Abstract
BRCA1 carboxyl-terminal (BRCT) motifs are present in a number of proteins involved in DNA repair and/or DNA damage signaling pathways. The BRCT domain-containing protein BRCTx has been shown to interact physically with RAD18, an E3 ligase involved in postreplication repair and homologous recombination repair. However, the physiological relevance of the interaction between RAD18 and BRCTx is largely unknown. In this study, we showed that RAD18 interacts with BRCTx in a phosphorylation-dependent manner and that this interaction, mediated via highly conserved serine residues on the RAD18 C terminus, is required for BRCTx accumulation at DNA damage sites. Furthermore, we uncovered critical roles of the RAD18-BRCTx module in UV-induced DNA damage repair but not PCNA mono-ubiquitination or homologous recombination. Thus, our results suggest that RAD18 has an additional function in the surveillance of the UV-induced DNA damage response signal.
Keywords: DNA damage, DNA repair, BRCT domain, RAD18
1. Introduction
Genomic DNA constantly suffers a variety of insults caused by its intrinsic instability, as well as by innumerable endogenous and environmental factors [1-4]. If unrepaired or repaired incorrectly, the resulting damage can cause cell death, mutations and chromosomal instability, which eventually lead to tumorigenesis [1-4]. Therefore, in response to DNA damage, cells elicit an elaborate signaling network, which is collectively known as the DNA damage response pathway (DDR) [5, 6]. Through a molecular cascade consisting of sensors, mediators, transducers, and effectors, the DDR coordinates several cellular processes, including cell cycle checkpoints, DNA repair, cellular senescence, and apoptosis [7-9].
The breast cancer gene 1 (BRCA1) carboxyl-terminal (BRCT) domain represents a class of phosphoprotein recognition motifs found on numerous DNA damage and checkpoint proteins across species [10-12]. The human genome encodes more than 20 gene products that contain BRCT domains, and many of these proteins have been implicated in DNA damage checkpoint regulation, DNA repair, and cell cycle control [13-15]. BRCTx, one of the BRCT domain-containing proteins, was originally identified in a database search for novel BRCT domain proteins that localize to regions of frequent rearrangement in cancer [16]. BRCTx was further conducted as a highly conserved RAD18-interacting protein in a yeast two-hybrid screen [16]. The E3 ligase RAD18 is well known for its function in DNA damage bypass and postreplication repair (PRR) in yeast and vertebrates via its ability to facilitate proliferating cell nuclear antigen (PCNA) mono-ubiquitination at stalled replication forks [17-20]. We recently found that, in addition to its traditional function in the DNA damage bypass pathway, RAD18 is also an integral component in translating the damage response signal to orchestrate homologous recombination repair (HRR) [21]. We showed that RAD18 promotes homologous recombination in a manner strictly dependent on its ability to be recruited to sites of DNA breaks and that this recruitment relies on the well-defined DNA damage signaling pathway mediated by another E3 ligase, RNF8 [21-24]. We further demonstrated that RAD18 functions as an adaptor to facilitate homologous recombination through direct interaction with the recombinase RAD51C [21]. Although BRCTx has been shown to interact with RAD18, the exact function of BRCTx in the DDR pathway and whether it fits into the well-defined RAD18-mediated DNA damage-signaling cascade remains unknown.
In this study, we provide evidence suggesting that BRCTx is a DNA damage response protein that acts downstream of RNF8-RAD18 in the DNA damage signal transduction cascade. We found that the RAD18-BRCTx interaction is mediated by the N-terminal tandem BRCT domains of BRCTx and requires the phosphorylation of the Ser442 and Ser444 residues on the RAD18 polypeptide. In addition, we showed that the specific interaction between RAD18 and BRCTx is important for RAD18 function in UV-induced DNA damage repair.
2. Materials and methods
2.1. Antibodies
Antibodies against pH2AX, RAD18, RAD51 and 53BP1 were described previously [21, 25]. Anti-GST and anti-myc antibodies were purchased from Santa Cruz Biotechnology. Anti-HA polyclonal and anti-Flag (M2) monoclonal antibodies were purchased from Abcam and Sigma, respectively.
