SUMMARY
Izumo, a sperm membrane protein, is essential for gamete fusion in the mouse. It has an Ig (Immunoglobulin) domain and an N-terminal domain for which neither the functions nor homologous sequences are known. In the present work we identified three novel proteins showing an N-terminal domain with significant homology to the N-terminal domain of Izumo. We named this region "Izumo domain", and the novel proteins “Izumo 2”,”Izumo 3” and “Izumo 4”, retaining “Izumo 1” for the first described member of the family. Izumo 1, 2 and 3 are transmembrane proteins expressed specifically in the testis, and Izumo 4 is a soluble protein expressed in the testis and in other tissues. Electrophoresis under mildly denaturing conditions, followed by Western blot analysis, showed that Izumo 1, 3 and 4 formed protein complexes on sperm, Izumo 1 forming several larger complexes and Izumo 3 and 4 forming a single larger complex. Studies using different recombinant Izumo constructs suggested the Izumo domain possesses the ability to form dimers, whereas the transmembrane domain or the cytoplasmic domain or both of Izumo 1 are required for the formation of multimers of higher order. Co-immunoprecipitation studies showed the presence of other sperm proteins associated with Izumo-1, suggesting Izumo 1 forms a multi-protein membrane complex. Our results raise the possibility that Izumo 1 might be involved in organizing or stabilizing a multi-protein complex essential for the function of the membrane fusion machinery.
Keywords: gamete fusion, sperm protein, fertilization
INTRODUCTION
Fertilization comprises a series of biological events leading to the formation of the embryo. Gamete fusion is the culminating step of fertilization, after which a single diploid cell, the zygote, is formed. The interaction between the gametes and the merging of their plasma membranes is believed to be a complex process involving the participation of multiple proteins on the surfaces of both the egg and the sperm (Rubinstein et al., 2006). To date, we know little about the molecular mechanisms of gamete fusion. In single-celled organisms, including the green alga Chlamydomonas and the malaria organism Plasmodium, both species-specific and highly conserved proteins are required for gamete fusion. Species-specific proteins bring about close adhesion of the plasma membranes of gametes, and members of the conserved HAP2/GCS1 family of proteins (first identified as a male sterile gene in Arabidopsis) function after membrane binding during the membrane fusion reaction (Liu et al., 2008).
In mouse and humans, the sperm protein Izumo has been shown to be essential for gamete fusion. The generation of knockout mouse models has provided a powerful tool to test the functional relevance of proteins proposed to have a role in mammalian fertilization. While otherwise normal, Izumo knockout male mice are infertile due to the specific failure of their sperm to fuse with the eggs (Inoue et al., 2005). In addition to being found in mouse, Izumo is present in humans, and the results of in vitro experiments suggest that Izumo also plays a role in human gamete fusion (Inoue et al., 2005). Izumo is an Immunoglobulin Super Family (IgSF) transmembrane protein containing a single immunoglobulin (Ig) domain in the extracellular domain and an N-terminal region absent in any previously described domain. Western blot analyses indicated that Izumo is expressed only in the testis and is present on mature sperm. The protein was detected by immunofluorescence on live cells only after acrosomal exocytosis, suggesting that it is a transmembrane acrosomal protein, exposed on the sperm surface after the acrosome reaction (AR).
Although it is absolutely essential for gamete fusion, the molecular function of Izumo is unknown and we know little about its relationship to other sperm proteins, its structure, or its expression on distinct sperm membranes. Here, we report that a ~ 150 amino acid residue region in the N-terminus of Izumo is present in three other novel proteins, thereby defining a new protein family, which we named “Izumo”. Results obtained with native and recombinant Izumo proteins suggest that the Izumo domain participates in formation of homo-dimers. Moreover, we show that Izumo 1 (the first described Izumo member) is associated with other non-Izumo proteins, suggesting Izumo 1 is part of a multi-protein membrane complex. Our findings raise the possibility that Izumo 1 plays a role in gamete fusion by organizing or stabilizing a molecular complex on the sperm membrane.
RESULTS
Identification of multiple Izumo domain-containing proteins
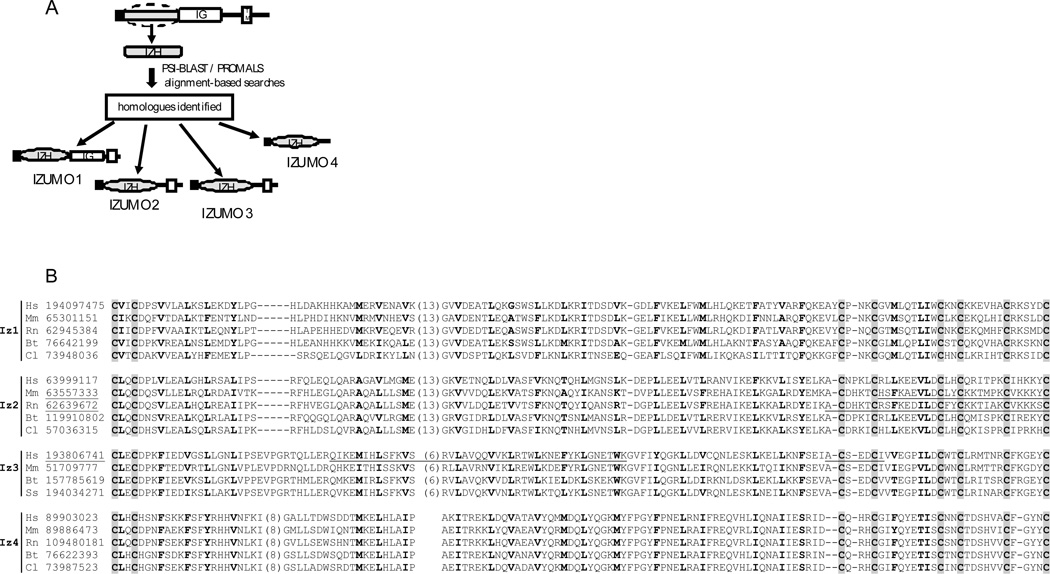
To search for proteins showing homology to the N-terminal region of Izumo, the sequence between amino acid residues 22 to 166 was used in a PSI-BLAST search (Altschul et al., 1997) and found homologs were aligned by Promals (Pei and Grishin, 2007). Three distinct novel proteins were identified (Fig 1A). Because they share homology with the N-terminal domain of Izumo, we propose naming this group of paralogs the “Izumo family”. Since the protein reported by Inoue et al (2005) was the first member of the family described, we refer to it as Izumo 1 (Mm.380445), assigning subsequent numbers for the novel members of the family (Izumo 2: Mm.273301, Izumo 3: Mm.55891, Izumo 4: Mm.35802). In addition to being present in mouse, each novel member of the Izumo family also has homologs in several mammalian species (Fig 1B), suggesting that this family is conserved among mammals. All members show a pattern of eight conserved cysteines (Fig 1B) and a similar predicted secondary structure consisting of four α helices in between the cysteine motifs (data not shown). Analysis of the sequence similarities between different species for each Izumo member shows Izumo 4 is the most conserved, followed by Izumo 3, 2 and 1 (Table 1). The sequence identities between the different Izumo members range between 16–18% (Table 1).
Figure 1.
A: Diagram of the approach used to identify novel Izumo homologues. The domain architecture of the novel proteins is also indicated. B. Sequence alignment for each Izumo member in several species. The two letters indicate the species (Hs: Homo sapiens, Mm: Mus musculus, Rn: Ratus norvergicus, Bt: Bos taurus, Cl: Canislupass, Ss: Sus scrofa). The numbers correspond to the Gi accession number. Positions of conserved cysteines are indicated by the shaded background. Positions with mainly hydrophobic residues are in bold letters. Three proteins with manually curated sequences have their gi numbers underlined. Also underlined are the regions that are absent from these sequences but are retrieved from tblastn searches or gene prediction results.
Table 1.
Sequence identity matrix.
| Izumo1 | Izumo 2 | Izumo 3 | Izumo 4 | |
|---|---|---|---|---|
| Izumo1 | 37.0 | 17.6 | 17.5 | 15.9 |
| Izumo 2 | 17.6 | 68.2 | 22.0 | 18.6 |
| Izumo 3 | 17.5 | 22.0 | 75.5 | 15.4 |
| Izumo 4 | 15.9 | 18.6 | 15.4 | 88.1 |
Analysis of ESTs (information from the UniGene database - National Center for Biotechnology Information) indicates that expression of mouse Izumo 2 and Izumo 3 is restricted to the adult testis whereas mouse Izumo 4 is present in a broader range of tissues (including testis). Similar expression patterns in the human are reported for Izumo 2 (Hs.414175) and Izumo 4 (Hs.424049), and information is unavailable for the expression of Izumo 3 (Hs.573530).
