Abstract
Background
Glioblastoma multiforme (GBM) is a highly aggressive brain tumor characterized by massive neovascularization, necrosis, and intense resistance to therapy. Deregulated Notch signaling has been implicated in the formation and progression of different malignancies. The present study attempted to investigate the activation status of Dll4-Notch signaling in primary human GBM and its association with vascular and clinical parameters in patients.
Methods
Major components of Dll4-Notch signaling were examined by real-time reverse-transcription polymerase chain reaction (PCR), Western blotting, and immunohistochemistry in GBM (n = 26) and control (n = 11) brain tissue. The vascular pattern (VP) and microvascular density (MVD) were analyzed after laminin immunostaining. O6-Methylguanine-methyltransferase (MGMT) promoter methylation in GBM samples was detected by methylation-specific PCR.
Results
The mRNA levels of Dll4, Jagged1, Notch1, Notch4, Hey1, Hey2, Hes1, and VEGF were 3.12-, 3.58-, 3.37-, 5.77-, 4.89-, 3.13-, 6.62-, and 32.57-fold elevated, respectively, in GBM samples, compared with the controls. Western blotting revealed a 4-, 3.7-, and 45.6-fold upregulation of Dll4, Notch1, and Hey1, respectively, accompanied by a downregulation of PTEN expression and an increase in the expression of p-Akt and VEGF. Immunostaining located the immunoreactivity of Dll4 and Notch1 in endothelial cells, microglia/macrophages, tumor cells, and astrocytes. Furthermore, the upregulation of Dll4-Notch signaling components was correlated to a low MVD and was potentially related to a classic VP, tumor edema, and MGMT promoter methylation.
Conclusions
The upregulation of Dll4-Notch signaling components was found in a subset of GBM samples and was associated with some angiogenic and clinical parameters. These findings highlight this signaling pathway as a potential therapeutic target for patients with GBM who show an activation of Dll4-Notch signaling.
Keywords: Dll4-Notch signaling, macrophage/microglia, microvascular density, primary glioblastoma multiforme, vascular pattern
Primary glioblastoma multiforme (GBM) is the most common and aggressive type of brain tumor, with an incidence of 5–6 cases/100 000 persons/year in European countries.1,2 This condition arises without clinical, radiological, or histopathological evidence of a less-malignant precursor lesion.3,4 GBM is characterized by rapid growth and diffuse invasiveness into the adjacent brain parenchyma. Surgical treatment can only control the solid component of the GBM, whereas the infiltrative component has to be treated by chemotherapy and radiotherapy.5 However, because of diffuse invasion, necrosis, anti-apoptosis, and microvascular proliferation, GBM appears to be resistant to current standard therapies and is ultimately incurable, with a median survival of 14.6 months.6,7
The Notch signaling pathway is an evolutionarily conserved intercellular signaling pathway affecting various cellular processes and the determination of cell fate during embryonic and postnatal development.8 Five transmembrane Notch ligands (Jagged1, Jagged2, Delta-like [Dll] 1, Dll3, and Dll4) and 4 Notch receptors (Notch 1–4) have been identified in mammalian cells. Binding of the ligand to a specific receptor activates Notch signaling by cleavage of the Notch intracellular domain (NICD) via intramembrane proteolysis by γ-secretase. The NICD then translocates to the nucleus, where it typically forms a complex with transcription factors and transcriptional coactivators to activate the transcription of target genes. The members of the Hey family (Hey1, 2, and L) and the Hairy/Enhancer of split family (Hes1-7) are well-characterized target genes of Notch signaling.9–12 Dll4 and Notch4 are specifically expressed in endothelial cells, whereas Notch1 is naturally expressed in extravascular tissues and in developing vasculature.10 This expression pattern suggests a pivotal role of Dll4 in the regulation of angiogenesis through the Notch1/4 receptors,12 whereas Jagged ligands have been shown to play a particular role in pericyte function.13
Notch receptors and Notch ligands are present or upregulated in several human malignancies.14,15 The activation of Notch signaling can be either oncogenic or tumor suppressive depending on the cellular and physiological context.16 Notch signaling is linked to several downstream targets, including the phosphatidylinositol-3-OH kinase (PI3K)-Akt pathway, which is a well-characterized signaling pathway regulating cell growth and cell survival. The activation of Notch signaling has been shown to lead to excessive PI3K/Akt activation, thereby promoting tumor progression.17–21
In addition to a direct involvement in tumor progression, the Dll4-Notch pathway has also been implicated in tumor angiogenesis.22 Expression of Dll4, Jagged1, Notch1, Notch4, Hes1, and Hey1 was upregulated in vivo after hypoxia, which in turn promoted tumor angiogenesis.23,24 Inhibition of Notch signaling by the Dll4 antibody was shown to significantly reduce tumor proliferation,25 and enhanced expression of Dll4 has been reported in tumor vessels of several cancers, including gliomas.26,27
Increasing evidence indicates the presence of deregulated Dll4-Notch signaling in patients with GBM,23,28 in whom the upregulation of Dll4-Notch is suggested to be oncogenic and to play a role in the progression of GBM. The inhibition of Notch signaling resulted in growth arrest and deregulated vasculature in a subset of investigated glial cells.29 Notch signaling also appeared to be involved in the activation of cancer stem cell self-renewal.29–31 Furthermore, the expression of Hey1, a target gene of Dll4-Notch, was found to be positively correlated with poor survival among patients with GBM.32
In the present study, we investigated the expression profile of the major components of the Dll4-Notch signaling pathway in patients with primary GBM. The expression pattern of these components was further correlated to vascular and clinical parameters in the patients.
Materials and Methods
Patients
Surgical specimens (n = 26) were consecutively collected from adult patients with primary GBM who were treated in the Department of Neurosurgery at the University Hospital of Essen from 2009 through 2010. The inclusion criterion was the histopathological diagnosis of primary GBM (according to World Health Organization classification, without the oligodendroglial component). Only tumors operated by gross total resection (GTR) were collected for further analyses. GTR was defined as a resection of at least 95% of the contrast-enhancing tumor mass, which was confirmed by postoperative magnetic resonance imaging (MRI). The tumor samples were immediately stored at –80°C. All experiments were performed with histopathologically confirmed tumor material taken from the core of the tumor and distant from the infiltration zone. The surgical specimens from patients who underwent anterior temporal lobe resections because of temporal lobe epilepsy were used as control tissue (n = 11). The study was performed in strict accordance with the Declaration of Helsinki and was approved by the local ethics committee of the University Hospital of Essen. Informed consent was obtained from all patients.
Grading of Edema in Preoperative MRI and Evaluation of the Karnofsky Performance Index (KPI)
The disruption of the blood-brain barrier in GBM leads to differentially expressed edema detected by MRI. Edema on MRI appears as a region of increased T2 signal intensity outside the contrast-enhanced area. Accordingly, edema was classified into 3 grades: grade 0 when the amount of edema was less than the amount of the tumor volume, grade 1 when the amount of edema was approximately equal to the amount of the tumor volume, and grade 2 when the amount of edema was greater than the amount of tumor volume.33
The KPI has been established as one of the major prognostic indicators for GBM survival. A KPI <70 has been shown to be significantly associated with a poor survival rate. All patients enrolled in the present study were evaluated with respect to a preoperative KPI above or below 70 according to the protocol described previously.34
O6-Methylguanine-Methyltransferase (MGMT) Promoter Methylation Analysis
For MGMT promoter methylation analysis, DNA was isolated from paraffin sections of GBM. MGMT promoter methylation was analyzed by methylation-specific polymerase chain reaction (PCR), as described previously.35,36 The primer sequences used to detect unmethylated MGMT promoter sequences were 5-TGT GTT TTT AGA ATG TTT TGT GTT TTG AT-3 and 5-CTA CCA CCA TCC CAA AAA AAA ACT CCA-3. The primer sequences used to detect methylated MGMT promoter sequences were 5-GTT TTT AGA ACG TTT TGC GTT TCG AC-3 and 5-CAC CGT CCC GAA AAA AAA CTC CG-3.37
Total RNA Extraction, cDNA Synthesis and Real-Time PCR
Total RNA was extracted from the surgical specimens of GBM and controls with use of the RNeasy Lipid Tissue kit (Qiagen; Hilden, Germany), and cDNA was synthesized using the iScript cDNA Synthesis Kit (Bio-Rad Laboratories GmbH; Munich, Germany) according to the manufacturer's instructions. The PCR mixture was prepared to a final volume of 15 μL, containing 6 μL of the cDNA sample (4 ng/μL), 7.5 μL of SYBR Green fluorescein mix (ABgene, Bio-Rad Laboratories GmbH; Munich, Germany), 0.3 μL each of the forward and reverse primers (10 μM), and 0.9 μL of RNase-free H2O. The primer sequences are shown in Table 1. Real-time PCR was performed using the following settings: initial denaturation at 95°C for 15 min, 40 cycles of amplification at 95°C for 30 s, 60°C for 30 s, and 72°C for 50 s. For the melting curve, the following settings were used: 72°C for 30 s, 95°C for 1 min, and 55–95°C with a heating rate increase of 0.5°C every 10 s. Relative mRNA expression (fold of change) was calculated according to the cycle threshold approach, normalized to the reference gene GAPDH. The specificity of amplification was monitored at the end of each reaction by melting curve analysis.38
Table 1.
