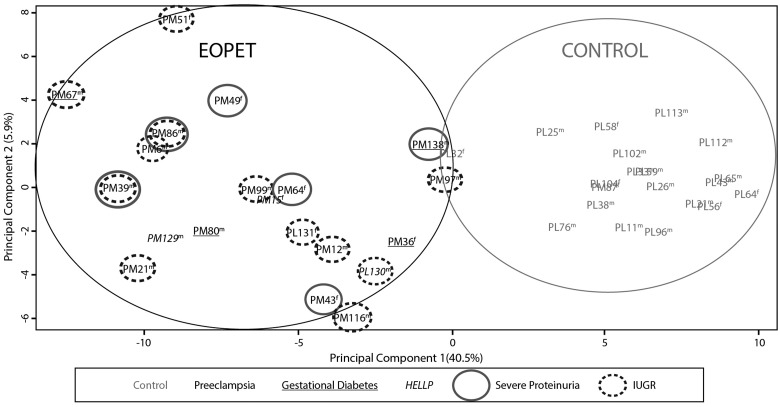
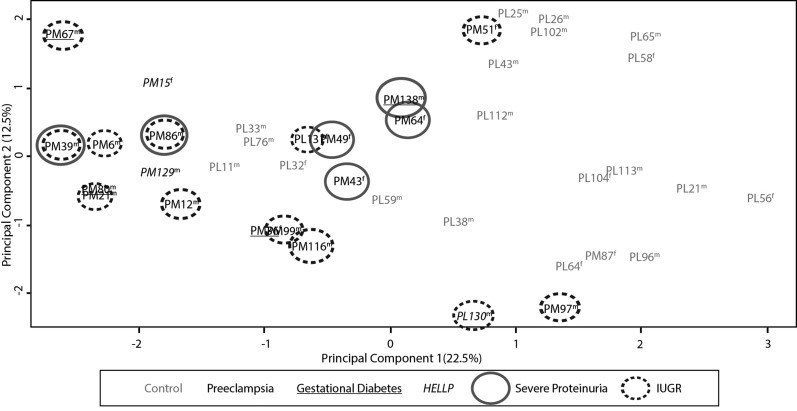
To assess the discriminatory power of the identified EOPET-associated DNA methylation changes, PCA was repeated on all samples using only the 282 candidate CpGs (Fig. 1). This revealed a distinct clustering of EOPET samples separate from controls along principal component 1 (40% of the variance). However, there was more variability among the EOPET samples than the controls. In order to determine if some of this variability was due to clinical variation in samples, we further coded samples by the presence or absence of IUGR, severe proteinuria, diabetes or HELLP syndrome. However, there were no distinct clusters of these clinical subtypes or sex using this set of probes (Fig. 1).
Figure 1.
Principle component analysis based upon values of the 282 significant differentially methylated probes in this study. EOPET and control samples form distinct clusters (circled), with no additional clustering based on the presence of IUGR, severe proteinuria, hemolysis, elevated liver enzymes and low platelet count (HELLP) or gestational diabetes. Sex of sample is indicated by m or f.
We also compared DNA methylation profiles between EOPET placentas with (n = 12) and without (n = 8) concurrent IUGR. No sites were found to be significant using modest criteria of an FDR ≤0.05 and Δβ ≥ 5%, in contrast to the large number of sites significant using such criteria when comparing EOPET versus controls. Separately comparing each subset of EOPET samples depending on the presence/absence of IUGR, against corresponding gestational age-matched controls produced similar sets of significant probes as the EOPET group as a whole (analysis not shown). Hence, we find no evidence for a major methylation difference between EOPET with and without associated IUGR.
Differentially methylated loci showed negative correlation with gene expression
A subset of placental samples run on the DNA methylation array (eight EOPET versus eight controls) was deemed of sufficient quality to be run on an Illumina HT-12v4 Expression BeadChip array. There were 195 probes representing 182 genes that were significantly changed using a cutoff of ≤0.05 FDR and ≥1.2–fold change (Supplementary data, Table SIV). Similarly, 17.4% of 282 candidate CpGs from the 450 K methylation array (87.9% of which were represented on the expression array) were associated with significantly altered gene expression using a nominal P-value of 0.05 (Supplementary data, Table SIII). There were multiple genes significantly over-expressed in this study that have been shown as over-expressed in other pre-eclampsia genome-wide expression studies despite differing sample criteria and methods (Louwen et al., 2012). These common genes include: INHA, LEP, PAPPA2, CGB, CRH, HTRA4, TREM1, CXCR6, EBI3, BTNL9, ULBP1, SIGLEC6 and QPCT.
In addition, 23 of the 282 candidate CpGs showed negative correlation between DNA methylation and gene expression (P < 0.05) (Supplementary data, Table SIII), although some of the associated genes did not reach genome-wide statistical significance for differential expression between cases and controls. These significant genes included EPAS1, FLT1 and FLNB, which are known to be important for angiogenesis. This further supports the involvement of DNA methylation in the regulation of a substantial number of genes that are associated with pre-eclampsia development. Small sample size likely caused an underestimation of altered methylation sites associated with altered expression. Changes in gene expression can also be regulated by other factors (e.g. availability of transcription factors) in addition to DNA methylation, and can change at term in response to labor and delivery, confounding these comparisons.