Expression on sperm
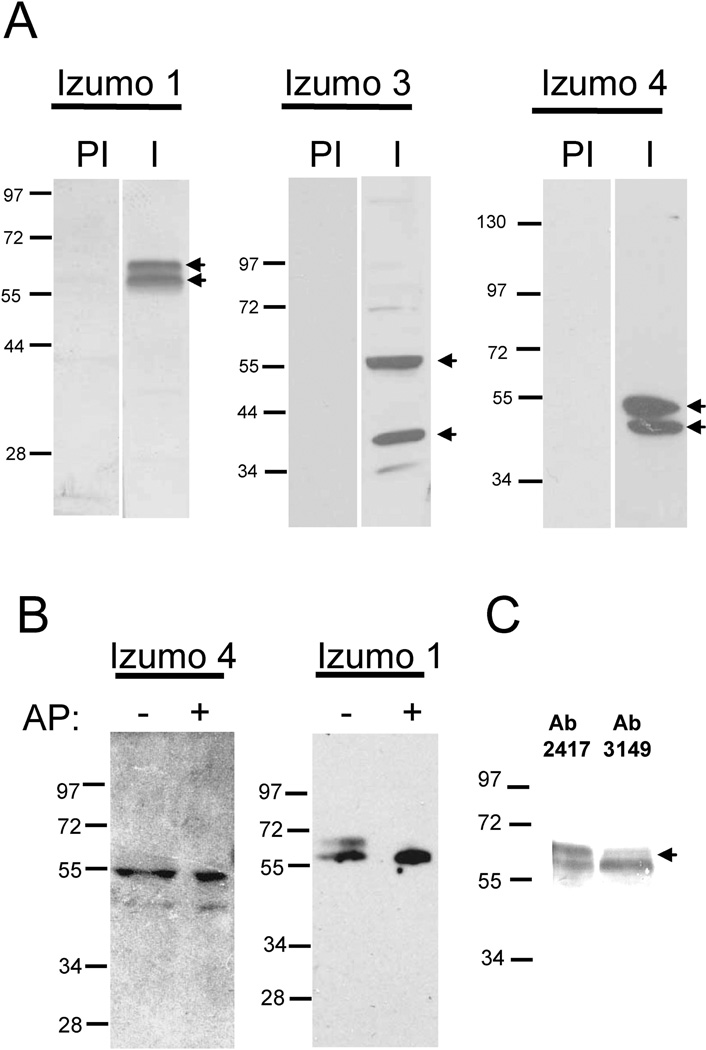
Because all three novel Izumo members are either testis-specific (Izumo 2 and 3) or expressed in various tissues including testis (Izumo 4), we tested for their presence on mature sperm. For this purpose we raised polyclonal antibodies against the recombinant mouse proteins. Immunization with Izumo 2 did not produce high-titer or high-specificity antibodies and therefore the study of this protein was not pursued further. Western blot experiments using all of the recombinant proteins confirmed that the specific antibodies did not cross-react with the other Izumo members (data not shown). In addition to detecting the predicted 56 kDa form (Inoue et al., 2005) with the Izumo I antibody, we also observed an additional ~60 kDa protein (Fig 2A). The antibody against Izumo 3 reacted mainly with two isoforms of ~55 kDa and ~37 kDa, whereas anti-Izumo 4 reacted with isoforms of ~50 kDa and ~43 kDa (Fig 2A). Two splice variants are predicted from the gene sequence for Izumo 3 and Izumo 4 but there are no predicted or reported splice variants for Izumo 1. The different isoforms observed for Izumo 3 and 4 could correspond to splice variants or to forms with different pos-translational modifications, whereas the two forms of Izumo 1 are more likely differences in pos-tranlational modifications. In order to evaluate if phosporylation accounts for the the two bands observed for Izumo 1 and Izumo 4 a sperm extract was treated or not with alkaline phosphatase and analyzed by SDS-PAGE and Western blot. Dephosphorylation with alkaline phosphatase did not affect the migration of Izumo 4 forms (Fig 2B), suggesting they are not phosphorylated proteins. In contrast, after the alkaline phosphatase treatment the ~60 kDa band of Izumo 1 was no longer observed whereas the ~56 kDa band became stronger (Fig 2B). These results suggest the ~56 kDa band correspond to the non-phosphorylated protein and the ~60 kDa band to the phosphorylated protein. The cytoplasmic tail of Izumo 1 contains several potential phosphorylation sites. We have previously raised an antibody (Ab 3149) against the sequence CDFNSDYSGDKSEATEN in the cytoplasmic tail of Izumo 1 (Stein et al., 2006) which encompass the phosphorylation site indicated with an underline. Comparing the reactivity of the two forms of Izumo 1 with Ab 3149 or the Ab against the extracellular domain (Ab 2417) it is observed that the non phosphorylated form reacts similarly with Ab 2417 and Ab 3149 but the phosphorylated form reacts more weakly with Ab 3149 than with Ab 2417 (Fig 2C), suggesting Izumo 1 is phosphorylated at the SYSGDK site.
Figure 2. Presence of Izumo 3 and 4 on sperm.
A: Acrosome-intact sperm were extracted with PFO and the proteins were separated by SDS-PAGE and transferred to a PVDF membrane. The membranes were probed with immune (I) or pre-immune (PI) sera specific for each protein at a 1:1000 dilution. B: Effect of treatment with alkaline phosphatase. Sperm extracts were treated (+) or not (−) with alkaline phosphatase (AP) and the effect on the MW of Izumo 1 and Izumo 4 was evaluated by SDS-PAGE and Western blot. C: Reactivity of the Izumo 1 forms with antibodies 2417 (against the extracellular domain) or 3149 (against a cytoplasmic region).
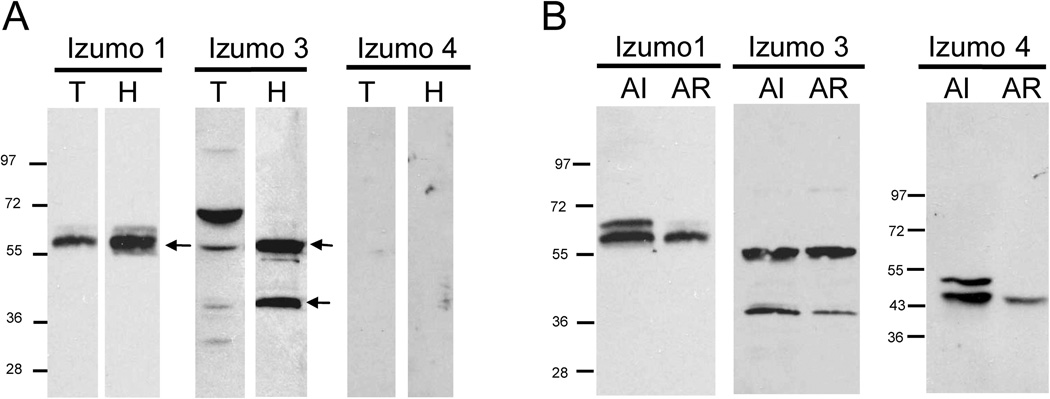
Having established the expression of Izumo 3 and Izumo 4 on sperm we then studied the localization of the Izumo members on sperm and their fate after the AR. Sperm were disrupted by sonication and fractionated by differential centrifugation and velocity sedimentation in sucrose gradients to yield sperm heads (80% pure) or sperm tails (75% pure). Analysis by immunoblotting showed that Izumo 1 was present in the head fraction with a stronger intensity than in the tail fraction (Fig 3A). Since Izumo 1 has been previously reported by immunofluorescence studies to be localized only in the sperm head (Inoue et al., 2005) it is possible that the signal in the tail preparation was due to the small percentage of contaminating sperm heads in the sample. Similarly, both forms of Izumo 3 were detected more strongly in the head preparation than in the tail preparation. In contrast, Izumo 4 was not detected in either the tail or head sperm sample, possibly because it was lost as a consequence of the sonication (see below).
Figure 3. Localization of Izumo members on sperm and fate after the AR.
A: Freshly isolated sperm were disrupted by sonication and head and tail fractions were isolated by velocity sedimentation on sucrose gradients. 2 × 106 isolated sperm heads or tails were loaded per lane and the presence of the Izumo members in the samples was analyzed by Western blot. B: Proteins from acrosome-intact (AI) and acrosome-reacted (AR) sperm were separated by SDS-PAGE and the presence of the different Izumo members was analyzed by Western blot. The percentage of acrosome –reacted cells was 84%.
