Abstract
Background
Because effective prediction of survival time can be highly beneficial for the treatment of glioblastoma patients, the relationship between survival time and multiple patient characteristics has been investigated. In this paper, we investigate whether the predictive power of a survival model based on clinical patient features improves when MRI features are also included in the model.
Methods
The subjects in this study were 82 glioblastoma patients for whom clinical features as well as MR imaging exams were made available by The Cancer Genome Atlas (TCGA) and The Cancer Imaging Archive (TCIA). Twenty-six imaging features in the available MR scans were assessed by radiologists from the TCGA Glioma Phenotype Research Group. We used multivariate Cox proportional hazards regression to construct 2 survival models: one that used 3 clinical features (age, gender, and KPS) as the covariates and 1 that used both the imaging features and the clinical features as the covariates. Then, we used 2 measures to compare the predictive performance of these 2 models: area under the receiver operating characteristic curve for the 1-year survival threshold and overall concordance index. To eliminate any positive performance estimation bias, we used leave-one-out cross-validation.
Results
The performance of the model based on both clinical and imaging features was higher than the performance of the model based on only the clinical features, in terms of both area under the receiver operating characteristic curve (P < .01) and the overall concordance index (P < .01).
Conclusions
Imaging features assessed using a controlled lexicon have additional predictive value compared with clinical features when predicting survival time in glioblastoma patients.
Keywords: glioblastoma, MRI, proportional hazards, survival analysis, VASARI
Glioblastoma (GBM) is the most commonly occurring type of malignant primary brain tumor and the second most common type of primary brain tumor in general.1 It is characterized by very poor survival rates: a 1-year survival rate of 35.2% and a 5-year survival rate of only 4.7%.1
Accurate prognosis for individual patients could be of high benefit to them, and thus multiple studies have been published examining the impact of various factors on time to death. Lacroix et al2 have shown that a high (≥98%) extent of tumor resection gives a significant survival advantage compared with a low (<98%) extent of resection. The dependence of survival on complete resection of the enhancing tumor was further confirmed by Stummer et al.3 Regarding clinical features, it has been demonstrated that age2,4 and Karnofsky Performance Status (KPS)2,4,5,6 are significant predictors of survival. Multiple recent studies focus on genomic predictors of survival in GBM patients. One among the most prominent studies is that of Verhaak et al,7 who found a gene expression–based classification for GBM patients that relates well to their clinical outcomes.
Although notably less attention has been given to the predictive value of pre- and postoperative medical imaging scans, some studies on the topic are available. Lacroix et al2 examined 7 different features based on pre- and postoperative MRI scans and showed that 4 of them were significant predictors of survival: tumor functional grade (proximity to eloquent brain), necrosis grade, edema grade, and enhancement grade. Pope et al8 evaluated the impact of 15 MRI variables on survival in GBM patients and found that noncontrast-enhancing tumor (nCET), edema, satellites, and multifocality were significant predictors of survival. Park et al6 identified that tumor involvement in prespecified eloquent brain regions and tumor volume were associated with poor postoperative survival. Recently, Zinn et al9 also showed that a model using KPS and age along with tumor volume (as determined by MRI) both predicts patient survival time well and correlates well with patient gene expression. These studies illustrate that particular features, or their combinations, are capable of classifying patients into groups that relate to survival. However, to our knowledge, there is limited scientific evidence in the literature10 that adding imaging features to popular clinical predictors such as KPS, age, and gender will improve the quality of survival predictions.
In this study, we evaluated the difference in performance (ie, how well survival can be predicted) of 2 models: one using only clinical features and one using both clinical and imaging features. In our study, we used a recently proposed set of controlled MRI features called VASARI (Visually Accessable REMBRANDT [Repository for Molecular Brain Neoplasia Data] Images). To date, only preliminary data on effectiveness of these features are available.