Differences in DNA methylation are regional and different in related disorders
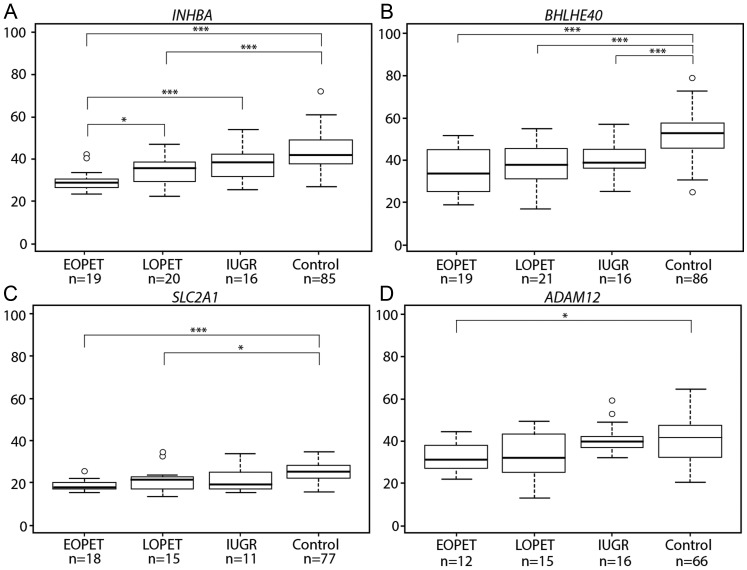
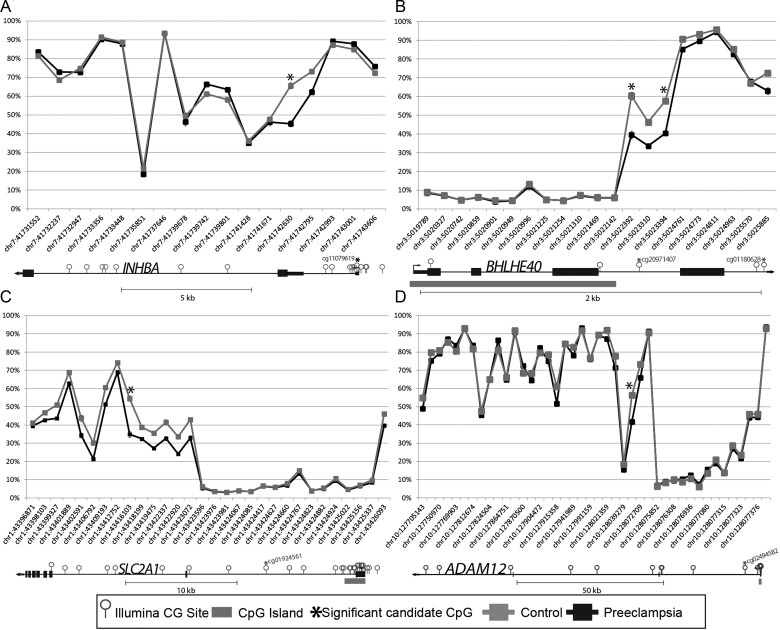
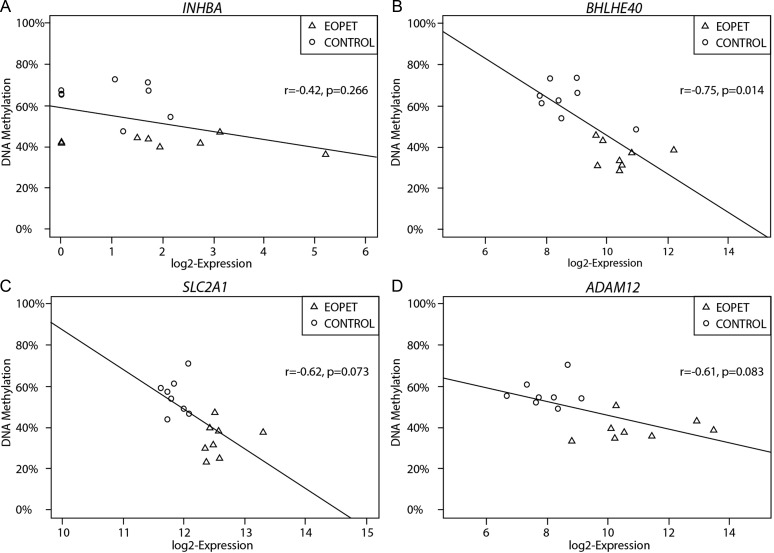
The ability to determine molecular differences specific to disease type is important for establishing accurate prediction methods and targeted treatment. Thus, bisulfite pyrosequencing of four altered CpG sites was performed on placentas from EOPET, LOPET, nIUGR and controls (Fig. 2). These four sites were chosen for further investigation due to their large Δβ and previous evidence that their expression can be affected by hypoxia (BHLHE40 (Chakrabarti et al., 2004) and SLC2A1 (Baumann et al., 2007)) or is relevant to prenatal screening (INHBA and ADAM12) (Kuc et al., 2011). While in some of these genes only one of the associated CpGs fulfilled the criteria of FDR < 0.01 and ≥12.5% Δβ, examining all array targets in these genes also showed DNA methylation differences between EOPET placentas and controls in nearby targets (Fig. 3). Comparing the DNA methylation at the significant CpG sites to expression in each gene revealed statistically significant negative correlation in BHLHE40, and trends in SLC2A1 and ADAM12 (Fig. 4). The increased expression reflects the expected association of decreased methylation near the promoter region of these genes.
Figure 2.
Boxplots from pyrosequencing follow-up of candidate CpGs associated with four genes. (A) INHBA (cg11079619); (B) BHLHE40 (cg20971407); (C) SLC2A1 (cg01924561) and (D) ADAM12 (cg02494582). Percent methylation is indicated on the Y-axis. Analysis of covariance (ANCOVA) adjusting for gestational age followed by Bonferroni corrected pair-wise post hoc analysis was used to determine significance between individual groups *P < 0.05, **P < 0.01 and ***P < 0.001.
Figure 3.
DNA methylation levels for cases and controls according to genomic location. Genomic order of CpGs is given on the X-axis. CpG islands are assigned according to UCSC Genome Browser delineations. Each plot represents one candidate gene: INHBA (A), BHLHE40 (B), SLC2A1 (C) and ADAM12 (D). Asterisk indicates candidate CpG site. Candidate CpGs fell into a variety of genomic regions relative to the gene structure.
Figure 4.
Methylation at candidate sites is compared with gene expression. Results for four genes are given: INHBA (A), BHLHE40 (B), SLC2A1 (C) and ADAM12 (D). The expected negative correlation in methylation and gene expression exists for BHLHE40, with similar trends in SLC2A1 and ADAM12.