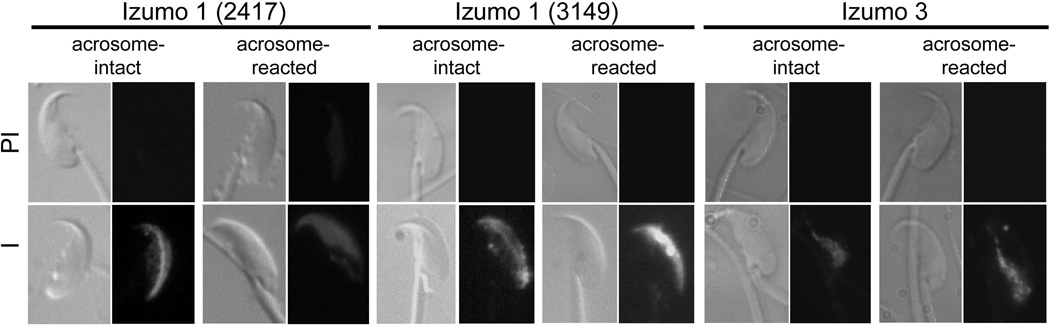
The fate of the Izumo proteins after the AR was analyzed by Western blot on protein extracts from acrosome-intact sperm or from sperm induced to undergo the AR by incubation in the calcium ionophore A23187. We found that that the ~55 kDa form of Izumo 1 was retained on the acrosome-reacted sperm, but the ~60 kDa form was greatly diminished (Fig 3B). In contrast, both forms of Izumo 3 were retained on the acrosome-reacted sperm. Analysis of Izumo 4, however, showed that both isoforms were reduced in acrosome-reacted sperm, with the more slowly migrating form being almost completely absent in samples from acrosome-reacted sperm. Because Izumo 4 is predicted to be a soluble protein, its loss after the AR suggests it could be an intra-acrosomal protein. Sonication affects the integrity of the acrosome and causes the release of its content. In this regard, it is interesting to note that the pattern observed for Izumo 1 in the sonicated sample (Fig 3A) resembled the one from the acrosome-reacted sperm (Fig 3B). Taken together, these results suggest Izumo 4 may be an intra-acrosomal protein and that Izumo 3 is expressed in the sperm head, although some expression in the sperm tail cannot be ruled out. As another approach to study the localization of Izumo family members on sperm we carried out immunofluorescence studies in non-capacitated and acrosome reacted permeabilized sperm. Both antibodies against Izumo 1, 2417 and 3149, stained non-capacitated sperm on the anterior acrosome but a weaker dotted staining was also observed on the equatorial region (Fig 4). However on acrosome-reacted sperm while antibody 3149 stained the anterior acrosome antibody 2417 stained the entire acrosomal region (including the equatorial segment) and the posterior edge of the post-acrosomal region (Fig 4). Izumo 3 was detected on the post-acrosomal region on both non-capacitated and acrosome-reacted sperm (Fig 4). Antibodies against Izumo 4 did not stain sperm in either condition (data not shown).
Figure 4. Immunofluorescence localization of Izumo 1 and Izumo 3 on sperm.
Acrosome-intact or acrosome-reacted sperm were fixed, permeabilized and incubated with antibodies 2417 or 3149 against Izumo 1 or antibodies against Izumo 3. I: immune sera, PI: pre-immune sera. The percentage of acrosome reaction was 77–81%.
Formation of protein complexes
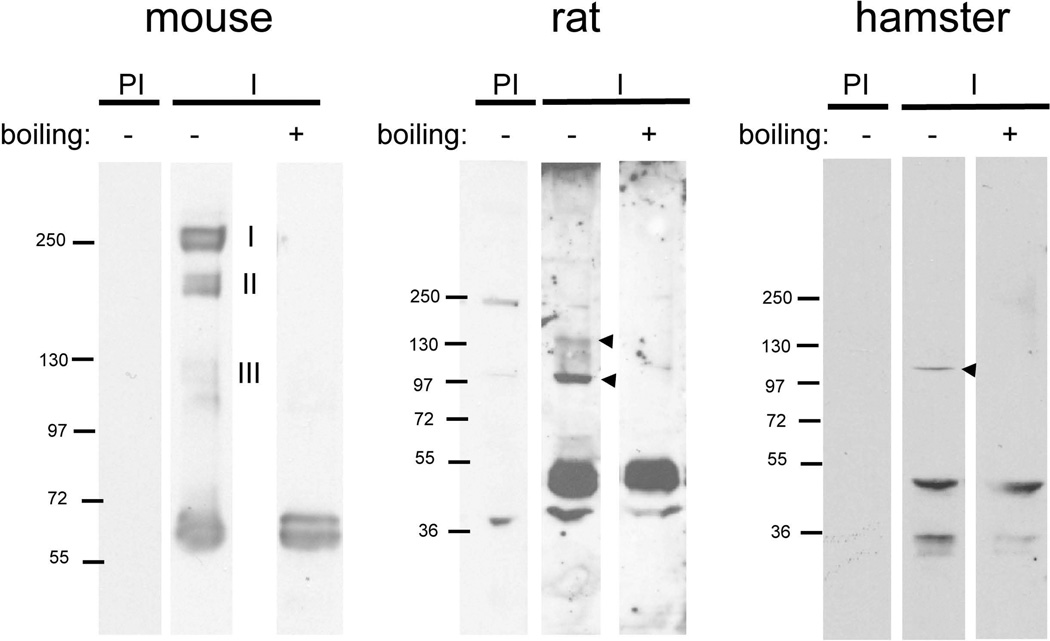
Because other sperm membrane proteins have been shown to exist as multimers (ADAMs, CatSper), we next analyzed whether members of the Izumo family also form complexes. For this purpose, sperm were incubated with perfluoro-octanoic acid (PFO), a weakly denaturing detergent capable of preserving certain high-affinity interactions (Ramjeesingh et al., 1999, Penna et al., 2008), centrifuged, and the supernatant was subjected to SDS-PAGE under “mildly denaturing” conditions. Samples were incubated at room temperature in electrophoresis running buffer containing 0.1% and analyzed by Western blotting. Under these conditions anti-Izumo 1 detected the monomeric forms of Izumo 1, and also three groups of more slowly migrating proteins called complex I (224–260 kDa), complex II (164–180 kDa) and complex III (110–120 kDa) (Fig 5A). If the protein extract was boiled in standard Laemmli (Laemmli, 1970) sample buffer containing 2% SDS before the electrophoresis, the ~60 kDa band was still visible but the high molecular weight bands were no longer observed. Because the Izumo molecular masses were close to multiples of 60 kDa (complex I: 4×, complex II: 3×, complex III: 2×) it was possible that they corresponded to Izumo 1 homo-tetramers, trimers and dimers, respectively. Formation of non-specific complexes of other sperm proteins (i.e. ADAM 2, ADAM 3 and basigin) was not observed (data not shown) consistent with previous work showing that PFO does not artifactually promote formation of complexes (Ramjeesingh et al., 1999) and supporting the model that Izumo 1 forms bona fide complexes. To determine whether Izumo 1 complex formation was specific to mouse or was also observed in other species, a similar analysis was carried out on rat and hamster sperm. Under denaturing conditions Izumo 1 was detected as a ~50 kDa band in both the rat and hamster sperm samples (Fig 5 B, C). Under mildly denaturing conditions two bands with molecular masses close to those expected for dimers (100 kDa) and trimers (150 kDa) were observed in the rat sample. In the hamster sample, however, only a single form corresponding to a putative dimer (100 kDa) was observed. Although the formation of complexes larger than 100 kDa was variable, these results suggest that the formation of Izumo 1 complexes is conserved in rodents, and potentially is relevant for Izumo 1 function in gamete fusion.
Figure 5. Multimerization of Izumo 1 in mouse, rat and hamster sperm.
Acrosome-intact mouse, rat or hamster sperm were extracted with PFO. The protein extracts were boiled in 2% SDS (boiling +) or left untreated (boiling −) followed by SDS-PAGE. Proteins were revealed by Western blot using the anti-Izumo-1 antibody (I) or the pre-immune serum as a control (PI).
