Abstract
Purpose
Gene promoter hypermethylation may be useful as a biomarker for cancer risk in histopathologically benign prostate specimens.
Materials and Methods
We performed a nested case-control study of gene promoter methylation status for 5 genes (APC, RARB, CCND2, RASSF1 and MGMT) measured in benign biopsy specimens from 511 prostate cancer case-control pairs. We estimated the overall and race stratified risk of subsequent prostate cancer associated with methylation status.
Results
On race stratified analysis RARB methylation was associated with a higher cancer risk in black American men (OR 2.18, 95% CI 1.39–3.44). APC methylation was associated with an increased risk of high grade tumors (OR 2.43, 95% CI 1.20–4.90), which was higher in black than in white men (OR 3.21 vs 2.04). In cases RARB and APC gene methylation in benign prostate samples persisted in matched malignant specimens. In black cases the combined risk associated with RARB and APC methylation (OR 3.04, 95% CI 1.44–6.42) was greater than the individual risk of each gene and significantly different from that in white cases (OR 1.14, 95% CI 0.56–2.30).
Conclusions
RARB gene methylation in histopathologically benign prostate samples was associated with a statistically significant increased risk of subsequent prostate cancer in black men. Methylation data on additional genes may improve risk stratification and clinical decision making algorithms for cancer screening and diagnosis.
Keywords: prostate, prostatic neoplasms, risk, DNA methylation, African Americans
Prostate carcinogenesis is associated with the silencing of tumor suppressor genes via promoter hypermethylation.1–3 For example, GSTP1 is almost universally methylated in prostate cancer and considered a highly specific marker for cancer in prostate tissue.4,5 Other genes are methylated less frequently but show signs of hypermethylation earlier in prostate carcinogenesis, making them potential markers of early cancer detection.3
Gene promoter methylation can also occur in prostate nonneoplastic cells before malignant transformation.6 For instance, RARB methylation in histopathologically negative tissue predicted prostate cancer in followup biopsies done within 24 months with 84% sensitivity.7 In a study of prostatectomy specimens RARB was hypermethylated in 72% of benign prostate adjacent to tumor.8 Similarly, in ex vivo core biopsies taken 1 to 4 mm from prostate cancer foci 4 of 11 specimens showed methylation in 1 or more of the tumor suppressor genes APC, RARB and RASSF1.9 Methylation of these genes in the preneoplastic state is consistent with a field defect that presages cancer in the human prostate and suggests the possibility of a biomarker for early detection before overt carcinogen-esis.10,11
Because DNA methylation is a common, consistent event that may develop early in carcinogenesis, it is an attractive biomarker for prostate cancer detection. Specific methylation events have also been linked to disease outcomes,12,13 suggesting that methylation biomarkers can be used to target treatment interventions toward the cases that would most benefit. A methylation based risk profile would also benefit the majority of men with negative prostate biopsies who are still considered at high risk due to increased PSA.14 Since many of these men have clinically indolent disease, aggressive clinical intervention would only decrease quality of life and not disease related mortality.15
Black American men experience a significantly higher incidence of prostate cancer and a more aggressive prostate cancer presentation.16 They are more likely to die of the disease17 and may have a higher incidence of cancer after negative biopsy than white men.18 Studies suggest that gene promoter methylation patterns in the prostate may differ by race.19,20 Characterizing the tumor suppressor genes inactivated by hypermethylation and the timeline of when such changes occur may improve early prostate cancer detection and targeted disease management, which could have a significant impact on these racial disparities.
To determine the significance of aberrant gene promoter methylation in early prostate carcinogenesis, we performed a matched case-control study nested in a large historical cohort of men with benign prostate tissue specimens to test whether gene promoter methylation in benign tissue is associated with incident prostate cancer and tumor aggressiveness, and whether these associations are race specific.
Methods
Study Sample
After obtaining approval from the Henry Ford Health System institutional review board, we identified a historical cohort of 6,692 men with a benign prostate specimen collected by needle core biopsy or TURP between January 1990 and December 2002. A nested case-control sample was drawn from this cohort based on study eligibility criteria, including PSA recorded within a year of cohort entry and no history of a prostate cancer diagnosis. Cohort entry date was defined as the date that the initial benign prostate specimen was harvested. Case diagnosis date was the date of the first cancer positive tissue specimen or the date that a clinician first reported a clinical diagnosis of prostate cancer. Patients diagnosed with prostate cancer less than 1 year from the cohort entry date were ineligible for study. We identified 808 potentially eligible cases diagnosed with prostate cancer before July 2007.
We used incidence density sampling to select controls with replacement from all cohort members at risk at the time of case occurrence. The reference date for controls was equal to the time between the case enrollment and diagnosis dates added to the control enrollment date. Matching criteria included age at cohort entry (±2 years), cohort entry date (±2 years), race (black or white) and specimen type (biopsy or TURP). We matched 802 of 808 potentially eligible cases. Further review decreased the final analytical sample to 574 case-control pairs.21 In the current study we ran methylation assays on the first 526 pairs enrolled and measured methylation levels for at least 1 gene in 511 of these pairs.
