Abstract
Background
CD44 is a molecular marker associated with cancer stem cell populations and treatment resistance in glioma. More effective therapies will result from approaches aimed at targeting glioma cells high in CD44.
Methods
Glioma-initiating cell lines were derived from fresh surgical glioblastoma samples. Expression of tissue transglutaminase 2 (TGM2) was attenuated through lentivirus-mediated short hairpin RNA knockdown. MTT assay [(3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] was used to evaluate the growth inhibition induced by TGM2 inhibitor. Terminal deoxynucleotidyl transferase deoxyuridine triphosphate nick end labeling was used to evaluate cell apoptosis following TGM2 inhibition. CD44+ glioma stem cells were sorted by flow cytometry. A nude mice orthotopic xenograft model was used to evaluate the in vivo effect of TGM2 inhibitor.
Results
TGM2 was highly expressed in CD44-high glioblastoma tissues and tumor-derived glioma-initiating cell lines. TGM2 knockdown impaired cell proliferation and induced apoptosis in CD44-high glioma-initiating cell lines. Further studies indicated that expression of inhibitor of DNA binding 1 protein (ID1) is regulated by TGM2 and might be an important mediator for TGM2-regulated cell proliferation in CD44-high glioma-initiating cell lines. TGM2 inhibitor reduces ID1 expression, suppresses cell proliferation, and induces apoptosis in CD44-high glioma-initiating cell lines. Furthermore, TGM2 is highly expressed in CD44+ glioma stem cells, while pharmacological inhibition of TGM2 activity preferentially eliminates CD44+ glioma stem cells. Consistently, TGM2 inhibitor treatment reduced ID1 expression and induced apoptosis in our orthotopic mice xenograft model, which can be translated into prolonged median survival in tumor-bearing mice.
Conclusions
TGM2 regulates ID1 expression in glioma-initiating cell lines high in CD44. Targeting TGM2 could be an effective strategy to treat gliomas with high CD44 expression.
Keywords: apoptosis, cell proliferation, glioma, tissue transglutaminase, tumor initiating cells
Despite progress in the study of the molecular aspects of malignant gliomas, the prognosis for these brain tumors, especially glioblastoma multiforme (GBM), continues to be dismal.1 GBM is associated with high mortality due to its aggressive growth and invasiveness. Interactions and functional cross-talk between tumor cells and their microenvironments are mediated by cell surface receptors that are responsible for cell–cell and cell–extracellular matrix (ECM) adhesion.2 Tissue transglutaminase (TGM2) is a unique multifunctional protein that, in addition to catalyzing calcium-dependent posttranslational modification of proteins in the ECM, can also catalyze calcium-independent protein disulfide isomerase, guanosine triphosphate/ATP hydrolase, and serine/threonine kinase functions.3,4 TGM2 is essential for integrin-mediated survival of bone marrow–derived mesenchymal stem cells.5 TGM2 expression is also elevated in multiple cancer cell types and is associated with poor drug response, increased metastatic potential, and poor patient survival.6–10 Recent studies have shown that TGM2 expression in epithelial cancer cells results in constitutive activation of cell growth and cell survival signaling pathways, transdifferentiation into mesenchymal cells, and acquisition of stem cell traits.11–13 Conversely, inhibition of TGM2 by small-molecule inhibitors or antisense, ribozyme, or small interfering RNAs inhibited tumor cell growth and invasiveness and rendered cancer cells sensitive to chemotherapy both in vitro and in animal models.14,15
The observation of increased TGM2 expression in glioblastomas has kindled great interest in understanding the contributions of this protein in glioma.16 ECM proteins, such as collagen, fibronectin (Fn), and laminin, are key structural components of the perivascular niche and regulate normal/tumor stem cell maintenance and migration.17 Some studies have demonstrated that disrupting the interaction between integrin and laminin impairs cancer stem cell maintenance and tumor initiation.17,18 TGM2 has been identified as an important factor that interacts with Fn in the ECM and integrins at the cell membrane, while abrogation of TGM2 activity disrupts Fn assembly in the ECM in glioma tissues.19 These findings underscore the potential role of ECM-integrin signaling on glioma cells and suggest that targeting TGM2 might be of great value in the treatment of glioma.
Cluster of differentiation (CD) 44 is a transmembrane glycoprotein expressed in glioma and serves as a surface receptor for components of the ECM such as hyaluronic acid.2 CD44 plays a critical role in efficient cell detachment from the hyaluronic acid substrate and promotes glioma cell migration.20,21 CD44 expression levels correlate with the histopathologic grade of gliomas.22 Recently, CD44 has been extensively used as a surface marker for isolating cancer stem cells from breast, prostate, pancreas, colorectal cancers, and glioma.23–26 In this study, we found that glioma-initiating cell lines high in CD44 express high levels of TGM2. Furthermore, TGM2 inhibition attenuates expression of inhibitor of DNA binding 1 protein (ID1) and suppresses cell proliferation in CD44-high glioma-initiating cell lines. These results may suggest that inhibition of TGM2 activity might be an effective strategy to combat CD44-high glioblastomas.
Materials and Methods
Reagents
The following were purchased: antibodies to phosphorylated (p)Akt (Ser473)/Akt and CD44 (Cell Signaling Technology); fluorescein isothiocyanate (FITC)–conjugated CD44 antibody (BD Biosciences); anti-TGM2 monoclonal antibody (Millipore); monoclonal antibody to β-actin and ID1 (Santa Cruz Biotechnology); horseradish peroxidase–conjugated goat anti-rabbit or anti-mouse antibodies (Bio-Rad); Dulbecco's modified Eagle's medium (DMEM)/F12, antibiotics, B27, epidermal growth factor (EGF), and basic fibroblast growth factor (bFGF) (Invitrogen); and monodansylcadaverine (MDC) and biotinylated cadaverine (Sigma Chemical).
Tumor Specimens and Glioma-Initiating Cell Culture
Eight primary glioblastoma specimens were obtained freshly from the operating room following protocols approved by the research ethics committee in the Cancer Center of Sun Yat-Sen University with informed consent obtained from all subjects. Glioma-initiating cell lines were isolated and subsequently cultured in tumorsphere medium as reported by Lee et al.27 Briefly, tumor samples were processed within 30 min after surgical resection. Minced pieces of human glioma samples were digested with 200 U/mL collagenase IV (Sigma) and 500 U/mL DNase I (Sigma) in phosphate buffered saline (PBS) for 2 h at 37°C with constant vigorous agitation. Single-cell suspension was filtered through a 70-μm cell strainer (BD Falcon) and washed with PBS. Glioma cells were resuspended and subsequently sphere cultured in tumorsphere medium. The tumorsphere medium consisted of DMEM/F12 supplemented with B27 (Gibco), l-glutamine (Gibco), penicillin/streptomycin, and growth factors (20 ng/mL EGF and 20 ng/mL bFGF). Normal mouse neural stem cells (mNSCs) were isolated from mouse embryonic cerebral cortex tissue (E12.5) and cultured as described previously.28
Lentiviral Construction and Transduction
Lentiviruses encoding short hairpin (sh)RNAs silencing TGM2 along with control scramble were purchased from Santa Cruz Biotechnology. ShRNA sequence silencing ID1 was reported previously.27 OmicsLink lentiviral ORF (open reading frame) expression clones for the TGM2 or ID1 gene were constructed by Genecopoeia. A site-directed mutation TGM2 inactive mutant (C277S) was created using the Quikchange Mutagenesis Kit (Agilent). Lentiviral particles were produced by transient cotransfection of packaging plasmids into the Lenti-Pac 293Ta cell line. Produced lentiviruses were concentrated by using a Centricon Plus-20 centrifugal filter device (Millipore).29 To ensure that the same number of lentiviruses were used in the same experiment, produced lentiviral stock was titered and stored at −80°C. For in vitro infection of cells with the lentivirus, cultured neurospheres were disaggregated prior to infection in order to increase infection efficiency and uniformity.
