Abstract
Long non-coding RNAs (ncRNAs) are emerging as important regulatory factors in mammalian genomics. A number of reports within the last 2 years have identified thousands of actively expressed long ncRNA transcripts with distinct properties. The long ncRNAs show differential expression patterns and regulation in a w ide variety of cells and tissues, adding significant complexity to the understanding of their biological role. Furthermore, genome-wide studies of transcriptional enhancers based on chromatin modifications and enhancer binding proteins have led to the identification of putative enhancers and provided insight into their tissue-specific regulation of gene expression. In an exciting turn of events, new evidence is indicating that long ncRNAs are associated with enhancer regions and that such non-coding transcription correlate with the increased activity of the neighboring genes. Moreover, additional experiments suggest that enhancer-function can be mediated through a transcribed long ncRNA and that this might be a common function for long ncRNAs. Here, we review recent advances made both in the genome-wide characterization of enhancers and in the identification of new classes of long ncRNAs, and discuss the functional overlap of these two classes of regulatory elements.
Introduction
Long non-coding RNAs (ncRNAs) are RNA transcripts of more than 200 nucleotides that do not encode proteins. Long ncRNAs have been implicated in the regulation of gene expression, dosage compensation and imprinting [1,2]. The functional role of long ncRNAs has long been underestimated, as protein-coding genes have taken center stage and been the primary focus of most research. Recent large-scale genome-wide sequencing studies have led to the conclusion that beyond the protein-coding genes, a large portion of the genome is being transcribed [3–5]. There are a number of loci of the mammalian genome that act as templates for the production of long ncRNAs. Indeed, the simplest classification of long ncRNAs could be based on their loci of origin. On the basis of such denomination long ncRNAs could be grouped into five separate categories: (1) transcripts that arise from the antisense strand of protein-coding genes, (2) transcripts that represent the introns of protein-coding genes, (3) transcripts that correspond to the promoters and/or 5′-untranslated or 3′-untranslated regions of protein-coding genes, (4) Independent transcripts that initiate within the protein-coding genes, and finally (5) transcripts originating from regions outside of protein-coding genes [5,6,7•,8•,9••,10,11,12].
In this review, we will focus on long ncRNAs that are transcribed from independent loci (category 5) of the mammalian genome. Moreover, we will discuss the genome-wide identification and characterization of transcriptional enhancers and the emerging evidence indicating that long ncRNAs are transcribed from and act as enhancers. Two classes of long ncRNAs will be discussed; one transcribed bidirectionally from promoters at activity-regulated enhancers in mouse neuronal cells showing association with enhancers and dynamic regulation termed enhancer-RNA (eRNAs) [13••], and the second, a set of long ncRNAs with enhancer-like functions expressed from regions independent of protein-coding genes [9••] called activating ncRNAs (ncRNA-a). Both examples suggest an active connection between classically defined enhancers and transcribed functional long ncRNAs.
Co-expression of long ncRNAs and protein-coding genes suggests regulatory functions
Recent advances in high throughput sequencing which have been combined with genome-wide mapping of chromatin modification signatures have resulted in the identification of a set of experimentally validated transcriptionally active long ncRNAs in multiple experimental systems. Through such efforts thousands of long ncRNAs displaying tissue specific expression have been identified [3,5,7•,8•,9••,11,13••,14]. Interestingly, while the conservation of protein-coding genes across species is relatively high, long ncRNAs display a lower degree of conservation [7•,8•,9••,15–17]. Multiple studies have been devoted to the analysis of expression patterns of long ncRNAs and the possible correlation of such patterns with the adjacent protein-coding genes.
In one such study identification of a subset of conserved long ncRNA transcripts in mouse brain and correlation of their expression levels and the neighboring protein-coding genes has been used to substantiate their functional importance [14]. Their analysis of sequencing data from mouse has identified 3122 putative long ncRNA transcripts encoded in between annotated protein-coding genes that exhibit evolutionary constraint. A subset of these long ncRNA transcripts (659) showed considerable evolutionary constraint and are expressed in the brain during development. This particular subset of long ncR NA is co-expressed with nearby genes encoding proteins that function in transcriptional regulation and nervous system development suggesting a cis mechanism of regulation by long ncRNAs [15].
