Abstract
NEIL1 [Nei (endonuclease VIII)-like protein 1], one of the five mammalian DNA glycosylases that excise oxidized DNA base lesions in the human genome to initiate base excision repair, contains an intrinsically disordered C-terminal domain (CTD; ∼100 residues), not conserved in its Escherichia coli prototype Nei. Although dispensable for NEIL1's lesion excision and AP lyase activities, this segment is required for efficient in vivo enzymatic activity and may provide an interaction interface for many of NEIL1's interactions with other base excision repair proteins. Here, we show that the CTD interacts with the folded domain in native NEIL1 containing 389 residues. The CTD is poised for local folding in an ordered structure that is induced in the purified fragment by osmolytes. Furthermore, deletion of the disordered tail lacking both Tyr and Trp residues causes a red shift in NEIL1's intrinsic Trp-specific fluorescence, indicating a more solvent-exposed environment for the Trp residues in the truncated protein, which also exhibits reduced stability compared to the native enzyme. These observations are consistent with stabilization of the native NEIL1 structure via intramolecular, mostly electrostatic, interactions that were disrupted by mutating a positively charged (Lysrich) cluster of residues (amino acids 355–360) near the C-terminus. Small-angle X-ray scattering (SAXS) analysis confirms the flexibility and dynamic nature of NEIL1's CTD, a feature that may be critical to providing specificity for NEIL1's multiple, functional interactions.
Keywords: DNA repair, intrinsically unstructured region, electrostatic interactions, protein stability, oxidative DNA damage
Introduction
Oxidatively damaged bases in the genome, induced both endogenously and by exogenous toxicants, are repaired by the evolutionary conserved base excision repair (BER) pathway,1,2 which is initiated by a DNA glycosylase (DG). DGs specific for oxidized DNA bases in mammalian cells have dual enzymatic functions: base excision followed by cleavage of the resulting abasic (AP) site via a lyase activity, generating a 1-nt gap at the lesion site. Five mammalian DGs responsible for repairing oxidized DNA bases belong to two major families: the Nth and Fpg/Nei, named after their bacterial prototypes endonuclease III (Nth) and endonuclease VIII (Nei), respectively. NEIL1 [Nei (endonuclease VIII)-like protein 1],3,4 NEIL2,5 and NEIL36 in the Fpg/Nei family are distinct from the Nth family members OGG1 and NTH1 with regard to both structure and AP lyase reaction,7 in spite of their overlapping substrate range.1 While NEIL1/2 generate 3′ phosphate (P) at the strand break via a βδ-lyase activity, OGG1 and NTH1 generate 3′ α,β unsaturated phosphoaldehyde (dRP) via β-elimination reaction. These 3′ blocking groups are removed by polynucleotide kinase 3′ phosphatase and AP endonuclease 1, respectively, in the next repair step, common with DNA single-strand break repair.7 The 1-nt gap at the resulting 3′OH-containing strand break is then filled in with the appropriate nucleotide by a DNA polymerase, followed by nick sealing by a DNA ligase.1,2
NEIL1 is unique among the oxidized base-specific DGs because of its S-phase-specific activation,3 ability to excise substrate bases in single-stranded DNA,8 and functional association with DNA replication proteins.9–12 All of these observations implicate NEIL1 in preferential repair of the replicating mammalian genome. NEIL1 interacts with downstream, conventional repair proteins, as well as noncanonical accessory proteins, invariably via a common interaction interface (amino acids 312–349) within its C-terminal (∼ 100 residues) domain (CTD).13 How such a short peptide is involved in more than 20 specific protein–protein interactions is unclear. The ∼78 C-terminal residues (amino acids 312–389) are dispensable for NEIL1's lesion excision and AP lyase activities but are required for efficient, overall repair both in cells and in vitro.13 The disordered structure of the NEIL1's CTD was predicted from PONDR analysis14 and likely was involved in the failure to crystallize the full-length protein.15 Notably, the crystal structure of the NEIL1 polypeptide lacking 56 C-terminal residues showed a lack of electron density for the 40 C-terminal residues that remained.15 Thus, no structural information is available for the CTD.
In apparent contrast to the data supporting an intrinsically disordered CTD, we surprisingly found decreased stability and distinct intrinsic fluorescence of the CTD-truncated NEIL1 polypeptide compared to that of the wild-type (WT) protein. As these findings are counter to the expectation of a disordered extension structurally dissociated from the folded core domain, we decided to carry out further investigation of the CTD structure. Based on combined biochemical, mutational, and biophysical studies, and solution structure analysis, we show here that the CTD folds back to interact with the globular domain in native NEIL1 where the dynamic association is driven by electrostatic interactions and that this intramolecular association in native NEIL1 is important for maintaining stability and structural integrity of the genome.
Results
Distinct fluorescence spectra of NEIL1 and its C-terminal deletion mutants
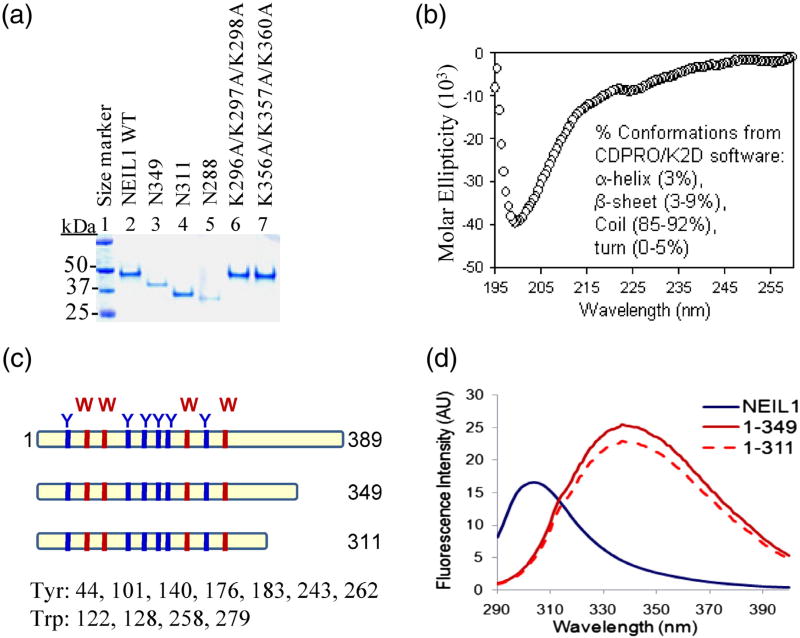
A series of studies from our laboratory demonstrated that NEIL1 interacts with downstream repair and other noncanonical accessory proteins via a 38-residue common interaction domain in the CTD. 9–13,16 We also showed recently that the CTD, dispensable for NEIL1's in vitro DG activity, is required for NEIL1-initiated complete repair and that deletion of the CTD significantly increases the sensitivity of human cells to oxidative stress.13 However, little information is available about the structure of this CTD region and its influence on the structure of NEIL1's core domain. Predominantly random-coil conformation of the CTD (78-residue segment), initially predicted by PONDR modeling,14 was confirmed by the circular dichroism (CD) spectrum of the purified C78 peptide (Fig. 1b). The purity of WT NEIL1 and its mutant polypeptides used in this study is shown in Fig. 1a.
Fig. 1.