Cell Sorting and Tumorsphere Formation Assay
CD44+ glioma stem cells were sorted as described previously.25 Tumorspheres were dissociated and individual cells were incubated for 15 min in blocking solution containing 10 μg/mL human immunoglobulin (Ig)G, followed by anti-CD44 antibody or the control IgG2b isotype, both FITC conjugated (BD Pharmingen). Cells were incubated for 20 min on ice protected from light and washed in PBS. Cells were then sorted (MoFlo, Dako) after staining with CD44-FITC or analyzed by flow cytometry (FACSCalibur, Becton Dickinson). Tumorsphere formation assay on CD44+/CD44− cell population was performed as described previously.25
Western Blot Analysis
Cell lysates were prepared with cell lysis buffer containing 1% Nonidet P40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), and protease inhibitor cocktail. After standard SDS–polyacrylamide gel electrophoresis and Western blotting procedures, proteins were visualized using an electrochemiluminescence system (Amersham Biosciences). Antibodies against TGM2, ID1, pAkt, Akt, and CD44 were used at 1:1000 dilution. Anti–β-actin was used at 1:5000 dilution. Blotting band intensities were quantitated densitometrically using ImageJ 1.37v software (National Institutes of Health).
Immunofluorescence Staining
Cells were cultured on glass coverslips precoated with polylysine in 6-well plates, rinsed thrice with PBS, fixed with 3.7% paraformaldehyde for 15 min, and blocked with 5% normal goat serum for 1 h. The cells were immunostained by using primary antibodies specific to various antigens. Goat anti-mouse IgG Alexa 488 or goat anti-rabbit IgG Alexa 546 was used as the secondary antibody. The stained coverslips were mounted on glass microscope slides in anti-fade Prolong mounting medium (Invitrogen). Images were taken under a Zeiss AxioCam fluorescence microscope using Zeiss AxioVision software. The percentage of CD44 or active caspase-3–positive cells in each section was determined at a magnification of 400× by counting 500 cells in a randomly selected field.
TUNEL Assay
We also determined the level of apoptosis using the terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate nick end labeling (TUNEL) method. For the TGM2 knockdown experiment, apoptotic cells were evaluated 5 days after shRNA lentivirus infection. For drug treatment, cells were treated with MDC at the indicated doses for 48 h. Thereafter, cells were fixed with 4% paraformaldehyde and then permeabilized with 0.1% Triton-X 100 in PBS. After being washed, cells were incubated with reaction mixture for 60 min at 37°C. Stained cells were mounted and analyzed under a fluorescent microscope (Zeiss). The percentage of cells identified by TUNEL in each section was determined at a magnification of 400× by counting 500 cells in a randomly selected field.
Cell Viability Assay
The antiproliferation effect of MDC on glioma cells was determined by using an MTT assay [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] . Briefly, 3 × 103 cells were plated in 100 μL of medium in 96-well microtiter plates precoated with polylysine and incubated for 24 h. Drugs were added and the cells were incubated for a further 3 days. MTT solution (20 μL) was added to each well, followed by incubation at 37°C for 2 h. Absorbance was then read at 570 nm with a 96-well plate reader. The viability of untreated cells was regarded as 100%. The concentration that caused 50% growth inhibition (GI50) was calculated using SPSS 10.0 statistical software. To determine the effect of TGM2 or ID1 knockdown on cell proliferation, cells were infected with scramble or shRNA-targeting lentivirus for 48 h, then dissociated into single cells and plated onto 6-well plates (1 × 104 cells/well) and incubated for 7 days. The formed tumorspheres were dissociated into single cells, and the viable cells in each group were counted with a hemocytometer using 0.2% trypan blue exclusion.
In situ Transglutaminase Activity
In situ transglutaminase activity was measured by the incorporation of biotinylated cadaverine into Fn.7,8 For this assay, 2 × 105 cells/mL were plated onto 96-well plates precoated with plasma Fn in 100 μL complete DMEM without serum in the presence of 0.1 mM biotinylated cadaverine. As a negative control, Fn-coated 96-well plates were incubated with 100 μL serum-free DMEM containing 0.1 mM biotinylated cadaverine alone. Cells were allowed to incubate for 1 h at 37°C and then were washed twice with PBS, pH 7.4, containing 3 mM EDTA in order to stop the reaction. A detergent solution (100 μL) consisting of 0.1% (weight/volume [w/v]) deoxycholate in PBS, pH 7.4, containing 3 mM EDTA was then added to each well, and the mixture was incubated with gentle shaking for 20 min. The supernatant was discarded and the remaining Fn layer washed 3 times with Tris-HCl, pH 7.4. Wells were then blocked with 3% (w/v) bovine serum albumin in Tris-HCl buffer for 30 min at 37°C and washed with buffer, and then incorporated biotinylated cadaverine was revealed with a 1:5000 dilution of ExtrAvidin peroxidase conjugate (Sigma), which was incubated for 1 h at 37°C using 3,3′,5,5′-tetramethylbenzidine as a substrate. Color development was stopped by adding 50 μL stop solution to each well. The resulting color was then read in an ELISA plate reader at 450 nm.
TGM2 enzyme activity was measured using a microscopic assay as previously described.30 Briefly, TGM2 activity was assessed by exposing mouse brain sections to 1 mM biotinylated cadaverine for 48 h. Sections were then fixed for 30 min in 3.7% (volume/volume [v/v]) formaldehyde in PBS and permeabilized with 0.2% (v/v) Triton X-100 in PBS for 30 min at room temperature. Sections were further incubated with an Alexa Fluor 488 streptavidin conjugate (1:1000, v/v) in PBS for 1 h at room temperature, washed, and mounted. The sections were observed under a fluorescent microscope (BX51, Olympus). TGM2 activity was seen as increased fluorescence labeling and quantified by ImageJ analysis software.
Immunohistochemistry
Immunohistochemical staining was performed as described previously.9 Briefly, frozen tissue sections were fixed with 4% paraformaldehyde, incubated with primary antibodies overnight at 4°C, and then incubated for 30 min each with horseradish peroxidase–labeled secondary antibody. Antigen/antibody reactions were detected by exposure to 3,3-diaminobenzidine and hydrogen peroxide chromogen substrate (Dako). Slides were counterstained with hematoxylin and mounted. The negative controls were incubated with nonimmune mouse IgG in place of the primary antibody. The immunostained slides were examined under a light microscope and scored by a double-blinded pathologist and laboratory researcher.
Animal Studies
Immunocompromised nude mice were obtained from the breeding facility at the animal center of Sun Yat-Sen University. All animal studies were performed in accordance with institutional ethical guidelines for experimental animal care. For subcutaneous (s.c.) xenograft study, 5 × 105 cells (0814) were inoculated into the flanks of nude mice, and tumors were allowed to establish for 10 days before initiation of treatments. Mice were injected intraperitoneally (i.p.) daily with either PBS or MDC (10 or 25 mg/kg/d) for 10 days. After drug treatment (day 21), tumors were removed and weighed to determine size.