Materials and Methods
Patient Population
In this study, we used data provided by The Cancer Genome Atlas (TCGA) that contained clinical as well as genomic information for patients. The data in the TCGA set were collected according to appropriate institutional review board approval (TCGA Research Network 2008).11 For the subset of the GBM patients from TCGA, the MRI exams were made available by The Cancer Imaging Archive (TCIA) through a collaborative effort between the National Cancer Institute (NCI) and multiple clinical institutions in the United States. For this study, out of these we identified 82 GBM subjects for whom both the clinical information of interest (age, gender, and KPS) and imaging features extracted from the MRI exams by radiologists were available. Each of these cases was scrutinized and assigned MRI features by a panel of radiologists using the standardized VASARI lexicon (http://cabig.cancer.gov/action/collaborations/vasari/). For each case, a consensus rating was established by summarizing the radiologists' ratings. Each case was assigned to at least 3 radiologists for rating, and 76/82 cases were in fact rated by at least 3 radiologists. For the majority (68/82), the consensus was based on ratings by exactly 3 radiologists per case. Of the remaining 14 cases, 5 were rated by 6 radiologists, 3 were rated by 4 radiologists, 3 were rated by 2 radiologists, and 3 were rated by 1 radiologist. The reason for a case being rated by fewer than 3 radiologists (6 total cases) is that, 1 or 2 radiologists were not able to identify all necessary exams in the database (eg, 1 of the 3 MRI modalities) or deemed the case exams unsuitable for rating. In such situations, an additional arbiter investigated the case to resolve the conflict. The image annotations were collected through an NCI-coordinated multi-institutional effort by members of the TCGA Glioma Phenotype Research Group and were made available to us by the group.
Patient Features
Each patient was characterized by a set of clinical and imaging (ie, VASARI) features. Specifically, the clinical features were age (in days), gender, and KPS. The VASARI lexicon for MRI annotation contains 26 imaging descriptors based on different MRI modalities, including T1 and T2/fluid attenuated inversion recovery (FLAIR). The exact description of all the features can be found at the National Cancer Institute's Cancer Imaging Archive (https://wiki.cancerimagingarchive.net/display/Public/VASARI+Research+Project).
The following MRI features were used: major axis length, minor axis length, tumor location, side of lesion center, eloquent brain, enhancement quality, proportion enhancing, proportion nCET, proportion necrosis, cysts, multifocal or multicentric, T1/FLAIR ratio, thickness of enhancing margin, definition of the enhancing margin, definition of the nonenhancing margin, proportion of edema, edema crosses midline, hemorrhage, diffusion characteristics, pial invasion, ependymal invasion, cortical involvement, deep white matter invasion, nCET tumor crosses midline, enhancing tumor crosses midline, satellites, and calvarial remodeling. Additionally, for each patient, time to death or time to last follow-up (for censored patients) was available.
Statistical Modeling
The modeling goal was to predict patients' time to death (dependent variable) based on their clinical and/or imaging features (independent variables). Two imaging features (tumor location and eloquent brain) were split into dummy binary variables representing each value of these features. To achieve this goal, we used a multivariate Cox proportional hazards regression model12 preceded by feature selection using univariate Cox proportional hazards regression models. Specifically, in the feature selection step, we first removed any features where the value of that feature across all subjects was constant. Such features have no predictive power. Then, we constructed a univariate Cox regression model for each remaining feature. Our feature selection algorithm, selected the features such that the P-value was significant for the feature in the univariate Cox regression model, which was P > .05. Finally, a multivariate Cox regression model was constructed using only the selected features. The survival prediction for each subject of interest was generated by a linear combination of the features, where the feature weights were determined by the multivariate Cox model.
For these calculations, we used the Cox proportional hazards regression function (coxphfit) in MATLAB's Statistical Toolbox.
Statistical Model Evaluation
To evaluate the models, we used leave-one-out cross-validation along with the receiver operating characteristic (ROC) methodology.13 Specifically, we divided the available dataset of subjects into a training set of all but 1 subject and a testing set containing the 1 remaining subject. Then, we conducted feature selection and model construction using only the training set and used the constructed model to calculate the survival prediction for the subject in the test set. The feature selection, model construction, and survival prediction are described in the Statistical Modeling section. Finally, we repeated this procedure multiple times so that each subject was excluded exactly once from training and received exactly 1 survival prediction.