DNA methylation for all four genes showed a small but significant negative correlation with increasing gestational age; therefore, all statistical tests were performed using gestational age as a co-variate. It should be noted that EOPET showed decreased DNA methylation at these loci, the opposite direction of that expected if gestational age influenced these changes. Each gene showed a statistically significant difference (P < 0.05) between groups, with EOPET being significantly different from the controls for all genes, confirming the array findings. LOPET showed significant alterations compared with controls for INHBA, BHLHE40 and SLC2A1, while only BHLHE40 showed a statistical difference between nIUGR and controls.
EOPET shows hypomethylation at loci that are hypermethylated with hypoxia exposure
EOPET is associated with initial placental hypoxia followed by re-oxygenation stress (Hung et al., 2002); thus, we wanted to assess whether sites altered by hypoxia exposure in culture would also be altered in pre-eclampsia. We recently used the 450 K array to assess DNA methylation changes in cultured trophoblast cells exposed to three oxygen levels (Yuen et al., 2013). Of the strictly defined subset of 282 altered CpGs associated with EOPET, only five overlapped with those identified as significant changes upon exposure of cells to hypoxia. However, 78.3% of probes identified as significant in the hypoxia study are considered significantly different between EOPET and control samples when using a less strict t-test P-value cutoff of 0.05. However, these CpGs all demonstrated increased DNA methylation upon exposure to hypoxia in cytotrophoblast, whereas these same loci showed decreased DNA methylation in pre-eclampsia. As many of these same loci are hypermethylated in syncytiotrophoblast as compared with cytotrophoblast (Yuen et al., 2013), these results could also reflect an alteration in trophoblast cell populations in EOPET placentas (i.e. reduced cytotrophoblast:syncytiotrophoblast ratio).
We further performed PCA using just the 147 hypoxia-associated loci to determine if EOPET and control placentas would cluster into separate groups using this subset of loci. A separation of EOPET and control placentas was achieved with few outliers (Fig. 5). Again, there was no further separation of cases based on clinical subclassification of EOPET.
Figure 5.
Principle component analysis using CpG sites that are significantly altered in cultured cytotrophoblast exposed to hypoxic conditions for 24 h (Yuen et al., 2013). Control and pre-eclampsia samples had a tendency to cluster apart from each other. Sex of sample is indicated by m or f.
Genomic regional and functional enrichments of differentially methylated CpG sites
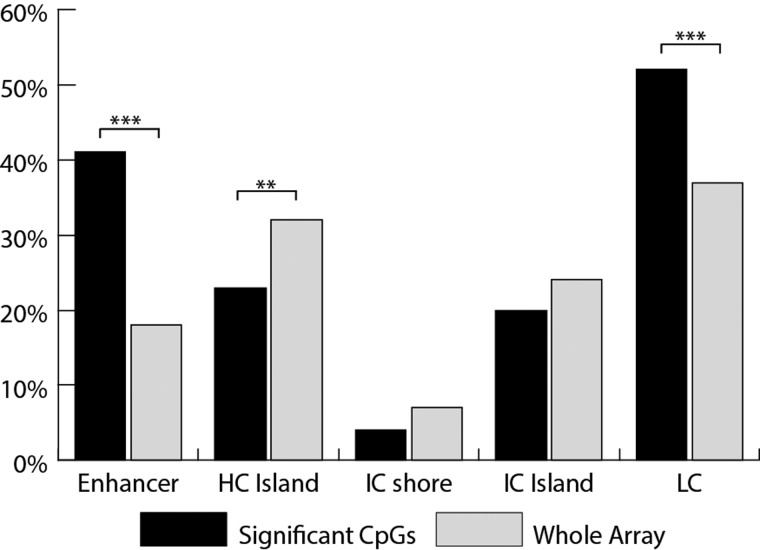
Investigating whether significant CpGs are located in certain genomic elements such as CpG islands or enhancers can help elucidate the nature of the biological processes that are disrupted in EOPET (Fig. 6). Significantly more probes fell into enhancer regions from among our candidate sites than expected at random from the array. There are also proportionally fewer probes falling into high-density CpG islands, typically unmethylated and associated with gene promoters, than non-CpG islands. Enhancers are often located outside of CpG islands, explaining the enrichment of significant probes in low CpG density areas. There is no significant difference from the expected ratios in probes that fall into intermediate density CpG islands or CpG island shores.
Figure 6.
Comparison of proportions of significant (candidate) CpGs relative to the array as a whole that fall into different genomic elements. The elements compared include high CpG density islands (HC), intermediate CpG density islands (IC), intermediate density CpG islands that are on the shore of an HC (ICshore), low CpG density regions (LC) and annotated genomic enhancers. **P < 0.01 and ***P < 0.001.
Of significant CpG sites, 29.8% are located in ‘intergenic regions’, which are defined using the Illumina annotation as being further than 2 kb from the transcription start or end site of a gene; however upstream enhancers could be located in such intergenic regions (Sanyal et al., 2012). As it is unclear without functional studies on placental tissue whether an upstream enhancer has an effect on expression or not, we associated all unannotated significant CpGs with the closest transcription start site to a RefSeq gene (Price et al., 2013) to expose potential transcriptionally important regions for future studies.
Gene ontology analysis for DNA methylation returned a list of enriched biological processes (Table III, Supplementary data, Table SV). The top significant gene ontology clusters were involved in regulation of gene transcription, implying an overall disregulation of transcription factors affecting gene expression in EOPET placentas. Consistent with this, invocation of the hypoxia response pathway targets many transcription factors (Schodel et al., 2011). Many of the genes associated with these gene ontology terms are affected by oxygen levels or directly involved in the hypoxia response pathway (e.g. EPAS1). Differentiation and morphogensis were also significant ontology terms, consistent with the observed altered development of trophoblast in pre-eclampsia (Gauster et al., 2009). Gene ontology analysis for gene expression returned only one hit of P < 0.01: ‘female pregnancy’ (GO:0007565). Consistent with this, many of the genes in this category have been shown to be over-expressed in pre-eclampsia as detectable in maternal serum (e.g. PAPPA) (Kuc et al., 2011).
Table III.