DNA Isolation and Methylation Specific PCR
Methylation studies were performed using DNA from benign prostate tissue acquired from needle biopsies and TURP, and from tumor specimens acquired from prostatectomy tissue. Sections (5 μ) of paraffin embedded prostate tissue were placed individually on untreated, uncharged glass microscope slides, air dried and placed in a 60C oven for 45 minutes to remove excess paraffin. Heated slides were immersed in xylene overnight, washed for 3 minutes in 100%, 95% and 80% ethanol, and air dried. All specimens were processed in 50 μl microdissection buffer composed of tris, KCl, MgCl2, Tween® 20 and water, followed by overnight digestion with Proteinase K (Sigma-Aldrich®) at 55C.
Extracted DNA was bisulfite treated using the EZ DNA Methylation-Gold™ Kit to convert unmethylated cytosines into uracils, while leaving methylated cytosines unchanged. Nested methylation specific PCR was used due to its ability to amplify degraded DNA associated with formalin fixation and long-term storage in paraffin.22 In addition, stage I primers were multiplexed to maximize DNA recovered from tissue (less than 200 ng for most biopsies). Stage I primers recognize bisulfite modified templates but do not discriminate methylated from unmethylated alleles. Thus, the stage I PCR product was diluted and subjected to repeat PCR (stage II) with primers specific to a methylated template located around the transcriptional start site, where methylation (when present) strongly correlates with gene silencing. To ensure that only methylated alleles were amplified, stage II PCR reactions were performed at annealing temperatures greater than the melting temperature of stage I primers. Sensitivity was approximately 1/500 even with the nested methylation specific PCR approach due to tissue degradation from formalin fixation and storage in paraffin. The supplementary table (http://jurology.com/) lists primer sequences. Supplementary figure 1 (http://jurology.com/) shows the gene region queried.
Statistical Analysis
We performed conditional logistic regression analysis to estimate prostate cancer incidence ORs during followup. Individual matching was controlled for age, race and specimen type. Models that assessed associations between the prostate cancer incidence and gene promoter methylation for each of the 5 study genes were tested with and without adjusting for PSA and the presence of HGPIN. Stratified models were compared using a conditional logistic regression model that included interaction terms with the stratified variable. We performed sensitivity analysis of the effect of shorter followup on risk estimates to determine whether cases diagnosed soonest (those with the highest probability of misclassification at cohort entry) may have had biased results.
Results
Demographics and Clinical Characteristics
The average age of cases was 65.9 years at cohort entry and 41% of the cases were black (table 1). Approximately 50% of the cases were diagnosed between 1 and 4 years after cohort entry. The remaining cases were diagnosed between 4 and 15 years after cohort entry. Cases had significantly higher PSA at cohort entry and higher PSA velocity between the time of cohort entry and diagnosis. They underwent an average of 2 more PSA tests during this period than controls. Of the cases 40% and 52% showed stage 1 and 2 tumors, respectively, and 30% had advanced tumor grade, defined as Gleason score 8 or greater, or 7 with a grade 4 primary tumor.
Table 1. Demographic and clinical characteristics of 511 case-control pairs.
| Controls | Cases | |
|---|---|---|
| No. race (%):* | ||
| White | 300 (59) | 300 (59) |
| Black | 211 (41) | 211 (41) |
| Mean ± SD age at cohort entry* | 65.9 ± 7.5 | 65.9 ± 7.5 |
| Median cohort entry date* | 1/95 | 11/94 |
| Median followup (yrs)* | 4.0 | 4.0 |
| No. benign specimen (%): | ||
| Needle core biopsy* | 481 (94) | 481 (94) |
| TURP | 30 (6) | 30 (6) |
| Mean ± SD PSA:† | ||
| Cohort entry (ng/ml) | 5.7 ± 5.7 | 7.8 ± 7.2 |
| No. tests cohort entry-diagnosis | 6.7 ± 4.7 | 8.7 ± 5.3 |
| Velocity (ng/ml/yr) | 0.003 ± 2.1 | 7.3 ± 97.7 |
| No. tumor stage (%): | — | |
| 1 | 202 (40) | |
| 2 | 264 (52) | |
| 3 | 37 (7) | |
| 4 | 6 (1) | |
| No. Gleason grade (%): | — | |
| 6 or Less | 228 (49) | |
| 7 (3 + 4) | 97 (21) | |
| 7 (4 + 3) | 49 (10) | |
| 8–10 | 95 (20) |
Matched variables
p <0.001
Associations
Gene promoter methylation and prostate cancer
Of the 5 genes assayed increased methylation of RARB had the strongest association with prostate cancer (OR 1.76, 95% CI 1.29–2.40, table 2). Adjusting for PSA at cohort entry and the presence of HGPIN moderately decreased this association (OR 1.59, 95% CI 1.15–2.21). When stratified by race, the association between RARB methylation and prostate cancer was significant only in black men (OR 2.18, 95% CI 1.39–3.44). The other significant race specific association was the adjusted risk estimate for methylation of the MGMT gene in white men (OR 0.50, 95% CI 0.27–0.95).