To calculate the performance of the models, we pooled the predictions for all the subjects and computed an ROC curve. In this calculation, binary ground truth labels were assigned to each patient based on his/her survival time. Patients with a survival time >1 year were labeled positive and those with a survival time ≤1 year were labeled negative. The censored subjects with a follow-up time of ≤1 year were excluded from the ROC calculation because it could not be determined whether such patients survived >1 year. The censored subjects with a last follow-up time >1 year were labeled positive in the calculation because it is known that they survived >1 year. For both models, we calculated the area under the ROC curve using the trapezoidal rule. We calculated the confidence intervals (CIs) according to Delong et al.14 To statistically compare the models, we used the nonparametric comparison method proposed by DeLong et al.14 The ROC analysis was performed in R using the popular pROC package.15 Specifically, we used the roc and auc functions to calculate the areas under the curves, the ci.auc function to calculate CIs, and the roc.test function (with the DeLong method) to compare the 2 areas under the curves.
Additionally, we compared the 2 models of interest using the concordance (C) index16,17 measure, which allows for comparing 2 models over all applicable time thresholds rather than 1 selected threshold, as is done in ROC analysis. The C index is the proportion of all usable pairs of subjects such that the prediction of survival time and the actual survival time are in agreement.13 The C index has been previously used in the context of brain tumor survival analysis.18,19 We used the Student t-test for dependent samples for statistical comparison of the 2 C indices.20
To calculate and compare C indices, we used the SurvComp package20 for R; specifically we used the function concordance.index (using the Noether method) to calculate C indices with CIs, and the function cindex.comp for statistical comparison of the C indices.
Finally, as an additional, exploratory analysis, we evaluated the individual predictive power of each of the variables by calculating the area under the ROC curve for a classifier where the variable is the predictor of the survival class (as noted, positive was >365 days, negative was ≤365 days). The ROC calculation for this analysis was also done in R using the pROC package. The variables that had the same value for all the patients were excluded as having no predictive power in this context.
Results
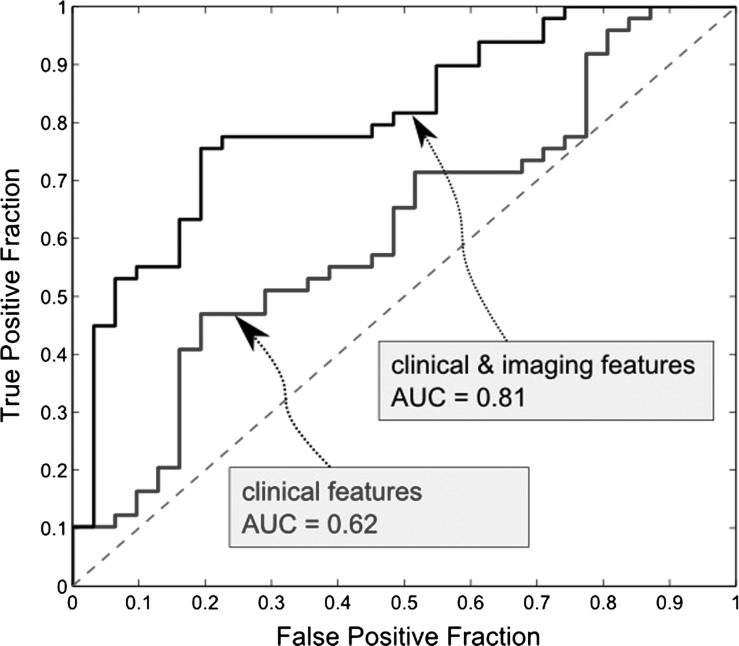
The ROC curves for the model using only clinical features and the model using both clinical and imaging features are presented in Fig. 1. The area under the ROC curve for the model using only clinical features was 0.62 (CI: 0.49–0.74). The area under the ROC curve for the model using clinical and imaging features was 0.81 (CI: 0.71–0.9). The area under the curve for the latter model was statistically significantly higher (P < .01), which demonstrates the added value of imaging descriptors for survival prognosis.