Gene Ontology top 10 hits for genes with altered DNA methylation.
| GO Number | Term | Count | P-value |
|---|---|---|---|
| GO:0006355a | regulation of transcription, DNA-dependent | 43 | 4.35E-05 |
| GO:0051252a | regulation of RNA metabolic process | 43 | 7.32E-05 |
| GO:0030182b | neuron differentiation | 16 | 5.05E-04 |
| GO:0010628a | positive regulation of gene expression | 18 | 1.26E-03 |
| GO:0045941a | positive regulation of transcription | 17 | 2.39E-03 |
| GO:0010604a | positive regulation of macromolecule metabolic process | 22 | 3.09E-03 |
| GO:0006928b | cell motion | 15 | 3.16E-03 |
| GO:0045449a | regulation of transcription | 49 | 3.93E-03 |
| GO:0000904b | cell morphogenesis involved in differentiation | 10 | 4.13E-03 |
| GO:0010557a | positive regulation of macromolecule biosynthetic process | 18 | 4.30E-03 |
aTerms that are related to gene transcription.
bTerms that are involved in cell differentiation.
MEME identified two recurrent motifs in proximity to the significant CpGs. These two motifs each occurred in 28 of 282 sites. To determine if they were associated with a transcription factor binding site, the sequences were submitted to TOMTOM, where the transcription factor databases JASPAR and uniprobe were searched for corresponding defined motifs. There were no significant occurrences of specific transcription factor motifs, consistent with there being a wide range of transcription factors that may be affected by the pre-eclamptic state.
Discussion
In this study, we demonstrate widespread alterations in DNA methylation in placentas from pregnancies complicated by EOPET. This expands upon and confirms our previous report of altered methylation in EOPET (Yuen et al., 2010) with much larger sample size and number of analyzed loci. EOPET cases can be distinguished from controls based on this methylation profile, regardless of the presence/absence of additional features (e.g. IUGR, severe proteinuria and HELLP). These findings support the concept of EOPET being a distinct clinical entity that includes various presentations including HELLP syndrome.
Clinical criteria for the definition of ‘severe’ pre-eclampsia vary (Steegers et al., 2010) with some based on a gestational age cutoff (i.e. EOPET), and others based on levels of proteinuria (e.g. >2 g/day) (Nishizawa et al., 2011). In our study, some cases classified as EOPET did not have documented proteinuria (i.e. diagnosis was based on the presence of hypertension in combination with other findings such as fetal IUGR as per Canadian guidelines (Magee et al., 2008)) while others had heavy proteinuria (>3 g/day); however, no relationship between proteinuria level and methylation profile was identified. Other conditions concurrent with EOPET in this study (IUGR, gestational diabetes and HELLP syndrome) did not cluster separately from other EOPET samples. Consistent with this observation, HELLP syndrome has been shown to have a similar transcription profile as pre-eclampsia (Varkonyi et al., 2011). Thus, there may be consistent molecular features of EOPET placentas despite variability in clinical symptoms; this variability may be reflective of other factors such as maternal health.
When comparing LOPET and nIUGR for a subset of loci, EOPET showed the largest differences from controls, but LOPET and nIUGR trended towards EOPET for these genes. In a separate study using these samples, but focusing on stress and other hormonal pathway genes, LOPET samples similarly tended to exhibit levels of methylation overlapping EOPET and control values, suggesting this may be a heterogeneous group with a subset of cases possibly fitting an ‘EOPET-like’ pattern and the remainder not (Hogg et al., 2013a, b). Furthermore, previous studies have shown that genes associated with altered DNA methylation or expression in EOPET show fewer and smaller changes in LOPET and/or nIUGR (Nishizawa et al., 2007; Yuen et al., 2010; Hogg et al., 2013a, b). It has been suggested that LOPET and EOPET have distinct etiologies (von Dadelszen et al., 2003; Rolfo et al., 2010), with EOPET having greater placental dysfunction (Chen et al., 2012). The present data support the concept of greater placental dysregulation in EOPET, though this needs to be followed up in a more comprehensive manner.
Detecting the future onset of pre-eclampsia early in pregnancy could lead to improved management and help mitigate the effects of the disorder. Ideally, the detection method would be non-invasive, likely investigating components of a first trimester maternal serum sample. Altered maternal serum concentrations of a number of proteins have been reported (PAPP-A, ADAM12, hCG, Inhibin, PP13, sFLT1, Endoglin, PGF, Leptin (Kuc et al., 2011; Kleinrouweler et al., 2012; Masoura et al., 2012; Hogg et al., 2013a, b)) prior to the onset of pre-eclampsia, with the placenta being the likely source of many of these. Expression of several genes coding for up-regulated proteins is increased in pre-eclampsia placentas from the 3rd trimester (ADAM12, LEP, CGB, PAPPA, INHA) as indicated in this and other studies (Louwen et al., 2012). Altered DNA methylation in many of the genes coding for these proteins was observed in the present study including ADAM12, CGA, FLT1, INHBA and, using a lower Δβ cutoff of 8%, PAPPA and LEP. We also found a correlation of DNA methylation and gene expression for most of these genes in this study. Hence, other genes we identified as showing altered DNA methylation, could perhaps offer candidates to investigate as potential biomarkers in maternal serum for the enhanced prediction of pre-eclampsia.
Furthermore, methylation differences could be utilized for early screening using an approach based on circulating cell-free fetal DNA (cffDNA), which is increased in mothers predisposed to pre-eclampsia (Zeybek et al., 2012). We previously suggested that methylation of TIMP3 was a candidate for DNA methylation-based diagnosis based upon the differential DNA methylation patterns between blood and placenta, with more extreme values in EOPET-associated placentas (Yuen et al., 2010); a finding confirmed by others (Xiang et al., 2012). Some sites showing similar properties to those in the present study include BHLHE40 and INHBA. Further evaluation of such sites for their ability to enhance the early detection of EOPET over simply quantifying the cffDNA fraction (Yuen et al., 2011) would be of interest.