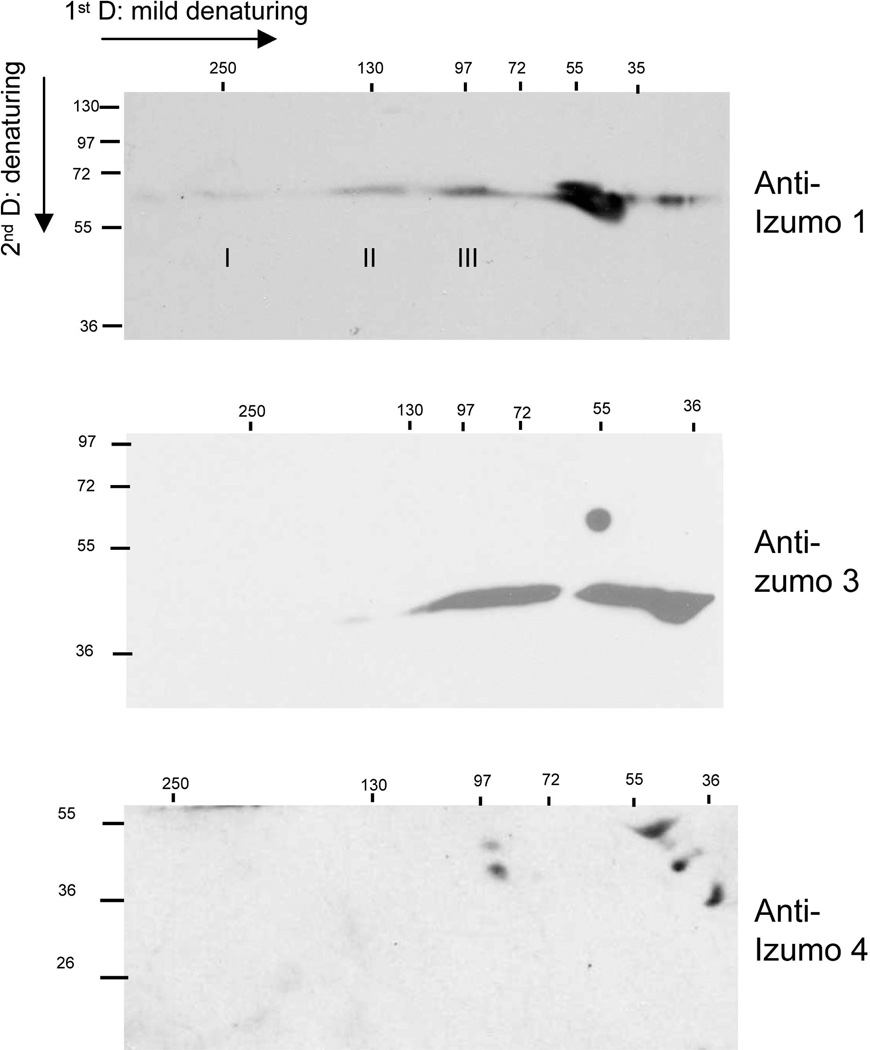
We used two dimensional (2D) electrophoresis to study the potential formation of complexes of Izumo 3 and Izumo 4. Mildly denaturing conditions were used in the first dimension and standard denaturing conditions in the second dimension. When Izumo 1 was analyzed by 2D, the monomer was detected at different regions along the horizontal axis (Fig 6), consistent with the existence of molecular complexes as described above. Analysis of Izumo 3 yielded different results for the ~37 kDa form compared to the ~55 kDa form. Although the ~55 kDa was observed as a monomer only, the ~37 kDa form of Izumo 3 was detected as two separate smears, one of 36–44 kDa (corresponding to the monomer), and one of 55–80 kDa (Fig 6). In turn, the two forms of Izumo 4 were detected in their corresponding monomeric positions, and also in forms originating from a complex migrating with an apparent molecular mass of of ~90 kDa (Fig 6). These results suggest the the ~55 kDa form of Izumo 3 fails to form complexes, and that the ~37 kDa forms of Izumo 3 and Izumo 4 are involved in the formation of protein complexes.
Figure 6. Presence of Izumo 3 and Izumo 4 in protein complexes.
Proteins from acrosome-intact sperm were extracted with PFO and separated by PAGE under mildly denaturing conditions followed by a second separation by denaturing SDS-PAGE. Proteins were detected by Western blot using the indicated, specific antibodies.
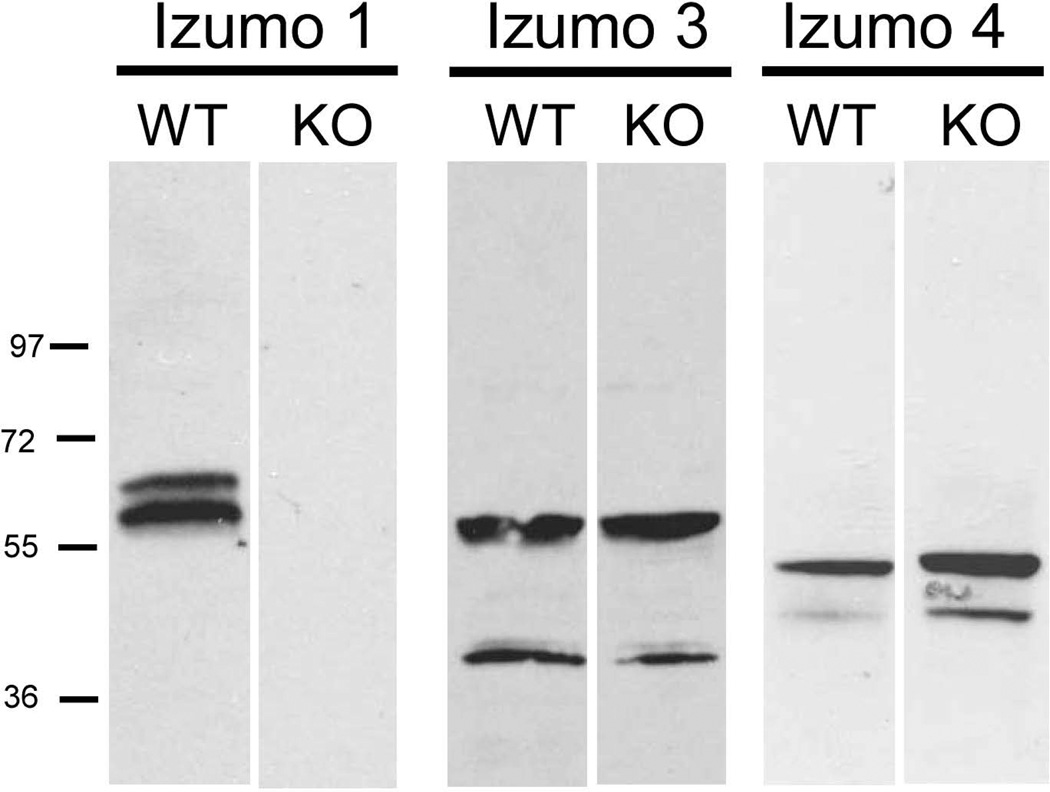
In other sperm surface protein families that exist as multimers, i.e. ADAM (Nishimura et al., 2001) and CatSper (Carlson et al., 2005), blocking expression of one of the subunits by targeted gene disruption reduces the expression levels of other members of the family. To analyze if the absence of Izumo 1 affected the expression of the other Izumo members, their presence on wild type or Izumo 1 null sperm was compared by Western blotting. As shown in Fig 7, both Izumo 3 and Izumo 4 were detected at comparable amounts in sperm from wild type or Izumo 1 knockout males (Fig 7. Thus, unlike other sperm membrane protein families that form complexes, expression of Izumo 3 and 4 does not depend on expression of Izumo 1.
Figure 7. Presence of Izumo members on Izumo 1-null sperm.
Acrosome-intact sperm from wild type or Izumo 1-null sperm were extracted and analyzed by Western blot for the presence of the different Izumo members.