For survival analysis, 1 × 105 cells (0814) were injected stereotactically into 4-week-old nude mouse cortex, following administration of general anesthesia. The injection coordinates were 3 mm to the left of the midline, 2 mm anterior to the lambdoid suture, and 3 mm deep. The incision was closed with wound clips, which were removed 4 days after inoculation. Animals that died, lost weight, or developed neurologic deficits within 24 h of cell injection were excluded. Drug treatment was started 5 days after inoculation. Mice were injected i.p. daily with either PBS or MDC (10 or 25 mg/kg/d) for 2 weeks. The animals were monitored daily until signs of neurologic deficit developed, at which time they were euthanized and their brains were removed.
For histopathologic analysis, the mouse brains or s.c. xenografts embedded in optimal cutting temperature compound were stored in liquid nitrogen overnight and then sectioned at 5-µm thickness on a MicromHM200 cryotome (Eryostar). Sections stained with hematoxylin and eosin (H&E) were evaluated for evidence of tumor.
The Cancer Genome Atlas Data Analysis
Array comparative genomic hybridization and mRNA and gene mutation data from GBM patients were downloaded from The Cancer Genome Atlas (TCGA) data portal (http://cancergenome.nih.gov/dataportal). Details on data processing and platforms are in the publication describing the GBM data analysis.31
Statistical Analysis
Statistical evaluations were carried out using SPSS 10.0 software. Error bars throughout the figures in this paper indicate standard deviation (SD). Student's t-test was used to compare means of 2 groups. A 1-way ANOVA was used to compare means of 3 or more groups. Kaplan–Meier curves of overall survival were drawn, and survival in the groups was compared using the log-rank test. For all tests, the level of statistical significance was P < .05. All statistical tests were 2-sided.
Results
TGM2 Is Highly Expressed in CD44-high GBM and Tumor-derived Glioma-initiating Cells
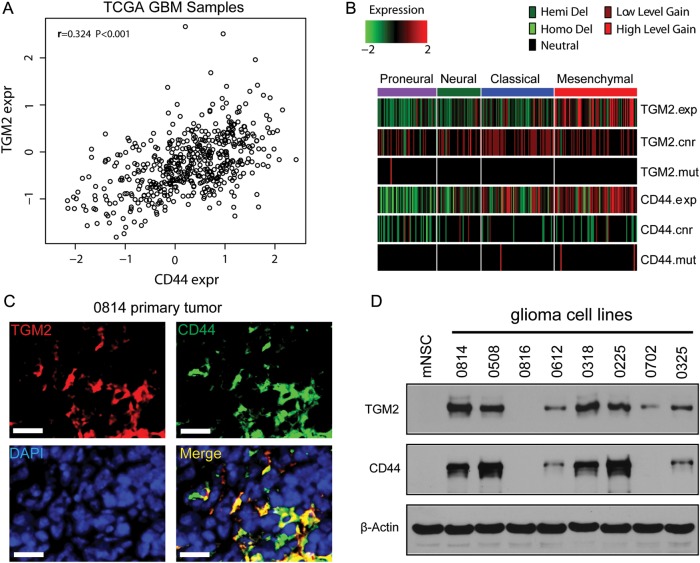
TCGA GBM profiling revealed that CD44 mRNA expression is significantly correlated with TGM2 expression (Pearson's r = 0.324, P < .001; Fig. 1A). Furthermore, higher TGM2 expression was consistent with higher CD44 expression in mesenchymal molecular subclasses (Fig. 1B). TGM2 expression was higher in some primary GBM tissues (0508) but not in normal brain tissues (Supplementary Fig. S1A). TGM2 immunoreactivity was also seen in CD44+ glioma cells in 0814 primary tumor sections (Fig. 1C). We established 8 glioma-initiating cell lines from surgical samples. Stem cell markers such as CD133 and CD44 were differentially expressed in these glioma-initiating cell lines (Supplementary Fig. S1B). Orthotopic transplantation of a small number of glioma-initiating cells into mice brains formed tumors (Supplementary Fig. S1C). Glioma-initiating cells grew as tumorspheres and expressed NSC markers nestin and Sox2 but showed only low expression for differentiation markers glial fibrillary acidic protein (GFAP) and neuron-specific class III beta-tubulin (TuJ1; Supplementary Fig. S2A, S2B). To better establish the correlation between CD44 and TGM2 in these cell lines, we evaluated the expression of CD44 and TGM2 in mNSCs and 8 glioma-initiating cell lines. Western blotting analysis showed that TGM2 protein expression was higher in all 4 CD44-high cell lines compared with mNSCs and 4 CD44-low cell lines (Fig. 1D). Therefore, TGM2 might exert an important function in CD44-high glioma-initiating cell lines.
Fig. 1.
TGM2 is highly expressed in CD44-high GBM and tumor-derived glioma-initiating cells. (A) Expression correlation between TGM2 and CD44 in TCGA GBM samples. (B) TGM2 is coexpressed with CD44 in TCGA GBM mesenchymal subclasses. (C) Immunostaining of TGM2 and CD44 in cryosection of 0814 primary tumor. TGM2 (red), CD44 (green), 4′,6′-diamidino-2-phenylindole (DAPI; blue). Bar, 50 μm. (D) Western blotting analysis on TGM2 and CD44 expression in mNSC and glioma-initiating cell lines. β-actin was used as loading control.
TGM2 Regulates the Cell Proliferation in CD44-high Glioma-initiating Cell Lines
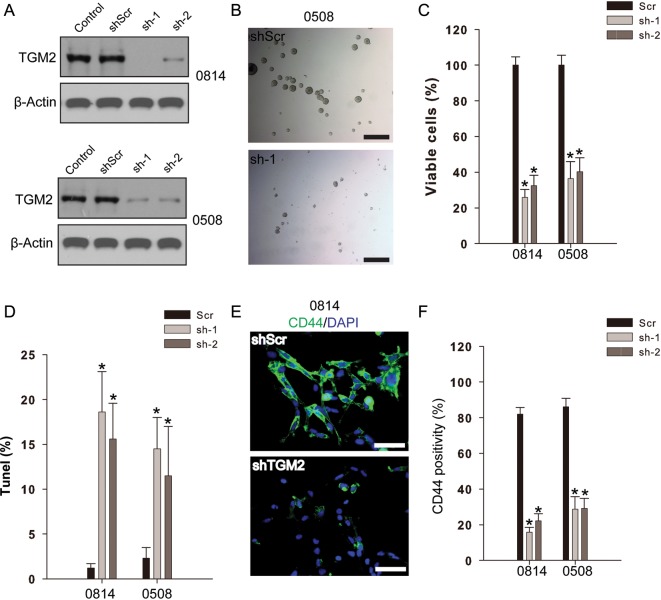
To interrogate the role of TGM2 in the regulation of cell proliferation in glioma-initiating cells, we targeted TGM2 expression by infection with lentivirus expressing shRNA specific to TGM2. The 2 shRNAs efficiently reduced TGM2 expression in both 0508 and 0814 cell lines compared with the control or scramble shRNA group (Fig. 2A). Consequently, TGM2 knockdown significantly suppressed cell proliferation in 0814 and 0508 cell lines, accompanied with less and smaller tumorsphere formation (Fig. 2B and C). Furthermore, TUNEL assays showed that TGM2 knockdown markedly induced apoptosis in both 0814 and 0508 cell lines compared with scramble controls (P < .01; Fig. 2D). We also examined CD44 expression in glioma cell lines following TGM2 knockdown. Our results indicated that 0814 and 0508 cell lines both exhibited high CD44 positivity (>80%). TGM2 knockdown considerably reduced the percentage of CD44+ cells in both cell lines compared with the shRNA scramble group (Fig. 2E and F). Therefore, TGM2 knockdown might regulate proliferation in CD44-high glioma-initiating cell lines.
Fig. 2.