There are several potential drivers for the altered DNA methylation observed in this study. One key driver is oxygen tension (hypoxia/re-oxygenation), an etiological factor in the development of pre-eclampsia (Hung et al., 2002). The hypoxia response pathway involves the expression of many genes, including the hypoxia-inducible factors (HIFs), transcription factors designed to activate genes whose expression can help reestablish viable oxygen levels. EPAS1, which encodes for HIF2α, is an early factor in the hypoxia response pathway and has been shown to be active in pre-clampsia placentas (Rajakumar et al., 2001). CpGs in the shore of the promoter CpG Island of this gene were significantly hypomethylated in pre-clampsia placentas in this study, indicating that there is increased binding of transcription factors such as RNA polymerase (Ogoshi et al., 2011). Supporting this, EPAS1 was over-expressed in this and a previous study (Jarvenpaa et al., 2007). Frequent targets of HIF2α are BHLHE40 and INHBA (Schodel et al., 2011), both of which code for proteins that can prevent trophoblast differentiation (Meinhardt et al., 2005; Ezashi et al., 2012). BHLHE40 was shown to have significantly decreased DNA methylation in the shore of the CpG island promoter and increased expression in pre-clampsia placentas, while INHBA showed decreased DNA methylation directly in the promoter region and has demonstrated increased expression in several studies (Enquobahrie et al., 2008; Sitras et al., 2009; Hoegh et al., 2010; Nishizawa et al., 2011; Tsai et al., 2011).
We previously demonstrated that a subset of genes underwent hypermethylation upon exposure of cytotrophoblast cells to 24 h of <1% oxygen (Yuen et al., 2013). Many of these same genes became hypomethylated upon differentiation of cytotrophoblast to syncytiotrophoblast, consistent with the observation that hypoxia can block the differentiation of cytotrophoblast cells into syncytiotrophoblast (Alsat et al., 1996). In the present study, we found that these hypoxia/differentiation sites tended to be hypomethylated in EOPET, which is the opposite of what would be expected due to hypoxic exposure. It may be that re-oxygenation stress, also proposed to occur in the development of pre-eclampsia, may result in hypomethylation of these same loci. It has been hypothesized that this sudden change in physiological state later in pregnancy can induce a stress response in the trophoblast (Hung et al., 2002), potentially changing the DNA methylation of the chorionic villi. It is also possible that the short-term effects of reduced oxygen level in culture do not accurately reflect the long-term effects in vivo. Reduced DNA methylation is in fact observed with long-term hypoxic exposure in some cancer tissues (Shahrzad et al., 2007).
Hypomethylation of these hypoxia-associated loci could be evidence of altered cell composition as many of these CpG sites were significantly hypomethylated in syncytiotrophoblast compared with cytotrophoblast; however, an excess of cytotrophoblast is generally observed (Redline and Patterson, 1995) in pre-eclampsia, making this explanation less likely. Other potential drivers of altered DNA methylation changes include effects of common treatments given to these women such as the prescription of anti-hypertensive medication or corticosteriod treatments, the latter given for fetal lung maturation before delivery. The effects of anti-hypertensive medicine on DNA methylation in the placenta are unstudied; however there are clear disruptions in the corticosteroid pathways in these samples (Hogg et al., 2013a, b), perhaps from the use of antenatal corticosteriods. As these are not individual events, but logically succeed one another, a mix of these factors driving the DNA methylation differences is plausible.
There are limitations to this study. To avoid confounding by gestational age—associated methylation changes (Novakovic et al., 2011), control placentas were also obtained from premature births. DNA methylation changes could theoretically be associated with pre-term labor or pre-term rupture of membranes, which were some causes of premature births in these controls. Furthermore, pre-eclampsia might have developed in some of these pregnancies had they continued further to term. Nonetheless, the tight clustering of control placentas relative to the EOPET ones, suggests they are a relatively homogenous group. Another limitation comes from the difficulty in obtaining high-quality RNA from placentas. We were only able to obtain quality RNA from 8 of the 20 EOPET samples, each matched to a control, resulting in reduced power when studying expression versus DNA methylation in this sample set.
By investigating the DNA methylation profile of pre-eclampsia placentas, this study has laid the groundwork for future studies. As many altered CpG sites located in upstream enhancer sites were identified, functional studies investigating DNA methylation-dependent regulation at these sites would help determine the biological significance of these findings. Furthermore, candidate genes could be investigated for altered expression in maternal blood or for properties appropriate for development of DNA methylation-based non-invasive prenatal screening methods. Additionally, a more thorough genome-wide investigation of DNA methylation differences in LOPET and nIUGR samples would help elucidate the nature of any potential alterations in placental methylation profile and the nature of the overlap with EOPET.
Supplementary data
Supplementary data are available at http://molehr.oxfordjournals.org/.
Authors’ roles
R.K.C.Y., W.P.R. and J.D.B. conceived and designed the study. D.E.F. and P.V.D. provided the samples and important expertise. J.D.B., R.K.C.Y. and B.L. performed the experiments and collected the data. J.D.B. performed the data analysis. J.D.B. and W.P.R. wrote the draft manuscript. All authors contributed to the final version of the manuscript.
Funding
This study was funded through a Canadian Institutes of Health Research grant (49520) to W.P.R. P.v.D. and W.P.R. receive salary support from the Child & Family Research Institute. Funding to pay the Open Access publication charges for this article was provided by Canadian Institutes of Health Research Operating Grant (WPR).
Conflict of interest
None declared.
Supplementary Material
Acknowledgements
We acknowledge Ruby Jiang for assistance with placental sampling, Kristal Louie and Joanna Mendel for patient recruitment and clinical data collection and Michael Kobor for use of the Pyromark Q96 instrument.