Primer sequences and annealing temperatures for real-time reverse-transcription PCR
| Primer | Sequence | Annealing temperature (°C) |
|---|---|---|
| DLL4 | ||
| for. | GCG GGG TAC CTT CTC GCTCAT CAT C | 60 |
| rev. | GCC TCC CCA GCC CTC ATC ACA AGT A | |
| Jagged1 | ||
| for. | TCG CTG TAT CTG TCC ACC TG | 60 |
| rev. | AGT CAC TGG CAC GGT TGT AG | |
| Notch1 | ||
| for. | CAG GCA ATC CGA GGA CTA TG | 60 |
| rev. | CAG GCG TGT TGT TCT CAC AG | |
| Notch4 | ||
| for. | TCC TGG GGC CCG GGC TGA AGA AAA G | 58 |
| rev. | ACG CCG GAT GAG CTG GAG GAC GAG A | |
| HEY1 | ||
| for. | CAGGCAATCCGAGGACTATG | 60 |
| rev. | CAGGCAATCCGAGGACTATG | |
| HEY2 | ||
| for. | GTA CCA TCC AGC AGT GCA TC | 60 |
| rev. | AGA GAA TTC AGT CAG GGC ATT T | |
| HES1 | ||
| for. | AGT GAA GCA CCT CCG GAA C | 60 |
| rev. | CGT TCA TGC ACT CGC TGA | |
| VEGF | ||
| for. | GAA GTG GTG AAG TTC ATG GAT GT | 60 |
| rev. | TGG AAG ATG TCC ACC AGG GTC | |
| GAPDH | ||
| for. | AGC CAC ATC GCT CAG ACA | 58 |
| rev. | GCC CAA TAC GAC CAA ATC C |
Abbreviations: for., forward; rev., reverse.
Western Blotting
The surgical specimens were sonicated in a lysis buffer containing 10% glycerol, 3% sodium dodecylsulfate, 0.05 mol/L Tris (pH 6.8), and 0.01% of protease and phosphatase inhibitor cocktails (Sigma-Aldrich; Seelze, Germany). The protein concentration was detected using a Micro BCA Kit (ThermoScientific; Schwerte, Germany). Samples containing an equal amount of total protein (50 μg) were loaded onto 10.0% sodium dodecyl sulfate–polyacrylamide gels. After electrophoresis, the proteins were transferred onto a nitrocellulose membrane. Unspecific binding was blocked by a buffer containing 0.1% Tween-20, 2% bovine serum albumin, and 5% nonfat dry milk in Tris-buffered saline. The blots were then incubated with different primary antibodies overnight at 4°C. The following primary antibodies were used: rabbit anti-Dll4 (1:1000, Cell Signaling Technology; Frankfurt, Germany), rabbit anti-Notch1 (1:1000; Cell Signaling Technology), mouse anti–phospho-Akt (1:1000; Cell Signaling Technology), rabbit anti-AKT (1:1000; Cell Signaling Technology), rabbit anti-VEGF (1:1000, Abcam; Cambridge, UK), rabbit anti-actin (1:1000; Sigma-Aldrich), mouse anti-PTEN (1:1000; Cell Signaling Technology), and rabbit anti-Hey1 (1:200; Sigma Aldrich). After the secondary antibody reaction, the signal was produced by enhanced chemiluminescence (Amersham Bioscience; Freiburg, Germany). For semi-quantification of the blot, the integrated optical density of the bands on the blots was measured using Image J software (http://rsbweb.nih.gov/ij/). The relative expression of the target protein in the GBM samples was calculated by determining the integrated optical density ratio of the target protein to the housekeeping protein actin and was expressed as a percentage of the control.
Immunofluorescence
After deparaffinization using standard graded ethanol, the sections underwent heat-induced epitope retrieval in a retrieval solution (DakoCytomation; Glostrup, Denmark). Unspecific binding was blocked by incubation of the sections with a blocking buffer containing 0.3% Triton X-100 (Sigma-Aldrich) and 5% goat serum in phosphate-buffered saline. For double staining, the sections were incubated with the following antibody mixtures: rabbit anti-Dll4 (1:100) and mouse anti-CD68 (1:100; DakoCytomation), rabbit anti-Dll4 (1:100), and mouse anti-GFAP (1:100; Sigma); rabbit anti-Notch1 (1:100) and mouse anti-CD68; and rabbit anti-Notch1 (1:100) and mouse anti-GFAP (1:100). The sections were then incubated overnight at 4°C with biotinylated goat anti-rabbit IgG and Texas red anti-mouse IgG (H + L) (Vector Laboratories; Dossenheim, Germany), followed by the substrate reaction with FITC-labeled avidin (DakoCytomation; Denmark). Counterstaining was performed with Hoechst-33258 (Invitrogen; Karlsruhe, Germany). The sections were analyzed using a fluorescence microscope (Olympus BX51). Images were acquired using an Olympus DP 70 camera and the associated Olympus cellF software.
Immunohistochemistry Staining
Paraffin-embedded sections were deparaffinized in xylene, dehydrated in graded alcohol, and rinsed in H2O for 15 min. After antigen retrieval, the sections were incubated with either laminin-specific polyclonal antibody (1:300, Sigma-Aldrich; L9393), rabbit anti-human Dll4 (1:100, AbD serotec; Düsseldorf, Germany; AHP1274), or rabbit anti-human Notch1 (1:100; Cell Signaling Technology; #3608) overnight at 4°C. The negative control sections were incubated with nonimmune rabbit/mouse IgG in equal concentrations to the primary antibody.
Evaluation of Microvascular Density (MVD) and Vascular Pattern (VP)
The hot-spot method, a widely used approach for vessel analysis in solid tumors, was performed to evaluate microvascular density.39,40 Laminin-immunostained tissue sections, taken from the tumor core, were scanned at a low magnification (40×), and then 3 tumor areas with the highest density of distinctly highlighted microvessels (“hot spots”) were selected. One microscopic field was identified in each hot spot at 200× magnification, providing a 0.81 mm2 field size. All individual microvessels were counted using an eye-grid. Microvascular density was defined as the number of manually counted vessels per square millimeter and presented as the mean of 3 hot spots.
Recent investigations have revealed the existence of distinct vascular patterns with prognostic impact in GBM.41,42 We therefore analyzed vascular patterns in GBM tumor core on laminin-immunostained tissue sections according to an algorithm described by Preusser et al.43 In brief, the presence of vascular clusters (score B1), vascular garlands (score B2), or glomeruloid vascular formations (score B3) was assessed (none = 0; few/discrete = 1; many/prominent = 2). Then, score B was calculated as the sum of the subscores (score B = score B1 + score B2 + score B3). The vascular pattern was determined using scores A and B and the following algorithm: if score A = 1 and score B ≤ 2, then the VP was classified as a classic vascular pattern; if score A = 0 or score B > 2, then the VP was classified as a bizarre vascular pattern. The tendency to be classified as a classic or bizarre VP was also evaluated by the sum of scores B1 to B3 in individual GBM samples, with a score ranging from 0 to 6. The larger score reflexes the greater tendency to bizarre VP in the tumor.
Statistical Analysis
The quantitative data are presented as the mean ± standard deviation. All statistical analyses were performed using the GraphPad Prism software, version 4.0, and SPSS, version 17.0 (SPSS). Student's t test with Welch's correction for data sets with unequal variances was performed for the analysis of the data from real-time PCR (fold change), Western blot, and the association of mRNA expression (fold change) with vascular measures (VP, MVD) and with clinical parameters (MGMT promoter methylation, edema, KPI). P < .05 was considered as a statistically significant difference between comparison groups.
Results
Patients
Twenty-six patients with primary GBM were enrolled in the study. Clinical data are summarized in Table 2. The mean age at the time of primary diagnosis was 59.4 ± 11.8 years, and the mean KPI score was 80 ± 24. There were 16 male (61.5%) and 10 (38.5%) female patients, which reflects the male predominance of patients with GBM. Twelve (46.2%) of 26 patients had a positive MGMT-promoter methylation status, and 21 (80.8%) of 26 patients received combined postoperative radio-chemotherapy.
Table 2.
Clinical data on patients with GBM
| Case Number | Age (year) | Sex | KPI (%) | MGMT | Edema (grade) |
|---|---|---|---|---|---|
| 1 | 62 | M | 60 | + | 2 |
| 2 | 27 | F | 100 | − | 1 |
| 3 | 69 | M | 100 | − | 0 |
| 4 | 61 | M | 80 | + | 2 |
| 5 | 48 | M | 90 | − | 2 |
| 6 | 79 | M | 100 | − | 2 |
| 7 | 67 | M | 80 | + | 2 |
| 8 | 60 | F | 100 | + | 2 |
| 9 | 71 | F | 20 | − | 0 |
| 10 | 57 | F | 100 | + | 2 |
| 11 | 57 | M | 100 | − | 1 |
| 12 | 63 | M | 60 | − | 2 |
| 13 | 56 | F | 90 | + | 2 |
| 14 | 69 | M | 100 | + | 2 |
| 15 | 63 | F | 70 | − | 2 |
| 16 | 51 | F | 70 | − | 2 |
| 17 | 63 | M | 40 | − | 1 |
| 18 | 43 | M | 70 | + | 1 |
| 19 | 59 | M | 100 | + | 1 |
| 20 | 42 | F | 70 | + | 2 |
| 21 | 64 | F | 30 | + | 0 |
| 22 | 66 | M | 100 | − | 2 |
| 23 | 66 | M | 100 | + | 0 |
| 24 | 77 | M | 100 | − | 2 |
| 25 | 40 | F | 100 | − | 0 |
| 26 | 65 | M | 70 | − | 2 |
Abbreviations: KPI, Karnofsky Performance Index; MGMT+/−, O6-methylguanine-methyltransferase (MGMT) promoter methylation positive/negative.