2.2. Cell culture and transfection
293T and HeLa Cells were maintained in RPMI 1640 supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin. SF9 insect cells were maintained in Grace's medium supplemented with 10% fetal bovine serum. Human cell lines were maintained in 37 °C incubator with 5% CO2, whereas insect cells were maintained at 27 °C. RAD18-/- MEFs were gifts from Dr. M Yamaizumi (Kumamoto University, Japan). Cell transfection was performed using Lipofectamine 2000 (Invitrogen), following the manufacture's protocol.
2.3. Plasmids
Full-length and deletion mutants of human RAD18 and mouse BRCTx were generated by PCR and subcloned into the pDONR201 vector using Gateway technology (Invitrogen). The corresponding fragments in entry vectors were transferred into a Gateway-compatible destination vector, which harbors an N-terminal triple-epitope tag (S-protein, Flag, and streptavidin-binding peptide; SFB), HA or Myc epitope tag for expression in mammalian cells.
2.4. Establishment of stable cell lines and affinity purification of SFB-tagged protein complexes
293T cells were transfected with plasmids encoding SFB-tagged proteins. Cell lines stably expressing tagged proteins were selected by culturing in medium containing puromycin (2 μg/ml) and confirmed by immunoblotting and immunostaining. For affinity purification, 293T cells stably expressing tagged proteins were lysed with NETN buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, and 0.5% Nonidet P-40) for 20 min. Crude lysates were removed by centrifugation at 14,000 rpm at 4°C for 10 min, and pellet was sonicated for 40 sec in high-salt solution (20 mM HEPES [pH 7.8], 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA, and protease inhibitor) to extract chromatin-bound protein fractions. The supernatants were cleared at 14,000 rpm to remove debris and then incubated with streptavidin-conjugated beads (Amersham Biosciences) for 2 h at 4°C. The beads were washed three times with NETN buffer, and then bead-bound proteins were eluted with NETN buffer containing 1 mg/ml biotin (Sigma). Eluted proteins were incubated with S protein beads (Novagen). The beads were again washed three times with NETN buffer and then subjected to SDS-PAGE. Protein bands were excised and digested, and the peptides were analyzed by mass spectrometry.
2.5. Co-immunoprecipitation and western blotting
For co-immunoprecipitation assays, constructs encoding SFB-tagged and Myc-tagged proteins were transiently co-transfected into 293T cells. Cells were lysed with NETN buffer containing 20 mM NaF, 1 μg /ml of pepstatin A, and 1 μg /ml aprotonin on ice for 20 min. After removal of cell debris by centrifugation, the soluble fractions were collected and incubated with S-protein beads for 2 h at 4°C. Beads were washed three times with NTEN buffer, boiled in 2 × SDS loading buffer, and resolved on SDS-PAGE. Membranes were blocked in 5% milk in TBST buffer and then probed with antibodies as indicated.
2.6. Immunofluorescence staining
To visualize damage-induced foci, cells cultured on coverslips were treated with gamma radiation (10 Gy) for 6 h. Cells were then washed with PBS and fixed using 3% paraformaldehyde solution for 10 min at room temperature and then extracted with buffer containing 0.5% Triton X-100 for 5 min. Samples were blocked with 5% goat serum and incubated with primary antibody for 20 min. Samples were washed and incubated with secondary antibody for 20 min. Cells were then counterstained with DAPI to visualize nuclear DNA.
2.7. Cell survival assays
Cells (1 × 103) were seeded onto 60-mm dishes in triplicates. At 24 h after seeding, cells were treated with UV radiation as indicated. The medium was replaced 24 h later and cells were then incubated for 14 days. Resulting colonies were fixed and stained with Coomassie blue and colonies were counted.
2.8. Gene conversion assay
Cells (1 × 106) were electroporated with 12 μg of DR-GFP plasmid together with 12 μg of pCBASce plasmid at 270 V, 975 μF using a BioRad genepulsar II as described previously [26]. Cells were plated onto 10 cm dishes and incubated in culture medium for 48 h before FACS analyses. Means were obstained from three independent experiments.
2.9. Chromatin fractionation
Chromatin fractions were prepared as described previously, with some modifications [21, 25, 27]. Briefly, 4 h after treatment with UV radiation (60 J/m2), cells were collected and washed once with PBS. Cell pellets were subsequently resuspended in NETN buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, and 0.5% Nonidet P-40 and protease inhibitors) and incubated on ice for 20 min. Crude lysates were removed by centrifugation at 14,000 rpm at 4 °C for 10 min, and pellet was recovered and resuspended in 0.2 M HCl for 20 min. The soluble fraction was then neutralized with 1 M Tris-HCl (pH 8.0) for further analysis.