Distinct intrinsic fluorescence emission spectra of WT NEIL1 and its mutants lacking the disordered C-terminus. (a) Coomassie-stained SDS-PAGE of purified WT NEIL1 and its deletion or point mutants. (b) The CD spectrum of NEIL1's C-terminal 78-residue peptide (amino acids 312–389; C78) reveals a prominently disordered structure with random-coil conformation. (c) Locations of four Trp and six Tyr residues within the N-terminal core domain in NEIL1's sequence. (d) Intrinsic fluorescence emission spectra of NEIL1 and its N349 and N311 C-terminal deletion mutants; excitation at 280 nm.
To gain further insight into the CTD structure in the context of the native protein, we analyzed the intrinsic fluorescence of the WT NEIL1 and its Cterminally truncated mutants. WT NEIL1 contains four Trp and six Tyr residues, all of which are located in its globular region (amino acids 1–311; Fig. 1c). Excitation at 280 nm (for both Tyr and Trp) showed typical Tyr emission spectrum of WT NEIL1 with λmax at 306 nm (Fig. 1d). However, the fluorescence of NEIL1 polypeptides with C-terminal deletions of 40 aa (N349) or 78 aa (N311) showed marked red shifts toward the Trp emission spectrum. The blue shift observed in the fluorescence of WT NEIL1 compared to its deletion mutants suggests a more nonpolar environment of these Trp residues, indicating that they are more shielded from the solvent than in the mutants. The fluorescence output of the N349 and N311 mutants was also about 2-fold higher than that of WT NEIL1. These unexpected observations suggested a profound impact of the C-terminal segment on the core structure, which could be due to intramolecular fold-back of the extended C-terminal tail on the globular core surface, causing solvent shielding of the Trp residues.
Propensity of NEIL1's CTD to form an ordered structure
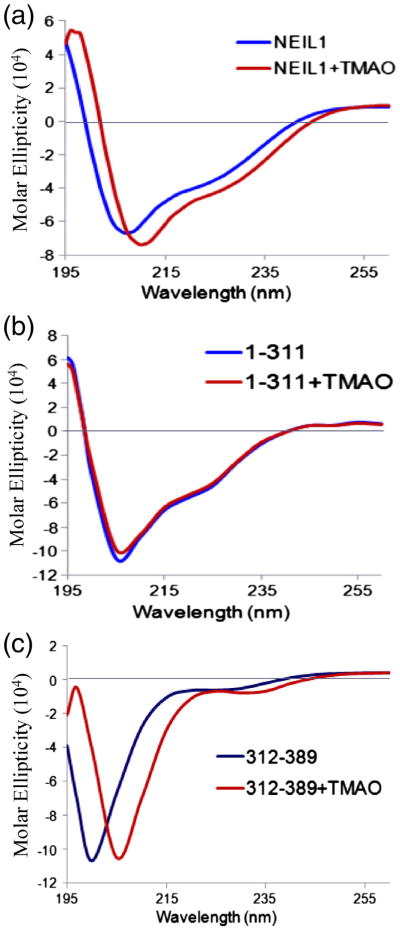
The specific interactions involving NEIL1's C-terminal region with multiple partner proteins suggest that the flexible CTD could lock into distinct conformations with the partners. To test whether the disordered region alone could form an ordered structure, we examined the effect of osmolytes trimethylamine-N-oxide (TMAO) and sarcosine, well-known protein-folding agents.17–19 While TMAO (3 M) induced folding in WT NEIL1 (Fig. 2a) and its C78 peptide (Fig. 2c) as monitored by the CD spectra, no change was observed for the N311 mutant lacking most of the disordered segment. Although the disordered segment extends to 100 C-terminal residues, the C78 peptide, the product of endoproteinase Asp-N,12 was studied. The inability of TMAO to induce additional folding in N311 suggested that the increased secondary structure in WT NEIL1 is attributable to the C-terminal segment. Sarcosine treatment produced similar results (data not shown).
Fig. 2.
Folding propensity of NEIL1's disordered C-terminal segment. CD spectral analysis of the effect of the osmolyte TMAO (3 M) on the secondary structure of WT NEIL1 (a) and its N311 mutant (b) and C78 peptide (c).
The CTD contributes to NEIL1's stability
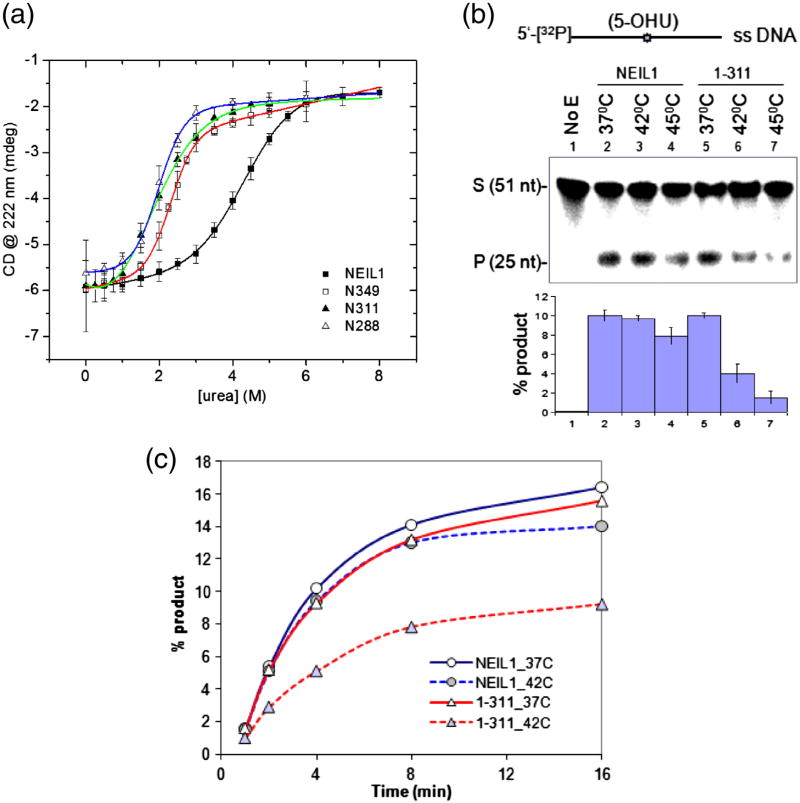
We reasoned that the NEIL1 CTD contributed to the structural stability of the WT protein, which would affect NEIL1's urea or heat denaturation. While the midpoint of transition from the native to denatured states for WT NEIL1 was observed at 4.3 M urea, 50% denaturation of the deletion mutants N349, N311, and N288 occurred at about 2 M urea (Fig. 3a). Furthermore, while the mutants exhibited an apparently cooperative single transition between the folded and unfolded states,19,20 the transition for the WT protein appears less cooperative, presumably due to the presence of multiple intermediate states. The Gibbs free energies (ΔG0) of unfolding were calculated to be 4.4 kJ/mol for WT NEIL1 and 2.9, 2.5, and 1.3 kJ/mol for the N349, N311, and N288 mutants, respectively.
Fig. 3.