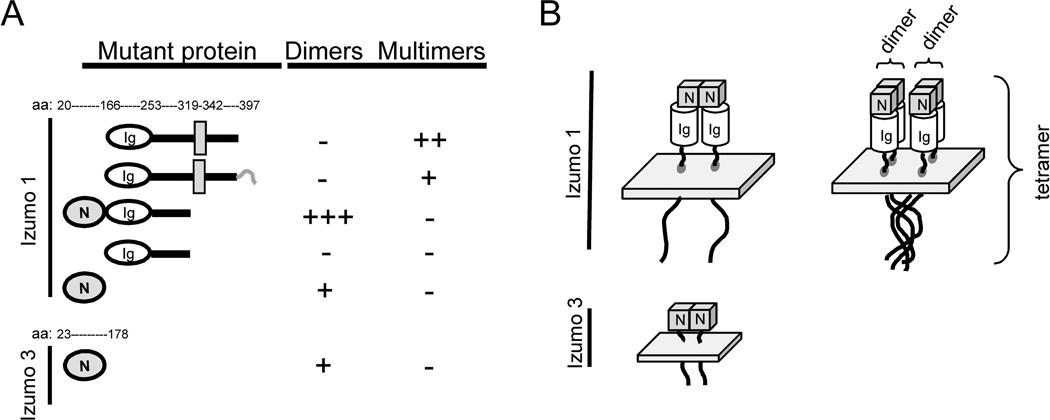
Izumo 1 domains and multimer formation
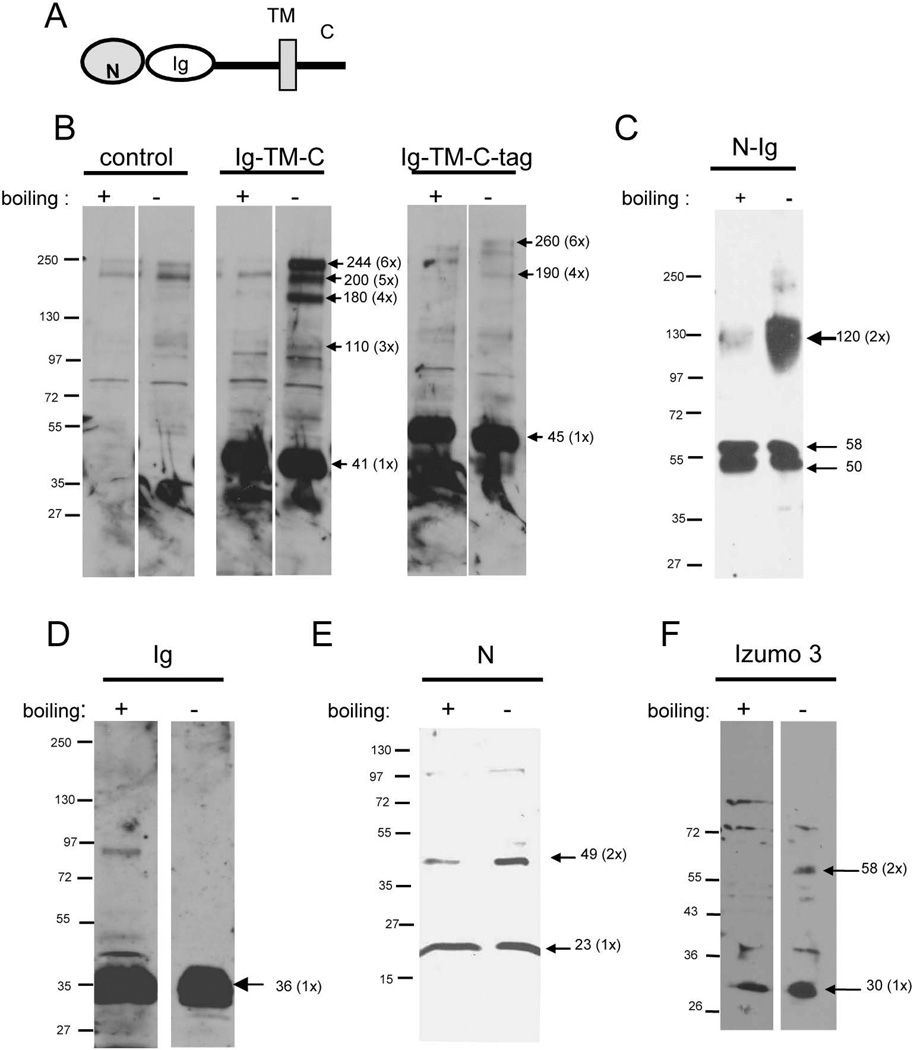
In order to gain insights into the domains of Izumo 1 involved in formation of multimers, several recombinant constructs were expressed in CHO cells and analyzed for their abilities to form complexes. The proteins were separated by SDS-PAGE with or without a prior denaturing incubation at 95°C for 5 min in 2% SDS to disrupt any complexes. A diagram of the different regions of Izumo 1 is shown in Fig 8A. A construct missing only the N-terminal domain formed large complexes, although putative dimers could not be identified (Fig 8B, Ig-TM-C). When a similar construct containing a His-tag at the C-terminal end of the cytoplasmic tail was used, large complexes were still observed but in a much lower intensity, suggesting that the His-tag interfered with the formation of complexes (Fig 8B Ig-TM-C-Tag). Expression of a construct corresponding to the entire extracellular domain yielded a strong band consistent with formation of a homodimer under mildly denaturing conditions, and with a greatly reduced intensity under denaturing conditions (Fig 8C). The result that significant amounts of large complexes failed to be observed for this construct suggested that the transmembrane domain or the cytoplasmic tail or both might be required for large complexes. On the other hand, the presence of a complete extracellular domain was sufficient for the formation of putative dimers. Thus, the formation of complexes of varying sizes appeared to rely on different regions of the molecule.
Figure 8. Analysis of the regions of Izumo 1 involved in complex formation.
A: Diagram showing the different regions of Izumo 1. Recombinant proteins corresponding to regions of Izumo 1 (B–E) or the extracellular domain of Izumo 3 (F) were expressed in CHO cells. They were analyzed for their ability to form complexes by PAGE under denaturing or mildlydenaturing conditions followed by Western blot using the specific antibodies. B: untransfected cells (control), construct lacking the N-terminal domain (N-Ig-TM-C) or with an additional His tag at the C-terminus (N-Ig-Tm-C-tag); C: full extracellular domain (N-Ig); C: Ig domain of Izumo 1 (Ig); E: N-terminal domain of Izumo 1 (N), F: N-terminal domain of Izumo 3 (Izumo 3).
To characterize the region responsible for formation of the putative dimers, we examined the complex-forming ability of distinct domains in the extracellular region of Izumo expressed in CHO cells. Because in many IgSF proteins the Ig domain is involved in homotypic interactions, we examined a soluble construct containing only the Ig domain. The construct, however, failed to form complexes (Fig 8D). On the other hand, the recombinant N-terminal domain was able to form complexes of a size consistent with dimmers (Fig 8E). In order to determine whether the ability to form complexes of putative dimers was restricted to the Izumo domain of Izumo 1, or on the contrary, was shared by other members of the family, the Izumo domain of Izumo 3 was also expressed in CHO cells and analyzed for its ability to form complexes. Results show that the recombinant protein was detected as a main band of ~30 kDa under denaturing conditions. Under mildly denaturing conditions an additional band of ~60 kDa was also observed, consistent with the presence of dimers (Fig 8F). A summary of the results is presented in Figure 9A. Our results suggest that Izumo 1 possesses two regions that participate in formation of complexes (Fig 9B): 1) a region in the N-terminal end, the Izumo domain, capable of forming complexes of the size of dimers that is probably conserved in other members of the family; and 2) a region between the transmembrane domain and the cytoplasmictail capable of forming larger complexes.
Figure 9.
A: Summary of the results on complex formation for the recombinant proteins. The second construct differs from the top one by the presence of the histidine tag (shown as a short hook) in the C-terminal end. The numbers indicate the aminoacid residues marking the boundaries of the different domains. B: Proposed model for the formation of multimers for Izumo 1 and Izumo 3. Dimers of Izumo 1 and Izumo 3 are stabilized by interactions in the Izumo domain. Formation of Izumo 1 multimers of higher order is proposed to be mediated by the interaction of the cytoplasmic tails and/or the transmembrane domains.