Fig. 1.
ROC curves for multivariate Cox proportional hazards regression model using clinical features only (blue), and both clinical and imaging features (black). The areas under these ROC curves (AUC) are significantly different (P < .01).
This result was confirmed using analysis based on the C index. The C index for the model combining clinical and imaging features was 0.69 (CI: 0.63–0.75). The C index for the model using clinical features only was 0.58 (CI: 0.5–0.66). The difference was statistically significant (P < .01). This confirms that imaging features improve the predictive power of survival models for GBM patients.
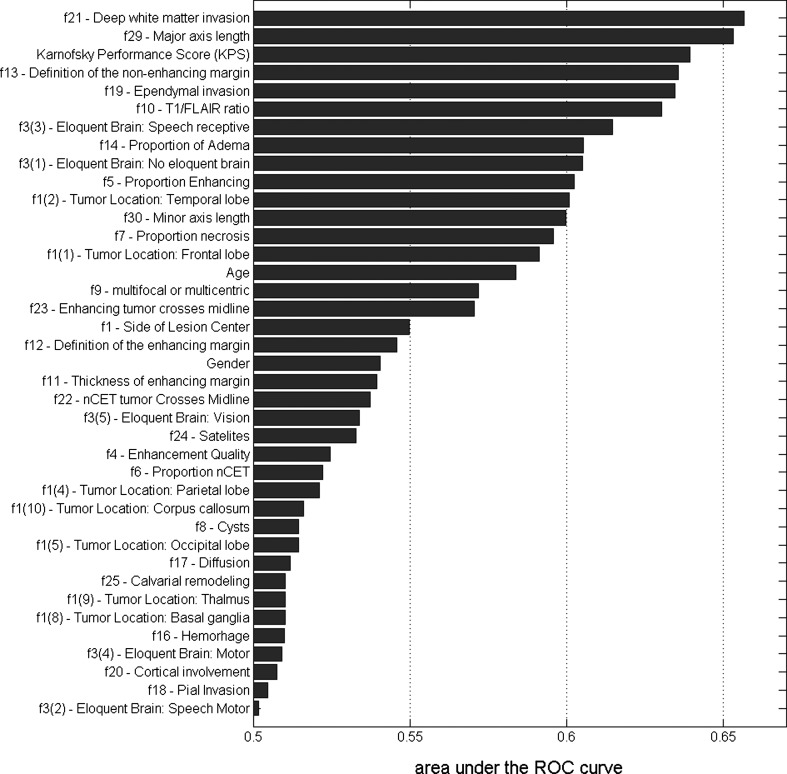
The results of our exploratory analysis regarding the importance of individual variables are presented in Fig. 2. Our analysis confirms the importance of some imaging features previously identified as important, such as tumor functional grade (proximity to eloquent brain), necrosis grade, edema grade, and enhancement grade, while identifying some imaging features that were previously not commonly identified as important (see Fig. 2. for details). Out of the 3 clinical features analyzed in this study, KPS was confirmed to have relatively high predictive power, while the power of age and gender in survival analysis is more limited.
Fig. 2.
Predictive power of individual features when predicting survival. The values of the bars are areas under the ROC curve where the individual feature is the predictor of the survival class (1-y threshold).
Discussion
Most previous studies on survival in GBM have focused on whether one or more patient features are a significant predictor of survival. Those studies, while undoubtedly important, do not offer insight into the added value of including additional covariates. Please note that even though presenting hazard ratios in univariate or multivariate Cox models is suggestive of the importance of individual covariates, most often it is insufficient for answering the question of whether adding a new set of features will improve the prediction of survival because such statistical analysis is simply not designed for that. Furthermore, detecting small improvements in predictive power of different sets of predictors is typically difficult and requires a large sample size.