The C-terminal region enhances NEIL1's stability. (a) Denaturation of WT and deletion mutants of NEIL1 by increasing amounts of urea, measured by CD spectra at 222 nm with a 1-min time scan. The sigmoidal transition curves were fitted for experimental points of molar ellipticity at 222 nm as a function of urea concentration using ORIGIN software. (b) Activity of WT NEIL1 and N311 mutant with 5′-32P-labeled 5-OHU-containing single-stranded DNA oligo at the indicated temperature. Change in activity as percent product is given as histogram (bottom panel). (c) Kinetics of activity.
To test whether the change in stability affects enzymatic activity, we compared the base excision/AP lyase activities of WT NEIL1 versus N311 mutant using a 5-OHU (an oxidative deamination product of C)-containing single-stranded deoxyoligonucleotide substrate as a function of temperature. While the activity decreased marginally (20%) for WT NEIL1 by raising the temperature from 37°C versus 45°C, the N311 mutant showed ∼80% loss of activity (Fig. 3b), strongly suggesting decreased thermal stability of the 311 mutant. Taken together, these data demonstrate that the CTD is critical for NEIL1's functional stability.
Intramolecular fold-back involves interactions between NEIL1's C-terminal and core domains
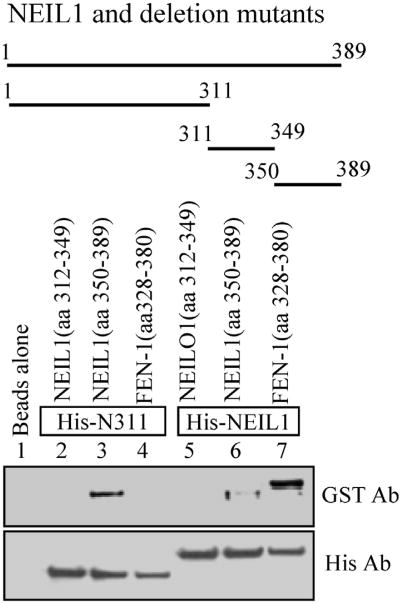
The ∼2-fold difference in the stability of WT NEIL1 versus its deletion mutants was surprising and is counter to the common perception of disordered terminal domains as being structurally independent of the core domain. To gain further structural insight about this observation and to experimentally test the intramolecular interaction between the C-terminal tail and the globular core surface, we monitored coelution of purified C-terminal peptides with Histagged WT NEIL1 and its N311 mutant bound to Ni-NTA beads (Fig. 4). The 40-residue C-terminal peptide (amino acids 350–389) but not an interior peptide (amino acids 312–349) stably bound to the mutant protein and, to a lesser extent, to the WT protein. The weak binding of the CTD also to the WT NEIL1 could be due to the dynamic nature of intramolecular fold-back, which was competitively disrupted by the exogenous peptide.
Fig. 4.
Binding of purified C-terminus to peptides truncated and full-length NEIL1. His-affinity co-elution of purified GST-tagged C-terminal peptides (amino acids 312–349 and amino acids 350–389) with His-tagged N311 and WT NEIL1 bound to N1-NTA beads. A GST-tagged FEN-1 C-terminal peptide9 that binds to NEIL1 via its C-terminal residues (312–349) was used as a control.
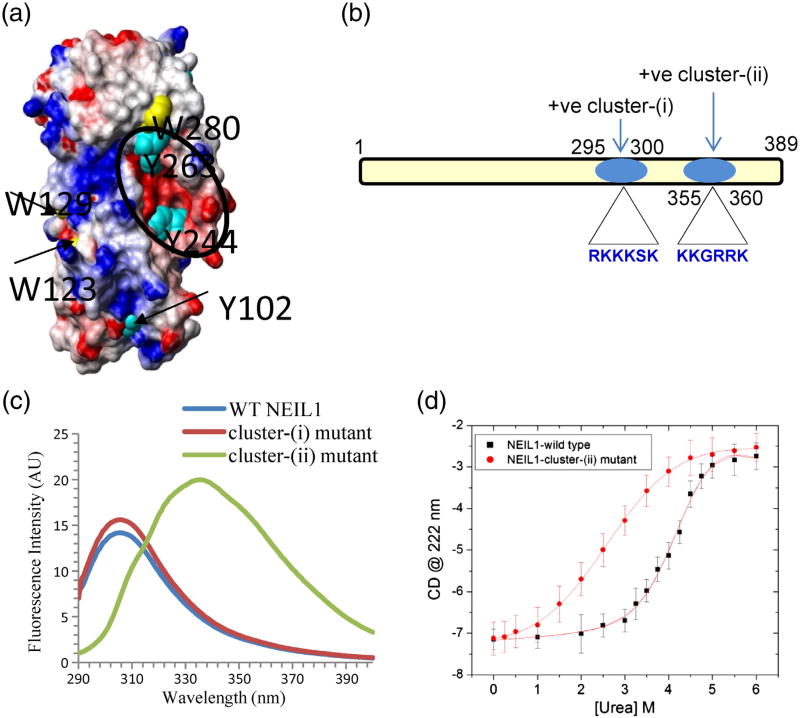
Identification of clustered C-terminal Lys residues involved in fold-back
The space-filling model of NEIL1's folded domain showed an acidic residue cluster (indicated in red in Fig. 5a), based on its crystal structure.15 Two clusters of Lys and Arg residues, cluster(i) containing amino acids 295–300 and cluster(ii) containing amino acids 355–360, are present in the C-terminal segment of NEIL1 (Fig. 5b). We purified two sitespecific point mutants of NEIL1 in which all of the Lys residues in these clusters were substituted with Ala. The fluorescence spectrum of the cluster(i) mutant (K296A/K297A/K298A) was similar to that of WT NEIL1, but the cluster(ii) mutant (K356A/K357A/K360A) exhibited Trp emission spectrum similar to that of the truncated NEIL1 (Fig. 5c). These results suggest that the basic residue cluster(ii) participates in intramolecular interaction with the acidic cluster in the core domain. Importantly, Trp280 is surface exposed in truncated NEIL1 (Fig. 5a), despite the blue-shifted emission spectrum for WT NEIL1. This suggests that the emission from Trp280 is likely masked due to the fold-back in the WT protein.
Fig. 5.
Contribution of electrostatic binding in NEIL1's intramolecular interaction involving a basic residue cluster at the C-terminus and a surface-exposed acidic residue cluster in the core domain. (a) Space-filling model of NEIL1's core domain based on crystal structure coordinates15 displaying a surface-exposed cluster of acidic amino acid residues (in red) within the circle. The Tyr/Trp residues partially or fully surface exposed are indicated. (b) Presence of two basic residue clusters at NEIL1's C-terminus. (c) Intrinsic fluorescence of cluster(i) and cluster(ii) mutants of NEIL1. (d) Urea denaturation of WT NEIL1 and cluster(ii) mutant.
Urea denaturation studies showed a significant decrease in the stability of the cluster(ii) mutant, compared to the WT NEIL1 (Fig. 5d). Although the transition midpoint and stability of the cluster(ii) mutant were comparable to those of the C-terminal deletion mutants, the denaturation pattern and transition characteristics were different. While the deletion mutants showed an apparent single phase transition (Fig. 3a), the cluster(ii) mutant showed a broad transition like WT NEIL1. These denaturation data suggest incomplete disruption of the intramolecular interactions in the cluster(ii) mutant. Taken together, these results underscore the contribution of electrostatic interactions in stabilizing the fold-back of the CTD.