Upregulation of mRNA Levels of the Components of Dll4-Notch Signaling in GBM
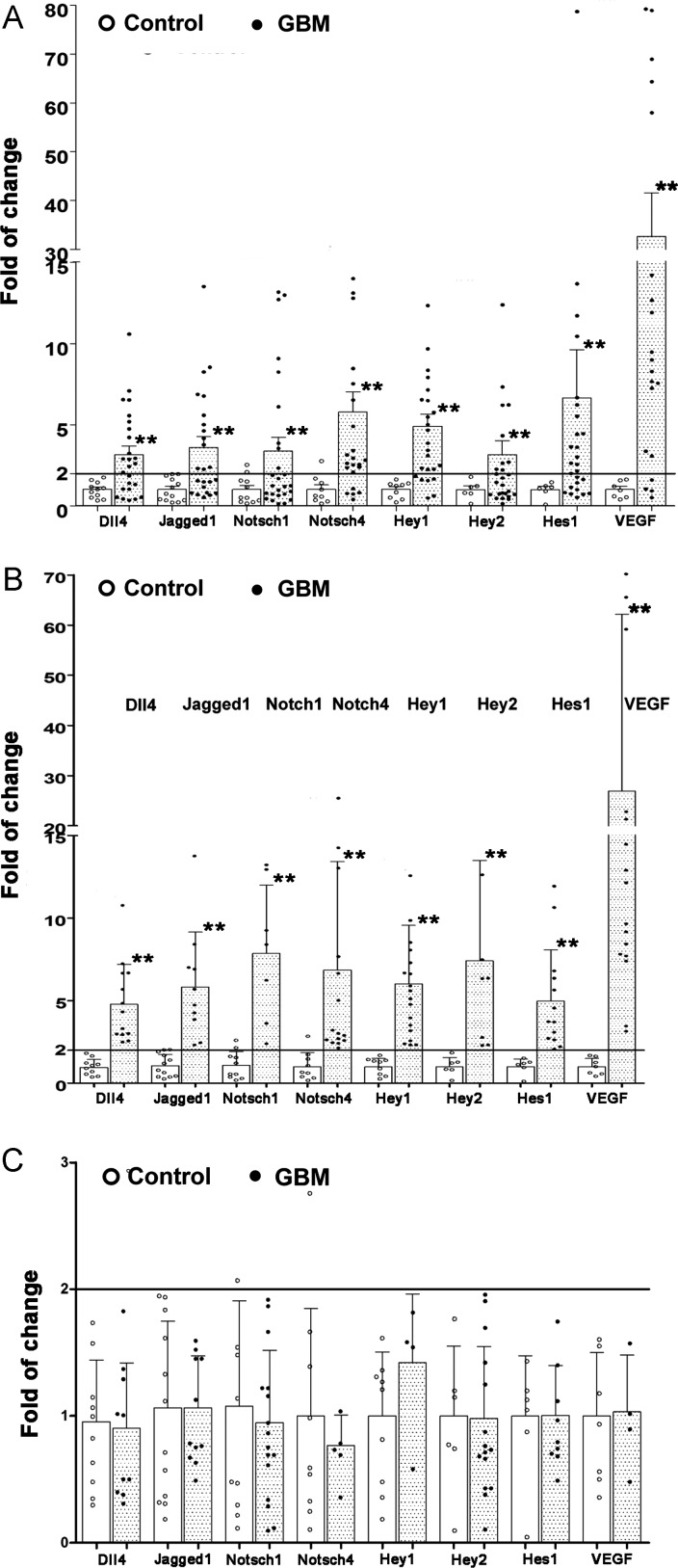
Real-time reverse-transcription PCR revealed a significant upregulation of all investigated components of Dll4-Notch signaling in GBM samples, compared with controls (Fig. 1A). The mean folds of change according to the cycle threshold approach from 26 tested GBM samples were 3.12, 3.58, 3.37, 5.77, 4.89, 3.13, 6.62, and 32.57 for Dll4 (P < .001), Jagged1 (P = .001), Notch1 (P = .01), Notch4 (P = .001), Hey1 (P < .001), Hey2 (P < .001), and Hes1 (P < .001), respectively, when the corresponding gene expression in the control was normalized to 1.00. The expression of Notch signaling target genes Hey1, Hey2, and Hes1 was significantly upregulated, indicating the activation of Notch signaling in GBM. On the other hand, the PCR data derived from all of the investigated GBM samples were heterogeneous. Therefore, we conducted additional analyses based on a fold of change > 2 or fold of change ≤ 2 in gene expression. Figure 1B shows the subgroups of GBM with a fold of change > 2. The mean folds of change in the individual genes were 4.50, 5.70, 8.00, 7.08, 6.02, 6.62, 5.52, and 38.57 for Dll4, Jagged1, Notch1, Notch4, Hey1, Hey2, Hes1, and VEGF, respectively. Moreover, if the fold of change > 2 was considered as upregulation, 61.5%, 54.2%, 33.3%, 76.9%, 76.9%, 42.3%, 65.4%, and 87.5% of the GBM samples showed an upregulation of these individual genes listed above. Another subgroup of GBM with a fold of change < 2 expressed these genes at a level similar to the controls (Fig. 1C).
Fig. 1.
The gene expression profiles of Notch signaling in GBM. Total RNA was extracted from the operative specimens of glioblastoma and normal brain control tissues. The mRNA levels of DLL4, Jagged1, Notch1, Notch4, Hey1, Hey2, Hes1, and VEGF were detected by real-time reverse-transcription PCR. The fold of change of individual genes was calculated according to the cycle threshold (Ct) approach. (A) Fold of change of the individual genes in all of the examined GBM (dots) and control (open cycles) samples. (B) Subgroup comparison of the expression of the individual genes in GBM samples with a fold of change > 2 (dots) and in control samples (open cycles). (C) Subgroup comparison of the expression of the individual genes in GBM samples with a fold change ≤ 2 (dots) and in control samples (open cycles). *P < .05 and **P < .01, compared with the control.
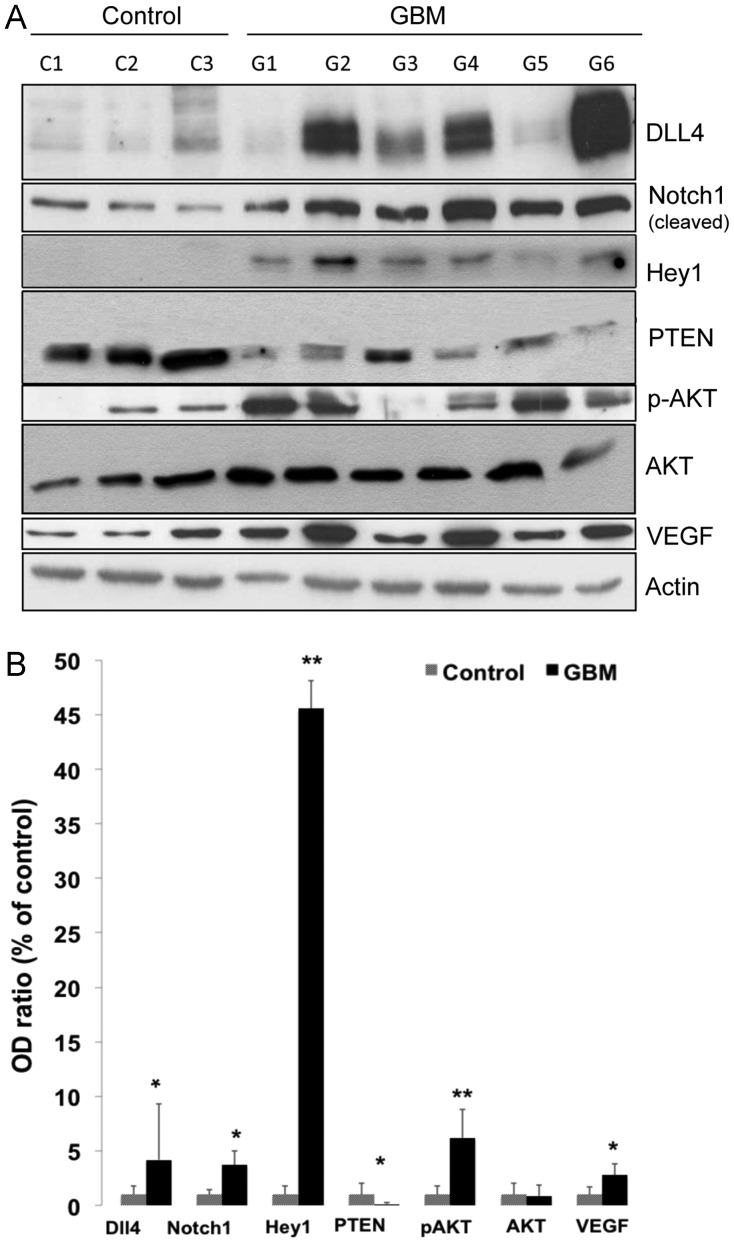
Upregulation of Protein Levels of Dll4, Notch1, and Hey1 was Concomitantly Accompanied by a Decreased PTEN Expression and an Increase in the Expression of Phospho-Akt (p-Akt) and VEGF
To confirm the activation of Dll4-Notch signaling, we detected the expression of Dll4, Notch1 (cleaved), and Hey1 by Western blotting in the subgroup of GBM with the fold of change > 2. Figure 2A shows a representative blot. Semiquantification of the blots revealed 4.2-, 3.7-, and 45.6-fold elevated protein levels of Dll4 (P = .04), cleaved Notch1 (P = .003), and Hey1 (P < .001), respectively, in the GBM samples, compared with the controls (Fig. 2B), confirming an activation of Dll4-Notch signaling in this subgroup of GBM samples. Meanwhile, this subgroup of GBM samples also showed a marked downregulation of PTEN expression (0.13-fold; P = .003) and a significant upregulation of p-Akt (P = .01) and VEGF (P = .004), whereas pan-Akt expression did not differ significantly between the GBM and control samples (Fig. 2B).