Similar correlations have been observed during the differentiation of human primary keratinocytes [9••]. Induced changes in long ncRNA expression tend to be in loci associated with keratinocyte differentiation. In addition, a correlation between the expression levels of long ncRNAs and their neighboring genes has been reported for activity-regulated neuronal enhancers in mouse [13••]. However, although co-regulated expression of non-coding transcripts and their neighboring protein-coding genes have been reported in mammalian cells [18], such data have been interpreted to suggest that the act of transcription through the non-coding locus rather than the transcript itself was the determining factor for the induction of the protein-coding genes. Overall, most genome-wide studies suggest a positive correlation between the expression of ncRNAs and the protein-coding genes surrounding them, which is in contrast to repressive effects described for ncRNAs involved in epigenetic silencing.
Genome-wide definition of enhancers
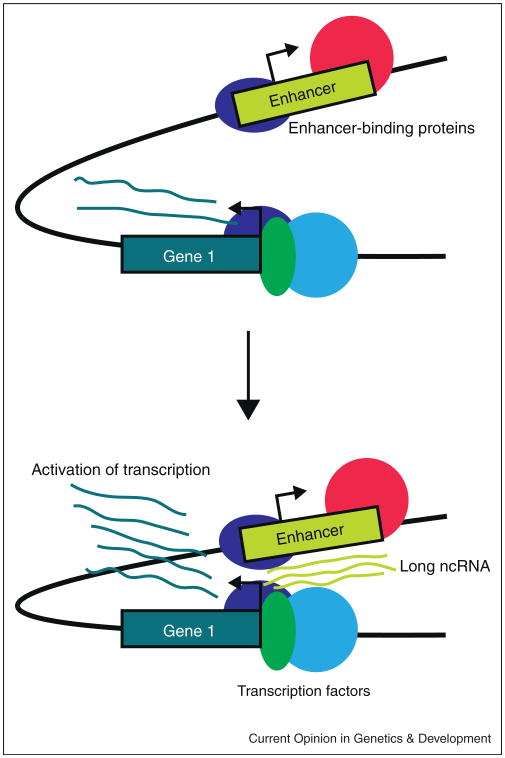
Enhancers are classically defined by their ability to potentiate transcription at a distance in a cis regulatory fashion and by their orientation independence with respect to the regulated gene [19]. Recent genome-wide analyses of chromatin modifications have suggested that enhancers and promoters may be distinguished from each other by their respective chromatin signatures [3,20]. While promoters can be identified by H3K4me2 and H3K4me3, enhancers can often be characterized by binding of the protein p300 and the presence of H3K4me1 and H3K4me2 marks, along with DNaseI hypersensitive sites [13••,21,22••,23,24]. Furthermore, RNA polymerase II (RNAPII) is reported to be associated with many predicted enhancers [13••,28•]. A recent study also identified Cohe-sin and Mediator as essential factors for the activity of enhancers regulating embryonic stem cell specific gene expression [25•]. Interestingly, such chromatin signatures of promoters and enhancers are overlapping to a great extent. Both promoters and enhancers display transcriptional co-activator occupancy such as p300/CBP binding as well as RNAPII, Mediator and cohesin occupancy. Indeed, it is tempting to speculate that many of the enhancer sites identified using such chromatin-modification and transcription factor-binding profiles may indeed correspond to the promoters of long ncRNAs (Figure 1).
Figure 1.
Enhancers are classically defined as distal regulatory genomic elements. Recent research suggests that several enhancers function through a long ncRNA. In panel A, an inactive enhancer is depicted. The DNA sequence is bound by enhancer-binding proteins, but does not communicate to the promoter of the gene that is regulated. In panel B, an active enhancer is depicted, where the functional molecule mediating the enhancing function is the ncRNA transcribed from the enhancer.