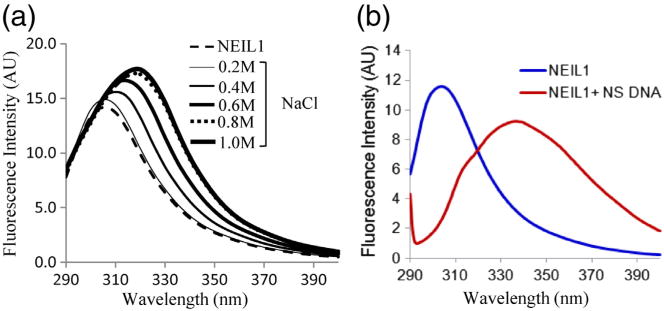
To get further insights into the involvement of electrostatic forces in the fold-back, we analyzed the salt dependence of NEIL1's intrinsic fluorescence spectrum, which revealed partial disruption of the fold-back interaction at 800 mM NaCl as indicated by a shift in fluorescence emission spectra (Fig. 6a). The higher-than-expected NaCl concentration required for disrupting the fold-back suggested that although the initial affinity was provided by electrostatic forces, the interaction may be further stabilized by hydrophobic forces, which would be enhanced with increasing salt concentration.
Fig. 6.
Disruption of NEIL1's intramolecular fold-back by salt or DNA binding. Intrinsic fluorescence emission spectra of WT NEIL1 (2 μM) in the presence of NaCl (a) or of equimolar 50-mer duplex DNA oligo (b).
DNA binding disrupts fold-back interaction to release the tail
Fluorescence analysis of NEIL1 in the presence of a 50-mer duplex DNA oligo showed partial shift from Tyr to Trp emission spectrum (Fig. 6b), suggesting the release of NEIL1's C-terminal tail. Interestingly, similar fold-back of the disordered C-terminal segment to the core and its release after DNA binding was observed for the Drosophila cryptochrome CRY protein that was attributed to DNA mimicry of the flexible tail.21 While a similar scenario may exist for NEIL1, it is also possible that the C-terminal region itself binds independently to DNA, leading to its release from the core. In support of the latter, our unpublished observations suggest that the C-terminal segment does bind to DNA and facilitate target DNA search via intra-strand transfer (J. Iwahara et al., unpublished results).
Small-angle X-ray scattering analysis indicates flexibility of NEIL1's C-terminal region
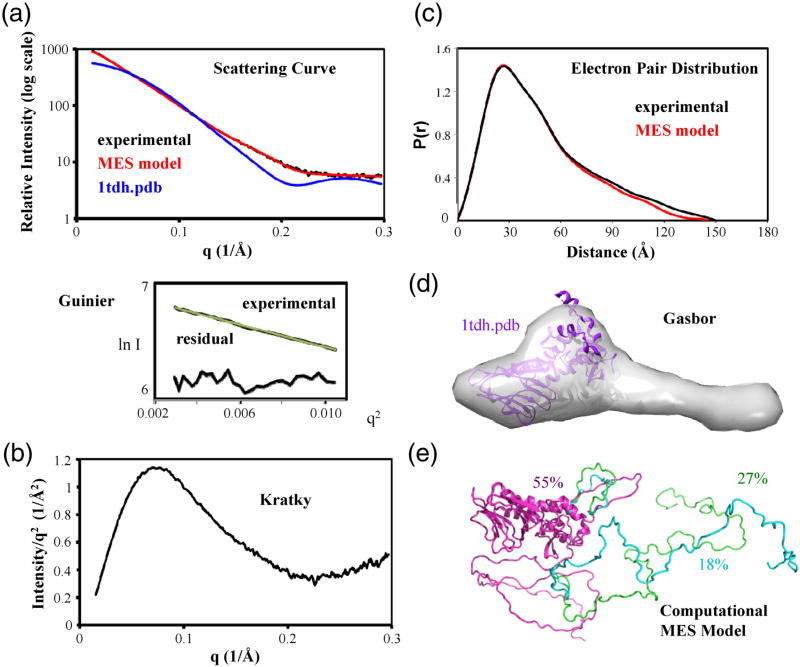
Small-angle X-ray scattering (SAXS) analysis of full-length NEIL1, combined with its crystal structure restraints and conformational analyses (Fig. 7), provided evidence for flexible conformation and ensembles of NEIL1 in solution. SAXS is a suitable technique for studying flexible proteins.22–26 The scattering curve of native NEIL1 was distinctly different from the crystal structure of the folded domain. The linearity of the Guinier plots confirmed lack of aggregation of NEIL1 samples. The parabolic curve and upward tail in the Kratky analysis, the tail at longer distances in the electron pair distribution or P(r) plot, and the ab initio shape prediction are all consistent with a folded domain and an unfolded tail. The radius of gyration, Rg, independent of sample concentration to provide an important constraint for possible solution structures,22 was 37.8 Å and 40.3 Å from the Guinier and P(r) calculations, respectively (Table 1). To objectively evaluate the SAXS results, we used the molecular dynamics CHARMM program implemented in the program BilboMD to generate ∼5000 atomic models where the catalytic NEIL1 core was fixed and the C-terminal region was allowed to move flexibly.27,28 CHARMM parameters set up in BilboMD are designed to explore conformational space. The predicted scattering curve of each BilboMD model was calculated using the program FOXS and compared to the experimental scattering curve.29 In a Minimal Ensemble Search (MES),27,30 random combinations of three predicted scattering curves were then compared to find the ensemble with the best fit to the scattering curve, as indicated by the Χ2 fit. Addition of the C-terminus improved the Χ2 fit from 15 for the catalytic core to 1.69 for the best single BilboMD model. The fit further improved to 1.07 for the ensemble. Although the conformations of NEIL1 are obviously not limited to three in solution, the improved fit of the ensemble to the experimental scattering data supports our conclusion that any single conformation cannot explain the experimental scattering data and that multiple conformations exist in solution.
Fig. 7.
SAXS analysis of WT NEIL1 is consistent with the structure of its catalytic core connected to a disordered C-terminus. (a) The SAXS scattering curve shows that the experimental data of the full-length protein do not match the scattering data calculated from the catalytic core crystal structure but match the scattering predicted from an ensemble of MES models of the catalytic core with a disordered C-terminus. The Guinier plot and residuals of the experimental data (below) indicate monodispersity of the sample. (b) The Kratky plot shows a parabolic curve with a rise in the curve at higher q, consistent with the presence of an ordered core and an additional disordered region. (c) The P(r) plot shows a bilobal shape with an extension, consistent with the crystal structure and an extended tail. The calculated P(r) of the MES models is shown for comparison to the experimental data. (d) The ab initio shape predicted from the experimental data shows not only a compact volume consistent with the crystal structure but also a volume extending from the C-terminal portion of the protein. (e) BilboMD models and their percentage representation in the population that were selected by the MES fit the experimental scattering data as an ensemble. The calculated scattering and P(r) from these MES models are shown in (a) and (c).