Table 2. Gene promoter methylation status and prostate cancer.
| Gene (No. pairs) | % Methylated | Crude Model | Adjusted Model* | |||
|---|---|---|---|---|---|---|
|
|
|
|
||||
| Controls | Cases | OR (95% CI) | p Value | OR (95% CI) | p Value | |
| Overall: | ||||||
| MGMT (414) | 11.8 | 8.5 | 0.68 (0.43–1.08) | 0.106 | 0.69 (0.42–1.13) | 0.139 |
| APC (453) | 22.1 | 24.5 | 1.15 (0.84–1.58) | 0.378 | 1.12 (0.80–1.56) | 0.502 |
| RARB (442) | 21.9 | 32.6 | 1.76 (1.29–2.40) | <0.001 | 1.59 (1.15–2.21) | 0.005 |
| CCND2 (404) | 8.7 | 8.4 | 0.96 (0.56–1.65) | 0.891 | 0.97 (0.55–1.69) | 0.908 |
| RASSF1 (426) | 22.8 | 19.2 | 0.80 (0.57–1.12) | 0.194 | 0.82 (0.57–1.17) | 0.280 |
| Black: | ||||||
| MGMT (166) | 9.0 | 8.4 | 0.93 (0.44–1.98) | 0.847 | 1.14 (0.51–2.51) | 0.754 |
| APC (180) | 24.4 | 30.0 | 1.36 (0.83–2.21) | 0.220 | 1.37 (0.83–2.26) | 0.224 |
| RARB (184) | 25.0 | 42.4 | 2.18 (1.39–3.44) | <0.001 | 2.09 (1.30–3.36) | 0.002 |
| CCND2 (160) | 5.6 | 10.0 | 2.00 (0.81–4.95) | 0.134 | 1.85 (0.73–4.67) | 0.192 |
| RASSF1 (165) | 20.6 | 16.4 | 0.74 (0.42–1.32) | 0.309 | 0.85 (0.47–1.56) | 0.611 |
| White: | ||||||
| MGMT (248) | 13.7 | 8.5 | 0.57 (0.31–1.03) | 0.061 | 0.50 (0.27–0.95) | 0.034 |
| APC (273) | 20.5 | 20.9 | 1.02 (0.67–1.55) | 0.916 | 0.95 (0.61–1.47) | 0.804 |
| RARB (258) | 19.8 | 25.6 | 1.43 (0.93–2.20) | 0.106 | 1.24 (0.78–1.96) | 0.356 |
| CCND2 (244) | 10.7 | 7.4 | 0.60 (0.29–1.23) | 0.162 | 0.62 (0.30–1.32) | 0.215 |
| RASSF1 (261) | 24.1 | 21.1 | 0.83 (0.54–1.27) | 0.389 | 0.80 (0.51–1.26) | 0.336 |
Adjusted for PSA and HGPIN at baseline.
To determine whether the association of prostate cancer with methylation status was modified by matched time variables or disease aggressiveness, we stratified analyses by the interval between cohort entry and diagnosis or end of followup in controls as well as by age and tumor grade (table 3). When stratified by time to diagnosis, the highest ORs were observed for RARB and APC methylation in black men diagnosed 4 or more years after cohort entry. In this group the methylation of either gene increased the risk estimate more than twofold (RARB methylation OR 2.53, 95% CI 1.30–4.91 and APC methylation OR 2.45, 95% CI 1.08–5.54). When stratified by age, we observed an increased risk estimate only for APC methylation in black men 65 years old or older (OR 2.08, 95% CI 0.97–4.46). Interestingly, age stratification also revealed an inverse association for MGMT methylation in older white men (OR 0.27, 95% CI 0.09–0.82). When cases were stratified by tumor grade (low vs high), the highest risk estimates were observed in black men. APC methylation was associated with high grade tumors (OR 3.21, 95% CI 1.06–9.75), while RARB methylation was more prominently associated with an increased risk of low grade tumors (OR 2.54, 95% CI 1.37–4.74).
Table 3. Gene promoter methylation status by matching factors and case characteristics.
| Gene | Overall | Black | White | |||
|---|---|---|---|---|---|---|
|
|
|
|
||||
| OR (95% CI) | p Value | OR (95% CI) | p Value | OR (95% CI) | p Value | |
| 1–4-Yr followup: | ||||||
| MGMT | 0.66 (0.32–1.38) | 0.270 | 0.90 (0.32–2.51) | 0.836 | 0.44 (0.15–1.29) | 0.134 |
| APC | 0.85 (0.53–1.38) | 0.515 | 0.83 (0.42–1.65) | 0.597 | 0.93 (0.47–1.86) | 0.841 |
| RARB | 1.74 (1.06–2.87) | 0.029 | 1.89 (0.93–3.84) | 0.078 | 1.68 (0.83–3.41) | 0.147 |
| CCND2 | 0.77 (0.34–1.75) | 0.528 | 2.14 (0.55–8.40) | 0.275 | 0.36 (0.11–1.25) | 0.107 |
| RASSF1 | 0.72 (0.42–1.24) | 0.239 | 0.44 (0.17–1.15) | 0.093 | 0.97 (0.49–1.92) | 0.938 |
| 4–15-Yr followup: | ||||||
| MGMT | 0.71 (0.36–1.38) | 0.312 | 1.44 (0.39–5.31) | 0.580 | 0.55 (0.25–1.23) | 0.148 |