While the chromatin state at promoters if reported to be largely invariant across diverse cell types, enhancers are reported to be associated with chromatin marks in a highly cell-type specific manner [22••]. More than 55,000 enhancers have been predicted in human cell lines, many of which are correlated with cell-type specific gene expression [22••]. Further evidence for the vast abundance of enhancers in mammalian genomes are provided by chromatin-immunoprecipitation with p300 and deep sequencing from mouse embryonic forebrain, midbrain and limb tissue [24]. Many of the enhancers identified are validated for tissue specific enhancer activity associated with tissue specific p300 binding. Interestingly, since long ncRNAs display an exquisite pattern of tissue expression, it is likely that the p300 occupancy and the chromatin signatures of their promoters may also display a tissue-specific and cell type-specific pattern of histone modifications. Therefore, it would be a provocative finding if the enhancer binding sites displaying cell type-specific and/or tissue-specific pattern of expression may turn out to be promoters of non-coding transcripts that are expressed in a temporal and cell-specific manner.
Although recent studies have detected some sequence conservation across species for a subset of the identified enhancers, strict conservation does not seem to be functionally crucial for enhancers in general [3,22••,26,27]. Enhancers are defined operationally as DNA elements that positively regulate transcription of their target genes from long distances and are likely to be composed of different functionally distinct classes. Therefore, although common characteristics of the genome-wide enhancer finding strategies such as the presence of H3K4me1 and especially p300 are strongly suggestive of enhancers, it is not clear whether all or only a subset of enhancers can be identified by such criteria.
Enhancers are transcribed and long ncRNAs act as enhancers
Several enhancer regions with associated RNAPII binding show evidence of transcription of long ncRNAs [13••,28•]. This association, along with binding of p300 and H3K4me1 marks at many enhancers has been applied in a study on neuronal activity-regulated enhancers in mouse [13••]. p300 binding sites with juxtaposed H3K4me1 domains of 2–4 kb were defined as enhancer domains and observed to bind transcription factors known to mediate activity-regulated gene expression, such as CREB, SRF and NPAS4. Transcription of non-polyadenylated long ncRNA was detected from 2000 of the predicted activity-regulated enhancers following membrane depolarization, extending in both directions from the CBP binding sites presumably from a shared bidirectional promoter. Transcription of an enhancer RNA (eRNA) derived from enhancer elements close to the Arc promoter was shown to be dependent on the presence of the Arc promoter. When the promoter was removed in a transgenic mouse, the eRNA was no longer transcribed, suggesting that such eRNA transcripts require an active promoter of its target gene. In contrast, the long ncRNAs arising from the HS2 enhancer of the beta-globin locus control region [29] have been shown to be transcribed independently and in the absence of the promoter they regulate [30]. Although such transcripts have not been implicated in the enhancer function thus far, these data are suggestive of an important role in transcriptional activation for such enhancer-derived transcripts.
Transcription from the Dlx-5/6 ultraconserved region has been shown to result in the expression of the long ncRNAs Evf-1 [31–33] and Evf-2 [34] in mouse. Evf-1 is a 3.8 kb developmentally regulated long ncRNA identified as a downstream target of Sonic hedgehog signaling [33]. Evf-1 is transcribed from an ultraconserved region overlapping with an intergenic enhancer upstream of the Dlx-6 gene. The Evf-2 long ncRNA is an alternatively spliced form of Evf-1, shown to cooperate with the home-odomain protein Dlx-2 to increase the transcriptional activity of the Dlx-5/6 enhancer [34], suggesting a transacting enhancer activity of the long ncRNA Evf-2. The RNA dependency of the enhancing effect is demonstrated by single-stranded sense RNA that is inhibited by the presence of anti-sense RNA. Furthermore, knockdown of Evf-2 with siRNAs inhibits the transcriptional enhancing activity in a dose-dependent manner. Another example of an activating long ncRNA is the LINoCR (LPS inducible ncRNA) identified in chicken cells. The transcription elongation of LINoCR from an enhancer region through a negative regulatory element has been shown to be required for the switching of a cis-regulatory region from a repressive to an active conformation [35].