TGM2 knockdown suppresses cell proliferation and induces apoptosis in CD44-high glioma-initiating cell lines. (A) Lentivirus shRNA mediated TGM2 knockdown in glioma-initiating cell lines. Glioma cells infected by lentivirus containing 2 shRNAs (sh-1, sh-2) for TGM2 or scramble (Scr) were subject to Western blotting analysis. (B) Light microscopy photographs from 0508 cells following TGM2 knockdown compared with scramble shRNA control. Bar, 500 µm. (C) TGM2 knockdown suppresses cell proliferation of CD44-high glioma-initiating cells. Data were calculated as percentage to scramble shRNA control. *P < .05 compared with scramble shRNA control (n = 4). (D) TGM2 knockdown induces apoptosis in CD44-high glioma-initiating cell lines. TUNEL assay was performed to detect apoptotic cells. *P < .05 compared with scramble shRNA control (n = 4). (E and F) TGM2 knockdown eliminates CD44-high glioma cells. *P < .05 compared with scramble shRNA control (n = 4). Bar, 100 µm.
TGM2 Regulates ID1 Expression in CD44-high Glioma-initiating Cell Lines
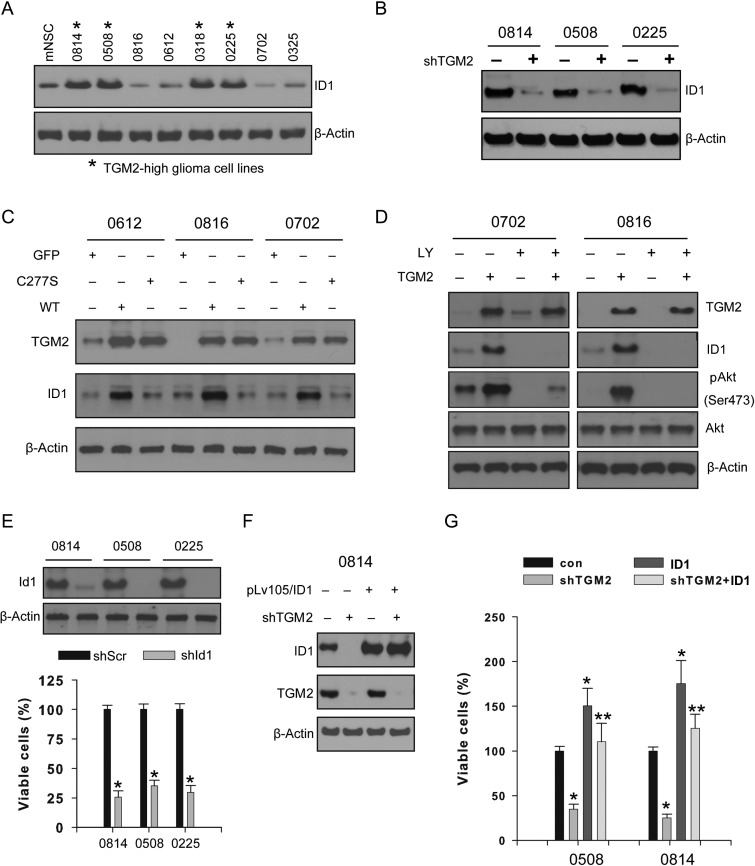
ID1 is a critical transcriptional factor closely linked to tumorigenesis as well as cell proliferation and survival.32 ID1 promotes cell proliferation and cell cycle progression through activation of growth-promoting pathways in certain cancer cells.33,34 Here we showed that TGM2-high glioma-initiating cell lines exhibited higher ID1 levels (Fig. 3A). TGM2 depletion (hereafter we used sh-1 shRNA for TGM2 knockdown) in 0814, 0508, and 0225 cell lines decreased ID1 levels compared with scramble control (Fig. 3B). In order to confirm TGM2 regulation on ID1 expression, we constitutively expressed TGM2 or its inactive mutant (C277S) in the 0612, 0816, and 0702 cell lines with low TGM2 expression. Wild-type TGM2, rather than the C277S mutant or green fluorescent protein (GFP) control, markedly increased ID1 expression (Fig. 3C). Importantly, TGM2 might regulate ID1 expression through activating the phosphoinositide-3 kinase (PI3K)/Akt pathway, as blocking the PI3K/Akt pathway by Ly294002 attenuated the TGM2 effect on ID1 expression in the 0702 and 0816 cell lines (Fig. 3D). Moreover, ID1 knockdown suppressed the cell proliferation of the 0814, 0508, and 0225 cell lines, suggesting that ID1 might mediate TGM2 signals to maintain cell proliferation (Fig. 3E). Our data also indicated that ID1 overexpression rescued cell proliferation following TGM2 knockdown in the 0814 and 0508 cell lines (Fig. 3F and G). Therefore, TGM2 might regulate ID1-mediated cell proliferation in CD44-high glioma-initiating cell lines.
Fig. 3.
TGM2 regulates ID1 expression in CD44-high glioma-initiating cell lines. (A) TGM2 is coexpressed with ID1 in glioma-initiating cell lines. (B) TGM2 knockdown reduces ID1 protein expression in CD44-high glioma-initiating cells. (C) TGM2 expression regulates ID1 expression in glioma-initiating cells. Lentivirus vector containing green fluorescent protein (GFP), wild-type (WT) TGM2, or TGM2 inactive mutant (C227A) was added into cell cultures for infection, and cell lysates were subjected to Western blotting analysis. (D) Constitutive TGM2 expression regulates ID1 expression through PI3K/Akt pathway. PI3K inhibitor Ly294002 (Ly) was added 48 h after lentiviral TGM2 infection and further incubated for 24 h. (E) ID1 knockdown attenuates cell proliferation in CD44-high glioma-initiating cells. Data were calculated as percentage to scramble shRNA control. *P < .05 compared with scramble shRNA control (n = 4). (F and G) Constitutive ID1 expression restores the cell proliferation inhibited by TGM2 knockdown. Lentivirus containing shRNA targeting TGM2 alone or expressing ID1 was used to infect glioma cells. Data were calculated as percentage to scramble shRNA control. *P < .05 compared with scramble shRNA control (n = 4). **P < .05 compared with TGM2 shRNA alone (n = 4).
TGM2 is Highly Expressed in the CD44+ Glioma Stem Cell Population
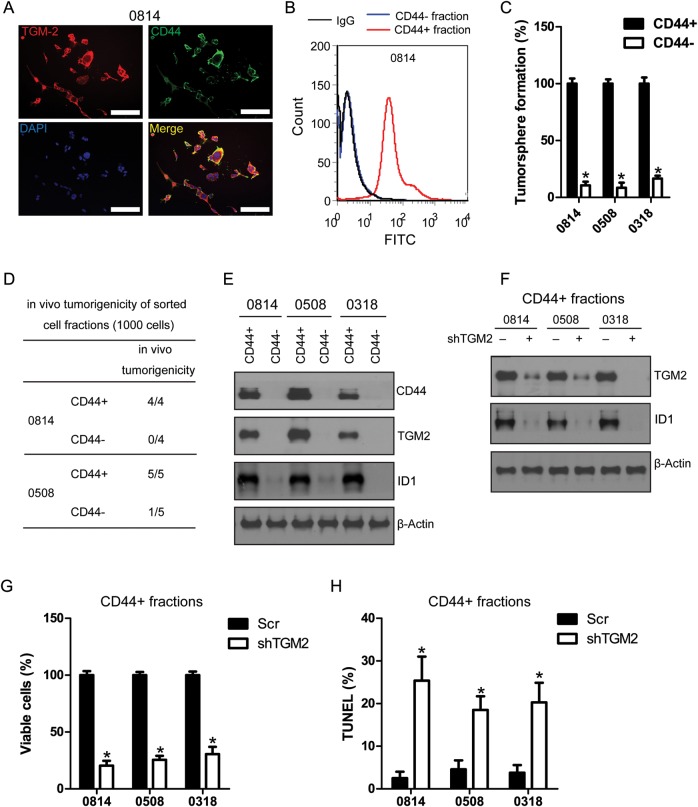
CD44 is a cell surface marker associated with cancer stem cell populations in glioma.25 Our immunostaining results showed that TGM2 was highly expressed in CD44+ glioma cells (Fig. 4A). Flow cytometry analysis confirmed the high CD44 expression in a sorted CD44+ glioma cell population (Fig. 4B). The sorted CD44+ glioma cell fractions met cancer stem cell criteria because they were highly self-renewable and tumorigenic (Fig. 4C and D). CD44+ glioma stem cells can differentiate into neural lineage cells with increased expression of glial fibrillary acidic protein, TuJ1, and 2-3-cyclic nucleotide 3-phosphodiesterase after induction with neural differentiation agents (Supplementary Fig. S2C). Importantly, we observed high expression of TGM2 and ID1 in CD44+ glioma stem cells sorted from 0814, 0508, and 0318 cell lines (Fig. 4E). TGM2 knockdown led to reduction of ID1 expression in CD44+ glioma stem cell fractions (Fig. 4F). Furthermore, loss of TGM2 expression led to significant growth inhibition and induction of apoptosis in CD44+ glioma stem cells (Fig. 4G and H).