Fig. 2.
Expression of Dll4, Notch1, Hey1, PTEN, p-AKT, pan-AKT, and VEGF proteins in GBM. Total protein was extracted from the operative specimens of GBM (n = 15) and control tissues (normal brain, n = 5). The expression of Dll4, Notch1, Hey1, PTEN, p-AKT, AKT, and VEGF was detected using specific antibodies. Actin was measured as a housekeeping protein to control the total protein loaded into each lane. (A) A representative Western blot of GBM (G1–G6) and control (C1–C3) samples. (B) Semiquantification of protein levels. The optical density (OD) of the bands on blots representing individual specific proteins and the housekeeping protein actin was measured by ImageJ software. The OD ratio of the target protein to actin was calculated, and data are presented as a percentage of the control. *P < .05 and **P < .01, compared with the control.
The Correlation of the Expression of Dll4-Notch Signaling Components with the VP and MVD
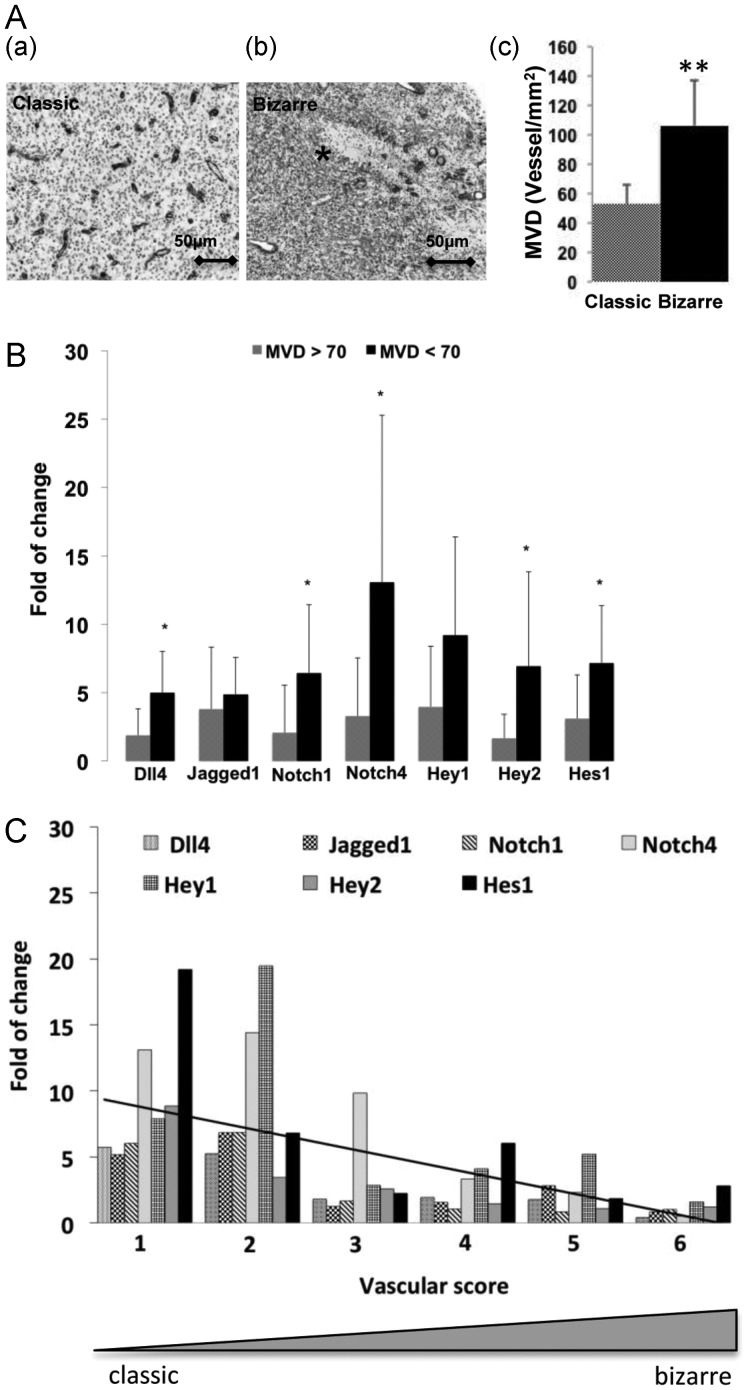
VP, comprising classic and bizarre patterns, and MVD are well-known parameters associated with angiogenesis and clinical outcomes in GBM.41,42 In the present study, VP and MVD were evaluated in GBM paraffin sections after staining with laminin. Although a quantitative evaluation method for judging a classic or bizarre VP was described previously,42,43 we found both classical and bizarre vascular patterns co-existing in one tumor, often with a predominance of one pattern. According to this evaluation method, the classic VP (Fig. 3A-a) was detected in 35% of investigated patients with GBM. There was no significant difference in age, sex, or KPI between the 2 types of vascular patterns (data not shown). Of interest, the bizarre VP was found frequently in the area containing extensive necrosis (Fig. 3A-b), whereas the classic VP was often located in the areas distant from the necrosis (Fig. 3A-a), reflecting nonproductive and productive vasculature for bizarre and classic VP, respectively. We also analyzed the MVD on these laminin-stained GBM sections and found that the mean MVD was 92 vessels/mm2. GBM that predominantly contained a classic VP showed a significantly lower MVD, compared with GBM with a bizarre VP (53 vs. 106 vessels/mm2; P < .001) (Fig. 3A-c).
Fig. 3.
Upregulation of Dll4-Notch signaling components was associated with the classic vascular pattern (VP) and lower microvascular density (MVD). The fold of change refers to the relative expression of the individual genes and was calculated based on the expression of the internal reference gene GAPDH and in the relation to the control samples (normal brain). (A) The association of VP with MVD in GBM. VP and MVD were evaluated in GBM sections (n = 26) after staining with laminin, as described in the Methods section. The typical classic (productive) and bizarre (nonproductive) VPs were frequently observed in the infiltrating (a) and necrotic (b, asterisk) areas of GBM tissue, respectively. The classic VP was associated with a lower MVD in GBM (c). **P < .01, compared with the classic vascular pattern. (B) The association of the expression of Dll4-Notch signaling components with MVD. *P < .05, compared with GBMs with a MVD >70 vessels/mm2. (C) The association of the expression of Dll4-Notch signaling components with VP. The tendency from a classic toward a bizarre VP was evaluated in individual GBM samples based on a score ranging from 0 to 6. A smaller score reflected the tendency toward a classic VP and was associated with a greater upregulation of Dll4-Notch signaling components.
Next, we determined the association of the expression of Dll4-Notch signaling components with MVD in all the tested GBM samples. The expression of the individual genes was presented as fold of change, which was calculated on the basis of the delta-delta Ct approach in relation to the internal reference gene GAPDH and to the control brain tissue. As shown in Fig. 3B, tumors with a lower MVD (<70 vessels/mm2) showed a significantly higher expression of Dll4 (P = .02), Notch1 (P = .04), Notch4 (P = .03), Hey2 (P = .03), and Hes1 (P = .05) (Fig. 3B), indicating that the activation of Notch signaling was inversely correlated to MVD in GBM tissue.
We also analyzed the potential association of Notch signaling component expression with VP. As mentioned above, classic and bizarre VPs coexisted in one tumor, which limits the use of judging a classic or bizarre VP by using the previously described mathematic evaluation method. Therefore, we modified the evaluation method by scoring the tendency from classic toward bizarre VP in individual GBM samples. A lower score (a score of 1 or 2) reflects a greater tendency toward classic VP. As shown in Fig. 3C, GBM samples with a score of 1 or 2 had an ∼5-fold upregulation of the majority of the Notch signaling components. GBM samples with a score of 3–6 also showed an upregulation of some of the components, but to a lesser extent than the groups with a score of 1–2. The regression analysis revealed a potential association of Notch signaling with VP in GBM tissue.