2.10. GST pull-down assay
GST fusion proteins were expressed in E. coli and purified as described previously [25]. GST fusion protein (2 μg) or GST alone was immobilized on glutathione Sepharose 4B beads and incubated with lysates prepared from cells that were transiently transfected with plasmids encoding indicated proteins.
2.11. Retrovirus production and infection
pDONR201-RAD18 constructs were transferred into a Gateway-compatible pEF1A-HA-FLAG or SFB-tagged retroviral vector. Virus supernatant was collected 48 h after the co-transfection of retroviral vectors and pcl-ampho into BOSC23 cells. MEFs were infected with viral supernatant in the presence of polybrene (8 μg/ml), and then Cells were selected in growth medium containing 2 μg/ml puromycin. Protein expression in transduced cells was confirmed by western blotting or immunofluorescence staining using anti-Flag antibodies.
3. Results and discussion
3.1. BRCTx is a DNA damage response protein
BRCT domains are evolutionarily conserved modules that exist in numerous prokaryotic and eukaryotic proteins [10-12]. Most BRCT domains function as protein-protein modules that recognize phosphorylated serine motifs, and interactions between BRCT domains and phosphorylated proteins are thought to have essential roles in the transduction of DNA damage signals; however, it is unclear whether and how the BRCT domain-containing protein BRCTx participates in mammalian DNA damage responses.
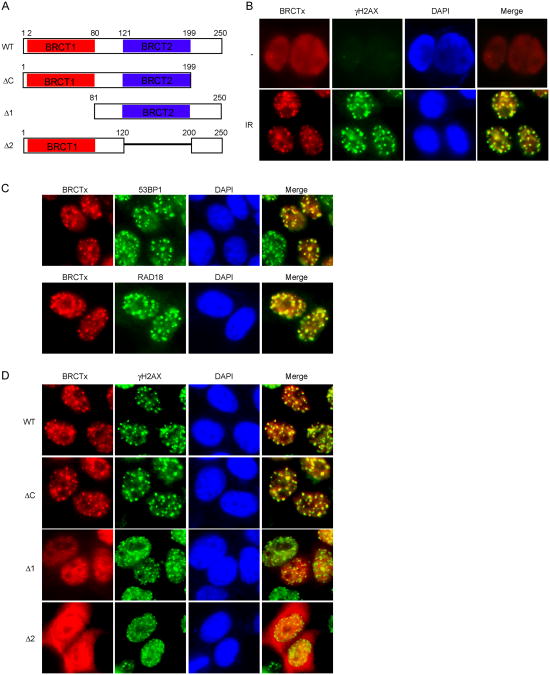
We first performed structural analysis of the BRCTx protein sequence. Interestingly, in contrast to a previous study, we found that not one but two BRCT domains are present in the BRCTx N terminus [16] (Fig. 1A). Because many BRCT domain-containing proteins form nuclear foci after DNA damage (e.g., MDC1 and BRCA1), we investigated the nuclear localization of BRCTx before and after DNA damage. BRCTx protein normally localizes diffusely in the nuclei (Fig. 1B). However, after gamma irradiation, BRCTx relocalized to nuclear foci that co-localized with phosphorylated H2AX (γH2AX), 53BP1 and RAD18 foci (Fig. 1B and C), suggesting that BRCTx relocalizes to sites of DNA damage and probably functions in the DNA damage response pathway.
Fig.1. BRCTx forms IR-induced foci.
(A) Schematic representation of BRCTx and its deletion mutants used in this study. (B) Localization of BRCTx in response to IR. 293T cells transfected with SFB-tagged BRCTx were treated with IR, fixed, and immunostained with anti-Flag and anti-pH2AX antibodies. (C) Colocalization of BRCTx with 53BP1 and RAD18. 293T cells transfected with SFB-tagged BRCTx were treated with IR, fixed, and immunostained with anti-Flag and anti-53BP1 or RAD18 antibodies. (D) The BRCT domains of BRCTx target it to IR-induced foci. 293T cells expressing indicated Flag-tagged proteins were treated with IR, fixed and immunostained with anti-Flag and anti-pH2AX antibodies.