Association of Izumo 1 with other proteins
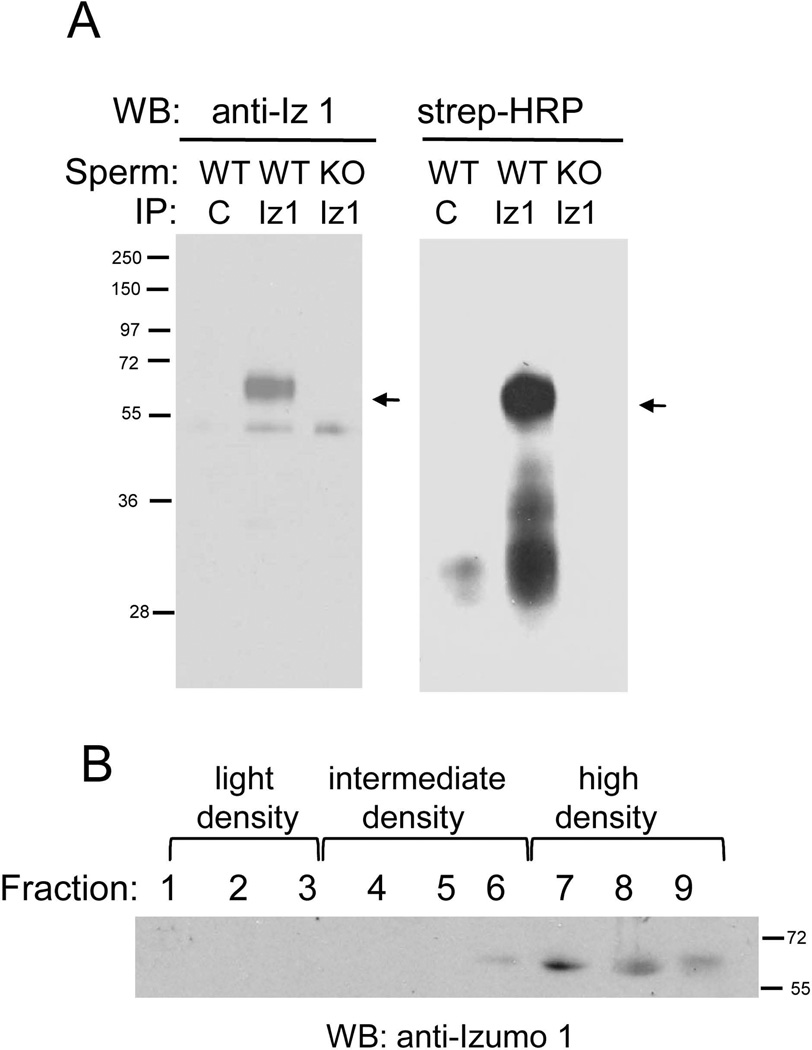
We also tested whether Izumo 1 formed complexes with other sperm surface proteins. Acrosome-reacted sperm were labeled with membrane impermeable biotin, proteins were extracted with the mild detergent dodecylmaltoside (DDM) and then immunoprecipitated with beads coupled to anti-Izumo 1 or to purified IgGs from normal rabbit serum as a control. Sperm from Izumo 1 knockout mice were also used as a control. We found that the anti-Izumo 1 antibody, but not the control antibody, was able to immunoprecipitate Izumo 1 from wild type sperm (Fig 10A). Incubation of the same membranes with Streptavidin-peroxidase indicated the presence of biotin-tagged proteins with molecular masses ranging from ~30 to ~45 kDa in the wild type sperm sample immunoprecipitated with anti-Izumo 1. Although a weak reaction was also observed in the sample immunoprecipitated with the control antibody, no biotinylated protein was observed in the Izumo-1 null sperm control (Fig 10A). These results suggested that Izumo 1 associates with other sperm proteins. Izumo 1 has been shown to be present in detergent-resistant microdomains (DRMs) in non capacitated sperm (Sleight et al., 2005). Thus it is possible that the anti-Izumo 1 antibodies precipitated proteins co-residing in the same DRMs but not necessarily forming a protein complex. In order to analyze whether in the conditions used for the co-immunprecipitation Izumo 1 is present in DRMs or not, acrosome-reacted sperm were extracted with DDM and the proteins were separated in a discontinues sucrose gradient. Western blot analysis of the different fractions of the sucrose gradient showed Izumo 1 is present mainly in the detergent soluble, high density fractions and marginally in the intermediate density fractions (Fig 10B), arguing against the notion that Izumo 1 is associated with DRMs in the conditions we used for the co-immunoprecipitation.
Figure 10. Proteins co-immunoprecipitating with Izumo 1.
A: Acrosome-reacted sperm from wild type (WT) or Izumo 1 knock out (KO) were biotinylated and extracted with dodecyl maltoside (DDM). The protein extract was immunoprecipitated with beads coupled to anti-Izumo 1 antibody (anti-Iz 1) or to IgGs from normal rabbit serum as a control (C). After separation by SDS-PAGE, Izumo 1 was visualized with the corresponding antibody. The biotinylated proteins co-immunoprecipitating with Izumo 1 were visualized by blotting the membrane with Streptavidine-peroxidase (Strep-HRP). The position of Izumo 1 is indicated by the arrows. B: Acrosome reacted sperm were ectracted with DDM and the proteins separated in a discontinues sucrose gradient. Fractions were collected form the top to the bottom of the gradient and the presence of Izumo 1 was analyzed by SDS-PAGE and Western blot.
DISCUSSION
Our goal in this study was to characterize domains of the IgSF sperm protein Izumo and identify paralogs, whose investigation might ultimately contribute to our knowledge of Izumo function in sperm-egg fusion. PSI-BLAST searches allowed us to identify novel proteins showing significant homology to the N-terminal domain of Izumo 1. Izumo 1, 2 and 3 have in common testis-specific expression and the presence of a transmembrane domain. On the other hand, Izumo 4 stands out from the other three members because it lacks a transmembrane domain and shows a wider tissue expression. Proteins that are testis-specific are reported to evolve more rapidly on average than proteins with maximal expression in other tissues (Turner et al., 2008). This seems to be the case for Izumo 1, 2 and 3, which are testis-specific and show a greater divergence than Izumo 4 which is predicted to be expressed in other tissues in addition to testis. Proteins involved in sperm-egg interactions also often evolve rapidly (Swanson and Vacquier 2002). That Izumo 1 has a required role in sperm-egg fusion and shows the lowest inter-species homology in the family is consistent with that idea. The other members show significantly higher inter-species homologies and their functions (if any) in fertilization remain to be analyzed.
Izumo was described by Inoue et al (2005) as a single 56 kDa band on mouse sperm. In this work using both an antibody against the whole recombinant extracellular domain and an antibody against a cytoplasmic peptide we detected a similar ~56 kDa band and an additional ~60 kDa band. Dephosphorylation experiments indicated the ~60 kDa protein is a phosphorylated form of Izumo 1. Both Izumo 3 and Izumo 4 were also detected as two different forms, which remained after treatment with alkaline phosphatase. The two different forms of each likely represent predicted splice variants. All three Izumo members are predicted to be N-glycosylated proteins, and Inoue et al (2008) recently demonstrated Izumo 1 indeed has N-linked carbohydrates. Whether the different forms of Izumo 3 and 4 correspond to splice variants, glycosylation variants, or other post-translational modifications is currently unknown.
Izumo 1 was shown by immunofluorescence studies to be localized exclusively on the sperm head (Inoue et al., 2005). As described previously by others (Miranda et al., 2009), we detected Izumo 1 on the anterior acrosome region of acrosome-intact sperm. However on acrosome-reacted sperm while Miranda et al (2009) observed Izumo 1 localized on regions adjacent to the equatorial segment we observed Izumo 1 localizing on either the entire acrosomal region or on the anterior acrosome. The different results on localization of Izumo 1 on acrosome-reacted sperm using different antibodies raise the possibility of existence of separate subpopulations of the molecule, but further studies are required to clarify the localization of Izumo 1 on fusion-competent sperm. The Western blot experiments performed on purified sperm heads suggest that Izumo 3 and Izumo 4 are are expressed on the sperm head and the immunofluorescence experiments confirmed the presence of Izumo 3 on the post-acrosomal region of the sperm head. Even though Izumo 1, 3 and 4 were all expressed in the sperm head, the fates of specific Izumo members after the AR were different: The two forms of Izumo 3 and the ~56 kDa form of Izumo 1 were unchanged by the AR, whereas Izumo 4 and the ~60 kDa form of Izumo 1 were greatly diminished after the AR. The specific loss of the phosphorylated (~60 kDa) form of Izumo 1 could have been due to its localization on the outer acrosomal membrane, which is lost as vesicles after the AR, or alternatively, to the dephosphorylation of this form.
Gene disruption studies have shown that expression of some sperm proteins influences the expression of others (Nishimura et al., 2001, Carlson et al., 2005). Our results that the amounts of Izumo 3 and Izumo 4 in sperm were similar in wild type and Izumo 1 knockout animals indicate that expression of Izumo 3 and Izumo 4 is independent of Izumo 1. The possibility that Izumo 1 forms protein complexes was analyzed by extraction of proteins with the detergent PFO followed by electrophoresis under mildly denaturing conditions. We found that Izumo 1 was present in several larger complexes. We favor the model that the complexes represent homo-multimers of the Izumo proteins. The molecular masses of the complexes in sperm were multiples of the monomers (55–60 kDa). And, Izumo 1 formed complexes not only in mouse sperm, but also in rat and hamster sperm. Moreover, the masses of the several complexes in CHO cells expressing the several recombinant forms of the Izumo family members supported the idea that the complexes were homo-multimers. Formation of putative multimers of higher order was variable among species, but the presence of putative dimers was observed in mouse, rat and hamster sperm, suggesting that this ability is conserved among rodents. Additional experiments will be required to determine if other proteins are present in the complexes and whether the complexes are essential for the function of Izumo 1 in gamete fusion.
The dissection of the regions required for complex formation of Izumo 1 suggests the presence of two distinct regions, each involved in separate Izumo 1 functions. According to this model, a region in the Izumo domain participates in formation of smaller complexes, possibly dimers. And, and a region in the transmembrane domain and/or the cytoplasmic tail participates in formation of larger complexes, possibly Izumo 1 multimers. The significant reduction in complex formation observed when a His-tag was added to the C-terminal end of the cytoplasmic tail suggests that the cytoplasmic tail plays an important role in the formation of mouse Izumo 1 putative trimers and tetramers.