In this study, for the first time, we present statistical evidence that imaging features improve the prediction of survival in GBM patients over clinical features alone. This has a potential of improving patient management. Our ability to detect this difference with only a moderate sample size (82 patients) was due to the high magnitude of the difference (ie, the effect of interest was very strong). Such a large improvement over the clinical features is potentially the result of using controlled imaging features (ie, VASARI). This hypothesis, however, needs additional experiments.
The benefit of using a controlled set of imaging features extends beyond the potential improvement in survival prediction. It also has a potential of reducing inter- and intraobserver variability. Additionally, with more controlled features, the results of different studies using MRI descriptors in GBM could be much more comparable to each other. This is very difficult in the current state where the definitions of imaging features are left to the interpretation of each individual reader. A controlled lexicon will in turn facilitate the creation of a more concrete body of knowledge regarding the relation of imaging features with clinical, genomic, and other features, rather than a set of loosely related hypotheses. A controlled lexicon can also improve the clinical management of GBM patients through improved interreader agreement and easier communication of results among clinicians. The benefits of controlled lexicons have been appreciated in breast imaging in the form of the Breast Imaging-Reporting and Data System, BI-RADS.21 While VASARI is only a very recent development, it has the potential of providing similar benefits to patient care and research.
There are some limitations of our study as well as potential future investigation that should be discussed. First, in this study we used the popular leave-one-out cross-validation technique for model evaluation, which generally provides an accurate estimate of model test performance. However, an independent dataset would be beneficial in order to further validate the hypothesis of this study.
Furthermore, this study focuses on the added value provided by presurgical imaging features when they are combined with standard clinical features. Therefore, it was important that our study includes 2 clinical features that are commonly studied and accepted as good predictors of survival: age and KPS. Gender was included because it was available in the dataset and is also a potentially useful feature. In the future, though, other less commonly studied but potentially predictive features could also be evaluated. Such features might include neurologic signs and symptoms, especially seizure history. Future studies could also include surgical features such as the extent of tumor resection, which has generally been shown to correlate with survival time. It also remains to be seen whether some relatively easy to obtain features might replace features more difficult to assess (eg, extent of tumor resection could replace some preoperative imaging features) and whether combining all these features could result in an even better survival model than the one presented in this paper.
Finally, due to the limited sample size available for this study, we limited the hypotheses tested in this paper to comparing the 2 multivariate Cox proportional hazards regression models with the features selected in a simple automatic feature selection step. To get some insight into the importance of individual features, we evaluated their predictive values, but no statistical analysis is presented for these additional exploratory results. Other combinations of features (eg, combinations of 2 features) could potentially yield models with improved predictive value or simplicity. However, a larger sample size would be needed for such analysis due to the necessity of repeated statistical tests (to statistically compare different combinations). Such analysis could be part of future work when more data of this type are available. Following the acceptance of this manuscript, another study10 was published using the VASARI feature set with a subset of features. Our study is complementary to10 in that our study focuses on cross validation-based comparison of the predictive power of a model that uses standard clinical variables and a model that uses both: clinical and imaging variables. The other study10, in addition to evaluating the association between some VASARI features and survival also investigates the relationship between imaging and genomic features in GBM patients.
Acknowledgments
The authors would like to thank the TCGA Glioma Phenotype Research Group for providing the reading data used in this study. We would also like to acknowledge The Cancer Imaging Archive and The Cancer Genome Atlas initiatives for making the imaging and the clinical data used in this study publicly available.
Conflict of interest statement. None declared.