Having shown that BRCTx accumulates at sites of DNA damage, we next sought to identify the region(s) within BRCTx important for its translocation to ionizing radiation-induced foci (IRIF). As shown in Figure 1D, whereas wild-type and C-terminal deletion mutant of BRCTx both localized to IRIF, deletion of either one of the two BRCT domains impaired BRCTx IRIF formation. These observations suggest that both BRCT domains are essential for targeting BRCTx to IRIF.
3.2. BRCTx interacts with RAD18 in a phosphorylation-dependent manner
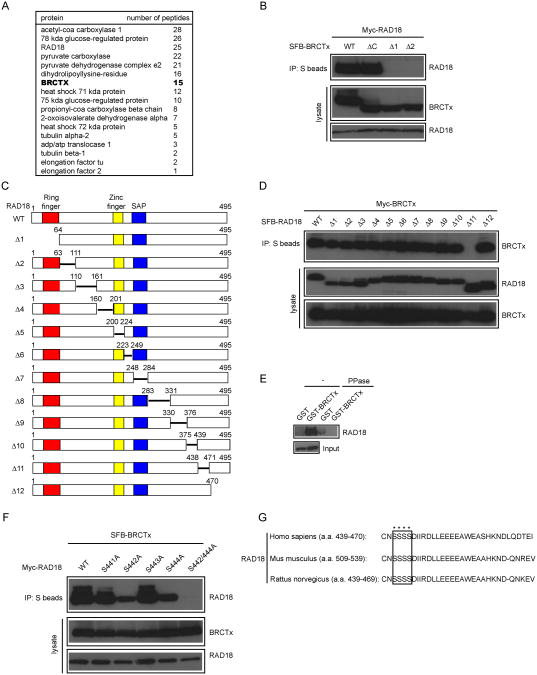
To better understand BRCTx functions in DNA damage response, we generated a human HEK-293T-derived cell line stably expressing a triple-tagged BRCTx for the identification of potential BRCTx-interacting proteins. Following a tandem affinity purification (TAP) scheme, proteins associated with BRCTx were identified by mass spectrometry analysis. We repeatedly found RAD18, a well known E3 ligase involved in postreplication repair and homologous recombination repair, as a major BRCTx binding partner (Fig. 2A). In fact, an interaction between BRCTx and RAD18 has been previously reported [16]. We performed co-immunoprecipitation experiments and confirmed an interaction between RAD18 and BRCTx, suggesting that these two proteins indeed associate with each other in vivo (Fig. 2B).
Fig.2. BRCTx interacts with RAD18.
(A) 293T cells stably expressing SFB-tagged-BRCTx were used for tandem affinity purification of protein complexes specifically from chromatin fractions. Tables are summaries of proteins identified by mass spectrometry analysis. Letters in bold indicate the bait proteins. (B and D) Mapping of the corresponding regions required for the BRCTx-RAD18 interaction. Immunoprecipitation reactions were performed using S-protein beads and then subjected to western blot analyses using antibodies as indicated. (C) Schematic representation of wild-type and deletion mutants of RAD18 used in this study. (E) Phosphorylation-dependent interaction between BRCTx and RAD18. GST or GST–BRCTx was incubated with cell lysates containing exogenously expressed Flag-tagged wild-type RAD18, with or without phosphatase (PPase). Bound RAD18 was analyzed by anti-Flag immunoblotting. Lower gel shows amounts of RAD18 protein used in these experiments. (F) Both the Ser442 and Ser444 residues of RAD18 are required for its binding to BRCTx. 293T cells were transfected with plasmids encoding SFB-tagged BRCTx together with plasmids encoding Myc-tagged wild-type or serine point mutants of RAD18. Cells were collected 24 h after transfection. Coimmunoprecipitation reactions were performed using S-protein beads, and cells were then subjected to immunoblotting using the antibodies as indicated. (G) Alignment of the BRCTx binding region of RAD18 from different species.
Next, we sought to identify the region(s) within BRCTx responsible for its interaction with RAD18 by using SFB-tagged wild-type BRCTx and a series of BRCTx deletion mutants (Fig. 1A). We showed that deletions of either the first or the second BRCT domain of BRCTx led to a dramatic decrease in BRCTx-RAD18 interaction, indicating that both the tandem BRCT domains of BRCTx are required for its binding to RAD18 (Fig. 2B). Conversely, using a series of overlapping RAD18 truncations and deletion mutants spanning its entire coding sequence, we mapped the minimal BRCTx-binding region to residues 439 to 470 of RAD18 (Fig. 2C and 2D).