Correlation of the Expression of Dll4-Notch Signaling Components with Clinical Parameters
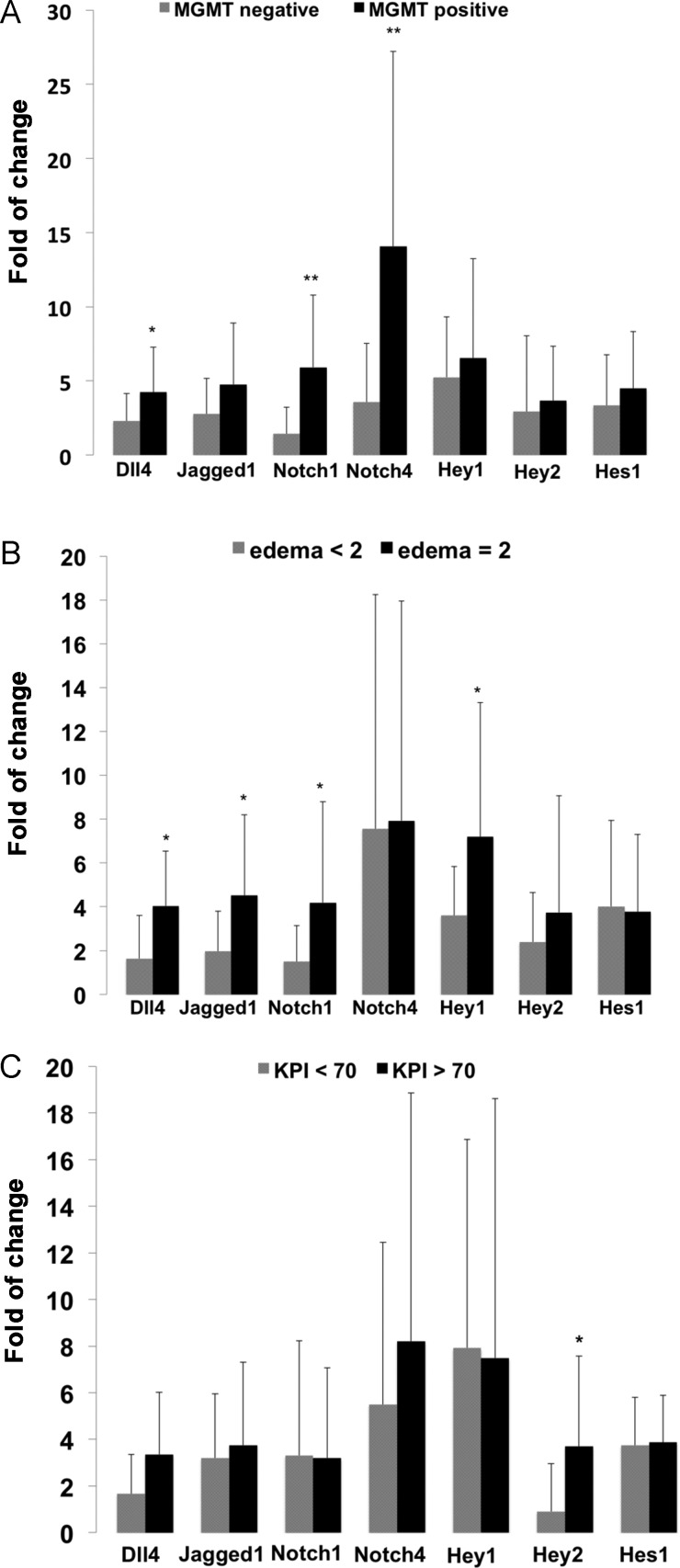
We elucidated a possible link between Dll4-Notch signaling component expression and important clinical parameters at the time of diagnosis, including MGMT-promoter methylation status, preoperative tumor edema detected on MRI, and the KPI in all tested GBM cases. As mentioned above, the fold of change referred to the relative expression of the individual genes and was calculated on the basis of the expression of the internal reference gene GAPDH in relation to the control (normal brain). GBM samples with a positive MGMT promoter methylation status presented with a significantly higher mRNA level of Dll4, Notch1, and Notch4 in comparison to the GBM with negative MGMT promoter methylation (Fig. 4A). Moreover, the degree of peritumoral edema detected by preoperative MRI was positively associated with the upregulation of Dll4, Jagged1, Notch1, and Hey1 (Fig. 4B). The expression of Hey2 was significantly upregulated in patients with GBM with a KPI >70, a parameter indicative of a better preoperative clinical condition, in comparison with the GBM with a KPI <70 (P = .002) (Fig. 4C).
Fig. 4.
Correlation of the expression of Dll4-Notch signaling components with clinical parameters. The fold of change refers to the relative expression of the individual genes and was calculated on the basis of the expression of the internal reference gene GAPDH and in the relation to the control samples (normal brain). MGMT positive/negative: O6-methylguanine-methyltransferase promoter methylation positive/negative status; KPI: Karnofsky performance index. A KPI >70 is indicative of a better preoperative condition of the patient. *P < .05 and **P < .01, compared with normal control brains.
Cellular Localization of Dll4 and Notch1 in GBM
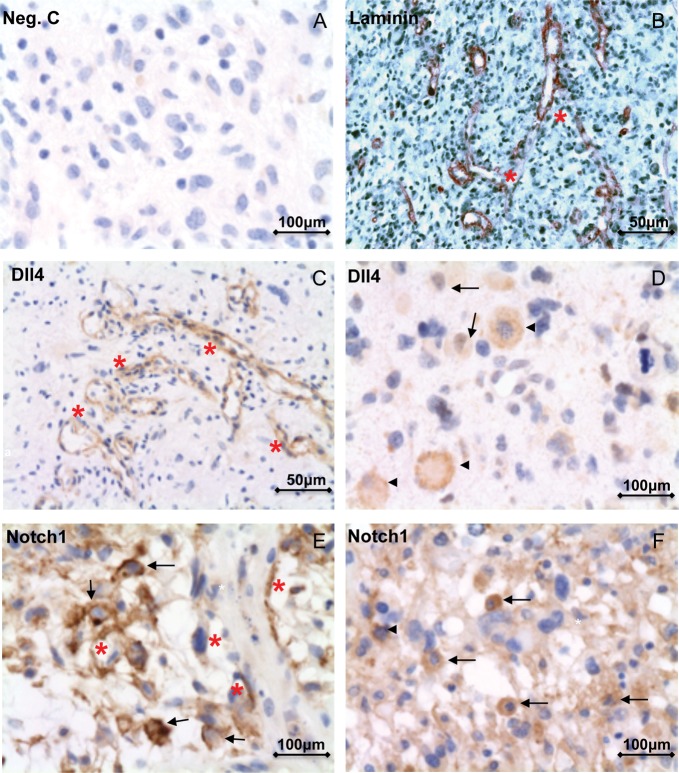
The control staining without the primary antibody showed negative staining (Fig. 5A). Immunostaining with laminin revealed the classic neovascularization pattern of multiple branching vessels in GBM (Fig. 5B). Of interest, the immunoreactivity of Dll4 and Notch1 was frequently detected in the endothelial cells of such classically patterned vessels (Fig. 5C and E), in the macrophage-like cells (Fig. 5D and F), and in some tumor cells (Fig. 5D–F). Of note, the Notch1 antibody applied during the immunostaining specifically detected cleaved Notch1. Therefore, the intensive immunoreactivity of Notch1 in tumor cells (Fig. 5E and F) and in endothelial cells of tumor vessels (Fig. 5E) reflected an activation of Notch signaling in these cells.
Fig. 5.
Cellular localization of immunoreactivity of Dll4 and cleaved Notch1 in GBM. (A) Negative control without the primary antibody for immunostaining did not show a signal. (B) Immunohistochemistry of laminin revealed the typical classic neovascularization pattern of branching vessels (red asterisks) in GBM samples. (C–F) Immunohistochemistry of Dll4 (C and D) and cleaved Notch1 (E and F) in GBM samples. The immunoreactivity of Dll4 and cleaved Notch1 was frequently detected in the endothelial cells of classically patterned vessels (red asterisks in C and E), in macrophage-like cells (arrowheads in D and F), and in some tumor cells (arrows in D, E, and F).
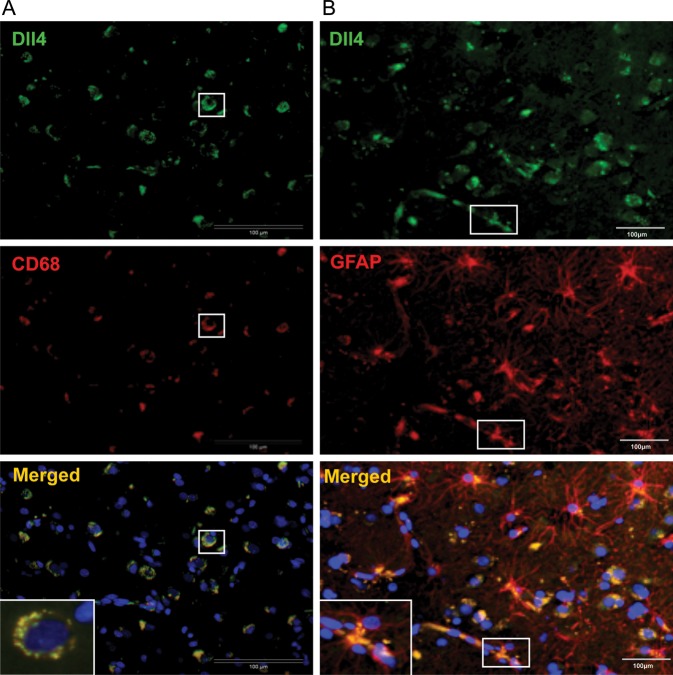
To confirm the macrophage expression of Dll4 and Notch1, double staining of Dll4/CD68 and Notch1/CD68 was performed. Figure 6A shows a representative GBM section stained for Dll4 and macrophage marker CD68. Colocalization of Dll4 and CD68 immunoreactivity is shown in the merged photo. A similar expression pattern was observed after Notch1/CD68 double staining of GBM tissue (data not shown).
Fig. 6.
