Abstract
Molecular chaperones of the Hsp70/40 family protect against the accumulation of mutated or misfolded proteins in part by facilitating their degradation. In the polyglutamine (polyQ) diseases, mutant proteins containing expanded polyQ repeats accumulate in intracellular inclusions and cause neurodegeneration. Although the ubiquitin–proteasome system and chaperones all help protect against accumulation of such toxic proteins, their precise roles are still unclear. Here we observed that the polyQ-expanded mutant ataxin-1 [82Q] was rapidly and selectively degraded in yeast while the wild-type protein [30Q] was stable. The selective degradation of the mutant ataxin-1 required proteasomes, but did not require Ydj1p, an Hsp40 homolog, which is involved in the disaggregation and/or breakdown of a number of misfolded proteins. However, another chaperone Hsp104 promoted degradation of mutant ataxin-1 without influencing the solubility or breakdown of short-lived cell proteins generally. Thus Hsp104-dependent degradation of mutant ataxin-1 may account for the ability of this chaperone to reduce toxicity caused by polyQ-repeat proteins.
Keywords: Polyglutamine, Ataxin-1, Proteasomes, Molecular chaperones, Hsp104
Introduction
Eukaryotic cells utilize multiple strategies to prevent the intracellular accumulation of unfolded mutant proteins: selective degradation by the ubiquitin–proteasome system (UPS), elimination of protein aggregates by autophagy and refolding by molecular chaperones [1]. Cytoplasmic chaperones, particularly Hsp70 and Hsp40 family members, also promote the rapid degradation of certain aberrant proteins [2,3]. Despite these protective mechanisms, in polyQ diseases such as Huntington disease (HD) and spinocerebellar ataxia (SCA), the mutant proteins containing expanded polyQ repeats accumulate in intracellular inclusions and cause neurodegeneration [4]. The inclusions in the affected neurons from patients with HD and SCA contain not only the mutant polyQ proteins but also ubiquitin, proteasome subunits and molecular chaperones. The presence of ubiquitin and proteasome subunits suggests either that the mutant proteins in the inclusions are being continually degraded or that there is a failure of the cells’ degradative apparatus to digest the mutant proteins [1,4].
There is growing evidence that over-expression of molecular chaperones can retard the pathogenesis of polyQ diseases and/or inclusion formation [4–10]. For example, over-expression of Hsp70 and/or Hsp40 can suppress the neurodegeneration and the aggregate formation caused by polyQ-repeat proteins [6,7]. Expression of another type of hexameric chaperone, Hsp104, a member of the AAA family of ATPases, can also modulate aggregation of polyQ proteins and reduce their toxicity in Caenorhabditis elegans and mammalian cells [8–10]. It is noteworthy that mammals do not contain Hsp104 or similar chaperones. Interestingly, the aggregation of polyQ domain of huntingtin in yeast is also dependent on the presence of prion proteins, whose propagation requires the function of Hsp104 [11]. By contrast Hsp104, together with Hsp70 family members (Ssa proteins) and an Hsp40 (Ydj1p), can reduce polyQ-induced toxicity in yeast presumably by facilitating the disaggregation of misfolded proteins [8,12]. In addition, Hsp104 deletion also is important for the ER-associated degradation of a model substrate in yeast [13].
These findings raised the possibility that Hsp104 not only influences the aggregation of polyQ proteins but also directly promotes the degradation of these misfolded proteins. To test this possibility, we have compared the stability of the wild-type [30Q] and polyQ-expanded mutant [82Q] forms of human ataxin-1 in yeast. Evidence is presented here that the proteasomal degradative pathway in yeast cells has the capacity to selectively hydrolyze the mutant ataxin-1 [82Q] proteins and that this process differs from the breakdown of most short-lived yeast proteins in requiring Hsp104 but not Ssa proteins or Ydj1p.