Fig. 4.
TGM2 is highly expressed in CD44+ glioma stem cells. (A) TGM2 coexpresses with CD44 in 0814 cell line. Bar, 50 µm. (B) Flow cytometry analysis on the expression of CD44 in sorted CD44+ and CD44− cell fractions (0814). FITC-labeled mouse isotype IgG was used as negative control. (C) CD44+ cell fractions exhibit higher self-renewal capability than matched CD44− cell fractions. *P < .05 compared with CD44+ cell fractions (n = 4). (D) CD44+ cell fractions are highly tumorigenic; 1000 cells from CD44+/CD44− glioma cells were orthopically transplanted into mouse brain. Mice were sacrificed 25 days after transplantation, and brains were collected for sectioning to examine the evidence of tumor. (E) CD44+ glioma stem cells express high TGM2 and ID1. (F) TGM2 knockdown reduces ID1 expression in CD44+ glioma stem cells. (G) TGM2 knockdown suppresses cell proliferation in CD44+ glioma stem cells. *P < .05 compared with scramble control (n = 4). (H) TGM2 knockdown induces apoptosis in CD44+ glioma stem cells. *P < .05 compared with scramble control (n = 4).
TGM2 Inhibition by MDC Reduces ID1 Expression and Suppresses Cell Proliferation in CD44-high Glioma-initiating Cells
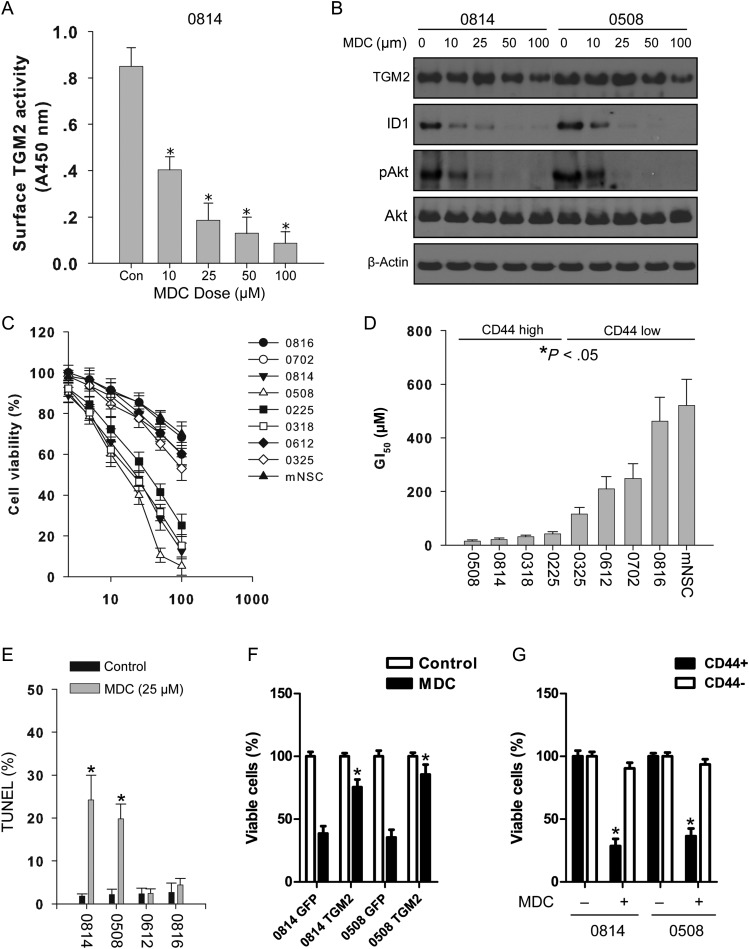
MDC, a primary alkyl amine with a fluorescent dansyl group attached at the end, has been used in a number of studies as a TGM2 inhibitor.35 We measured the TGM2 activity in 0814 cells with the in situ cadaverine incorporation assay. The findings showed that increasing doses of MDC decreased TGM2 activity in 0814 cells (Fig. 5A). Consistently, Western blotting of 0814 and 0508 cells treated with escalating doses of MDC showed attenuated TGM2 activity, accompanied by reduced pAkt and ID1 levels (Fig. 5B). Glioma-initiating cell lines treated with MDC were also assessed for growth inhibition with the MTT assay. MDC treatment significantly suppressed cell proliferation in a dose-dependent manner (Fig. 5C). The calculated GI50 for CD44-high glioma cell lines was significantly lower than that for glioma stem cells with low CD44 levels (P < .05; Fig. 5D). MDC displayed mild cytotoxicity on normal NSCs in the tested dose range, suggesting that TGM2 inhibition might be selective on CD44-high glioma cell lines rather than on normal NSCs. Furthermore, MDC also attenuated clonogenesis in vitro and induced apoptosis in CD44-high glioma cell lines (Fig. S3A, Fig. 5E). We also did a rescue experiment to observe the effect of overexpression of TGM2 on MDC-induced growth inhibition. We showed that overexpression of TGM2 partially reversed the antiproliferation effect induced by MDC treatment (Fig. S3C, Fig. 5F). MDC treatment also exhibited a differential effect on CD44+ and CD44− glioma cell fractions, with CD44+ glioma stem cells more sensitive to MDC treatment than CD44− glioma cells (Fig. 5G). Therefore, targeting TGM2 might be an effective strategy to eliminate glioma cell lines/glioma cells with high CD44 expression.
Fig. 5.