Double staining of Dll4-CD68 and Dll4-GFAP in GBM. (A) Double staining of Dll4/CD68 in GBM. GBM sections were stained for Dll4 (green) and macrophage marker CD68 (red). The colocalization of Dll4 and CD68 immunoreactivity is shown in the merged photo (yellow). The white box is an enlarged view of the double-labeled cells. (B) Double staining of Dll4/GFAP in GBM. GBM sections were stained for Dll4 (green) and glial cell marker GFAP (red). The colocalization of Dll4 and GFAP immunoreactivity is shown in the merged photo (yellow). Of note, both the glial and tumor cells were stained with GFAP, and these cells often appeared to be Dll4-positive (yellow-colored cells in the merged photo). The white box is an enlarged view of the direct contact of glial cells with the tumor vessel.
Positive staining of GBM tumor cells and glial cells with GFAP is one of the histomorphological features of GBM. We therefore performed double staining of Dll4/GFAP and Notch1/GFAP on GBM sections (Fig. 6B). As predicted, both glial and tumor cells stained positive for GFAP. Furthermore, these cells often appeared to be Dll4-positive. Colocalization of Dll4 and GFAP can be clearly viewed in the merged photo. The direct contact of glial cells with the tumor vessel was observed in GBM tissue. The double staining of Notch1/GFAP revealed a similar expression pattern (data not shown).
Discussion
The Association of Dll4-Notch Signaling Activation and Angiogenic Parameters
Increasing evidence has implicated Notch signaling in the formation and progression of tumors.15,44–46 The activation of Dll4-Notch signaling is suggested to be oncogenic in GBM and to contribute to the transformation of glial cells and to glioma growth.15,28,29,31 Experimental overexpression and knockout of Notch-receptors/ligands promotes and inhibits tumor growth, respectively.25,47 Of note, activation of the Notch signaling pathway has been recently associated with the prognosis in patients with GBM.32
The present study investigated major components of Notch signaling in patients with GBM. In addition to demonstrating a significant upregulation of the mRNA levels of the ligands (Dll4 and Jagged1) and receptors (Notch1 and Notch4), we also showed an enhanced expression of the target genes, comprising Hey1, Hey2, and Hes1. Moreover, markedly elevated protein levels of cleaved Notch1 and Hey1 highlighted an activation of the Notch signaling pathway in GBM tissue. On the other hand, detailed analysis of our mRNA expression data revealed very heterogeneous expression of these genes in individual GBM samples. If a fold of change > 2 was considered to be biologically meaningful,48 one subset of tested GBM samples showed an upregulation of mRNA of the individual Dll4-Notch signaling components (Fig. 1B), whereas another group of GBM samples expressed the components of Notch signaling at a level similar to the controls (Fig. 1C). Therefore, GBM can be classified according to the difference in the activation status of Dll4-Notch signaling, supporting previously observed heterogeneous gene expression in human GBM tissue.28,29
PTEN is a well-defined tumor suppressor. Loss of PTEN has been found frequently in GBM, which causes a sustained activation of Akt and a subsequent upregulation of VEGF expression because of the stabilization of HIF-1α via the mTOR pathway.49 Upregulation of Dll4 expression in tumor vessels has been suggested to be directly regulated by VEGF.50–52 In addition, increasing evidence indicates a link between the Notch and PI3K pathways via PTEN.17,18,20 Endothelial Dll4 has been shown to inhibit lung cancer cell growth through upregulation of PTEN.53 Recent studies have demonstrated that upregulation of the Notch target gene Hes1 directly repressed PTEN expression, leading to PI3K activation in T cell leukemia20 and in thymocytes.54 Furthermore, Notch1-mediated activation of the PI3K-Akt pathway has been shown to promote glioma cell migration and invasion.21 In the present study, we found a downregulation of PTEN and an activation of Akt accompanied by an upregulation of VEGF in patients with GBM with activated Notch signaling. Our findings support a possible feedback regulation loop comprising the PTEN/PI3k-Akt/VEGF/Dll4-Notch signaling pathways in a subgroup of GBM samples. The present work was performed with patients with GBM, and the results were obtained from whole extracts of the tumor tissues and, therefore, reflect a cumulative alteration of oncogenic and angiogenic signaling pathways at the tissue level. It will be very interesting to explore the mechanistic link between Notch signaling and p-Akt and VEGF at the cellular level in future studies.
Considering the crucial role of angiogenesis in promoting the progression of solid tumors, anti-angiogenic therapy has become a new strategy to significantly prolong progression-free survival among patients with certain tumors.55,56 Anti-angiogenic VEGF therapy has also been applied to patients with GBM. However, only some patients with GBM respond to anti-angiogenic VEGF therapy, and even this population becomes resistant at a later stage.56 Activation of Dll4-Notch signaling is thought to mediate resistance to anti-VEGF therapy52. The present study demonstrated an upregulation of VEGF at both the mRNA (Fig. 1B) and the protein levels (Fig. 2) in patients with GBM with activated Notch signaling. Whether activation of Dll4-Notch signaling in GBM serves as a mechanism accounting for the resistance to anti-angiogenic VEGF therapy needs to be further elucidated. If this mechanism is proven, evaluation of the activation status of Dll4-Notch signaling in patients with GBM could be helpful for designing personalized therapy in the future.
VP and MVD have been defined as parameters associated with angiogenesis in different clinical malignancies,42,43 and the assessment of GBM neovascularization by evaluating MVD and VP may have clinical relevance.57,58 However, the association between VP and MVD is controversial,41,42 most likely because of the limitation of the method for detecting classic versus bizarre VPs. In fact, GBM tissue usually contains both classic and bizarre VPs, despite the predominance of one pattern. Nevertheless, according to this evaluation method, we found that a classic VP correlated to a low MVD in patients with GBM. In addition, we observed that the bizarre VP (Fig. 3A-b) was often located in the necrotic area of the tumor core, whereas the classic VP was more frequently seen in the area of infiltration that was distant from the necrosis (Fig. 3A-a). We therefore assumed that the bizarre and classic VPs may resemble nonproductive and productive vasculature in GBM tissue, respectively. Of interest, upregulation of Dll4-Notch signaling components was associated with a lower MVD but a higher tendency toward a classic VP in GBM tissue. Moreover, the immunoreactivity of Dll4 and Notch1 was frequently and intensively detected in the endothelial cells of tumor vessels with a predominately classic VP (Fig. 5C and E). These findings are in accordance with the current angiogenic concept of the role of the Dll4-Notch signaling pathway in regulating tumor angiogenesis. It has been previously shown that overexpression of Dll4 in a glioma xenograft model improved vascular perfusion and reduced intratumoral hypoxia and necrosis despite a decrease in vessel density.59 In contrast, blocking Dll4 induced nonproductive angiogenesis accompanied with hypoxia and necrosis. Activation of Dll4-Notch signaling in tumors has been shown to promote better perfusion in blood vessels (productive vessels), thereby stimulating tumor growth despite a decreased vessel density.50 Furthermore, these productive vessels, induced by the activation of Dll4-Notch signaling, appeared to be insensitive to VEGF-therapy.52 Anti–Dll4-Notch signaling for cancer therapy was thus directed to blocking the formation of functional vessels, but of interest, promoted the formation of nonproductive vessels.50,59 To our knowledge, the present study established for the first time the association of Dll4-Notch signaling with VP and MVD in patients with GBM, supporting the unique role of Notch signaling in promoting the formation of a more productive vascular pattern.
It is well known that gliomas are infiltrated by astrocytes and microglia/macrophages and that the extent of their infiltration positively correlates with malignancy.60 Microglia/macrophages promote glioma growth by secreting proteolytic enzymes and multiple angiogenic factors, such as matrix metalloproteinases (MMPs) and VEGF, and by affecting nuclear factor kappa B (NF-kB).5,61–63 NF-kB and MMPs are critically involved in tumor cell invasion and tumor angiogenesis and have been shown to be inactivated by inhibition of Notch1.64 The present study revealed the immunoreactivity of Dll4 and Notch1 in the endothelia of tumor vessels (Fig. 5), in macrophages, and in GFAP-positive cells (e.g. astrocytic/microglial and tumor cells). Of note, a cell-cell contact network was established frequently through astrocytic/microglial cells, which was essential for intercellular Dll4-Notch signaling transduction. We therefore hypothesize that a direct interaction among tumor cells, microglia/macrophages, and endothelial cells may determine the activation status of Dll4-Notch signaling, thereby specifying a predominantly classic or bizarre vascular pattern in GBM.
The Association of Dll4-Notch Signaling Activation and Prognostic Clinical Parameters
Several genetic and clinical parameters have been associated with the prognosis in patients with GBM.65–67 The present study demonstrated the correlation of the upregulation of multiple Dll4-Notch signaling components with enhanced peritumoral edema and with a higher rate of positive MGMT promoter methylation. The higher grade of edema observed in patients with GBM with upregulated Notch signaling may be attributable to the elevated VEGF level and, at least in part, to the better perfusion of vessels in tumors with a classic VP. Despite the increased blood flow, these vessels have an elevated permeability associated with blood-brain barrier disruption, which results in vasogenic cerebral edema.68 This explanation is in line with the well-known clinical finding that anti-angiogenic therapy dramatically reduces peritumoral edema.69 MGMT promoter methylation is an important prognostic factor for GBM-related survival,70 and novel findings highlight the MGMT gene as a critical upstream regulator of angiogenesis in GBM.71 However, whether the components of Dll4-Notch signaling in GBM are regulated by the MGMT gene requires further investigation. In addition, because of incomplete survival information for the tested patients with GBM, we were unable to conduct survival analyses.