Materials and methods
Yeast strains and plasmids
Yeast strains used in this study were W303a (MATa, ade2-1, can1–100, his3–11,15, leu2–3,112, trp1-1, ura3-1, [psi+], ssd1-d), YS483 (MATa, ade2-1, can1–100, his3–11,15, leu2–3,112, trp1-1, ura3-1, [psi+], ssd1-d, hsp104:b:LEU2), W303-1b (MATα, ade2-1, can1–100, his3–11,15, leu2–3,112, trp1-1, ura3-1), ACY17b (MATα, ade2-1, can1–100, his3–11,15, leu2–3,112, trp1-1, ura3-1, ydj1–2::HIS3, LEU2::ydj1–151) and JN284 (MATa, his7, leu2, ura3, ise1). The yeast expression plasmids carrying human ataxin-1 (wild-type and mutant) under the control of GAL1 promoter – provided by Prof. Huda Zoghbi (Baylor College of Medicine) – were constructed by inserting human ATXN1 (SCA1) coding region into SpeI and SalI sites of p416 Gal1 or p423 Gal1 vector (ATCC). The yeast expression plasmid carrying wild-type Hsp104 under the control of GAL1 promoter was a gift from Prof. Susan Lindquist (Whitehead Institute, MIT).
Measurement of ataxin-1 degradation in yeast
The degradation of ataxin-1 in yeast was determined either by radiolabeling and following the loss of labeled proteins or by the promoter shut-off assay. To label cell proteins, rapidly growing yeast cells (induced to express ataxin-1 for 6–10 h) were labeled for 15 min with 200 µCi of 35S-methionine (Easy-Tag EXPRESS: NEN), washed twice with fresh medium and then further incubated for 3 h in the chase medium containing cycloheximide and methionine (0.5 mg/ml). Aliquots of cells collected at different times during the chase period were re-suspended in IP buffer (50 mM Tris–HCl, pH 7.5; 100 mM NaCl; 5 mM EDTA; 1% Triton X-100) plus protease inhibitors (1× Complete-Mini™, 1 mM PMSF and 5 mM NEM). Cells were then disrupted by vortexing with an equal volume of glass-beads and centrifuged gently at 500g for 10 min to remove unbroken cells and debris. Aliquots of extract were further centrifuged at 20,000g for 30 min to separate into the soluble (supernatant) and the particulate (pellet) fractions. The pellets were re-suspended in the IP buffer plus 0.1% SDS and briefly sonicated. The resulting extract (or fractions) was incubated with an anti-ataxin-1 antibody 11750 (provided by Prof. Huda Zoghbi) for 2 h and then with protein A/G-agarose (Oncogene) for an additional 90 min. The immunoprecipitated ataxin-1 was separated by 8% SDS–PAGE and then analyzed with a PhosphorImager.
For the promoter shut-off assay, yeast cells carrying ataxin-1 were first induced with 2% galactose for 6–10 h to express the protein and then transferred to the chase medium containing 2% glucose and cycloheximide (0.5 mg/ml) to shut off its expression. The cells were further incubated for 3 h and an aliquot of cells was collected at every 90 min during this chase period. The content of ataxin-1 in the cell extract (or fractions) was measured by immunoblotting with the anti-ataxin-1 antibody 11750.
Measurement of breakdown of other proteins in yeast
The degradation of Ub-Pro-β-gal and the breakdown of short-lived proteins (pulse-labeled with 200 µCi of 35S-methionine for 10 min and then further incubated for up to 30 min in the chase medium containing cycloheximide and methionine) in the wild-type and Δhsp104 mutant yeast cells was measured as described previously [14].