TGM2 inhibition suppresses cell proliferation and triggers apoptosis in glioma-initiating cells. (A) TGM2 activities are reduced following TGM2 treatment in glioma-initiating cells. 0814 cells were treated with indicated dose of MDC for 4 h. Dimethyl sulfoxide (DMSO) was used as vehicle control. *P < .05 compared with no treatment control (n = 4). (B) TGM2 inhibition by MDC reduces pAkt (Ser473) and ID1 expression. 0814 and 0508 cells were treated with indicated dose of MDC for 24 h, then cells were collected and cell lysates were subject to Western blotting analysis. DMSO was used as vehicle control. (C) Effects of MDC on cell proliferation of glioma-initiating cell lines. Glioma cells (3 × 103) were plated per well of 96-well culture plate precoated with poly-l-ornithine and incubated with MDC for 72 h. Cell viability in DMSO control was treated as 100%. Results were from 3 independent experiments. (D) GI50 values of glioma cell lines treated with MDC. *P < .05 compared with CD44-low cell lines. (E) TGM2 inhibition triggers apoptosis in CD44-high glioma-initiating cell lines. Two CD44-high glioma cell lines (0814, 0508) and 2 CD44-low glioma cell lines (0612, 0816) were incubated with MDC (25 μM) for 72 h, then TUNEL assay was performed. *P < .05 compared with vehicle control (n = 4). (F) Overexpression of TGM2 partially restored cell proliferation inhibited by MDC. *P < .05 compared with green fluorescent protein control (n = 4). (G) MDC preferentially targets CD44+ glioma stem cells. *P < .05 compared with matched CD44− cell fraction after MDC treatment (n = 4).
TGM2 Inhibitor Suppresses Tumor Growth and Eliminates CD44-high Glioma Cells In vivo
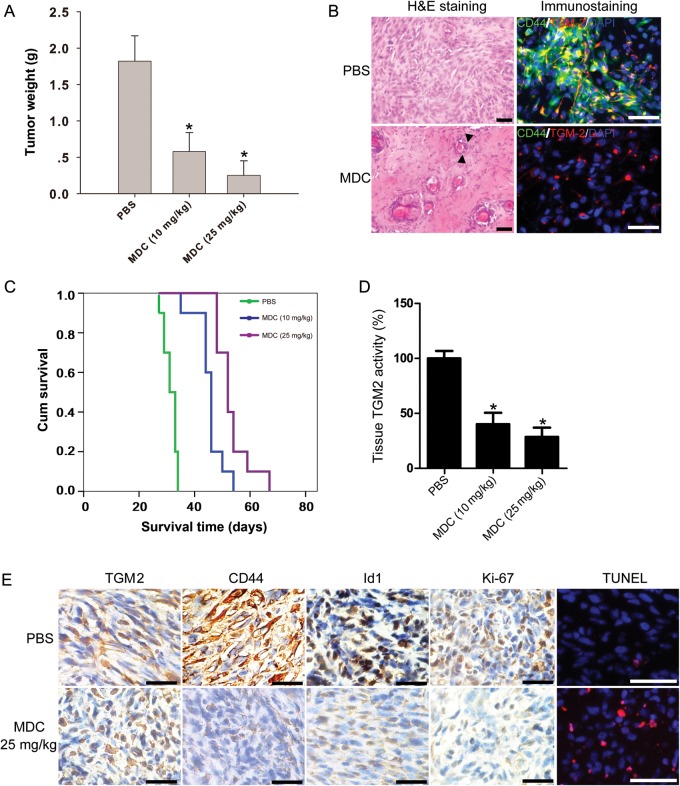
In vitro findings that treatment with the TGM2 inhibitor MDC suppressed cell proliferation and induced apoptosis in CD44-high glioma cell lines were extended to observations with in vivo studies. The ability of MDC to inhibit tumor growth in glioblastoma cells was examined in mice with 0814 glioblastoma s.c. xenografts. There were no mice deaths in the groups treated with 10 or 25 mg/kg of MDC. The group of mice treated with PBS developed tumors weighing 1.54 ± 0.35 g (n = 4). The tumors from the group treated with MDC 10 mg/kg weighed 0.48 ± 0.26 g (n = 4). The group treated with MDC 25 mg/kg developed tumors that weighed 0.25 ± 0.22 g (n = 4) (Fig. 6A). The tumor weights in 2 MDC-treated groups were significantly lower than that in the PBS-treated group (P < .05). H&E staining revealed several areas of severe necrosis scattered within the tumor following MDC treatment compared with PBS control (Fig. 6B). TGM2 inhibition also decreased the percentage of CD44+ glioma cells, as shown by immunostaining on cryosections (Fig. 6B).
Fig. 6.
TGM2 inhibition suppresses tumor growth and prolongs survival time in tumor-bearing mice (0814). (A) Tumor weights from the s.c. xenograft groups following MDC treatment. The data show that groups of mice treated with MDC (10 or 25 mg/kg) had tumors that were significantly smaller than those treated with PBS only. *P < .05 compared with PBS control (n = 4). (B) Photographs show representative staining on s.c. tumor cryosections from mice treated with PBS or MDC 25 mg/kg. Left panel, H&E staining. Tumors were composed of heterogeneous spindled to gemistocytic glial cells with frequent mitoses in the PBS group. Solid triangles: necrotic cells. Bar, 50 μm. Right panel, immunostaining of TGM2 (red) and CD44 (green) on tumor cryosections from mice treated with PBS or MDC. Bar, 50 μm. (C) Kaplan–Meier survival curves from mice bearing i.c. 0814 tumors. Mice were treated with PBS or MDC (10 or 25 mg/kg). Log-rank analysis showed the combination group had an increased survival compared with PBS group or MDC (n = 10), *P < .05 compared with PBS control. (D) MDC treatment reduces TGM2 activity in orthotopic xenografts (n = 3), *P < .05 compared with PBS control. (E) Immunostaining of the i.c. transplanted tumors that were treated with PBS or MDC alone of mice killed 15 days after the treatment started. The tissue sections were incubated with antibodies against TGM2, CD44, ID1, and Ki67 (dilution 1:200). Diaminobenzidine was used as a chromogen, followed by counterstaining with hematoxylin. Incidence of apoptosis was determined with TUNEL staining. Bar, 100 μm.
TGM2 Inhibitor Prolongs Survival Time in an Orthotopic Mouse Xenograft Model
We next evaluated the antitumor effect of MDC in the i.c. xenograft model. The animals were treated with MDC (10 or 25 mg/kg) after inoculation of 0814 glioma cells (n = 13). The primary endpoint was to evaluate animal survival (n = 10). In addition, we examined the effects of TGM2 inhibition in vivo in the context of MDC treatment (n = 3). The median duration of survival of the control mice (PBS-treated) was 31 days (95% confidence interval [CI] = 28.5–33.4 d). The median duration of survival for animals treated with MDC 10 mg/kg was 46 days (95% CI = 44.7–47.2 d). Importantly, in the group of animals treated with MDC 25 mg/kg, 60% of the animals survived >50 days (P < .01; Fig. 6C). Next, we investigated whether ID1 was reduced by the effect of the drug in vivo. For this purpose, tumor-bearing mice from each group (n = 3) were sacrificed 15 days after treatment started. Xenografts treated with MDC 25 mg/kg showed decreased TGM2 activity (Fig. 6D). CD44, Ki67, and ID1 expression in the tumors was also decreased after MDC treatment (Fig. 6E). Importantly, TUNEL analyses identified elevated apoptotic cells in the MDC-treated tumors (Fig. 6E). Taken together, our results proved the antitumoral efficacy of TGM2 inhibitor in eliminating CD44-high glioma cells.