Long ncRNAs with enhancer-like functions have been demonstrated in several human cell lines [9••]. In a recent report, we used the GENCODE annotation of the human genome [36] to identify long ncRNAs transcribed from regions residing outside of protein-coding loci [9••]. Using siRNA-mediated knock-down of individual ncRNA-acti-vating (ncRNA-a) transcripts, we demonstrated that long ncRNA-a transcripts increased expression of specific genes in their immediate surroundings. This effect was demonstrated to be RNA dependent, as knock-down of the ncRNA-a transcripts abolished the enhancing activity observed, both in vivo and in reporter assays [9••]. A similar effect of RNA dependent positive regulation of gene expression was observed in X-inactivation, where the Jpx long ncRNA was reported to induce Xist on the inactive X-chromosome [37•].
Expression of TAL1 is shown to be enhanced by ncRNA-a3 in human cells [9••]. Studies in the mouse have identified an enhancer element mapping to genomic region syngenic to the ncRNA-a3 locus in human [38]. Interestingly, this locus in mouse displays evidence of transcription of what could be the mouse homologue of ncRNA-a3. More detailed examination of the TAL1 locus in human has revealed a complex pattern of predicted promoters, insulators and enhancers based on chromatin signatures, supporting the findings in mice [39•].
The future of enhancers and RNA
The fact that transcripts arise from enhancers has been known for many years [40]. However, a number of recent genome-wide analyses of enhancers and their targets have begun to paint a picture suggesting that for many enhancers their transcribed RNAs are functional and are the mediators of the enhancing activity. Many enhancers and long ncRNAs share common characteristics in their tissue-specific expression and modest conservation rate across species [7•,8•,9••,15–17]. Moreover, several enhancers arising from the p300 and H3K4me1 prediction studies show evidence of transcription of long, spliced ncRNAs according to HAVANA annotation of the human genome [3,36] (Ulf Ørom and Ramin Shiekhattar, unpublished), suggesting that a large fraction of transcriptional enhancers could be working through the act of a transcribed long ncRNA. Further functional analysis of the large number of identified enhancers and expression profiling of the relevant genomic loci in several cell lines is required to fully address this observation, and might provide valuable insight into the function of enhancer-encoded transcripts.
Conclusions
The precise mechanism by which enhancer-like RNAs mediate their action is not known. One of the working models for enhancer function contends that the enhancer region may be looped to contact the promoter of the gene they regulate. Such a looping could be facilitated by transcriptional activators and co-activators recruited to the induced locus [19]. Indeed, the binding of transcription factors has been shown to be critically involved in mediating such interactions [41]. It is conceivable that ncRNAs transcribed from the enhancer mediate such chromatin looping by providing a structural scaffold facilitating transcriptional assembly of activators and coactivators. Another hypothesis contends that a class of enhancer-like RNAs may be endowed with enzymatic activities to modify chromatin or DNA This model is akin to transcriptional coactivators with histone acetyl-transferase or methyl-transferase activities. An important question that remains to be answered is the exact nature of the structural and/or domain requirements for enhancer-like RNAs that are required to mediate their function. Future structure/function studies will reveal whether a specific domain(s) of the ncRNA is critical for its activating function and whether such a domain is conserved among other ncRNAs evolutionarily.
Several studies have reported the identification of large sets of long ncRNAs [3,5,7•,8•,9••,11,13••,14], and it is becoming increasingly clear that long ncRNAs play important roles in cellular processes. Concomitantly, large-scale identification of transcriptional enhancers [13••,22••,23,24] and the functional overlap of long ncRNAs and enhancers add increasing intricacy to the understanding of gene expression and regulatory elements. Genome-wide functional validation assays, and approaches for studying the 3-dimensional structure of the genome such as Chromosome Conformation Capture (3C) [42], for the thousands of identified regulatory elements and long ncRNAs could provide insightful information of their functions and is one of the important next steps for understanding the impact of the intersection of these two expanding areas of molecular biology.