References
- Alsat E, Wyplosz P, Malassine A, Guibourdenche J, Porquet D, Nessmann C, Evain-Brion D. Hypoxia impairs cell fusion and differentiation process in human cytotrophoblast, in vitro. J Cell Physiol. 1996;168:346–353. doi: 10.1002/(SICI)1097-4652(199608)168:2<346::AID-JCP13>3.0.CO;2-1. doi:10.1002/(SICI)1097-4652(199608)168:2<346::AID-JCP13>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Bahado-Singh RO, Akolekar R, Mandal R, Dong E, Xia J, Kruger M, Wishart DS, Nicolaides K. First-trimester metabolomic detection of late-onset preeclampsia. Am J Obstet Gynecol. 2012;25:1840–1847. doi: 10.1016/j.ajog.2012.11.003. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. doi:10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann MU, Zamudio S, Illsley NP. Hypoxic upregulation of glucose transporters in BeWo choriocarcinoma cells is mediated by hypoxia-inducible factor-1. Am J Physiol Cell Physiol. 2007;293:C477–C485. doi: 10.1152/ajpcell.00075.2007. doi:10.1152/ajpcell.00075.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, Delano D, Zhang L, Schroth GP, Gunderson KL, et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–295. doi: 10.1016/j.ygeno.2011.07.007. doi:10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- Chakrabarti J, Turley H, Campo L, Han C, Harris AL, Gatter KC, Fox SB. The transcription factor DEC1 (stra13, SHARP2) is associated with the hypoxic response and high tumour grade in human breast cancers. Br J Cancer. 2004;91:954–958. doi: 10.1038/sj.bjc.6602059. doi:10.1038/sj.bjc.6602059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Huang Y, Jiang R, Teng Y. Syncytiotrophoblast-derived microparticle shedding in early-onset and late-onset severe pre-eclampsia. Int J Gynaecol Obstet. 2012;119:234–238. doi: 10.1016/j.ijgo.2012.07.010. doi:10.1016/j.ijgo.2012.07.010. [DOI] [PubMed] [Google Scholar]
- Cote AM, Firoz T, Mattman A, Lam EM, von Dadelszen P, Magee LA. The 24-hour urine collection: gold standard or historical practice? Am J Obstet Gynecol. 2008;199:625.e1–625.e6. doi: 10.1016/j.ajog.2008.06.009. doi:10.1016/j.ajog.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Cox B, Sharma P, Evangelou AI, Whiteley K, Ignatchenko V, Ignatchenko A, Baczyk D, Czikk M, Kingdom J, Rossant J, et al. Translational analysis of mouse and human placental protein and mRNA reveals distinct molecular pathologies in human preeclampsia. Mol Cell Proteomics. 2011;10:M111.012526. doi: 10.1074/mcp.M111.012526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross JC. The genetics of pre-eclampsia: a feto-placental or maternal problem? Clin Genet. 2003;64:96–103. doi: 10.1034/j.1399-0004.2003.00127.x. doi:10.1034/j.1399-0004.2003.00127.x. [DOI] [PubMed] [Google Scholar]
- Davis S, Du P, Bilke S. methylumi: Handle Illumina Methylation Data. 2011. R package version 2.0.13 http://www.bioconductor.org/ . [Google Scholar]
- Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, Lin SM. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. doi:10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enquobahrie DA, Meller M, Rice K, Psaty BM, Siscovick DS, Williams MA. Differential placental gene expression in preeclampsia. Am J Obstet Gynecol. 2008;199:566.e1–566.e11. doi: 10.1016/j.ajog.2008.04.020. doi:10.1016/j.ajog.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudero C, Bertoglia P, Hernadez M, Celis C, Gonzalez M, Aguayo C, Acurio J. Impaired A(2A) adenosine receptor/nitric oxide/VEGF signaling pathway in fetal endothelium during late- and early-onset preeclampsia. Purinergic Signal. 2013;9:215–226. doi: 10.1007/s11302-012-9341-4. doi:10.1007/s11302-012-9341-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezashi T, Telugu BP, Roberts RM. Model systems for studying trophoblast differentiation from human pluripotent stem cells. Cell Tissue Res. 2012;349:809–824. doi: 10.1007/s00441-012-1371-2. doi:10.1007/s00441-012-1371-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauster M, Moser G, Orendi K, Huppertz B. Factors involved in regulating trophoblast fusion: potential role in the development of preeclampsia. Placenta. 2009;30(Suppl A):S49–S54. doi: 10.1016/j.placenta.2008.10.011. doi:10.1016/j.placenta.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Ghulmiyyah L, Sibai B. Maternal mortality from preeclampsia/eclampsia. Semin Perinatol. 2012;36:56–59. doi: 10.1053/j.semperi.2011.09.011. doi:10.1053/j.semperi.2011.09.011. [DOI] [PubMed] [Google Scholar]
- Hoegh AM, Borup R, Nielsen FC, Sorensen S, Hviid TV. Gene expression profiling of placentas affected by pre-eclampsia. J Biomed Biotechnol. 2010;2010:787545. doi: 10.1155/2010/787545. doi:10.1155/2010/787545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg K, Blair JD, McFadden DE, von Dadelszen P, Robinson WP. Early onset pre-eclampsia is associated with altered DNA methylation of cortisol-signalling and steroidogenic genes in the placenta. PloS One. 2013a;8:e62969. doi: 10.1371/journal.pone.0062969. doi:10.1371/journal.pone.0062969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg K, Blair JD, von Dadelszen P, Robinson WP. Hypomethylation of the LEP gene in placenta and elevated maternal leptin concentration in early onset pre-eclampsia. Mol Cell Endocrinol. 2013b;367:64–73. doi: 10.1016/j.mce.2012.12.018. doi:10.1016/j.mce.2012.12.018. [DOI] [PubMed] [Google Scholar]