Conclusions
GBM is a heterogeneous type of tumor; therefore, personalized therapies may be desirable. The present study demonstrated an activation of Dll4-Notch signaling in a subset of GBM samples, which was associated with some angiogenic and clinical parameters. These findings suggest this pathway as a potential therapeutic target for a subgroup of primary human GBM cases. Notch signaling plays a crucial role in the production of functional vasculature, which is associated with the mechanism of resistance to anti-VEGF treatment; therefore, it is of interest to further study the association of the activation status of Dll4-Notch signaling and the failure of anti-angiogenic VEGF therapy in patients with GBM.
Funding
None.
Acknowledgments
We thank Chao You, Rita Haase, and Eva Kusch for technical assistance, and Mike Sucker for assistance with the preparation of the graphs.
Conflict of interest statement. None declared.
References
- 1.Kleihues P, Louis DN, Scheithauer BW, et al. The WHO classification of tumors of the nervous system. J Neuropathol Exp Neurol. 2002;61(3):215–225. doi: 10.1093/jnen/61.3.215. discussion 226–219. [DOI] [PubMed] [Google Scholar]
- 2.Sant M, van der Sanden G, Capocaccia R. Survival rates for primary malignant brain tumours in Europe. Eur J Cancer. 1998;34(14):2241–2247. doi: 10.1016/s0959-8049(98)00336-0. [DOI] [PubMed] [Google Scholar]
- 3.Ohgaki H, Dessen P, Jourde B, et al. Genetic pathways to glioblastoma: a population-based study. Cancer Res. 2004;64(19):6892–6899. doi: 10.1158/0008-5472.CAN-04-1337. [DOI] [PubMed] [Google Scholar]
- 4.Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol. 2005;109(1):93–108. doi: 10.1007/s00401-005-0991-y. [DOI] [PubMed] [Google Scholar]
- 5.Demuth T, Berens ME. Molecular mechanisms of glioma cell migration and invasion. J Neurooncol. 2004;70(2):217–228. doi: 10.1007/s11060-004-2751-6. [DOI] [PubMed] [Google Scholar]
- 6.Roth W, Wagenknecht B, Klumpp A, et al. APRIL, a new member of the tumor necrosis factor family, modulates death ligand-induced apoptosis. Cell Death Differ. 2001;8(4):403–410. doi: 10.1038/sj.cdd.4400827. [DOI] [PubMed] [Google Scholar]
- 7.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 8.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284(5415):770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 9.Gray GE, Mann RS, Mitsiadis E, et al. Human ligands of the Notch receptor. Am J Pathol. 1999;154(3):785–794. doi: 10.1016/S0002-9440(10)65325-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iso T, Hamamori Y, Kedes L. Notch signaling in vascular development. Arterioscler Thromb Vasc Biol. 2003;23(4):543–553. doi: 10.1161/01.ATV.0000060892.81529.8F. [DOI] [PubMed] [Google Scholar]
- 11.Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194(3):237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 12.Shutter JR, Scully S, Fan W, et al. Dll4, a novel Notch ligand expressed in arterial endothelium. Genes Dev. 2000;14(11):1313–1318. [PMC free article] [PubMed] [Google Scholar]
- 13.Regan JN, Majesky MW. Building a vessel wall with notch signaling. Circ Res. 2009;104(4):419–421. doi: 10.1161/CIRCRESAHA.109.194233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jang MS, Zlobin A, Kast WM, Miele L. Notch signaling as a target in multimodality cancer therapy. Curr Opin Mol Ther. 2000;2(1):55–65. [PubMed] [Google Scholar]
- 15.Bolos V, Grego-Bessa J, de la Pompa JL. Notch signaling in development and cancer. Endocr Rev. 2007;28(3):339–363. doi: 10.1210/er.2006-0046. [DOI] [PubMed] [Google Scholar]
- 16.Koch U, Radtke F. Notch and cancer: a double-edged sword. Cell Mol Life Sci. 2007;64(21):2746–2762. doi: 10.1007/s00018-007-7164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gutierrez A, Look AT. NOTCH and PI3K-AKT pathways intertwined. Cancer Cell. 2007;12(5):411–413. doi: 10.1016/j.ccr.2007.10.027. [DOI] [PubMed] [Google Scholar]
- 18.Fitzgerald K, Harrington A, Leder P. Ras pathway signals are required for notch-mediated oncogenesis. Oncogene. 2000;19(37):4191–4198. doi: 10.1038/sj.onc.1203766. [DOI] [PubMed] [Google Scholar]
- 19.Palomero T, Ferrando A. Oncogenic NOTCH1 control of MYC and PI3K: challenges and opportunities for anti-NOTCH1 therapy in T-cell acute lymphoblastic leukemias and lymphomas. Clin Cancer Res. 2008;14(17):5314–5317. doi: 10.1158/1078-0432.CCR-07-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palomero T, Dominguez M, Ferrando AA. The role of the PTEN/AKT Pathway in NOTCH1-induced leukemia. Cell Cycle. 2008;7(8):965–970. doi: 10.4161/cc.7.8.5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Chen T, Zhang J, et al. Notch1 promotes glioma cell migration and invasion by stimulating beta-catenin and NF-kappaB signaling via AKT activation. Cancer Sci. 2012;103(2):181–190. doi: 10.1111/j.1349-7006.2011.02154.x. [DOI] [PubMed] [Google Scholar]
- 22.Thurston G, Noguera-Troise I, Yancopoulos GD. The Delta paradox: DLL4 blockade leads to more tumour vessels but less tumour growth. Nat Rev Cancer. 2007;7(5):327–331. doi: 10.1038/nrc2130. [DOI] [PubMed] [Google Scholar]
- 23.Stockhausen MT, Kristoffersen K, Poulsen HS. The functional role of Notch signaling in human gliomas. Neuro Oncol. 2010;12(2):199–211. doi: 10.1093/neuonc/nop022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poellinger L, Lendahl U. Modulating Notch signaling by pathway-intrinsic and pathway-extrinsic mechanisms. Curr Opin Genet Dev. 2008;18(5):449–454. doi: 10.1016/j.gde.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 25.Li JL, Sainson RC, Shi W, et al. Delta-like 4 Notch ligand regulates tumor angiogenesis, improves tumor vascular function, and promotes tumor growth in vivo. Cancer Res. 2007;67(23):11244–11253. doi: 10.1158/0008-5472.CAN-07-0969. [DOI] [PubMed] [Google Scholar]
- 26.Mailhos C, Modlich U, Lewis J, Harris A, Bicknell R, Ish-Horowicz D. Delta4, an endothelial specific notch ligand expressed at sites of physiological and tumor angiogenesis. Differentiation. 2001;69(2–3):135–144. doi: 10.1046/j.1432-0436.2001.690207.x. [DOI] [PubMed] [Google Scholar]
- 27.Kuhnert F, Kirshner JR, Thurston G. Dll4-Notch signaling as a therapeutic target in tumor angiogenesis. Vasc Cell. 2011;3(1):20. doi: 10.1186/2045-824X-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Purow BW, Haque RM, Noel MW, et al. Expression of Notch-1 and its ligands, Delta-like-1 and Jagged-1, is critical for glioma cell survival and proliferation. Cancer Res. 2005;65(6):2353–2363. doi: 10.1158/0008-5472.CAN-04-1890. [DOI] [PubMed] [Google Scholar]
- 29.Kanamori M, Kawaguchi T, Nigro JM, et al. Contribution of Notch signaling activation to human glioblastoma multiforme. J Neurosurg. 2007;106(3):417–427. doi: 10.3171/jns.2007.106.3.417. [DOI] [PubMed] [Google Scholar]
- 30.Jubb AM, Browning L, Campo L, et al. Expression of vascular Notch ligands Delta-like 4 and Jagged-1 in glioblastoma. Histopathology. 2012;60(5):740–747. doi: 10.1111/j.1365-2559.2011.04138.x. [DOI] [PubMed] [Google Scholar]
- 31.Gursel DB, Berry N, Boockvar JA. The contribution of Notch signaling to glioblastoma via activation of cancer stem cell self-renewal: the role of the endothelial network. Neurosurgery. 2012;70(2):N19–N21. doi: 10.1227/01.neu.0000410937.38828.6f. [DOI] [PubMed] [Google Scholar]
- 32.Gaetani P, Hulleman E, Levi D, et al. Expression of the transcription factor HEY1 in glioblastoma: a preliminary clinical study. Tumori. 2010;96(1):97–102. doi: 10.1177/030089161009600116. [DOI] [PubMed] [Google Scholar]
- 33.Hammoud MA, Sawaya R, Shi W, Thall PF, Leeds NE. Prognostic significance of preoperative MRI scans in glioblastoma multiforme. J Neurooncol. 1996;27(1):65–73. doi: 10.1007/BF00146086. [DOI] [PubMed] [Google Scholar]
- 34.Stark AM, van de Bergh J, Hedderich J, Mehdorn HM, Nabavi A. Glioblastoma: clinical characteristics, prognostic factors and survival in 492 patients. Clin Neurol Neurosurg. 2012;114(7):840–845. doi: 10.1016/j.clineuro.2012.01.026. [DOI] [PubMed] [Google Scholar]
- 35.Takai D, Yagi Y, Wakazono K, et al. Silencing of HTR1B and reduced expression of EDN1 in human lung cancers, revealed by methylation-sensitive representational difference analysis. Oncogene. 2001;20(51):7505–7513. doi: 10.1038/sj.onc.1204940. [DOI] [PubMed] [Google Scholar]