Acknowledgments
We thank Glenda Harris and Jeppe Falsig for comments on the manuscript. UAO is supported by a personal grant from the Danish Research Council. R.S. was supported by a grant from NIH, GM 079091.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5(4):e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wan LB, Bartolomei MS. Regulation of imprinting in clusters: noncoding RNAs versus insulators. Adv Genet. 2008;61:207–223. doi: 10.1016/S0065-2660(07)00007-7. [DOI] [PubMed] [Google Scholar]
- 3.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308(5725):1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 5.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316(5830):1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 6.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322(5909):1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7•.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. doi: 10.1038/nature07672. This paper demonstrates the identification of transcribed domains through chromatin modification patterns. Using the assumption that transcripts intervening known protein coding genes are non-coding, a catalogue of long non-coding RNAs in the mouse is established. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8•.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106(28):11667–11672. doi: 10.1073/pnas.0904715106. This paper extends the approach of Guttmann et al. to include a catalogue of long non-coding RNAs in human. The association to chromatin-modifying complexes with non-coding RNAs is suggested to be involved in regulation of gene expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9••.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143(1):46–58. doi: 10.1016/j.cell.2010.09.001. This paper uses the HAVANA annotation of the human genome to identify a set of long non-coding RNAs. Using knock-down approaches and reporter assays long non-coding RNAs are demonstrated to mediate RNA-dependent enhancer-like effects. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322(5909):1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 11.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322(5909):1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13••.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465(7295):182–187. doi: 10.1038/nature09033. This paper uses larges-scale approaches to identify numerous putative enhancers in the mouse. Several of these enhancers are shown to be transcribed into RNAs with suggested transcriptional regulatory functions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ponjavic J, Oliver PL, Lunter G, Ponting CP. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009;5(8):e1000617. doi: 10.1371/journal.pgen.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chodroff RA, Goodstadt L, Sirey TM, Oliver PL, Davies KE, Green ED, Molnar Z, Ponting CP. Long noncoding RNA genes: conservation of sequence and brain expression among diverse amniotes. Genome Biol. 2010;11(7):R72. doi: 10.1186/gb-2010-11-7-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marques AC, Ponting CP. Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness. Genome Biol. 2009;10(11):R124. doi: 10.1186/gb-2009-10-11-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 18.Ebisuya M, Yamamoto T, Nakajima M, Nishida E. Ripples from neighbouring transcription. Nat Cell Biol. 2008;10(9):1106–1113. doi: 10.1038/ncb1771. [DOI] [PubMed] [Google Scholar]
- 19.Bulger M, Groudine M. Enhancers: the abundance and function of regulatory sequences beyond promoters. Dev Biol. 2010;339(2):250–257. doi: 10.1016/j.ydbio.2009.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koch CM, Andrews RM, Flicek P, Dillon SC, Karaoz U, Clelland GK, Wilcox S, Beare DM, Fowler JC, Couttet P, et al. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 2007;17(6):691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, et al. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42(4):343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22••.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:7243, 108–112. doi: 10.1038/nature07829. This paper uses large-scale chromatin immunoprecipitation to define histone tail modifications associated with active enhancers in the human genome across a number of cell lines. Enhancers are suggested to be the basis of cell-type specific gene expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39(3):311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 24.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25•.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467(7314):430–435. doi: 10.1038/nature09380. This paper demonstrates an important role for Mediator and Cohesin in mediating enhancer function in embryonic stem cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Margulies EH, Cooper GM, Asimenos G, Thomas DJ, Dewey CN, Siepel A, Birney E, Keefe D, Schwartz AS, Hou M, et al. Analyses of deep mammalian sequence alignments and constraint predictions for 1% of the human genome. Genome Res. 2007;17(6):760–774. doi: 10.1101/gr.6034307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruvinsky I, Ruvkun G. Functional tests of enhancer conservation between distantly related species. Development. 2003;130(21):5133–5142. doi: 10.1242/dev.00711. [DOI] [PubMed] [Google Scholar]