Izumo 3 and Izumo 4 were also observed to form complexes. Although a smear rather than a defined spot was observed for Izumo 3, the molecular masses of Izumo 3 and Izumo 4 complexes were in the range expected for the corresponding dimers (~74 kDa Izumo 3, 86–110 kDa for Izumo 4). Moreover, the recombinant Izumo domain of Izumo 3 was able to form complexes of the mass of dimers. Taken together, these results support the model that the Izumo domain possesses a conserved protein-protein interaction motif. The absence of a transmembrane domain in Izumo 4 coupled with its loss after the AR and its presence in several somatic tissues suggest that this member of the Izumo family does not have a central role in in sperm-egg fusion. The generation of knockout animal models will help elucidate the functions of the novel Izumo members, which perhaps might be in spermatogenesis or earlier steps in fertilization.
The co-immunoprecipitation studies suggested Izumo 1 associates with other, as of yet unidentified sperm surface proteins. Although Izumo 1 has been shown to be present in DRM in non-capacitated sperm (Sleight et al., 2005), under the conditions we performed the co-immunoprecipitation experiments (acrosome-reacted sperm, detergent extraction peformed at RT) Izumo 1 did not partition in the detergent-insoluble, light buoyant fractions suggesting the co-precipitating proteins rather than linked to Izumo 1 by a lipid microdomain are part of a a multi-protein complex. Although the importance of Izumo 1 in gamete fusion is well established, its precise molecular function is unknown. We reported earlier that the tetraspanin CD9, an egg protein essential for gamete fusion (Kaji et al., 2000; Le Naour et al., 2000; Miyado et al., 2000), is able to bind directly to pSG17, a member of the IgSF, raising the possibility that it could also bind other sperm IgSF members (Ellerman et al., 2003). Since Izumo 1 has an Ig domain, it is conceivable that it could bind directly to CD9. We assayed all of the recombinant Izumo 1 constructs described here for their ability to bind to the oocyte and affect sperm-egg fusion in vitro, with negative results (data not shown). This argues against the idea that Izumo 1 interacts directly with a protein in the oocyte.
We favor the model that rather than directly mediating the interaction between the sperm and the egg, Izumo 1 is essential for formation of complexes in cis with other sperm proteins that are required for gamete fusion. In other biological systems involving fusion between membranes, the participation of membrane protein complexes is well established. In intracellular vesicle fusion, SNARE proteins localized in the vesicles and in the target membrane zipper into an alpha-helical bundle that pulls the two membranes tightly together to exert the force required for fusion. Many other proteins interact with this SNARE complex to control its assembly and/or membrane fusion (Rizo and Rosenmund, 2008; Sudhof and Rothman, 2009). In addition, fusion of Herpes simplex with host cells requires four glycoproteins, gB, gD, gH and gL, and recent data indicate these proteins are part of a single complex (Atanasiu et al., 2007; Avitabile et al., 2007). One model is that the complex is required for the fusogen molecule to adopt the fusion-active configuration, or alternatively that the proteins are biologically active only when they are assembled into a complex (Campadelli-Fiume et al., 2007). Given that Izumo 1 is essential for gamete fusion and that it is part of a protein complex, it is possible that the absence of Izumo 1 influences the organization or stability of a protein complex essential for gamete fusion. The identification of the sperm proteins associated with Izumo 1 will allow exploration of this possibility.
Material and Methods
Animals
Animals were kept at the UC Davis animal facility, and used in accordance to the UC Davis Institutional Animals Care and Use Committee.
Antibodies
The whole extracellular domain of Izumo 1 (amino acids 22 to 319) was expressed using the pTrcHis (Invitrogen, Carlsbad, CA) vector as a His-tagged protein. The protein was expressed in an insoluble form. It was solubilized in 6M guanidinium and purified using its binding to nickel beads. Izumo 2 and Izumo 3 were quickly proteolyzed when expressed in the pTrc. Thus the extracellular domain of Izumo 2 (amino acids 21 to 158), Izumo 3 (amino acids 23 to 178) or the entire Izumo 4 were expressed using the pMAL-p vector (New England Biolabs, Ipswich, MA). This overcame the degradation problem but some of the constructs showed a very low affinity to the amylose column. Therefore a histidine tag was added at the C-terminal end of the proteins. The proteins were purified using a nickel column (Invitrogen, Carlsbad, CA), following the instructions of the manufacturer. The purified proteins were used to immunize rabbits at the UC Davis polyclonal antibody production facility.
Expression of recombinant proteins in CHO cells
The constructs coding for the different recombinant proteins were generated by PCR and cloned into the pSecTag2 expression vector (Invitrogen) which carries a myc tag. CHO cells were transfected with the different constructs using PolyFect (Quiagen, Germantown, MD) and 48 h later the cells were re-plated in the presence of 80 µg/ml hygromycin (Invitrogen). Individual clones were isolated using cloning discs (Sigma, St Louis, MO) and amplified. Clones showing the highest expression levels by Western blot using either anti-myc antibodies or the specific antibodies were selected for further analysis. Soluble recombinant proteins were purified from the spent media by affinity purification using nickel beads. To analyze recombinant transmembrane proteins, a 10 cm dish grown to confluency was washed with PBS and then cells were lysed in 500 µl of perfluorooctanoic acid (PFO, Sigma) sample buffer (Ramjeesingh et al. 1999) in the presence of protease inhibitors (Sigma).
Sperm collection
Sperm were collected from the caudae epididymides of adult mice, rats or hamsters by performing a few cuts in each epididymis with microdissection scissors. The sperm were allowed to disperse in PBS at 37°C. For induction of the acrosome reaction, mouse sperm were collected in medium M199 containing 0.4% BSA and incubated in 5% CO2 at 37°C for 1 h. A23187, calcium ionophore, (Sigma) was added to 10 µM final concentration and sperm were incubated for 30 min. The percentage of acrosome reaction was determined by peanut agglutinin staining and ranged between 78–91%. Sperm were harvested by centrifugation and washed in PBS. Purification of sperm heads and tails was done as described previously (Stein et al. 2006). The isolated sperm heads contained a 20% of sperm tails and the isolated tails had 25% of sperm heads.
Immunoprecipitation
The purified IgGs of the anti-Izumo 1 antibody were coupled to Sepharose beads using the AminoLink kit (Thermo Scientific, Rockford, IL) according to the manufacturer’s instructions. Purified IgGs from normal rabbit serum were coupled to the beads as a control. Acrosome-reacted sperm (~2× 107 cells) were biotinylated as described elsewhere (Nishimura et al., 2007), incubated in 200 µl of 2% dodecylmaltoside (DDM, Sigma) in PBS in the presence of a protease inhibitors cocktail (Sigma P8340) for 30 min at RT, and centrifuged at 20,000 × g for 10 min at 4°C. Half of the DDM sperm extract was incubated with 50 µl of control beads and the other half of the supernatant with beads conjugated to anti-Izumo 1 antibodies. After 1 h incubation at RT the beads were washed five times in 0.2% Tween 20 in PBS. The beads were eluted with 200 µl of low pH elution buffer (Thermo Scientific), the eluate was neutralized with 20 µl of 1M Tris, pH 8 and concentrated using a Microcon Y10 (Millipore Corp., Bedford, MA).
Sucrose density fractionation
Acrosome-reacted sperm were extracted in 2% DDM as described above. The detergent extract was processed and separated in a discontinuous sucrose gradient as described by Miranda et al (2009). Nine fractions of 200 µl each were collected carefully starting from the top to the bottom of the gradient.
Phosphatase alkaline treatment
Sperm (~4 × 107) were extracted in 1% NP40, 50 mM Tris (pH 7.4), 150 mM NaCl in the presence of protease inhibitors (Sigma) and centrifuged at 20,000 × g for 10 min at 4°C. An aliquot equivalent to 1 × 106 sperm was treated with 1U of alkaline phosphatase (New England Biolabs, Ipswich, MA) for 1 h at 37°C. The samples were then treated with non reducing Laemmli buffer and subjected to SDS-PAGE and Western blot analysis.