- 36.Mollemann M, Wolter M, Felsberg J, Collins VP, Reifenberger G. Frequent promoter hypermethylation and low expression of the MGMT gene in oligodendroglial tumors. Int J Cancer. 2005;113(3):379–385. doi: 10.1002/ijc.20575. [DOI] [PubMed] [Google Scholar]
- 37.Schaich M, Kestel L, Pfirrmann M, et al. A MDR1 (ABCB1) gene single nucleotide polymorphism predicts outcome of temozolomide treatment in glioblastoma patients. Ann Oncol. 2009;20(1):175–181. doi: 10.1093/annonc/mdn548. [DOI] [PubMed] [Google Scholar]
- 38.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 39.Weidner N. Current pathologic methods for measuring intratumoral microvessel density within breast carcinoma and other solid tumors. Breast Cancer Res Treat. 1995;36(2):169–180. doi: 10.1007/BF00666038. [DOI] [PubMed] [Google Scholar]
- 40.Weidner N, Semple JP, Welch WR, Folkman J. Tumor angiogenesis and metastasis—correlation in invasive breast carcinoma. N Engl J Med. 1991;324(1):1–8. doi: 10.1056/NEJM199101033240101. [DOI] [PubMed] [Google Scholar]
- 41.Onguru O, Gamsizkan M, Ulutin C, Gunhan O. Cyclooxygenase-2 (Cox-2) expression and angiogenesis in glioblastoma. Neuropathology. 2008;28(1):29–34. doi: 10.1111/j.1440-1789.2007.00828.x. [DOI] [PubMed] [Google Scholar]
- 42.Birner P, Piribauer M, Fischer I, et al. Vascular patterns in glioblastoma influence clinical outcome and associate with variable expression of angiogenic proteins: evidence for distinct angiogenic subtypes. Brain Pathol. 2003;13(2):133–143. doi: 10.1111/j.1750-3639.2003.tb00013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Preusser M, Birner P, Hainfellner JA. Algorithm for the standardized assessment of vascular patterns in glioblastoma specimens. Clin Neuropathol. 2004;23(5):238–240. [PubMed] [Google Scholar]
- 44.Jubb AM, Soilleux EJ, Turley H, et al. Expression of vascular notch ligand delta-like 4 and inflammatory markers in breast cancer. Am J Pathol. 2010;176(4):2019–2028. doi: 10.2353/ajpath.2010.090908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jubb AM, Turley H, Moeller HC, et al. Expression of delta-like ligand 4 (Dll4) and markers of hypoxia in colon cancer. Br J Cancer. 2009;101(10):1749–1757. doi: 10.1038/sj.bjc.6605368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patel NS, Dobbie MS, Rochester M, et al. Up-regulation of endothelial delta-like 4 expression correlates with vessel maturation in bladder cancer. Clin Cancer Res. 2006;12(16):4836–4844. doi: 10.1158/1078-0432.CCR-06-0285. [DOI] [PubMed] [Google Scholar]
- 47.Chen J, Kesari S, Rooney C, et al. Inhibition of Notch signaling blocks growth of glioblastoma cell lines and tumor neurospheres. Genes Cancer. 2010;1(8):822–835. doi: 10.1177/1947601910383564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delic S, Lottmann N, Jetschke K, Reifenberger G, Riemenschneider MJ. Identification and functional validation of CDH11, PCSK6 and SH3GL3 as novel glioma invasion-associated candidate genes. Neuropathol Appl Neurobiol. 2012;38(2):201–212. doi: 10.1111/j.1365-2990.2011.01207.x. [DOI] [PubMed] [Google Scholar]
- 49.Zundel W, Schindler C, Haas-Kogan D, et al. Loss of PTEN facilitates HIF-1-mediated gene expression. Genes Dev. 2000;14(4):391–396. [PMC free article] [PubMed] [Google Scholar]
- 50.Noguera-Troise I, Daly C, Papadopoulos NJ, et al. Blockade of Dll4 inhibits tumour growth by promoting non-productive angiogenesis. Nature. 2006;444(7122):1032–1037. doi: 10.1038/nature05355. [DOI] [PubMed] [Google Scholar]
- 51.Patel NS, Li JL, Generali D, Poulsom R, Cranston DW, Harris AL. Up-regulation of delta-like 4 ligand in human tumor vasculature and the role of basal expression in endothelial cell function. Cancer Res. 2005;65(19):8690–8697. doi: 10.1158/0008-5472.CAN-05-1208. [DOI] [PubMed] [Google Scholar]
- 52.Li JL, Sainson RC, Oon CE, et al. DLL4-Notch signaling mediates tumor resistance to anti-VEGF therapy in vivo. Cancer Res. 2011;71(18):6073–6083. doi: 10.1158/0008-5472.CAN-11-1704. [DOI] [PubMed] [Google Scholar]
- 53.Ding XY, Ding J, Wu K, et al. Cross-talk between endothelial cells and tumor via delta-like ligand 4/Notch/PTEN signaling inhibits lung cancer growth. Oncogene. 2012;31(23):2899–2906. doi: 10.1038/onc.2011.467. [DOI] [PubMed] [Google Scholar]
- 54.Wong GW, Knowles GC, Mak TW, Ferrando AA, Zuniga-Pflucker JC. HES1 opposes a PTEN-dependent check on survival, differentiation, and proliferation of TCRbeta-selected mouse thymocytes. Blood. 2012;120(7):1439–1448. doi: 10.1182/blood-2011-12-395319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Norden AD, Drappatz J, Wen PY. Antiangiogenic therapy in malignant gliomas. Curr Opin Oncol. 2008;20(6):652–661. doi: 10.1097/CCO.0b013e32831186ba. [DOI] [PubMed] [Google Scholar]
- 56.Norden AD, Drappatz J, Wen PY. Antiangiogenic therapies for high-grade glioma. Nat Rev Neurol. 2009;5(11):610–620. doi: 10.1038/nrneurol.2009.159. [DOI] [PubMed] [Google Scholar]
- 57.Preusser M, Heinzl H, Gelpi E, et al. Histopathologic assessment of hot-spot microvessel density and vascular patterns in glioblastoma: Poor observer agreement limits clinical utility as prognostic factors: a translational research project of the European Organization for Research and Treatment of Cancer Brain Tumor Group. Cancer. 2006;107(1):162–170. doi: 10.1002/cncr.21973. [DOI] [PubMed] [Google Scholar]
- 58.Verhoeff JJ, van Tellingen O, Claes A, et al. Concerns about anti-angiogenic treatment in patients with glioblastoma multiforme. BMC Cancer. 2009;9:444. doi: 10.1186/1471-2407-9-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ridgway J, Zhang G, Wu Y, et al. Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis. Nature. 2006;444(7122):1083–1087. doi: 10.1038/nature05313. [DOI] [PubMed] [Google Scholar]
- 60.Roggendorf W, Strupp S, Paulus W. Distribution and characterization of microglia/macrophages in human brain tumors. Acta Neuropathol. 1996;92(3):288–293. doi: 10.1007/s004010050520. [DOI] [PubMed] [Google Scholar]
- 61.Zhai H, Heppner FL, Tsirka SE. Microglia/macrophages promote glioma progression. Glia. 2011;59(3):472–485. doi: 10.1002/glia.21117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Demuth T, Rennert JL, Hoelzinger DB, et al. Glioma cells on the run - the migratory transcriptome of 10 human glioma cell lines. BMC Genomics. 2008;9:54. doi: 10.1186/1471-2164-9-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alterman RL, Stanley ER. Colony stimulating factor-1 expression in human glioma. Mol Chem Neuropathol. 1994;21(2–3):177–188. doi: 10.1007/BF02815350. [DOI] [PubMed] [Google Scholar]
- 64.Wang Z, Li Y, Banerjee S, et al. Down-regulation of Notch-1 and Jagged-1 inhibits prostate cancer cell growth, migration and invasion, and induces apoptosis via inactivation of Akt, mTOR, and NF-kappaB signaling pathways. J Cell Biochem. 2010;109(4):726–736. doi: 10.1002/jcb.22451. [DOI] [PubMed] [Google Scholar]
- 65.Sure U, Ruedi D, Tachibana O, et al. Determination of p53 mutations, EGFR overexpression, and loss of p16 expression in pediatric glioblastomas. J Neuropathol Exp Neurol. 1997;56(7):782–789. [PubMed] [Google Scholar]
- 66.Frenel JS, Botti M, Loussouarn D, Campone M. [Prognostic and predictive factors for gliomas in adults] Bull Cancer. 2009;96(4):357–367. doi: 10.1684/bdc.2008.0776. [DOI] [PubMed] [Google Scholar]
- 67.Ushio Y, Kochi M. [Prognostic factors in malignant gliomas] Gan To Kagaku Ryoho. 1996;23(5):643–648. [PubMed] [Google Scholar]
- 68.Rees JH, Smirniotopoulos JG, Jones RV, Wong K. Glioblastoma multiforme: radiologic-pathologic correlation. Radiographics. 1996;16(6):1413–1438. doi: 10.1148/radiographics.16.6.8946545. quiz 1462–1413. [DOI] [PubMed] [Google Scholar]
- 69.Jain RK, Tong RT, Munn LL. Effect of vascular normalization by antiangiogenic therapy on interstitial hypertension, peritumor edema, and lymphatic metastasis: insights from a mathematical model. Cancer Res. 2007;67(6):2729–2735. doi: 10.1158/0008-5472.CAN-06-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 71.Chahal M, Xu Y, Lesniak D, et al. MGMT modulates glioblastoma angiogenesis and response to the tyrosine kinase inhibitor sunitinib. Neuro Oncol. 2010;12(8):822–833. doi: 10.1093/neuonc/noq017. [DOI] [PMC free article] [PubMed] [Google Scholar]