- 28•.De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8(5):e1000384. doi: 10.1371/journal.pbio.1000384. This paper demonstrates the expression of transcripts corresponding to enhancers which are correlated to the expression levels of their neighboring genes. They show that the increase in the levels of enhancer transcripts preceeds that of the protein-coding genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ling J, Ainol L, Zhang L, Yu X, Pi W, Tuan D. HS2 enhancer function is blocked by a transcriptional terminator inserted between the enhancer and the promoter. J Biol Chem. 2004;279(49):51704–51713. doi: 10.1074/jbc.M404039200. [DOI] [PubMed] [Google Scholar]
- 30.Ling J, Baibakov B, Pi W, Emerson BM, Tuan D. The HS2 enhancer of the beta-globin locus control region initiates synthesis of non-coding, polyadenylated RNAs independent of a cis-linked globin promoter. J Mol Biol. 2005;350(5):883–896. doi: 10.1016/j.jmb.2005.05.039. [DOI] [PubMed] [Google Scholar]
- 31.Faedo A, Quinn JC, Stoney P, Long JE, Dye C, Zollo M, Rubenstein JL, Price DJ, Bulfone A. Identification and characterization of a novel transcript down-regulated in Dlx1/Dlx2 and up-regulated in Pax6 mutant telencephalon. Dev Dyn. 2004;231(3):614–620. doi: 10.1002/dvdy.20152. [DOI] [PubMed] [Google Scholar]
- 32.Kohtz JD, Baker DP, Corte G, Fishell G. Regionalization within the mammalian telencephalon is mediated by changes in responsiveness to Sonic Hedgehog. Development. 1998;125(24):5079–5089. doi: 10.1242/dev.125.24.5079. [DOI] [PubMed] [Google Scholar]
- 33.Kohtz JD, Fishell G. Developmental regulation of EVF-1, a novel non-coding RNA transcribed upstream of the mouse Dlx6 gene. Gene Expr Patterns. 2004;4(4):407–412. doi: 10.1016/j.modgep.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 34.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20(11):1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lefevre P, Witham J, Lacroix CE, Cockerill PN, Bonifer C. The LPS-induced transcriptional upregulation of the chicken lysozyme locus involves CTCF eviction and noncoding RNA transcription. Mol Cell. 2008;32(1):129–139. doi: 10.1016/j.molcel.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harrow J, Denoeud F, Frankish A, Reymond A, Chen CK, Chrast J, Lagarde J, Gilbert JG, Storey R, Swarbreck D, et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7(Suppl 1S4):1–9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37•.Tian D, Sun S, Lee JT. The long noncoding RNA, jpx, is a molecular switch for x chromosome inactivation. Cell. 2010;143(3):390–403. doi: 10.1016/j.cell.2010.09.049. This paper shows that Xist transcription is positively regulated by a long non-coding RNA expressed from a nearby location on the X-chromo-some. The mechanism is suggested to be trans-acting, as autosomally expressed Jpx RNA can rescue the X-linked Jpx null defects. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Delabesse E, Ogilvy S, Chapman MA, Piltz SG, Gottgens B, Green AR. Transcriptional regulation of the SCL locus: identification of an enhancer that targets the primitive erythroid lineage in vivo. Mol Cell Biol. 2005;25(12):5215–5225. doi: 10.1128/MCB.25.12.5215-5225.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39•.Dhami P, Bruce AW, Jim JH, Dillon SC, Hall A, Cooper JL, Bonhoure N, Chiang K, Ellis PD, Langford C, et al. Genomic approaches uncover increasing complexities in the regulatory landscape at the human SCL (TAL1) locus. PLoS One. 2010;5(2):e9059. doi: 10.1371/journal.pone.0009059. This paper provides an extensive analysis of the genomic landscape of the TAL1 locus in human. It is shown that a complex collection of regulatory factors exists in the locus offering the possibility of sophisticated transcriptional regulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kong S, Bohl D, Li C, Tuan D. Transcription of the HS2 enhancer toward a cis-linked gene is independent of the orientation, position, and distance of the enhancer relative to the gene. Mol Cell Biol. 1997;17(7):3955–3965. doi: 10.1128/mcb.17.7.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nolis IK, McKay DJ, Mantouvalou E, Lomvardas S, Merika M, Thanos D. Transcription factors mediate long-range enhancer-promoter interactions. Proc Natl Acad Sci U S A. 2009;106(48):20222–20227. doi: 10.1073/pnas.0902454106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295(5558):1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]