Table 1. SAXS data collection parameters.
| SAXS data collection parameters | |
| Beamline | 12.3.1 Advanced Light Source, Lawrence Berkeley National Laboratory |
| Wavelength (Å) | 1.12 |
| Q range (Å−1) | 0.0154–0.29699 |
| Exposure time (s) | 7 |
| Concentration (mg/ml) | 10 |
| Temperature (°C) | 4 |
| Structural parameters | |
| I(0) from P(r) (arbitrary units) | 1029 |
| Rg (Å) from P(r) | 40.3 |
| I(0) from Guinier (arbitrary units) | 1017 |
| Rg (Å) from Guinier | 37.8 |
| Dmax (Å) | 150 |
Discussion
Structures of BER proteins have proven particularly useful for characterizing structural elements associated with specific activities31 and general mechanisms.32 As structure determination is often precluded for proteins with flexible regions, such as NEIL1, SAXS and computational analysis together with other biophysical methods have provided important information about structure–function relationships including interactions.23,33 Human NEIL1 interacts binarily with most downstream repair proteins, including Polβ, Liglllα, and XRCC1, which act in the single nucleotide incorporation (SN)-BER pathway.7,13 NEIL1 also interacts with the replication-associated long-patch (LP)-BER proteins including flap endonuclease 1 (FEN-1),9 PCNA,10 and RPA.11 Furthermore, NEIL1 stably interacts with several noncanonical repair proteins, for example, Werner protein (WRN), a RecQ helicase,12 and hnRNP-U.16 These multiple interactions have been suggested to play various roles, including stabilizing the interacting proteins and selecting repair sub-pathways.13 Surprisingly, all of these interactions involve NEIL1's unstructured CTD that includes a 38-residue common interaction interface. This CTD is absent in NEIL1's prototype Nei in Escherichia coli and is likely to have been added during mammalian evolution, presumably to cope with higher complexity and regulation in mammalian BER relative to E. coli.1,13,14 However, how NEIL1 interacts with at least 20 partner proteins by using a short common interaction segment is not understood. Furthermore, no structural information was available for this C-terminal extension, which needed to be removed for its crystallization.15
The distinct fluorescence spectra of WT NEIL1 versus its deletion mutants suggested tight association between the core domain and CTD and a strong influence of the C-terminal region on NEIL1's structure. Based on biophysical, biochemical, and SAXS analysis of native NEIL1 and its mutants, we propose that NEIL1's C-terminal segment folds back to interact with the core domain in the native protein in the absence of its partners. The intramolecular fold-back was also supported by our preliminary MD simulation analysis (N.O., unpublished results). The intramolecular interaction between the basic amino acid cluster near the C-terminus and the acidic cluster in the core domain significantly enhances the stability of free NEIL1. This fold-back was partially disrupted in the presence of a DNA oligo as indicated by the change in its intrinsic fluorescence (Fig. 6b). The intramolecular interaction in native NEIL1 may regulate its half-life in vivo and prevent the CTD from nonspecific interactions, which could lead to aggregation, and susceptibility to proteolysis. Similar fold-back interaction of the N-terminal region may regulate interactions of tyrosyl-DNA phosphodiesterase 2.34
The flexibility of the C-terminal segment, suggested by SAXS analysis, may also be critical for NEIL1's binding to multiple partners. Our recent studies have demonstrated the role of NEIL1's C-terminal region in its multiple interactions with other repair and noncanonical proteins and its functional role in repair.9–14,16 While it is intriguing how a short, ∼38-residue segment in NEIL1's C-terminus provides binding interface for at least 20 partner proteins with high affinity and specificity, we predict that the flexibility of the C-terminus allows specific induced-fit conformation with each partner and thus facilitates its multiple interactions. However, further investigations are needed to experimentally establish this. SAXS has revealed important functional roles for unstructured regions in other DNA repair proteins, such as NBS1 where the N-terminal domains are folded but the CTD is unstructured but required for binding to its nuclease partner Mre11.35 The ability of the CTD to exist in multiple states with subtle change in conformation could help lock in distinct conformations specific for each partner. FEN-1 evidently employs analogous local and substrate-specific folding of its active-site gateway and cap regions to aid specific binding of DNA flaps.36 As seen for FEN-1, the initial affinity for an interacting partner may induce a subtle conformational change in NEIL1 required to stabilize the interaction. The propensity of the C-terminal segment to fold in the presence of osmolytes supports this scenario.
Intrinsic disorder and allosteric coupling
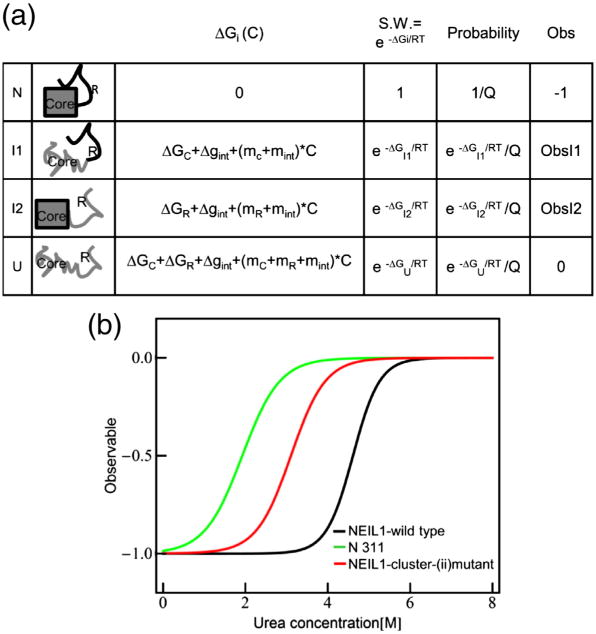
Intrinsically disordered proteins have emerged as an important class of polypeptides37–42 that adopt multiple structures after interaction with various ligands, suggesting that coupled folding and binding facilitate biological functions ranging from catalysis to signal transduction.43,44 By exploiting region-specific order-to-disorder transitions, as observed for the CTD of NEIL1, proteins with multiple partners could fine-tune the affinity for the ligand. In the case of NEIL1, the coupling between the core and the C-terminal amino acids [named as the core and the C-terminal region (R)] is schematically illustrated in Fig. 8a. Each segment is depicted to have intrinsic stability (ΔGR and ΔGC); the coupling free energy (Δgint) describes the quantitative influence of the unfolding of one domain on the stability of the other. In principle43 and in practice, this coupling energy can be positive45 or negative.46 For NEIL1, the fact that the stability of the full-length protein is higher than that of the truncated variants indicates positive coupling energy. This is further evident when comparing the simulated unfolding curve of a two-domain protein (Fig. 8b) with the experimental data for NEIL1 (Fig. 3a). For all scenarios wherein the R region is disordered (ΔGR < 0) and the core is folded (ΔGC > 0), destabilization of the protein through truncation or mutation (Fig. 8b) can only be facilitated with when Δgint > 0. Although it is clear that the intrinsically disordered C-terminal segment stabilizes NEIL1 and protects it from both thermal and chemical inactivation, what role this coupling plays in controlling NEIL1 function is at present unknown. However, the fact that the equilibrium appears poised at a point where both structured and unstructured states exist in measurable quantities suggests that these states are functionally relevant.43
Fig. 8.