- Horiuchi A, Hayashi T, Kikuchi N, Hayashi A, Fuseya C, Shiozawa T, Konishi I. Hypoxia upregulates ovarian cancer invasiveness via the binding of HIF-1alpha to a hypoxia-induced, methylation-free hypoxia response element of S100A4 gene. Int J Cancer. 2012;131:1755–1767. doi: 10.1002/ijc.27448. doi:10.1002/ijc.27448. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. doi:10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hung TH, Skepper JN, Charnock-Jones DS, Burton GJ. Hypoxia-reoxygenation: a potent inducer of apoptotic changes in the human placenta and possible etiological factor in preeclampsia. Circ Res. 2002;90:1274–1281. doi: 10.1161/01.res.0000024411.22110.aa. doi:10.1161/01.RES.0000024411.22110.AA. [DOI] [PubMed] [Google Scholar]
- Huppertz B. Placental pathology in pregnancy complications. Thromb Res. 2011;127(Suppl 3):S96–S99. doi: 10.1016/S0049-3848(11)70026-3. doi:10.1016/S0049-3848(11)70026-3. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. doi:10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvenpaa J, Vuoristo JT, Savolainen ER, Ukkola O, Vaskivuo T, Ryynanen M. Altered expression of angiogenesis-related placental genes in pre-eclampsia associated with intrauterine growth restriction. Gynecol Endocrinol. 2007;23:351–355. doi: 10.1080/09513590701350291. doi:10.1080/09513590701350291. [DOI] [PubMed] [Google Scholar]
- Jia RZ, Zhang X, Hu P, Liu XM, Hua XD, Wang X, Ding HJ. Screening for differential methylation status in human placenta in preeclampsia using a CpG island plus promoter microarray. Int J Mol Med. 2012;30:133–141. doi: 10.3892/ijmm.2012.983. [DOI] [PubMed] [Google Scholar]
- Kleinrouweler CE, Wiegerinck MM, Ris-Stalpers C, Bossuyt PM, van der Post JA, von Dadelszen P, Mol BW, Pajkrt E EBM CONNECT Collaboration. Accuracy of circulating placental growth factor, vascular endothelial growth factor, soluble fms-like tyrosine kinase 1 and soluble endoglin in the prediction of pre-eclampsia: a systematic review and meta-analysis. BJOG. 2012;119:778–787. doi: 10.1111/j.1471-0528.2012.03311.x. doi:10.1111/j.1471-0528.2012.03311.x. [DOI] [PubMed] [Google Scholar]
- Kuc S, Wortelboer EJ, van Rijn BB, Franx A, Visser GH, Schielen PC. Evaluation of 7 serum biomarkers and uterine artery Doppler ultrasound for first-trimester prediction of preeclampsia: a systematic review. Obstet Gynecol Surv. 2011;66:225–239. doi: 10.1097/OGX.0b013e3182227027. doi:10.1097/OGX.0b013e3182227027. [DOI] [PubMed] [Google Scholar]
- Louwen F, Muschol-Steinmetz C, Reinhard J, Reitter A, Yuan J. A lesson for cancer research: placental microarray gene analysis in preeclampsia. Oncotarget. 2012;3:759–773. doi: 10.18632/oncotarget.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee LA, Helewa M, Moutquin JM, von Dadelszen P Hypertension Guideline Committee and Strategic Training Initiative in Research in the Reproductive Health Sciences (STIRRHS) Scholars. Diagnosis, evaluation, and manage-ment of the hypertensive disorders of pregnancy. J Obstet Gynaecol Can. 2008;30:S1–48. [Google Scholar]
- Maksimovic J, Gordon L, Oshlack A. SWAN: Subset-quantile Within Array Normalization for Illumina Infinium HumanMethylation450 BeadChips. Genome Biol. 2012;13:R44. doi: 10.1186/gb-2012-13-6-r44. doi:10.1186/gb-2012-13-6-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoura S, Kalogiannidis IA, Gitas G, Goutsioulis A, Koiou E, Athanasiadis A, Vavatsi N. Biomarkers in pre-eclampsia: a novel approach to early detection of the disease. J Obstet Gynaecol. 2012;32:609–616. doi: 10.3109/01443615.2012.709290. doi:10.3109/01443615.2012.709290. [DOI] [PubMed] [Google Scholar]
- Meinhardt G, Husslein P, Knofler M. Tissue-specific and ubiquitous basic helix-loop-helix transcription factors in human placental trophoblasts. Placenta. 2005;26:527–539. doi: 10.1016/j.placenta.2004.09.005. doi:10.1016/j.placenta.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Mousa AA, Archer KJ, Cappello R, Estrada-Gutierrez G, Isaacs CR, Strauss JF, 3rd, Walsh SW. DNA methylation is altered in maternal blood vessels of women with preeclampsia. Reprod Sci. 2012;19:1332–1342. doi: 10.1177/1933719112450336. doi:10.1177/1933719112450336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa H, Pryor-Koishi K, Kato T, Kowa H, Kurahashi H, Udagawa Y. Microarray analysis of differentially expressed fetal genes in placental tissue derived from early and late onset severe pre-eclampsia. Placenta. 2007;28:487–497. doi: 10.1016/j.placenta.2006.05.010. doi:10.1016/j.placenta.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Nishizawa H, Ota S, Suzuki M, Kato T, Sekiya T, Kurahashi H, Udagawa Y. Comparative gene expression profiling of placentas from patients with severe pre-eclampsia and unexplained fetal growth restriction. Reprod Biol Endocrinol. 2011;9:107. doi: 10.1186/1477-7827-9-107. doi:10.1186/1477-7827-9-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novakovic B, Yuen RK, Gordon L, Penaherrera MS, Sharkey A, Moffett A, Craig JM, Robinson WP, Saffery R. Evidence for widespread changes in promoter methylation profile in human placenta in response to increasing gestational age and environmental/stochastic factors. BMC Genomics. 2011;12:529. doi: 10.1186/1471-2164-12-529. doi:10.1186/1471-2164-12-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogoshi K, Hashimoto S, Nakatani Y, Qu W, Oshima K, Tokunaga K, Sugano S, Hattori M, Morishita S, Matsushima K. Genome-wide profiling of DNA methylation in human cancer cells. Genomics. 2011;98:280–287. doi: 10.1016/j.ygeno.2011.07.003. doi:10.1016/j.ygeno.2011.07.003. [DOI] [PubMed] [Google Scholar]
- Price EM, Cotton AM, Lam LL, Farre P, Emberly E, Brown CJ, Robinson WP, Kobor MS. Additional annotation enhances potential for biologically-relevant analysis of the Illumina Infinium HumanMethylation450 BeadChip array. Epigenetics Chromatin. 2013;6:4. doi: 10.1186/1756-8935-6-4. doi:10.1186/1756-8935-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. 2011. http://www.bioconductor.org/ .