| APC | 1.47 (0.91–2.37) | 0.119 | 2.45 (1.08–5.54) | 0.032 | 1.13 (0.61–2.11) | 0.698 |
| RARB | 1.66 (1.05–2.60) | 0.029 | 2.53 (1.30–4.91) | 0.006 | 1.06 (0.56–2.03) | 0.856 |
| CCND2 | 1.19 (0.54–2.61) | 0.658 | 1.46 (0.41–5.26) | 0.562 | 1.05 (0.38–2.90) | 0.920 |
| RASSF1 | 0.90 (0.55–1.47) | 0.679 | 1.42 (0.61–3.32) | 0.414 | 0.73 (0.39–1.34) | 0.304 |
| Age less than 65: | ||||||
| MGMT | 0.82 (0.40–1.65) | 0.572 | 0.80 (0.21–2.97) | 0.736 | 0.83 (0.36–1.91) | 0.665 |
| APC | 1.02 (0.62–1.65) | 0.950 | 0.84 (0.40–1.79) | 0.658 | 1.15 (0.61–2.20) | 0.663 |
| RARB | 1.73 (1.10–2.74) | 0.018 | 2.17 (1.15–4.10) | 0.017 | 1.35 (0.69–2.62) | 0.378 |
| CCND2 | 0.46 (0.18–1.19) | 0.110 | 0.82 (0.17–4.04) | 0.807 | 0.32 (0.09–1.10) | 0.070 |
| RASSF1 | 0.88 (0.53–1.46) | 0.621 | 0.85 (0.36–2.00) | 0.713 | 0.89 (0.47–1.67) | 0.712 |
| Age 65 or greater: | ||||||
| MGMT | 0.63 (0.32–1.25) | 0.188 | 1.35 (0.50–3.69) | 0.554 | 0.27 (0.09–0.82) | 0.020 |
| APC | 1.20 (0.75–1.92) | 0.447 | 2.08 (0.97–4.46) | 0.059 | 0.82 (0.43–1.54) | 0.529 |
| RARB | 1.52 (0.93–2.47) | 0.096 | 2.15 (1.00–4.60) | 0.049 | 1.15 (0.59–2.22) | 0.680 |
| CCND2 | 1.29 (0.60–2.76) | 0.510 | 2.67 (0.71–10.01) | 0.144 | 0.83 (0.30–2.29) | 0.715 |
| RASSF1 | 0.80 (0.47–1.35) | 0.402 | 1.00 (0.41–2.44) | 0.998 | 0.73 (0.37–1.43) | 0.357 |
| Low grade tumor: | ||||||
| MGMT | 0.75 (0.43–1.30) | 0.301 | 1.51 (0.56–4.07) | 0.418 | 0.53 (0.26–1.06) | 0.074 |
| APC | 0.85 (0.58–1.26) | 0.419 | 1.05 (0.58–1.90) | 0.866 | 0.71 (0.42–1.21) | 0.210 |
| RARB | 1.63 (1.09–2.42) | 0.017 | 2.54 (1.37–4.73) | 0.003 | 1.16 (0.67–1.99) | 0.597 |
| CCND2 | 1.10 (0.56–2.13) | 0.788 | 1.55 (0.55–4.34) | 0.409 | 0.85 (0.35–2.10) | 0.730 |
| RASSF1 | 0.85 (0.56–1.28) | 0.436 | 0.91 (0.47–1.76) | 0.774 | 0.80 (0.47–1.36) | 0.412 |
| High grade tumor: | ||||||
| MGMT | 0.56 (0.21–1.54) | 0.262 | 0.65 (0.16–2.74) | 0.561 | 0.44 (0.10–1.83) | 0.258 |
| APC | 2.43 (1.20–4.90) | 0.014 | 3.21 (1.06–9.75) | 0.040 | 2.04 (0.80–5.18) | 0.133 |
| RARB | 1.55 (0.88–2.74) | 0.132 | 1.64 (0.76–3.52) | 0.207 | 1.40 (0.59–3.31) | 0.439 |
| CCND2 | 0.73 (0.25–2.09) | 0.558 | 3.87 (0.41–36.35) | 0.237 | 0.29 (0.06–1.38) | 0.118 |
| RASSF1 | 0.73 (0.35–1.50) | 0.389 | 0.67 (0.16–2.92) | 0.597 | 0.85 (0.37–1.95) | 0.707 |
*Adjusted for PSA and HGPIN at baseline.
Because cases with the shortest time to diagnosis had the highest probability of misclassification at cohort entry, we also performed sensitivity analysis to determine whether missed diagnoses at cohort entry may have biased results (supplementary fig. 2, http://jurology.com/). Pairs were removed sequentially from the sample, starting with the pair with the shortest followup. We then repeatedly re-estimated the OR to determine the effect of the earliest diagnosed cases on risk estimates. Results suggested that misclassifying controls at cohort entry would not bias results away from the null.
Clinical risk factors and gene methylation
Since HGPIN and increased PSA are independently associated with an increased prostate cancer risk, we tested for associations between these 2 factors and gene promoter methylation status (table 4). The presence of HGPIN was significantly higher in benign prostate with RARB methylation independent of race (p <0.0001). However, no other significant associations were found between HGPIN or PSA and methylation status. To determine whether HGPIN or increased PSA significantly modified the prostate cancer risk associated with methylation, we next tested a series of interaction models. We found no significant interaction of HGPIN and PSA with any of the 5 study genes.