(a) Schematic representation of the ensemble model of allosteric regulation in native NEIL1 containing the core domain and the regulatory region (R). Each region can be in native state or fully unfolded, resulting in four possible states (i.e., N, I1,I2, and U). ΔGi (C) is the free energy of each state in the presence of urea. ΔGC and ΔGR are the free energies of the fully unfolded state of the core domain and R region in the absence of urea, with the native state as reference. Δgint is the interaction energy between the coupled core domain and R region in absence of urea. mC, mR, and mint are the slopes of the free-energy dependence on urea concentration for the core domain, R region, and interaction interface, respectively. S.W. is the statistical weight of each state. Q is the partition function, which is the sum of the statistical weights of all the states in the ensemble. Obs is the observable for each state used in the simulation. (b) Simulated urea unfolding curve for NEIL1 WT (black line) composed of the core and regulatory region, N311 composed of only the core domain, and NEIL1-cluster(ii) mutant composed of the core domain and mutated regulatory region. The parameters used in generating the simulated curve are as follows: ΔGC = 2.5 kcal/mol, ΔGR = 1 kcal/mol, Δgint (WT) = 6 kcal/mol, Δgint [cluster(ii) mutant] = 2 kcal/mol, mC = −1.3 kcal/mol−1 K−1, mR = −0.1 kcal/mol−1 K−1, mint(WT) =−0.5 kcal/mol−1 K−1, mint[cluster(ii) mutant] =−0.1 kcal/mol−1 K−1, Obs1 =0, Obs2 = 0.8.
Intrinsically unstructured regions and electrostatic forces are known to be generally important for molecular interactions.40,47 Flexibility appears to favor sequence-based recognition, consistent with the NEIL1 CTD, as observed from the recognition of intact proteins by anti-peptide antibodies.48 Furthermore, unstructured regions present in FEN1 and DNA ligase are involved in their interaction with the PCNA partner.49,50 An unstructured motif promotes Rad51 assembly into multimers.51 Furthermore, electrostatic interactions could provide substrate guidance,52 orient proteins for functional interactions,53 and regulate DNA repair proteins by DNA mimicry.54 However, the role of electrostatic interaction in promoting stability of an intrinsically unstructured region as observed for NEIL1 is unprecedented, to the best of our knowledge. Thus, this study provides significant insight into the dynamic nature of NEIL1's intrinsically disordered CTD, which may be important for functional and structural roles in multiprotein interactions in general.13,14
Materials and Methods
Expression and purification of NEIL1 and its mutant polypeptides
Recombinant untagged WT NEIL1 and its truncated polypeptides 1–349 and 1–288 were purified to homogeneity from E. coli BL21 RIPL-enriched cells harboring corresponding expression plasmids.3,9 The 1–311 mutant clone of NEIL1 was generated by introducing stop codons after the 311th amino acid position in a NEIL1 expression plasmid (pET22b)3 using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) and purified as previously described.13 The glutathione S-transferase (GST)-fused NEIL1 CTDs 312–389, 312–349, and 350–389 were expressed in E. coli and purified from the cell extracts via glutathione-agarose affinity chromatography and eluting the bound proteins with 10 mM reduced glutathione.9,16 The proteins were digested with thrombin to cleave the GST tag, followed by removal of GST by chromatography on SP-Sepharose. The purity of the protein preparations was confirmed by SDS-PAGE analysis.
The clustered point mutant constructs for NEIL1, K296A/K297A/K298A, and K356A/K357A/K360A were generated using the QuikChange Site-Directed Mutagenesis Kit as above. Mutagenesis reactions were performed using a pET16(a) vector harboring NEIL1 cDNA, using appropriate primers. Mutations were confirmed by direct sequencing. After expression in E. coli transfected with the plasmid, the recombinant untagged proteins were purified according to the protocol used for WT NEIL1.
Fluorescence spectroscopy
The intrinsic fluorescence of NEIL1 and its mutants was measured in a SPEX Fluoromax spectrofluorimeter (Horiba Jobin Yvon). The protein solutions in 10 mM phosphate-buffered saline (PBS) buffer (pH 7.5) were excited at 280 nm at 25 °C, and emission was monitored at 300– 450 nm. The average spectrum was obtained from triplicate measurements.
CD spectroscopy
The CD spectra (195–260 nm) of the proteins (2 μM) in 10 mM PBS buffer (pH 7.5) in the absence or presence of osmolytes TMAO or sarcosine were taken in an AVIV CD spectrometer model 215 using a 1-mm cuvette.55 Four repetitions of the CD spectra were used for averaging and then corrected for the contribution from the buffer alone. The osmolytes TMAO and sarcosine were dissolved in pure water at 6 M, treated with granulated activated carbon, and filtered with 0.2 μM filter. The final concentration of the osmolyte solutions was calculated from the refractive index.19
Protein denaturation studies
Urea (>99.9% purity, Sigma) was dissolved in 10 mM PBS (pH 7.5) to 8 M, treated with activated charcoal, and the final concentration of the filtered solution was calculated from the refractive index.19 To analyze the effect of urea on NEIL1's folding, we diluted the protein to 2 μM into both urea and plain PBS buffer. The two solutions were then mixed at various proportions to yield increasing urea concentrations while maintaining the 2-μM protein concentration and used for CD measurements at 222 nm using time scan (1 min).
DG/AP lyase assay
To produce radiolabeled substrate, we labeled the singlestranded 5-OHU-containing deoxyoligonucleotide sequence 5′-GCTTAGCTTGGAATCGTATCATGTA(50HU) ACTCGTGTGCCGTGTAGACCGTGCC-3′ at the 5′-terminus with [Υ-32P]-ATP using T4-PNK (New England Biolabs). The labeled substrate was then separated from unincorporated radioactivity by chromatography on Sephadex G25. The strand cleavage of substrate DNA by NEIL1 was analyzed after incubation of 5′ 32P-5-OHU-containing substrate at the indicated temperatures for 5 min in a 10-μl reaction mixture containing 40 mM Hepes–KOH, pH 7.5, 50 mM KCI, 1 mM MgCI2, 100 μg/ml bovine serum albumin, and 5% glycerol. The reaction was stopped with the formamide dye mix (80% formamide, 20 mM NaOH, 20 mM ethylenediaminetetraacetic acid, 0.05% bromophenol blue, and 0.05% xylene cyanol). The products were separated by denaturing gel electrophoresis in 20% polyacrylamide containing 8 M urea, in 1× Tris/borate–ethylenediaminetetraacetic acid buffer, pH 8.4.9 The radioactivity was quantitated in a Phosphorlmager using Image Quant software (Amersham Biosciences).
His-affinity co-elution analysis
His-affinity pull-down assays were carried out as previously described.9,10 Briefly, NEIL1's C-terminal peptides 312–349 and 349–389 (untagged, 20 pmol) were mixed with His-tagged WT NEIL1, or its N-terminal polypeptide 1–311 (40 pmol), prebound with His-select magnetic nickel beads (Sigma) and incubated for 1 h at 4 °C with constant rotation in a buffer containing Trisbuffered saline, 5% bovine serum albumin, and 10% glycerol. After washing the beads with Tris-buffered saline containing 0.1% Triton X-100 and 400 mM NaCI, the bound proteins were eluted with SDS sample dye and tested for the presence of the C-terminal peptides by immunoblotting.