As BRCT is a phosphoprotein-binding domain, we reasoned that the N-terminal tandem BRCT domains of BRCTx might bind specifically to phosphorylated RAD18, and pull-down assays confirmed this hypothesis (Fig. 2E). We then generated several serine point mutants within the BRCTx-binding region of RAD18. Indeed, in contrast to wild-type and S441A and S443A mutants of RAD18, the S442A and S444A mutants had highly reduced binding abilities to BRCTx (Fig. 2F). Moreover, the serine double mutant of RAD18 (S442/444A) totally lost its binding ability to BRCTx, suggesting that residues Ser442 and Ser444 of RAD18 are critical for the BRCTx-RAD18 interaction (Fig. 2F). Interestingly, the Ser442 and Ser444 residues of RAD18 are highly evolutionarily conserved, suggesting that they may carry out an important function of RAD18 (e.g., an interaction with BRCTx) (Fig. 2G).
3.3. RAD18 acts upstream of BRCTx
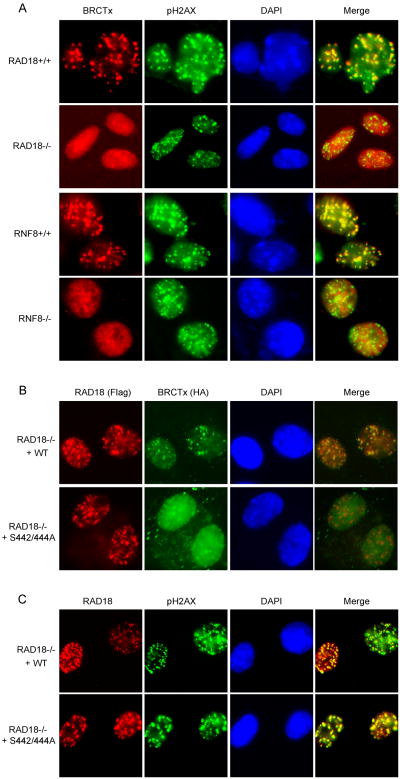
Our previous studies have shown that RAD18 proteins are recruited to sites of DNA breaks and rely on the well-defined DNA damage signaling pathway mediated by RNF8 [21-24]. To define where BRCTx fits into this established DNA damage signaling cascade, we examined DNA damage-induced BRCTx foci formation in a panel of MEF cell lines defective in various components known to be involved in this DDR pathway. No IR-induced BRCTx foci formation was observed in RAD18- and RNF8-deficient MEFs, in sharp contrast to their wild-type counterparts (Fig. 3A). Moreover, RAD18-S442/444A mutant failed to restore BRCTx foci formation in RAD18-/- cells (Fig. 3B). Conversely, both wild-type RAD18, and its S442/444A mutant, which is impaired in binding to BRCTx, could still relocalize to γ-H2AX containing foci (Fig. 3C). These data suggest that BRCTx acts downstream of RNF8 and RAD18 in the known DNA damage signal transduction pathway.
Fig.3. RAD18 is required for BRCTx foci formation.
(A) Genetic dependence of BRCTx relocalization after IR treatment. RAD18- and RNF8-deficient MEFs and their wild-type counterparts infected with HA-Flag-tagged BRCTx were irradiated, fixed, and immunostained using anti-pH2AX and anti-Flag antibodies. (B) Binding to RAD18 is required for BRCTx foci formation. RAD18-deficient MEFs infected with SFB-tagged wild-type RAD18 or the serine double mutant were transfected with HA-tagged BRCTx and then were irradiated, fixed, and immunostained using anti-Flag monoclonal and anti-HA polyclonal antibodies. (C) Binding to BRCTx is not required for RAD18 foci formation. RAD18-deficient MEFs infected with HA-Flag-tagged wild-type RAD18 or the serine double mutant were irradiated, fixed, and immunostained using anti-pH2AX and anti-Flag antibodies.