Discussion
While GBM has a poor prognosis overall, molecular subsets of tumors exist with differential outcomes to initial treatment. Recent TCGA GBM molecular subtyping demonstrated that the mesenchymal GBM subtype appears to have worse prognosis compared with tumors with a proneural signature.36 CD44 is a cell surface marker associated with tumor progression and treatment resistance in glioma. Phillips et al37 showed that CD44 is a typical biomarker for mesenchymal-subtype glioma. Lottaz et al38 divided 17 glioma stem cell lines into 2 groups, one expressing the “proneural” gene signature resembling normal fetal brain stem cells and the other expressing a mesenchymal gene signature including CD44, which is more similar to adult NSCs.38 In breast cancer, transforming growth factor (TGF)–β has been shown to increase the CD44-high cell population enriched for cancer-initiating cells through the induction of an epithelial-mesenchymal transition.39 Recently, Anido et al25 revealed the regulatory role of TGF-β signaling in the maintenance of CD44+ glioma stem cells. In this study, we found that TGM2 was highly expressed in CD44-high GBM and tumor-derived glioma-initiating cell lines. Knockdown of TGM2 expression suppressed cell proliferation and induced apoptosis. Importantly, we observed high TGM2 expression in CD44+ glioma stem cell populations, suggesting that TGM2 might be an important regulator for the maintenance of CD44+ glioma stem cells. As both TGM2 activity and expression were upregulated by TGF-β stimulation,40 it is reasonable to propose that TGM2 might be part of TGF-β signaling to maintain CD44-high glioma-initiating cells in a proliferative state.
ID1 is an important basic helix-loop-helix transcriptional factor regulating the stem cell maintenance of embryonic stem cells and B1-type adult NSCs.41,42 Soroceanu et al43 showed that ID1 expression levels positively correlate with glioma cell invasiveness in culture and with histopathologic grades in patient biopsies and that genetic knockdown of ID1 leads to a significant increase in survival in an orthotopic model of human GBM, suggesting that ID1 represents a novel and promising target for improving the therapy and outcome of GBM patients. Anido et al25 also identified a cell population enriched for glioma stem cells that expresses high levels of CD44 and ID1. The inhibition of the TGF-β pathway decreased the CD44-high/ID1-high stem cell population through repression of ID1 levels, therefore inhibiting the capacity of cells to initiate tumors.25 Our findings showed that TGM2 knockdown reduced ID1 expression, while constitutive TGM2 expression enhanced ID1 expression in glioma-initiating cells. Similarly, loss of TGM2 expression in sorted CD44+ glioma stem cells reduced ID1 expression. Importantly, overexpression of ID1 restored the cell proliferation suppressed by TGM2 knockdown, suggesting that ID1 might be a downstream mediator for TGM2-regulated cell proliferation in CD44-high glioma-initiating cells. We also showed that TGM2 might regulate ID1 expression through activating the PI3K/Akt pathway, suggesting that the PI3K/Akt pathway could be an alternative target in order to suppress ID1-mediated cell proliferation in CD44-high glioma.
Various TGM2 inhibitors have been studied in experimental therapeutics on glioblastoma.19,35 These studies identify TGM2 inhibitors as a potential new class of agents to enhance chemotherapy in glioblastoma. Some more specific TGM2 inhibitors, such as ZM39923 and Tyrphostin 47, were discovered and may serve as valuable lead compounds for the development of orally active TGM2 inhibitors to treat human diseases.44 In the present study, the finding that TGM2 regulated the cell proliferation of CD44-high glioma-initiating cells/CD44+ glioma stem cells sparked our interest to test the effect of a TGM2 inhibitor on glioma models. Our results indicated that CD44-high glioma-initiating cell lines/CD44+ glioma stem cells were more sensitive to TGM2 inhibitor MDC treatment. Treatment with MDC suppresses cell proliferation and induces apoptosis in CD44-high glioma-initiating cell lines/CD44+ glioma stem cells. The significance of these in vitro observations of inhibition on cell proliferation and induction of apoptosis in CD44-high glioma-initiating cell lines treated with TGM2 inhibitor were evaluated with an in vivo mouse model. Mice with s.c. xenografts treated with MDC compared with those treated with PBS showed significantly decreased tumor size. Furthermore, the benefits of TGM2 inhibitor treatment were confirmed in vivo, with more of the mice in the MDC groups surviving long-term than in the group that had received PBS. Treatment of animals bearing glioma xenografts with MDC also resulted in reduced CD44, ID1, and Ki67 positivity and induced apoptosis. These results identify TGM2 inhibitors as a new class of agents that may improve the treatment of CD44-high GBM.
Supplementary Material
Funding
This work was supported by a grant from the National Natural Science Foundation of China (no. 30772551).
Supplementary Material
Acknowledgments
We thank Dr Janusz Tucholski (Psychiatry and Behavioral Neurobiology, University of Alabama at Birmingham) for kindly providing us the TGM2 plasmid construct.
Conflict of interest statement. None declared.
References
- 1.Tabatabai G, Hegi M, Stupp R, Weller M. Clinical implications of molecular neuropathology and biomarkers for malignant glioma. Curr Neurol Neurosci Rep. 2012;12:302–307. doi: 10.1007/s11910-012-0263-x. [DOI] [PubMed] [Google Scholar]
- 2.Merzak A, Koocheckpour S, Pilkington GJ. CD44 mediates human glioma cell adhesion and invasion in vitro. Cancer Res. 1994;54:3988–3992. [PubMed] [Google Scholar]
- 3.Lesort M, Tucholski J, Miller ML, Johnson GV. Tissue transglutaminase: a possible role in neurodegenerative diseases. Prog Neurobiol. 2000;61:439–463. doi: 10.1016/s0301-0082(99)00052-0. [DOI] [PubMed] [Google Scholar]
- 4.Akimov SS, Krylov D, Fleischman LF, Belkin AM. Tissue transglutaminase is an integrin-binding adhesion coreceptor for fibronectin. J Cell Biol. 2000;148:825–838. doi: 10.1083/jcb.148.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song H, Chang W, Lim S, et al. Tissue transglutaminase is essential for integrin-mediated survival of bone marrow–derived mesenchymal stem cells. Stem Cells. 2007;25:1431–1438. doi: 10.1634/stemcells.2006-0467. [DOI] [PubMed] [Google Scholar]
- 6.Singer CF, Hudelist G, Walter I, et al. Tissue array-based expression of transglutaminase-2 in human breast and ovarian cancer. Clin Exp Metastasis. 2006;23:33–39. doi: 10.1007/s10585-006-9015-0. [DOI] [PubMed] [Google Scholar]
- 7.Fok JY, Ekmekcioglu S, Mehta K. Implications of tissue transglutaminase expression in malignant melanoma. Mol Cancer Ther. 2006;5:1493–1503. doi: 10.1158/1535-7163.MCT-06-0083. [DOI] [PubMed] [Google Scholar]
- 8.Antonyak MA, Li B, Regan AD, Feng Q, Dusaban SS, Cerione RA. Tissue transglutaminase is an essential participant in the epidermal growth factor–stimulated signaling pathway leading to cancer cell migration and invasion. J Biol Chem. 2009;284:17914–17925. doi: 10.1074/jbc.M109.013037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park KS, Kim HK, Lee JH, et al. Transglutaminase 2 as a cisplatin resistance marker in non–small cell lung cancer. J Cancer Res Clin Oncol. 2010;136:493–502. doi: 10.1007/s00432-009-0681-6. [DOI] [PubMed] [Google Scholar]