SAXS analysis
SAXS data for NEIL1 were collected at the SIBYLS 12.3.1 beamline at the Advanced Light Source, Lawrence Berkeley National Laboratory.26,56 Scattering measurements were performed on 20 μl of NEIL1 at 10 mg/ml dialyzed in PBS buffer and 1 mM DTT at 4 °C, 1.5 m from the Mar165 detector. Sequential exposures (7, 7, 35, and 7 s) were taken at 11,111 eV. Data from the 10-mg/ml solution were consistent with those for NEIL1 purified by gel filtration, indicating a lack of aggregation in the concentrated sample. No radiation-induced aggregation was observed, and the first and third exposures were merged. Data were analyzed using the ATSAS program suite57 and BilboMD.27 BilboMD models were generated with Rg between 36 and 44 Å. The envelopes and overlaid pdb were done in Chimera58 and BilboMD models were done in PyMOL (Shrodinger).
Acknowledgments
The research was supported by U.S. Public Health Service grants R01 CA81063, CA158910 (S.M.), and P01 CA92854 (J.A.T. and S.M.); R01 GM046312 (J.A.T.) and R01 GM 63747 (V.J.H.); University of Texas Medical Branch National Institute of Environmental Health Sciences Center and pilot grants P30 ES006676 (S.M. and M.L.H.); and Alzheimer's Association grant NIRG-12-242135 (M.L.H.). SAXS data were collected at the SIBYLS beamline 12.3.1 (Advanced Light Source, IDAT, Contract DE-AC02-05CH11231). CD and fluorescence experiments were performed at the biophysical core facility at the University of Texas Medical Branch. We thank Dr. David Konkel for carefully editing the manuscript.
Abbreviations used
- BER
base excision repair
- SAXS
small angle X-ray scattering
- NEIL1
Nei (endonuclease VIII)-like protein 1
- WT
wild type
- TMAO
trimethylamine-N-oxide
- CTD
C-terminal domain
- DG
DNA glycosylase
- MES
Minimal Ensemble Search
- FEN-1
flap endonuclease 1
- GST
glutathione S-transferase
- PBS
phosphate-buffered saline
References
- 1.Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedberg EC, Aguilera A, Gellert M, Hanawalt PC, Hays JB, Lehmann AR, et al. DNA repair: from molecular mechanism to human disease. DNA Repair (Amst) 2006;5:986–996. doi: 10.1016/j.dnarep.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, et al. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc Natl Acad Sci USA. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair (Amst) 2002;1:517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- 5.Hazra TK, Kow YW, Hatahet Z, Imhoff B, Boldogh I, Mokkapati SK, et al. Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J Biol Chem. 2002;277:30417–30420. doi: 10.1074/jbc.C200355200. [DOI] [PubMed] [Google Scholar]
- 6.Liu M, Bandaru V, Bond JP, Jaruga P, Zhao X, Christov PP, et al. The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. Proc Natl Acad Sci USA. 2010;107:4925–4930. doi: 10.1073/pnas.0908307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, et al. AP endonuclease-independent DNA base excision repair in human cells. Mol Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 8.Dou H, Mitra S, Hazra TK. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J Biol Chem. 2003;278:49679–9684. doi: 10.1074/jbc.M308658200. [DOI] [PubMed] [Google Scholar]
- 9.Hegde ML, Theriot CA, Das A, Hegde PM, Guo Z, Gary RK, et al. Physical and functional interaction between human oxidized basespecific DNA glycosylase NEIL1 and flap endonuclease 1. J Biol Chem. 2008;283:27028–27037. doi: 10.1074/jbc.M802712200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dou H, Theriot CA, Das A, Hegde ML, Matsumoto Y, Boldogh I, et al. Interaction of the human DNA glycosylase NEIL1 with proliferating cell nuclear antigen. The potential for replication-associated repair of oxidized bases in mammalian genomes J Biol Chem. 2008;283:3130–3140. doi: 10.1074/jbc.M709186200. [DOI] [PubMed] [Google Scholar]
- 11.Theriot CA, Hegde ML, Hazra TK, Mitra S. RPA physically interacts with the human DNA glycosylase NEIL1 to regulate excision of oxidative DNA base damage in primer-template structures. DNA Repair (Amst) 2010;9:643–652. doi: 10.1016/j.dnarep.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das A, Boldogh I, Lee JW, Harrigan JA, Hegde ML, Piotrowski J, et al. The human Werner syndrome protein stimulates repair of oxidative DNA base damage by the DNA glycosylase NEIL1. J Biol Chem. 2007;282:26591–26602. doi: 10.1074/jbc.M703343200. [DOI] [PubMed] [Google Scholar]
- 13.Hegde ML, Hegde PM, Arijit D, Boldogh I, Mitra S. Human DNA glycosylase NEIL1's interactions with downstream repair proteins is critical for efficient repair of oxidized DNA base damage and enhanced cell survival. Biomolecules. 2012;2:564–578. doi: 10.3390/biom2040564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hegde ML, Hazra TK, Mitra S. Functions of disordered regions in mammalian early base excision repair proteins. Cell Mol Life Sci. 2010;67:3573–3587. doi: 10.1007/s00018-010-0485-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doublie S, Bandaru V, Bond JP, Wallace SS. The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zincless finger motif required for glycosylase activity. Proc Natl Acad Sci USA. 2004;101:10284–10289. doi: 10.1073/pnas.0402051101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hegde ML, Banerjee S, Hegde PM, Bellot LA, Hazra TK, Boldogh I, Mitra S. Enhancement of NEIL1-initiated oxidized DNA base excision repair by heterogeneous nuclear ribonucleoprotein U (hnRNP-U) via direct interaction. J Biol Chem. 2012;287:34202–34211. doi: 10.1074/jbc.M112.384032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, Bolen DW. The peptide backbone plays a dominant role in protein stabilization by naturally occurring osmolytes. Biochemistry. 1995;34:12884–12891. doi: 10.1021/bi00039a051. [DOI] [PubMed] [Google Scholar]
- 18.Hegde ML, Rao KS. DNA induces folding in alpha-synuclein: understanding the mechanism using chaperone property of osmolytes. Arch Bio-chem Biophys. 2007;464:57–69. doi: 10.1016/j.abb.2007.03.042. [DOI] [PubMed] [Google Scholar]
- 19.Holthauzen LM, Auton M, Sinev M, Rosgen J. Protein stability in the presence of cosolutes. Methods Enzymol. 2011;492:61–125. doi: 10.1016/B978-0-12-381268-1.00015-X. [DOI] [PubMed] [Google Scholar]
- 20.Lumry R, Biltonen R. Validity of the “two-state” hypothesis for conformational transitions of proteins. Biopolymers. 1966;4:917–944. doi: 10.1002/bip.1966.360040808. [DOI] [PubMed] [Google Scholar]