Table 4. Gene promoter methylation status by HGPIN and PSA.
| Methylated Gene (No. pairs) | % HGPIN | Mean ± SD PSA (ng/ml) | ||
|---|---|---|---|---|
|
|
|
|||
| Pos/Neg Methylation | p Value | Pos/Neg Methylation | p Value | |
| Overall: | ||||
| MGMT (753) | 6.9/9.0 | 0.5 | 6.8 ± 5.3/6.9 ± 6.9 | 0.9 |
| APC (826) | 10.3/7.8 | 0.4 | 6.8 ± 5.6/6.8 ± 6.9 | 0.9 |
| RARB (806) | 14.0/5.3 | <0.0001 | 7.3 ± 7.2/6.7 ± 6.3 | 0.3 |
| CCND2 (772) | 8.6/8.0 | 0.9 | 6.5 ± 5.0/6.8 ± 6.5 | 0.7 |
| RASSF1 (790) | 9.8/8.5 | 0.6 | 6.2 ± 5.2/7.0 ± 6.9 | 0.09 |
| Black: | ||||
| MGMT (300) | 3.9/9.1 | 0.7 | 7.6 ± 6.0/7.5 ± 7.7 | 0.9 |
| APC (324) | 7.8/8.6 | >0.99 | 6.9 ± 4.8/7.8 ± 8.2 | 0.3 |
| RARB (322) | 13.1/4.2 | 0.005 | 8.2 ± 8.2/7.2 ± 6.7 | 0.3 |
| CCND2 (303) | 3.7/8.0 | 0.7 | 7.3 ± 5.4/7.3 ± 7.3 | >0.99 |
| RASSF1 (303) | 6.0/8.7 | 0.8 | 6.8 ± 5.8/7.9 ± 7.9 | 0.2 |
| White: | ||||
| MGMT (453) | 8.5/8.9 | >0.99 | 6.3 ± 4.8/6.4 ± 6.2 | 0.9 |
| APC (502) | 12.5/7.3 | 0.1 | 6.7 ± 6.2/6.3 ± 5.9 | 0.5 |
| RARB (484) | 14.8/6.0 | 0.0005 | 6.4 ± 6.0/6.4 ± 6.1 | 0.97 |
| CCND2 (469) | 11.6/8.0 | 0.4 | 6.1 ± 4.7/6.4 ± 5.9 | 0.7 |
| RASSF1 (487) | 11.5/8.3 | 0.4 | 6.0 ± 4.9/6.4 ± 6.0 | 0.4 |
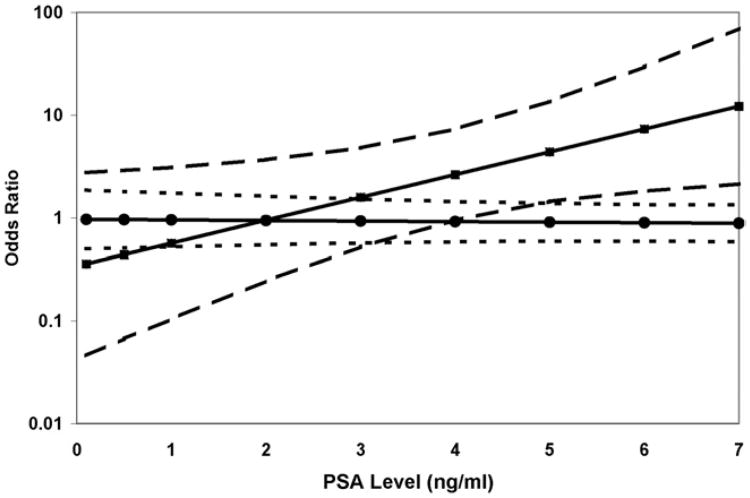
However, when the case sample was restricted to high grade tumors, the estimated OR of the interaction between PSA and APC methylation was statistically significant and consistent across race groups (p = 0.01). In a logistic regression model the OR of high grade prostate cancer associated with APC methylation increased from 0.63 (95% CI 0.19–2.16) at PSA 2 ng/ml to 3.67 (95% CI 1.50–9.01) at PSA 6 ng/ml. The risk estimate for low grade cancer was not affected by PSA level (fig. 1).
Figure 1.
Logistic regression model estimate of ORs of low (circles) and high (squares) grade prostate cancer associated with positive APC methylation status by PSA at cohort enrollment. Dashed lines represent 95% CI of risk estimates.
Gene Methylation
Joint effects of APC and RARB
Because APC and RARB were frequently methylated and associated with prostate cancer, we examined whether methylation of these genes had joint effects on prostate cancer risk estimates (table 5). In the full sample little additional cancer risk was associated with APC methylation when the RARB gene was also methylated. However, in black men APC methylation appeared to increase the risk associated with RARB methylation, particularly after adjusting for PSA and the presence of HGPIN (OR 3.04, 95% CI 1.44–6.42). This was not true in white men, in whom the joint OR of RARB and APC methylation was only slightly increased (OR 1.14), which differed significantly from that in black men (p = 0.01).
Table 5. Joint effects on prostate cancer risk of gene promoter methylation status at RARB and APC.