3.4. RAD18-BRCTx interaction is required for efficient repair of UV-induced DNA damage
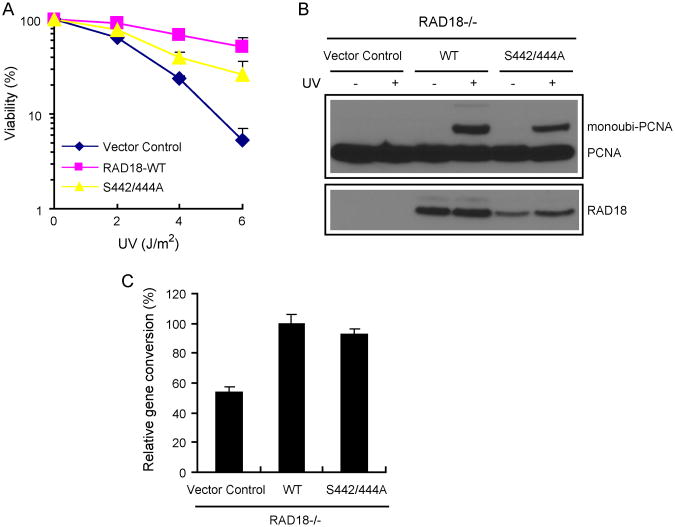
To explore the physiological relevance of the highly conserved serine residues of RAD18, which are required for its binding to BRCTx, we first set out to determine whether the RAD18 S442/444A mutant would be defective in restoring cell survival after DNA damage. RAD18-/- MEFs reconstituted with wild-type RAD18, the RAD18-S442/444A mutant, or vector alone were treated with various doses of UV radiation. Clonogenic assays indicated that only the cells reconstituted with wild-type RAD18, but not those reconstituted with the RAD18 S442/444A mutant, restored cell survival after UV treatment (Fig. 4A). These results suggest that the specific interaction between RAD18 and BRCTx is critical for RAD18 function in the UV-induced damage repair pathway. In agreement with this hypothesis, BRCTx-null MEFs display increased sensitivity to UV irradiation [16].
Fig. 4. RAD18-BRCTx interaction is required for efficient repair of UV-induced DNA damage.
(A) The S442/444A mutant of RAD18 is defective in restoring cell survival after UV treatment. RAD18-deficient cells were transduced with a control virus or a virus expressing HA-Flag-tagged wild-type RAD18 or the serine double mutant (S442/444A), which does not bind to BRCTx. Cell survival assays were performed as described in Materials and Methods. Date are presented as means and SD (error bars) from three independent experiments. (B) Binding to BRCTx is not required for RAD18 function in UV-induced PCNA monoubiquitination. RAD18-deficient cells were transduced with a control virus or a virus expressing HA-Flag-tagged wild-type RAD18 or the serine double mutant (S442/444A), which does not bind to BRCTx. Chromatin fractions were isolated (see Materials and Methods) and immunoblotted with the indicated antibodies. (C) Binding to BRCTx is not required for RAD18 function in homologous recombination. Gene conversion assays were performed as described in Materials and Methods. The percentage of GFP-positive cells was determined by flow cytometry 48 h after electroporation. Data shown are Means and SD (error bars) from three independent experiments.
The known functions of RAD18 in vivo are to facilitate PCNA mono-ubiquitination, which is required for DNA damage bypass in response to UV-induced lesions, and accumulation of RAD51C at damaged chromatin, which is required for homologous recombination in response to IR-induced lesions [21]. We first tested whether the binding of RAD18 to BRCTx is required for RAD18 function in promoting UV-induced PCNA mono-ubiquitination. Unexpectedly, both wild-type RAD18 and the RAD18 S442/444A mutant restored UV-induced PCNA mono-ubiquitination in RAD18-deficient MEFs (Fig. 4B). Moreover, the S442/444A mutant also restored HRR, indicating that the ability of RAD18 to bind to BRCTx is also not required for the homologous recombination function of RAD18 (Fig. 4C), consistently, Brctx-null MEFs are not overtly sensitive to ionizing radiation [16]. Thus, RAD18 might participate in UV-induced damage repair through a yet-to-be-identified pathway via its interaction with BRCTx.
4. Conclusions
In this study, we reported that RAD18 interacts with BRCTx in a phosphorylation-dependent manner. Whereas this interaction appears to be largely dispensable for RAD18 function in PCNA mono-ubiquitination and homologous recombination, it is clearly necessary for the full function of RAD18 in promoting cell survival after UV-induced DNA damage. Therefore, it is likely that RAD18, via its interaction with BRCTx, functions through a yet-to-be-identified pathway in the surveillance of the UV-induced DNA damage response signal.
Highlights.
> BRCTx is a DNA damage response protein. > BRCTx interacts with RAD18 in a phosphorylation-dependent manner. > RAD18-BRCTx interaction is required for UV-induced DNA damage repair.