- 21.Zoltowski BD, Vaidya AT, Top D, Widom J, Young MW, Crane BR. Structure of full-length Drosophila cryptochrome. Nature. 2011;480:396–399. doi: 10.1038/nature10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rambo RP, Tainer JA. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod–Debye law. Biopolymers. 2011;95:559–571. doi: 10.1002/bip.21638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rambo RP, Tainer JA. Bridging the solution divide: comprehensive structural analyses of dynamic RNA, DNA, and protein assemblies by small-angle X-ray scattering. Curr Opin Struct Biol. 2010;20:128–137. doi: 10.1016/j.sbi.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 25.Tsutakawa SE, Hura GL, Frankel KA, Cooper PK, Tainer JA. Structural analysis of flexible proteins in solution by small angle X-ray scattering combined with crystallography. J Struct Biol. 2007;158:214–223. doi: 10.1016/j.jsb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 26.Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL, 2nd, Tsutakawa SE, et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat Methods. 2009;6:606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pelikan M, Hura GL, Hammel M. Structure and flexibility within proteins as identified through small angle X-ray scattering. Gen Physiol Biophys. 2009;28:174–189. doi: 10.4149/gpb_2009_02_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammel M, Yu Y, Mahaney BL, Cai B, Ye R, Phipps BM, et al. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J Biol Chem. 2012;285:1414–1423. doi: 10.1074/jbc.M109.065615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneidman-Duhovny D, Hammel M, Sali A. FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res. 2010;38:W540–W544. doi: 10.1093/nar/gkq461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hilser VJ. Biochemistry. An ensemble view of allostery Science. 2010;327:653–654. doi: 10.1126/science.1186121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hitomi K, Iwai S, Tainer JA. The intricate structural chemistry of base excision repair machinery: implications for DNA damage recognition, removal, and repair. DNA Repair (Amst) 2007;6:410–28. doi: 10.1016/j.dnarep.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 32.Huffman JL, Sundheim O, Tainer JA. DNA base damage recognition and removal: new twists and grooves. Mutat Res. 2005;577:55–76. doi: 10.1016/j.mrfmmm.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 33.Perry JJ, Cotner-Gohara E, Ellenberger T, Tainer JA. Structural dynamics in DNA damage signaling and repair. Curr Opin Struct Biol. 2010;20:283–294. doi: 10.1016/j.sbi.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi K, Kurahashi K, Gao R, Tsutakawa SE, Tainer JA, Pommier Y, Aihara H. Structural basis for recognition of 5′-phosphotyrosine adducts by Tdp2. Nat Struct Mol Biol. 2012;19:1372–1377. doi: 10.1038/nsmb.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams RS, Dodson GE, Limbo O, Yamada Y, Williams JS, Guenther G, et al. Nbs1 flexibly tethers Ctp1 and Mre11–Rad50 to coordinate DNA double-strand break processing and repair. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsutakawa SE, Classen S, Chapados BR, Arvai AS, Finger LD, Guenther G, et al. Human flap endonuclease structures, DNA doublebase flipping, and a unified understanding of the FEN1 superfamily. Cell. 2011;145:198–211. doi: 10.1016/j.cell.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tantos A, Han KH, Tompa P. Intrinsic disorder in cell signaling and gene transcription. Mol Cell Endocrinol. 2012;348:457–465. doi: 10.1016/j.mce.2011.07.015. [DOI] [PubMed] [Google Scholar]
- 38.Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK. Intrinsic disorder in transcription factors. Biochemistry. 2006;45:6873–6888. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 40.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 41.Uversky VN. Intrinsically disordered proteins from A to Z. Int J Biochem Cell Biol. 2011;43:1090–1103. doi: 10.1016/j.biocel.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 42.Hilser VJ, Wrabl JO, Motlagh HN. Structural and energetic basis of allostery. Annu Rev Biophys. 2012;41:585–609. doi: 10.1146/annurev-biophys-050511-102319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hilser VJ, Thompson EB. Intrinsic disorder as a mechanism to optimize allosteric coupling in proteins. Proc Natl Acad Sci USA. 2007;104:8311–8315. doi: 10.1073/pnas.0700329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 45.Schrank TP, Bolen DW, Hilser VJ. Rational modulation of conformational fluctuations in adenylate kinase reveals a local unfolding mechanism for allostery and functional adaptation in proteins. Proc Natl Acad Sci USA. 2009;106:16984–16989. doi: 10.1073/pnas.0906510106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J, Motlagh HN, Chakuroff C, Thompson EB, Hilser VJ. Thermodynamic dissection of the intrinsically disordered N-terminal domain of human glucocorticoid receptor. J Biol Chem. 2012;287:26777–26787. doi: 10.1074/jbc.M112.355651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang C, Iwahara J, Clore GM. Visualization of transient encounter complexes in protein–protein association. Nature. 2006;444:383–386. doi: 10.1038/nature05201. [DOI] [PubMed] [Google Scholar]
- 48.Tainer JA, Getzoff ED, Alexander H, Houghten RA, Olson AJ, Lerner RA, Hendrickson WA. The reactivity of anti-peptide antibodies is a function of the atomic mobility of sites in a protein. Nature. 1984;312:127–134. doi: 10.1038/312127a0. [DOI] [PubMed] [Google Scholar]
- 49.Hosfield DJ, Mol CD, Shen B, Tainer JA. Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity. Cell. 1998;95:135–146. doi: 10.1016/s0092-8674(00)81789-4. [DOI] [PubMed] [Google Scholar]
- 50.Pascal JM, Tsodikov OV, Hura GL, Song W, Cotner EA, Classen S, et al. A flexible interface between DNA ligase and PCNA supports conformational switching and efficient ligation of DNA. Mol Cell. 2006;24:279–291. doi: 10.1016/j.molcel.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 51.Shin DS, Pellegrini L, Daniels DS, Yelent B, Craig L, Bates D, et al. Full-length archaeal Rad51 structure and mutants: mechanisms for RAD51 assembly and control by BRCA2. EMBO J. 2003;22:4566–4576. doi: 10.1093/emboj/cdg429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Getzoff ED, Tainer JA, Weiner PK, Kollman PA, Richardson JS, Richardson DC. Electrostatic recognition between superoxide and copper, zinc superoxide dismutase. Nature. 1983;306:287–290. doi: 10.1038/306287a0. [DOI] [PubMed] [Google Scholar]
- 53.Roberts VA, Freeman HC, Olson AJ, Tainer JA, Getzoff ED. Electrostatic orientation of the electron-transfer complex between plastocyanin and cytochrome c. J Biol Chem. 1991;266:13431–13441. [PubMed] [Google Scholar]
- 54.Putnam CD, Shroyer MJ, Lundquist AJ, Mol CD, Arvai AS, Mosbaugh DW, Tainer JA. Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coll uracil-DNA glycosylase. J Mol Biol. 1999;287:331–346. doi: 10.1006/jmbi.1999.2605. [DOI] [PubMed] [Google Scholar]
- 55.Hegde ML, Hegde PM, Holthauzen LM, Hazra TK, Rao KS, Mitra S. Specific inhibition of NEIL-initiated repair of oxidized base damage in human genome by copper and iron: potential etiological linkage to neurodegenerative diseases. J Biol Chem. 2010;285:28812–28825. doi: 10.1074/jbc.M110.126664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Classen S, Rodic I, Holton J, Hura GL, Hammel M, Tainer JA. Software for the high-throughput collection of SAXS data using an enhanced Blu-lce/DCS control system. J Synchrotron Radial. 2010;17:774–781. doi: 10.1107/S0909049510028566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Petoukhov MV, Konarev PV, Kikhney AG, Svergun DI. ATSAS 2.1—towards automated and web-supported small-angle scattering data analysis. J Appl Crystallogr. 2007;40:s223–s228. [Google Scholar]
- 58.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]