| Combined Gene Methylation Status | % Methylated | Crude Model | Adjusted Model* | |||
|---|---|---|---|---|---|---|
|
|
|
|
||||
| Controls | Cases | OR (95% CI) | p Value | OR (95% CI) | p Value | |
| Overall: | ||||||
| APC−/RARB− | 63.1 | 54.2 | 1 | 1 | ||
| APC+/RARB− | 14.6 | 12.4 | 0.99 (0.65–1.53) | >0.9 | 0.96 (0.61–1.49) | 0.8 |
| APC−/RARB+ | 14.1 | 20.0 | 1.69 (1.14–2.51) | 0.009 | 1.45 (0.96–2.18) | 0.08 |
| APC+/RARB+ | 8.2 | 13.4 | 1.91 (1.19–3.08) | 0.008 | 1.85 (1.12–3.05) | 0.02 |
| Black: | ||||||
| APC−/RARB− | 59.3 | 42.6 | 1 | 1 | ||
| APC+/RARB− | 16.0 | 11.7 | 1.06 (0.51–2.22) | 0.9 | 1.00 (0.47–2.12) | >0.9 |
| APC−/RARB+ | 15.4 | 25.9 | 2.19 (1.22–3.93) | 0.009 | 1.88 (1.02–3.46) | 0.04 |
| APC+/RARB+ | 9.3 | 19.8 | 2.96 (1.44–6.05) | 0.003 | 3.04 (1.44–6.42) | 0.004 |
| White: | ||||||
| APC−/RARB− | 65.7 | 62.0 | 1 | 1 | ||
| APC+/RARB− | 13.6 | 12.8 | 0.99 (0.58–1.68) | >0.9 | 0.96 (0.55–1.69) | 0.9 |
| APC−/RARB+ | 13.2 | 16.1 | 1.34 (0.77–2.33) | 0.3 | 1.19 (0.67–2.11) | 0.6 |
| APC+/RARB+ | 7.4 | 9.1 | 1.29 (0.67–2.49) | 0.4 | 1.14 (0.56–2.30) | 0.7 |
Adjusted for PSA and HGPIN
Correlation in benign and malignant tissue
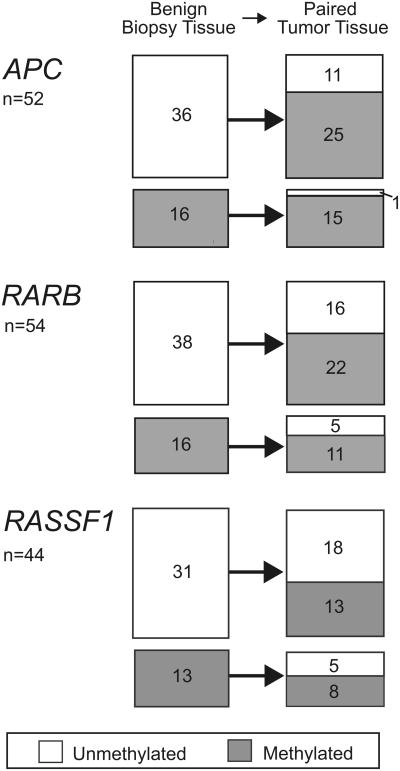
The association of RARB and APC methylation in benign tissue with incident prostate cancer suggests a generalized field defect that should be retained in the primary tumor. Therefore, in the 58 cases with available prostatectomy specimens we compared the methylation profile of all 5 genes in benign vs prostatectomy tissues (table 6). These cases were evenly distributed between black and white men. Cohort entry PSA, followup duration and the percent of methylation were comparable to those of the larger case sample. Prostatectomy cases were slightly younger (mean ± SD age 60.2 ±6.1 years) and had tumors of more advanced stage (15.4%) and grade (39.7%). As expected, the overall prevalence of methylation increased dramatically from the benign to malignant state for 4 of the 5 study genes with the percent methylation of only the MGMT gene lower in prostate cancer than in benign specimens (14.6% vs 4.2%). Figure 2 shows the change in methylation status from the benign to the malignant state in paired benign-tumor prostate specimens for the 3 genes most frequently methylated in benign prostate (APC, RARB and RASSF1). The APC gene was most likely to stay methylated in tumor when it was methylated in the benign prostate (100% to 93% methylated), or change to the methylated state when it was unmethylated in the benign prostate (0% to 70% methylated).
Table 6. Methylation status in histopathologically benign and matched prostate tumor specimens in 58 cases.
| Specimen | ||
|---|---|---|
|
|
||
| Benign | Tumor | |
| No. race (%): | ||
| White | 31 (54) | — |
| Black | 27 (47) | — |
| Cohort entry: | ||
| Mean ± SD cohort entry age | 60.2 ± 6.1 | 3.6 ± 2.0 |
| Median cohort entry date | 9/7/94 | 4/27/99 |
| Mean ± SD PSA cohort entry-diagnosis (ng/ml) | 7.7 ± 4.6 | 14.2 ± 12.9 |
| % Tumor stage: | — | |
| 2A | 26 | |
| 2B | 40 | |
| 2C | 19 | |
| 3A | 9 | |
| 3B | 5 | |
| 3C | 2 | |
| % Gleason grade: | — | |
| 6 or Less | 47 | |
| 7 (3 + 4) | 14 | |
| 7 (4 + 3) | 12 | |
| 8–10 | 28 | |
| % Pos methylation (No. pts): | ||
| MGMT (53) | 13 | 6 |
| APC (52) | 31 | 77 |
| RARB (54) | 30 | 61 |
| CCND2 (49) | 4 | 39 |
| RASSF1 (44) | 30 | 48 |
Figure 2.
Gene promoter methylation status of APC, RARB and RASSF1 genes in case paired benign and tumor specimens. In APC and RARB genes, for which biopsy tissue methylation was associated with prostate cancer risk, unmethylated benign tissue frequently became methylated in tumor tissue. Benign tissue methylation tended to predict tumor tissue methylation.
Discussion
RARB gene methylation in benign prostate biopsy tissue was associated with an increased risk of subsequent cancer. Moreover, this increased risk associated with RARB methylation alone or in the presence of APC methylation was only statistically significant in black men.