Western blotting
Acrosome-intact or acrosome-reacted sperm, or isolated sperm heads and tails were extracted in PFO sample buffer (Ramjeesingh et al., 1999) in the presence of protease inhibitors (Sigma) for 30 min at RT with end-to-end agitation. After centrifuging the samples at 20,000 ×g for 10 min at 4°C, the supernatants were prepared for PAGE and Western blotting. For samples prepared under denaturing conditions, bromophenol blue, glycerol, and SDS were added to the sperm samples to yield final concentrations of 0.25%, 6% and 2% respectively, and the samples were heated at 96°C for 5 min. Samples prepared under mildly denaturing conditions were prepared with the same concentrations of dye and glycerol. The proteins in the samples not treated in denaturing conditions were exposed to 4% PFO present in the extraction buffer and to 0.1% SDS present in the running buffer, thus these samples are referred to as treated in “mildly denaturing conditions”. 2×106 sperm equivalents were loaded per lane. For two dimensional analysis, PFO sperm extracts were first separated by SDS-PAGE under mild denaturing conditions. After the electrophoresis was complete a strip of the gel containing the proteins was cut out and incubated for 30 min in 5% SDS in PBS in the presence of protease inhibitors at 75 °C. The strip of gel was placed on top of a second SDS-polyacrylamide gel and proteins were separated by SDS- electrophoresis. Proteins were trasnferred onto Immobilon-P transfer membranes (Millipore). After blocking, the membranes were probed with either the pre-immune or immune sera at a 1:1000 dilution over night at 4°C, followed by horse radish-peroxidase-conjugated secondary antibody. If not stated differently, anti-Izumo 1 referes to antibody 2417. For detection of the biotinylated proteins in the co-immunoprecipitation experiment, the membrane was incubated with Streptavidin-horseradish peroxides 2% SDS at 96°C for 5 min e (1:1000) for 1 hr at RT. Signal was detected using the SuperSignal WestPico reagent (Thermo Scientific).
Immunofluorescence
Non- capacitated sperm or acrosome-reacted sperm were fixed in 2% formaldehyde in PBS for 10 min at RT. After washes sperm were permeabilized in 0.1% Triton X-100 in PBS for 30 min, washed and incubated ON at 4°C with the primaries antibodies. Sperm were washed in 1 ml of 3% BSA in PBS and incubated with the secondary antibody conjugated to Alexa Fluor 488 (Molecular Probes). After washing cells were mounted using 90% glycerol in PBS and observed in an epi-fluorescence microscope.
Acknowledgements
We thank Dr Masaru Okabe (Osaka University) for providing us with Izumo 1 knockout mice. This work was supported by NIH-GM 56778 to WJS and NIH 16580 and NIH U54-29125 to PP and DM.
REFERENCES
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasiu D, Whitbeck JC, Cairns TM, Reilly B, Cohen GH, Eisenberg RJ. Bimolecular complementation reveals that glycoproteins gB and gH/gL of herpes simplex virus interact with each other during cell fusion. Proc Natl Acad Sci U S A. 2007;104(47):18718–18723. doi: 10.1073/pnas.0707452104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avitabile E, Forghieri C, Campadelli-Fiume G. Complexes between herpes simplex virus glycoproteins gD, gB, and gH detected in cells by complementation of split enhanced green fluorescent protein. J Virol. 2007;81(20):11532–11537. doi: 10.1128/JVI.01343-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campadelli-Fiume G, Amasio M, Avitabile E, Cerretani A, Forghieri C, Gianni T, Menotti L. The multipartite system that mediates entry of herpes simplex virus into the cell. Rev Med Virol. 2007;17(5):313–326. doi: 10.1002/rmv.546. [DOI] [PubMed] [Google Scholar]
- Carlson AE, Quill TA, Westenbroek RE, Schuh SM, Hille B, Babcock DF. Identical phenotypes of CatSper1 and CatSper2 null sperm. J Biol Chem. 2005;280(37):32238–32244. doi: 10.1074/jbc.M501430200. [DOI] [PubMed] [Google Scholar]
- Ellerman DA, Ha C, Primakoff P, Myles DG, Dveksler GS. Direct binding of the ligand PSG17 to CD9 requires a CD9 site essential for sperm-egg fusion. Mol Biol Cell. 2003;14(12):5098–5103. doi: 10.1091/mbc.E03-04-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue N, Ikawa M, Isotani A, Okabe M. The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature. 2005;434(7030):234–238. doi: 10.1038/nature03362. [DOI] [PubMed] [Google Scholar]
- Inoue N, Ikawa M, Okabe M. Putative sperm fusion protein IZUMO and the role of N-glycosylation. Biochem Biophys Res Commun. 2008;377(3):910–914. doi: 10.1016/j.bbrc.2008.10.073. [DOI] [PubMed] [Google Scholar]
- Kaji K, Oda S, Shikano T, Ohnuki T, Uematsu Y, Sakagami J, Tada N, Miyazaki S, Kudo A. The gamete fusion process is defective in eggs of Cd9-deficient mice. Nat Genet. 2000;24(3):279–282. doi: 10.1038/73502. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Le Naour F, Rubinstein E, Jasmin C, Prenant M, Boucheix C. Severely reduced female fertility in CD9-deficient mice. Science. 2000;287(5451):319–321. doi: 10.1126/science.287.5451.319. [DOI] [PubMed] [Google Scholar]
- Liu Y, Tewari R, Ning J, Blagborough AM, Garbom S, Pei J, Grishin NV, Steele RE, Sinden RE, Snell WJ, Billker O. The conserved plant sterility gene HAP2 functions after attachment of fusogenic membranes in Chlamydomonas and Plasmodium gametes. Genes Dev. 2008;22(8):1051–1068. doi: 10.1101/gad.1656508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda PV, Allaire A, Sosnik J, Visconti PE. Localization of low-density detergent-resistant membrane proteins in intact and acrosome-reacted mouse sperm. Biol Reprod. 2009;80(5):897–904. doi: 10.1095/biolreprod.108.075242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyado K, Yamada G, Yamada S, Hasuwa H, Nakamura Y, Ryu F, Suzuki K, Kosai K, Inoue K, Ogura A, Okabe M, Mekada E. Requirement of CD9 on the egg plasma membrane for fertilization. Science. 2000;287(5451):321–324. doi: 10.1126/science.287.5451.321. [DOI] [PubMed] [Google Scholar]
- Nishimura H, Cho C, Branciforte DR, Myles DG, Primakoff P. Analysis of loss of adhesive function in sperm lacking cyritestin or fertilin beta. Dev Biol. 2001;233(1):204–213. doi: 10.1006/dbio.2001.0166. [DOI] [PubMed] [Google Scholar]
- Nishimura H, Myles DG, Primakoff P. Identification of an ADAM2-ADAM3 complex on the surface of mouse testicular germ cells and cauda epididymal sperm. J Biol Chem. 2007;282(24):17900–17907. doi: 10.1074/jbc.M702268200. [DOI] [PubMed] [Google Scholar]
- Pei J, Grishin NV. PROMALS: towards accurate multiple sequence alignments of distantly related proteins. Bioinformatics. 2007;23(7):802–808. doi: 10.1093/bioinformatics/btm017. [DOI] [PubMed] [Google Scholar]
- Penna A, Demuro A, Yeromin AV, Zhang SL, Safrina O, Parker I, Cahalan MD. The CRAC channel consists of a tetramer formed by Stim-induced dimerization of Orai dimers. Nature. 2008;456(7218):116–120. doi: 10.1038/nature07338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramjeesingh M, Huan LJ, Garami E, Bear CE. Novel method for evaluation of the oligomeric structure of membrane proteins. Biochem J. 1999;342(Pt 1):119–123. [PMC free article] [PubMed] [Google Scholar]
- Rizo J, Rosenmund C. Synaptic vesicle fusion. Nat Struct Mol Biol. 2008;15(7):665–674. doi: 10.1038/nsmb.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein E, Ziyyat A, Wolf JP, Le Naour F, Boucheix C. The molecular players of sperm-egg fusion in mammals. Semin Cell Dev Biol. 2006;17(2):254–263. doi: 10.1016/j.semcdb.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Stein KK, Go JC, Lane WS, Primakoff P, Myles DG. Proteomic analysis of sperm regions that mediate sperm-egg interactions. Proteomics. 2006;6(12):3533–3543. doi: 10.1002/pmic.200500845. [DOI] [PubMed] [Google Scholar]
- Sleight SB, Miranda PV, Plaskett NW, Maier B, Lysiak J, Scrable H, Herr JC, Visconti PE. Isolation and proteomic analysis of mouse sperm detergent-resistant membrane fractions: evidence for dissociation of lipid rafts during capacitation. Biol Reprod. 2005;73(4):721–729. doi: 10.1095/biolreprod.105.041533. [DOI] [PubMed] [Google Scholar]
- Stein KK, Go JC, Lane WS, Primakoff P, Myles DG. Proteomic analysis of sperm regions that mediate sperm-egg interactions. Proteomics. 2006;6(12):3533–3543. doi: 10.1002/pmic.200500845. [DOI] [PubMed] [Google Scholar]
- Sudhof TC, Rothman JE. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323(5913):474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD. The rapid evolution of reproductive proteins. Nat Rev Genet. 2002;3(2):137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- Turner LM, Chuong EB, Hoekstra HE. Comparative analysis of testis protein evolution in rodents. Genetics. 2008;179(4):2075–2089. doi: 10.1534/genetics.107.085902. [DOI] [PMC free article] [PubMed] [Google Scholar]