References
- 1. Central Brain Tumor Registry of the United States. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2004–2008. http://www.cbtrus.org/2012-NPCR-SEER/CBTRUS_Report_2004-2008_3-23-2012.pdf. (accessed December 18, 2012) [Google Scholar]
- 2.Lacroix M, Abi-Said D, Fourney DR, et al. A multivariate analysis of 416 patients with glioblastoma multiforme: prognosis, extent of resection, and survival. J Neurosurg. 2001;95(2):190–198. doi: 10.3171/jns.2001.95.2.0190. [DOI] [PubMed] [Google Scholar]
- 3.Stummer W, Reulen H-J, Meinel T, et al. Extent of resection and survival in glioblastoma multiforme: identification of and adjustment for bias. Neurosurgery. 2008;62(3):564–576. doi: 10.1227/01.neu.0000317304.31579.17. [DOI] [PubMed] [Google Scholar]
- 4.Carson KA, Grossman SA, Fisher JD, et al. Prognostic factors for survival in adult patients with recurrent glioma enrolled onto the new approaches to brain tumor therapy CNS consortium phase I and II clinical trials. J Clin Oncol. 2007;25:2601–2606. doi: 10.1200/JCO.2006.08.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Filippini G, Falcone C, Boiardi A, et al. Prognostic factors for survival in 676 consecutive patients with newly diagnosed primary glioblastoma. Neuro-oncology. 2008;10:79–87. doi: 10.1215/15228517-2007-038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park JK, Hodges T, Arko L, et al. Scale to predict survival after surgery for recurrent glioblastoma multiforme. J Clin Oncol. 2010;28(24):3838–3843. doi: 10.1200/JCO.2010.30.0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verhaak RGW, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pope WB, Sayre J, Perlina A, Villablanca JP, Mischel PS, Cloughesy TF. MR imaging correlates of survival in patients with high-grade gliomas. Am J Neuroradiol. 2005;26(10):2466–2474. [PMC free article] [PubMed] [Google Scholar]
- 9.Zinn PO, Sathyan P, Mahajan B, et al. A novel volume-age-KPS (VAK) glioblastoma classification identifies a prognostic cognate microRNA-gene signature. PLoS ONE. 2012;7(8):e41522. doi: 10.1371/journal.pone.0041522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gutman DA, Cooper LAD, Hwang SN, et al. MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Radiology. 2013 doi: 10.1148/radiol.13120118. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McLendon R, Friedman A, Bigner D, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox DR. Regression models and life-tables. J Roy Statist Soc B (Methodological) 1972;34(2):187–220. [Google Scholar]
- 13.Swets JA, Pickett RM. Evaluation of Diagnostic Systems: Methods from Signal Detection Theory. New York: Academic Press;; 1982. [Google Scholar]
- 14.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–845. [PubMed] [Google Scholar]
- 15.Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12(1):77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrell FE, Jr., Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15(4):361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 17.Pencina MJ, D'Agostino RB. Overall C as a measure of discrimination in survival analysis: model specific population value and confidence interval estimation. Stat Med. 2004;23(13):2109–2123. doi: 10.1002/sim.1802. [DOI] [PubMed] [Google Scholar]
- 18.van den Bent MJ, Gravendeel LA, Gorlia T, et al. A hypermethylated phenotype in anaplastic oligodendroglial brain tumors is a better predictor of survival than MGMT methylation in anaplastic oligodendroglioma: a report from EORTC study 26951. Clin Cancer Res. 2011;17(22):7148–7155. doi: 10.1158/1078-0432.CCR-11-1274. [DOI] [PubMed] [Google Scholar]
- 19.Quillien V, Lavenu A, Karayan-Tapon L, et al. Comparative assessment of 5 methods (methylation-specific polymerase chain reaction, methylight, pyrosequencing, methylation-sensitive high-resolution melting, and immunohistochemistry) to analyze O6-methylguanine-DNA-methyltranferase in a series of 100 glioblastoma patients. Cancer. 118(17):4201–4211. doi: 10.1002/cncr.27392. [DOI] [PubMed] [Google Scholar]
- 20.Haibe-Kains B, Desmedt C, Sotiriou C, Bontempi G. A comparative study of survival models for breast cancer prognostication based on microarray data: does a single gene beat them all? Bioinformatics. 2008;24(19):2200–2208. doi: 10.1093/bioinformatics/btn374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D’Orsi CJ, Mendelson EB, Ikeda DM. Breast Imaging Reporting and Data System: ACR BI-RADS–Breast imaging Atlas. Reston, VA: American College of Radiology; 2003. [Google Scholar]