Acknowledgments
We would like to thank Dr. M. Yamaizumi for providing RAD18-/- MEFs and all our colleagues in the Huang laboratory for insightful discussions and technical assistance. This work was supported in part by grants from the National Natural Science Foundation of China (grant 31071243), the Natural Science Foundation of Zhejiang Province (grant R2110569) and the China's Fundamental Research Funds for the Central Universities. J.C. is a recipient of an Era of Hope Scholar award from the Department of Defense (W81XWH-05-1-0470) and a member of MD Anderson Cancer Center (CA016672).
Abbreviations
- BRCT
BRCA1 carboxyl-terminal
- DDR
DNA damage response
- HRR
homologous recombination repair
- IRIF
ionizing radiation-induced foci
- MEFs
mouse embryonic fibroblasts
- PRR
postreplication repair
- PCNA
proliferating cell nuclear antigen
- SFB
S-protein, Flag, and streptavidin-binding peptide
Footnotes
Conflict of interest: None declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Friedberg EC. DNA damage and repair. Nature. 2003;421:436–440. doi: 10.1038/nature01408. [DOI] [PubMed] [Google Scholar]
- 2.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 3.McKinnon PJ, Caldecott KW. DNA strand break repair and human genetic disease. Annu Rev Genomics Hum Genet. 2007;8:37–55. doi: 10.1146/annurev.genom.7.080505.115648. [DOI] [PubMed] [Google Scholar]
- 4.Thompson SL, Bakhoum SF, Compton DA. Mechanisms of chromosomal instability. Curr Biol. 20:R285–295. doi: 10.1016/j.cub.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 8.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 9.Lombard DB, Chua KF, Mostoslavsky R, Franco S, Gostissa M, Alt FW. DNA repair, genome stability, and aging. Cell. 2005;120:497–512. doi: 10.1016/j.cell.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 10.Caldecott KW. Cell signaling. The BRCT domain: signaling with friends? Science. 2003;302:579–580. doi: 10.1126/science.1091463. [DOI] [PubMed] [Google Scholar]
- 11.Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- 12.Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 13.Mohammad DH, Yaffe MB. 14-3-3 proteins, FHA domains and BRCT domains in the DNA damage response. DNA Repair (Amst) 2009;8:1009–1017. doi: 10.1016/j.dnarep.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mesquita RD, Woods NT, Seabra-Junior ES, Monteiro AN. Tandem BRCT domains: DNA's Praetorian Guard. Genes Cancer. 1:1140–1146. doi: 10.1177/1947601910392988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez MC, Songyang Z. BRCT domains: phosphopeptide binding and signaling modules. Front Biosci. 2008;13:5905–5915. doi: 10.2741/3125. [DOI] [PubMed] [Google Scholar]
- 16.Adams DJ, van der Weyden L, Gergely FV, Arends MJ, Ng BL, Tannahill D, Kanaar R, Markus A, Morris BJ, Bradley A. BRCTx is a novel, highly conserved RAD18-interacting protein. Mol Cell Biol. 2005;25:779–788. doi: 10.1128/MCB.25.2.779-788.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tateishi S, Sakuraba Y, Masuyama S, Inoue H, Yamaizumi M. Dysfunction of human Rad18 results in defective postreplication repair and hypersensitivity to multiple mutagens. Proc Natl Acad Sci U S A. 2000;97:7927–7932. doi: 10.1073/pnas.97.14.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tateishi S, Niwa H, Miyazaki J, Fujimoto S, Inoue H, Yamaizumi M. Enhanced genomic instability and defective postreplication repair in RAD18 knockout mouse embryonic stem cells. Mol Cell Biol. 2003;23:474–481. doi: 10.1128/MCB.23.2.474-481.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 20.Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang J, Huen MS, Kim H, Leung CC, Glover JN, Yu X, Chen J. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat Cell Biol. 2009;11:592–603. doi: 10.1038/ncb1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, Pelletier L, Jackson SP, Durocher D. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 25.Liu T, Ghosal G, Yuan J, Chen J, Huang J. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science. 329:693–696. doi: 10.1126/science.1192656. [DOI] [PubMed] [Google Scholar]
- 26.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang J, Gong Z, Ghosal G, Chen J. SOSS complexes participate in the maintenance of genomic stability. Mol Cell. 2009;35:384–393. doi: 10.1016/j.molcel.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]