- Rajakumar A, Whitelock KA, Weissfeld LA, Daftary AR, Markovic N, Conrad KP. Selective overexpression of the hypoxia-inducible transcription factor, HIF-2alpha, in placentas from women with preeclampsia. Biol Reprod. 2001;64:499–506. doi: 10.1093/biolreprod/64.2.499. [DOI] [PubMed] [Google Scholar]
- Redline RW, Patterson P. Pre-eclampsia is associated with an excess of proliferative immature intermediate trophoblast. Hum Pathol. 1995;26:594–600. doi: 10.1016/0046-8177(95)90162-0. doi:10.1016/0046-8177(95)90162-0. [DOI] [PubMed] [Google Scholar]
- Redman CW, Sargent IL. Immunology of pre-eclampsia. Am J Reprod Immunol. 2010;63:534–543. doi: 10.1111/j.1600-0897.2010.00831.x. doi:10.1111/j.1600-0897.2010.00831.x. [DOI] [PubMed] [Google Scholar]
- Rolfo A, Many A, Racano A, Tal R, Tagliaferro A, Ietta F, Wang J, Post M, Caniggia I. Abnormalities in oxygen sensing define early and late onset preeclampsia as distinct pathologies. PLoS One. 2010;5:e13288. doi: 10.1371/journal.pone.0013288. doi:10.1371/journal.pone.0013288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. doi:10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schodel J, Oikonomopoulos S, Ragoussis J, Pugh CW, Ratcliffe PJ, Mole DR. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood. 2011;117:e207–e217. doi: 10.1182/blood-2010-10-314427. doi:10.1182/blood-2010-10-314427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwender H. Siggenes: Multiple Testing using SAM and Efron's Empirical Bayes Approaches. 2011. R package version 1.28.0 http://www.bioconductor.org/ [Google Scholar]
- Shahrzad S, Bertrand K, Minhas K, Coomber BL. Induction of DNA hypomethylation by tumor hypoxia. Epigenetics. 2007;2:119–125. doi: 10.4161/epi.2.2.4613. doi:10.4161/epi.2.2.4613. [DOI] [PubMed] [Google Scholar]
- Sitras V, Paulssen RH, Gronaas H, Leirvik J, Hanssen TA, Vartun A, Acharya G. Differential placental gene expression in severe preeclampsia. Placenta. 2009;30:424–433. doi: 10.1016/j.placenta.2009.01.012. doi:10.1016/j.placenta.2009.01.012. [DOI] [PubMed] [Google Scholar]
- Steegers EA, von Dadelszen P, Duvekot JJ, Pijnenborg R. Pre-eclampsia. Lancet. 2010;376:631–644. doi: 10.1016/S0140-6736(10)60279-6. doi:10.1016/S0140-6736(10)60279-6. [DOI] [PubMed] [Google Scholar]
- Tal R. The Role of Hypoxia and Hypoxia-Inducible Factor 1 Alpha in Preeclampsia Pathogenesis. Biol Reprod. 2012;87:134. doi: 10.1095/biolreprod.112.102723. doi:10.1095/biolreprod.112.102723. [DOI] [PubMed] [Google Scholar]
- Tsai S, Hardison NE, James AH, Motsinger-Reif AA, Bischoff SR, Thames BH, Piedrahita JA. Transcriptional profiling of human placentas from pregnancies complicated by preeclampsia reveals disregulation of sialic acid acetylesterase and immune signalling pathways. Placenta. 2011;32:175–182. doi: 10.1016/j.placenta.2010.11.014. doi:10.1016/j.placenta.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi T, Nagy B, Fule T, Tarca AL, Karaszi K, Schonleber J, Hupuczi P, Mihalik N, Kovalszky I, Rigo J, Jr, et al. Microarray profiling reveals that placental transcriptomes of early-onset HELLP syndrome and preeclampsia are similar. Placenta. 2011;32(Suppl):S21–S29. doi: 10.1016/j.placenta.2010.04.014. doi:10.1016/j.placenta.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Dadelszen P, Magee LA, Roberts JM. Subclassification of preeclampsia. Hypertens Pregnancy. 2003;22:143–148. doi: 10.1081/PRG-120021060. doi:10.1081/PRG-120021060. [DOI] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. doi:10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Zhang X, Li Q, Xu J, Zhou X, Wang T, Xing Q, Liu Y, Wang L, He L, et al. Promoter hypomethylation of TIMP3 is associated with pre-eclampsia in a Chinese population. Mol Hum Reprod. 2012;19:153–159. doi: 10.1093/molehr/gas054. doi:10.1093/molehr/gas054. [DOI] [PubMed] [Google Scholar]
- Yuen RK, Avila L, Penaherrera MS, von Dadelszen P, Lefebvre L, Kobor MS, Robinson WP. Human placental-specific epipolymorphism and its association with adverse pregnancy outcomes. PLoS One. 2009;4:e7389. doi: 10.1371/journal.pone.0007389. doi:10.1371/journal.pone.0007389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen RK, Penaherrera MS, von Dadelszen P, McFadden DE, Robinson WP. DNA methylation profiling of human placentas reveals promoter hypomethylation of multiple genes in early-onset preeclampsia. Eur J Hum Genet. 2010;18:1006–1012. doi: 10.1038/ejhg.2010.63. doi:10.1038/ejhg.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen RK, Manokhina I, Robinson WP. Are we ready for DNA methylation-based prenatal testing? Epigenomics. 2011;3:387–390. doi: 10.2217/epi.11.62. doi:10.2217/epi.11.62. [DOI] [PubMed] [Google Scholar]
- Yuen RK, Chen B, Blair JD, Robinson WP, Nelson DM. Hypoxia alters the epigenetic profile in cultured human placental trophoblasts. Epigenetics. 2013;8:192–202. doi: 10.4161/epi.23400. doi:10.4161/epi.23400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeybek YG, Gunel T, Benian A, Aydinli K, Kaleli S. Clinical evaluations of cell-free fetal DNA quantities in pre-eclamptic pregnancies. J Obstet Gynaecol Res. 2012;39:632–640. doi: 10.1111/j.1447-0756.2012.02011.x. doi:10.1111/j.1447-0756.2012.02011.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.