- 10.Mann AP, Verma A, Sethi G, et al. Overexpression of tissue transglutaminase leads to constitutive activation of nuclear factor-kappaB in cancer cells: delineation of a novel pathway. Cancer Res. 2006;66:8788–8795. doi: 10.1158/0008-5472.CAN-06-1457. [DOI] [PubMed] [Google Scholar]
- 11.Verma A, Guha S, Wang H, et al. Tissue transglutaminase regulates focal adhesion kinase/AKT activation by modulating PTEN expression in pancreatic cancer cells. Clin Cancer Res. 2008;14:1997–2005. doi: 10.1158/1078-0432.CCR-07-1533. [DOI] [PubMed] [Google Scholar]
- 12.Kubo H, Shimizu M, Taya Y, et al. Identification of mesenchymal stem cell (MSC)–transcription factors by microarray and knockdown analyses, and signature molecule-marked MSC in bone marrow by immunohistochemistry. Genes Cells. 2009;14:407–424. doi: 10.1111/j.1365-2443.2009.01281.x. [DOI] [PubMed] [Google Scholar]
- 13.Obinata A, Osakabe K, Yamaguchi M, et al. Tgm2/Gh, Gbx1 and TGF-beta are involved in retinoic acid–induced transdifferentiation from epidermis to mucosal epithelium. Int J Dev Biol. 2011;55:933–943. doi: 10.1387/ijdb.113326ao. [DOI] [PubMed] [Google Scholar]
- 14.Srivastava M, Khurana P, Sugadev R. Lung cancer signature biomarkers: tissue specific semantic similarity based clustering of digital differential display (DDD) data. BMC Res Notes. 2012;5:617– . doi: 10.1186/1756-0500-5-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ai L, Skehan RR, Saydi J, et al. Ataxia-telangiectasia, mutated (ATM)/nuclear factor κ light chain enhancer of activated B cells (NFκB) signaling controls basal and DNA damage-induced transglutaminase 2 expression. J Biol Chem. 2012;287:18330–18341. doi: 10.1074/jbc.M112.339317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang R, Tremblay TL, McDermid A, Thibault P, Stanimirovic D. Identification of differentially expressed proteins in human glioblastoma cell lines and tumors. Glia. 2003;42:194–208. doi: 10.1002/glia.10222. [DOI] [PubMed] [Google Scholar]
- 17.Lathia JD, Gallagher J, Heddleston JM, et al. Integrin alpha 6 regulates glioblastoma stem cells. Cell Stem Cell. 2010;6:421–432. doi: 10.1016/j.stem.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang D, Anderson JC, Gladson CL. The role of the extracellular matrix in angiogenesis in malignant glioma tumors. Brain Pathol. 2005;15:318–326. doi: 10.1111/j.1750-3639.2005.tb00117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan L, Siegel M, Choi K, et al. Transglutaminase 2 inhibitor, KCC009, disrupts fibronectin assembly in the extracellular matrix and sensitizes orthotopic glioblastomas to chemotherapy. Oncogene. 2007;26:2563–2573. doi: 10.1038/sj.onc.1210048. [DOI] [PubMed] [Google Scholar]
- 20.Knüpfer MM, Poppenborg H, Hotfilder M, Kühnel K, Wolff JE, Domula M. CD44 expression and hyaluronic acid binding of malignant glioma cells. Clin Exp Metastasis. 1999;17(1):71–76. doi: 10.1023/a:1026425519497. [DOI] [PubMed] [Google Scholar]
- 21.Wiranowska M, Ladd S, Moscinski LC, et al. Modulation of hyaluronan production by CD44 positive glioma cells. Int J Cancer. 2010;127:532–542. doi: 10.1002/ijc.25085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshida T, Matsuda Y, Naito Z, Ishiwata T. CD44 in human glioma correlates with histopathological grade and cell migration. Pathol Int. 2012;62:463–470. doi: 10.1111/j.1440-1827.2012.02823.x. [DOI] [PubMed] [Google Scholar]
- 23.Wang C, Xie J, Guo J, et al. Evaluation of CD44 and CD133 as cancer stem cell markers for colorectal cancer. Oncol Rep. 2012;28:1301–1308. doi: 10.3892/or.2012.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ponti D, Costa A, Zaffaroni N, et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65:5506–5511. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- 25.Anido J, Sáez-Borderías A, Gonzàlez-Juncà A, et al. TGF-β receptor inhibitors target the CD44(high)/Id1(high) glioma-initiating cell population in human glioblastoma. Cancer Cell. 2010;18:655–668. doi: 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 26.Ushio K, Hashimoto T, Kitamura N, Tanaka T. Id1 is down-regulated by hepatocyte growth factor via ERK-dependent and ERK-independent signaling pathways, leading to increased expression of p16INK4a in hepatoma cells. Mol Cancer Res. 2009;7:1179–1188. doi: 10.1158/1541-7786.MCR-08-0289. [DOI] [PubMed] [Google Scholar]
- 27.Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 28.Ahlenius H, Kokaia Z. Isolation and generation of neurosphere cultures from embryonic and adult mouse brain. Methods Mol Biol. 2010;633:241–252. doi: 10.1007/978-1-59745-019-5_18. [DOI] [PubMed] [Google Scholar]
- 29.Bao S, Wu Q, Li Z, et al. Targeting cancer stem cells through L1CAM suppresses glioma growth. Cancer Res. 2008;68:6043–6048. doi: 10.1158/0008-5472.CAN-08-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tovar-Vidales T, Roque R, Clark AF, Wordinger RJ. Tissue transglutaminase expression and activity in normal and glaucomatous human trabecular meshwork cells and tissues. Invest Ophthalmol Vis Sci. 2008;49:622–628. doi: 10.1167/iovs.07-0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruzinova MB, Benezra R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 2003;13:410–418. doi: 10.1016/s0962-8924(03)00147-8. [DOI] [PubMed] [Google Scholar]
- 33.Lee TK, Man K, Ling MT, et al. Over-expression of Id-1 induces cell proliferation in hepatocellular carcinoma through inactivation of p16INK4a/RB pathway. Carcinogenesis. 2003;24:1729–1736. doi: 10.1093/carcin/bgg145. [DOI] [PubMed] [Google Scholar]
- 34.Cheng YJ, Tsai JW, Hsieh KC, et al. Id1 promotes lung cancer cell proliferation and tumor growth through Akt-related pathway. Cancer Lett. 2011;307:191–199. doi: 10.1016/j.canlet.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 35.Yuan L, Choi K, Khosla C, et al. Tissue transglutaminase 2 inhibition promotes cell death and chemosensitivity in glioblastomas. Mol Cancer Ther. 2005;4:1293–1302. doi: 10.1158/1535-7163.MCT-04-0328. [DOI] [PubMed] [Google Scholar]
- 36.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 38.Lottaz C, Beier D, Meyer K, et al. Transcriptional profiles of CD133+ and CD133– glioblastoma-derived cancer stem cell lines suggest different cells of origin. Cancer Res. 2010;70:2030–2040. doi: 10.1158/0008-5472.CAN-09-1707. [DOI] [PubMed] [Google Scholar]
- 39.Shipitsin M, Campbell LL, Argani P, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 40.Kumar A, Xu J, Brady S, et al. Tissue transglutaminase promotes drug resistance and invasion by inducing mesenchymal transition in mammary epithelial cells. PLoS One. 2010;5:e13390. doi: 10.1371/journal.pone.0013390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romero-Lanman EE, Pavlovic S, Amlani B, Chin Y, Benezra R. Id1 maintains embryonic stem cell self-renewal by up-regulation of Nanog and repression of Brachyury expression. Stem Cells Dev. 2012;21:384–393. doi: 10.1089/scd.2011.0428. [DOI] [PubMed] [Google Scholar]
- 42.Nam HS, Benezra R. High levels of Id1 expression define B1 type adult neural stem cells. Cell Stem Cell. 2009;5:515–526. doi: 10.1016/j.stem.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soroceanu L, Murase R, Limbad C, et al. Id-1 is a key transcriptional regulator of glioblastoma aggressiveness and a novel therapeutic target. Cancer Res. 2013;73:1559–1569. doi: 10.1158/0008-5472.CAN-12-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lai T-S, Liu Y, Tucker T, et al. Identification of chemical inhibitors to human tissue transglutaminase by screening existing drug libraries. Chem Biol. 2008;15:969–978. doi: 10.1016/j.chembiol.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.