Racial differences in methylation patterns and prostate cancer were noted previously. Woodson et al found a higher frequency of CD44 methylation in black than in white men.20 Another study showed that GSTP1 methylation was more strongly related to prostate cancer in black than in white or Asian men.23 More recently, Kwabi-Addo et al observed significant differences between black and white men in the benign prostate tissue methylation levels of several genes that were highly methylated in prostate tumor tissue, including RARB.19
We found that men with RARB methylation in benign prostate were at increased risk for subsequent prostate cancer, which persisted almost 10 years after the initial benign biopsy. This increased risk was more than double in black men. This is consistent with the findings of Troyer et al, who reported that methylated RARB was the most sensitive marker for a subsequent diagnosis of prostate cancer within 24 months of an initial negative biopsy.7 However, RARB was also highly methylated in tissue from black controls. While Troyer et al implicitly assumed that cancer was present at the initial negative biopsy, we excluded cases diagnosed within a year of cohort entry to enrich for incident prostate cancer. Given the age range of our cohort and the high prevalence of undiagnosed prostate cancer in older men,24 synchronous prostate cancer was likely missed in some men in our cohort due to biopsy sampling error. However, considering the high rate of negative initial prostate biopsies among all prostate biopsies performed25 and the fact that RARB methylation likely indicates a prostate cancer field defect,8,9 our findings are generalizable to men with histopathologically benign biopsy findings.
Similar to RARB, APC methylation likely indicates a generalized prostate cancer field defect but APC is not as highly methylated in benign tissue adjacent to tumors.8,9 While APC methylation in benign prostate biopsy with HGPIN is strongly associated with a later cancer diagnosis,26 we found that HGPIN more strongly correlated with methylation in RARB than in APC. In our sample there was a slightly higher percent of HGPIN in men with APC methylation, which became more pronounced and approached statistical significance in white patients after stratifying by race (table 4). While there were similarities between the APC DNA sequences probed in in our study and that by Trock et al, they did not report the racial distribution of their sample.26 In our study APC methylation was more strongly associated with high grade tumors. This finding is consistent with several reports that APC methylation is an independent predictor of poor prostate cancer prognosis.12,13 Interestingly, in our series the association of APC methylation with high grade tumors increased with increasing PSA, suggesting that combining PSA with APC methylation status could have the potential to better stratify patient risk.
Our study has several limitations. The recovery of small amounts of DNA from biopsy specimens limited our analysis to 5 genes and DNA degradation limited our ability to amplify specific gene promoters with methylated or unmethylated stage II primers. This led to a relatively high rate of missing methylation data. Of all genes analyzed data on any 1 specific gene was missing in 7% to 16% of individuals, resulting in the removal of 11% to 21% of pairs from analysis due to the matched study design. Missing data were not related to case-control status or associated with potential confounders, such as HGPIN or PSA. Because our study was observational, cohort members were not uniformly followed after cohort entry. Cases underwent significantly more PSA tests between cohort entry and diagnosis than controls. The frequency of PSA tests in our study sample was greater than current screening recommendations even in controls but adjusting for the number of PSA tests did not substantively change our results.
Approximately 1 million prostate biopsies are performed annually in the United States, of which 700,000 are negative for cancer.14 The concept of using a measurement of epigenetic field defects to detect early stage prostate cancer is becoming more accepted as a potential clinical modality to decrease the high rate of unnecessary prostate biopsies and treatment.10 Methylation risk profiles may also assist physicians in selecting patients who would benefit most from aggressive followup, while sparing others unnecessary procedures and distress. This study is a step toward that goal. The race specific nature of our findings also highlights the need for additional molecular studies in under studied populations to better understand potential biological differences in prostate cancer progression. In conclusion, if measuring gene methylation status in prostate cancer is to have clinical usefulness, we must better understand methylation in early carcinogenesis and its implications in populations of different racial and ethnic backgrounds.
Supplementary Material
Acknowledgments
Travis Wheeler microdissected prostate specimens. Loren Muirhead assisted with early manuscript drafts.
Supported by National Institutes of Health 5R01-ES011126.
Abbreviations and Acronyms
- HGPIN
high grade prostatic intraepithelial neoplasia
- PCR
polymerase chain reaction
- PSA
prostate specific antigen
- TURP
transurethral prostate resection
References
- 1.Hoque MO. DNA methylation changes in prostate cancer: current developments and future clinical implementation. Expert Rev Mol Diagn. 2009;9:243. doi: 10.1586/erm.09.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yegnasubramanian S, Kowalski J, Gonzalgo ML, et al. Hypermethylation of CpG islands in primary and metastatic human prostate cancer. Cancer Res. 2004;64:1975. doi: 10.1158/0008-5472.can-03-3972. [DOI] [PubMed] [Google Scholar]
- 3.Nelson WG, Yegnasubramanian S, Agoston AT, et al. Abnormal DNA methylation, epigenetics, and prostate cancer. Front Biosci. 2007;12:4254. doi: 10.2741/2385. [DOI] [PubMed] [Google Scholar]
- 4.Brooks JD, Weinstein M, Lin X, et al. CG island methylation changes near the GSTP1 gene in prostatic intraepithelial neoplasia. Cancer Epidemiol Biomarkers Prev. 1998;7:531. [PubMed] [Google Scholar]
- 5.Millar DS, Ow KK, Paul CL, et al. Detailed methylation analysis of the glutathione S-transferase pi (GSTP1) gene in prostate cancer. Oncogene. 1999;18:1313. doi: 10.1038/sj.onc.1202415. [DOI] [PubMed] [Google Scholar]
- 6.Hanson JA, Gillespie JW, Grover A, et al. Gene promoter methylation in prostate tumor-associated stromal cells. J Natl Cancer Inst. 2006;98:255. doi: 10.1093/jnci/djj051. [DOI] [PubMed] [Google Scholar]
- 7.Troyer DA, Lucia MS, de Bruine AP, et al. Prostate cancer detected by methylated gene markers in histopathologically cancer-negative tissues from men with subsequent positive biopsies. Cancer Epidemiol Biomarkers Prev. 2009;18:2717. doi: 10.1158/1055-9965.EPI-09-0068. [DOI] [PubMed] [Google Scholar]
- 8.Steiner I, Jung K, Schatz P, et al. Gene promoter methylation and its potential relevance in early prostate cancer diagnosis. Pathobiology. 2010;77:260. doi: 10.1159/000318017. [DOI] [PubMed] [Google Scholar]
- 9.Mehrotra J, Varde S, Wang H, et al. Quantitative, spatial resolution of the epigenetic field effect in prostate cancer. Prostate. 2008;68:152. doi: 10.1002/pros.20675. [DOI] [PubMed] [Google Scholar]
- 10.Truong M, Yang B, Livermore A, et al. Using the epigenetic field defect to detect prostate cancer in biopsy negative patients. J Urol. 2013;189:2335. doi: 10.1016/j.juro.2012.11.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nonn L, Ananthanarayanan V, Gann PH. Evidence for field cancerization of the prostate. Prostate. 2009;69:1470. doi: 10.1002/pros.20983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richiardi L, Fiano V, Vizzini L, et al. Promoter methylation in APC, RUNX3, and GSTP1 and mortality in prostate cancer patients. J Clin Oncol. 2009;27:3161. doi: 10.1200/JCO.2008.18.2485. [DOI] [PubMed] [Google Scholar]
- 13.Henrique R, Ribeiro FR, Fonseca D, et al. High promoter methylation levels of APC predict poor prognosis in sextant biopsies from prostate cancer patients. Clin Cancer Res. 2007;13:6122. doi: 10.1158/1078-0432.CCR-07-1042. [DOI] [PubMed] [Google Scholar]
- 14.Welch HG, Fisher ES, Gottlieb DJ, et al. Detection of prostate cancer via biopsy in the Medicare-SEER population during the PSA era. J Natl Cancer Inst. 2007;99:1395. doi: 10.1093/jnci/djm119. [DOI] [PubMed] [Google Scholar]
- 15.Vickers AJ, Roobol MJ, Lilja H. Screening for prostate cancer: early detection or overdetection? Annu Rev Med. 2012;63:161. doi: 10.1146/annurev-med-050710-134421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Powell IJ, Bock CH, Ruterbusch JJ, et al. Evidence supports a faster growth rate and/or earlier transformation to clinically significant prostate cancer in black than in white American men, and influences racial progression and mortality disparity. J Urol. 2010;183:1792. doi: 10.1016/j.juro.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jemal A, Ward E, Thun M. Declining death rates reflect progress against cancer. PLoS ONE. 2010;5:e9584. doi: 10.1371/journal.pone.0009584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yanke BV, Salzhauer EW, Colon I. Is race a positive predictor of cancer on repeat prostate biopsy? J Urol. 2006;176:1114. doi: 10.1016/j.juro.2006.04.041. [DOI] [PubMed] [Google Scholar]
- 19.Kwabi-Addo B, Wang S, Chung W, et al. Identification of differentially methylated genes in normal prostate tissues from African American and Caucasian men. Clin Cancer Res. 2010;16:3539. doi: 10.1158/1078-0432.CCR-09-3342. [DOI] [PubMed] [Google Scholar]
- 20.Woodson K, Hayes R, Wideroff L, et al. Hypermethylation of GSTP1, CD44, and E-cadherin genes in prostate cancer among US Blacks and Whites. Prostate. 2003;55:199. doi: 10.1002/pros.10236. [DOI] [PubMed] [Google Scholar]
- 21.Kryvenko ON, Jankowski M, Chitale DA, et al. Inflammation and preneoplastic lesions in benign prostate as risk factors for prostate cancer. Mod Pathol. 2012;25:1023. doi: 10.1038/modpathol.2012.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herman JG, Graff JR, Myohanen S, et al. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Enokida H, Shiina H, Urakami S, et al. Ethnic group-related differences in CpG hypermethylation of the GSTP1 gene promoter among African-American, Caucasian and Asian patients with prostate cancer. Int J Cancer. 2005;116:174. doi: 10.1002/ijc.21017. [DOI] [PubMed] [Google Scholar]
- 24.Sakr WA, Grignon DJ, Crissman JD, et al. High grade prostatic intraepithelial neoplasia (HGPIN) and prostatic adenocarcinoma between the ages of 20–69: an autopsy study of 249 cases. In Vivo. 1994;8:439. [PubMed] [Google Scholar]
- 25.Djavan B, Mazal P, Zlotta A, et al. Pathological features of prostate cancer detected on initial and repeat prostate biopsy: results of the prospective European Prostate Cancer Detection study. Prostate. 2001;47:111. doi: 10.1002/pros.1053. [DOI] [PubMed] [Google Scholar]
- 26.Trock BJ, Brotzman MJ, Mangold LA, et al. Evaluation of GSTP1 and APC methylation as indicators for repeat biopsy in a high-risk cohort of men with negative initial prostate biopsies. BJU Int. 2012;110:56. doi: 10.1111/j.1464-410X.2011.10718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.