Results and discussion
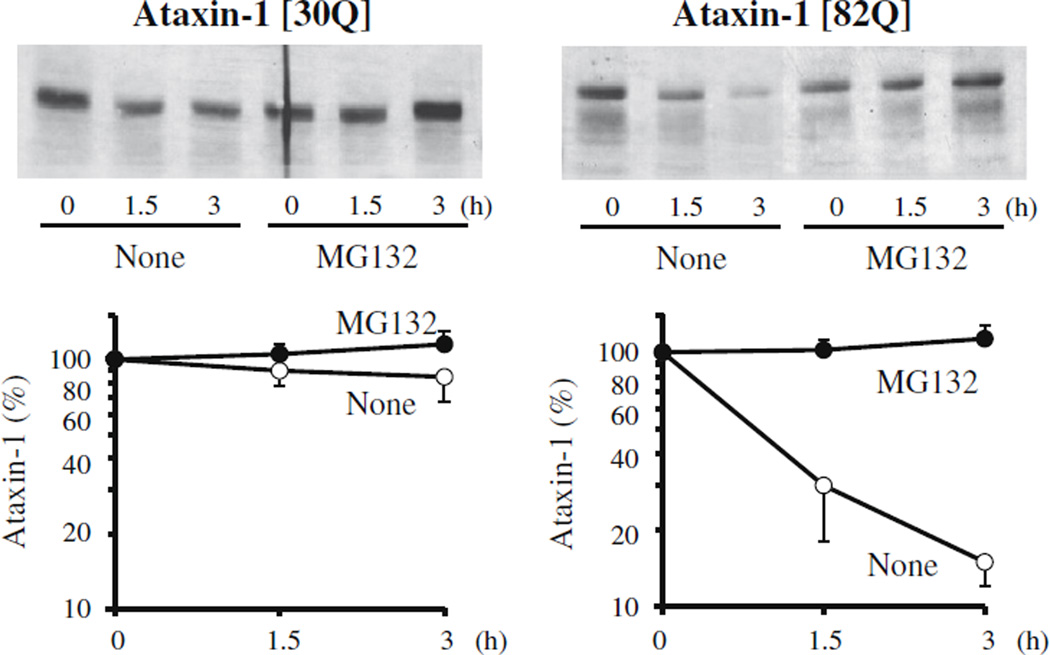
To measure the rates of degradation of human ataxin-1 in yeast, the protein was radiolabeled and the loss of radioactive protein was analyzed with time. After a 15-min pulse-labeling with 35S-methionine, the yeast cells expressing ataxin-1 were incubated in the chase medium containing cycloheximide and a large excess of methionine to prevent the re-incorporation of radioactive methionine. After cell lysis and centrifugation at low speeds (500g) to remove unbroken cells, the radiolabeled ataxin-1 in the extract was immunoprecipitated with the anti-ataxin-1 antibody. Wild-type ataxin-1 [30Q] was quite stable and underwent no significant degradation several hours after synthesis. By contrast, the mutant ataxin-1 [82Q] was rapidly degraded with a half-life of 1–1.5 h (Fig. 1). Next we tested if the ubiquitin–proteasome system is responsible for the turnover of mutant ataxin-1 in yeast cells. Treatment with 50 µM of MG132, a selective inhibitor of proteasomes, completely blocked the degradation of the mutant ataxin-1 confirming that the proteasome system in yeast recognizes and rapidly degrades the polyQ-expanded ataxin-1 (Fig. 1). This complete inhibition by MG132 is noteworthy because in several cellular models, the degradation of polyQ-expanded huntingtin is largely through autophagy [1,4,7].
Fig. 1.
Mutant human ataxin-1 with expanded polyglutamine repeats (82Q) is selectively degraded by the proteasome in yeast. Yeast cells (JN284) were transfected with the plasmids carrying human ataxin-1 (30Q or 82Q) whose expression is under the control of the GAL1 promoter. Cells were grown in the minimal medium containing 2% raffinose until mid-log phase (OD600 = 0.5–1.0) and then transferred to the induction medium containing 2% galactose for 6–8 h. The degradation of ataxin-1 in these cells was measured in the presence or absence of MG132 (50 µM) either by pulse-chase analysis and immunoprecipitation with the anti-ataxin-1 antibody 11750 or by the promoter shut-off assay. Similar results were obtained in three independent experiments.
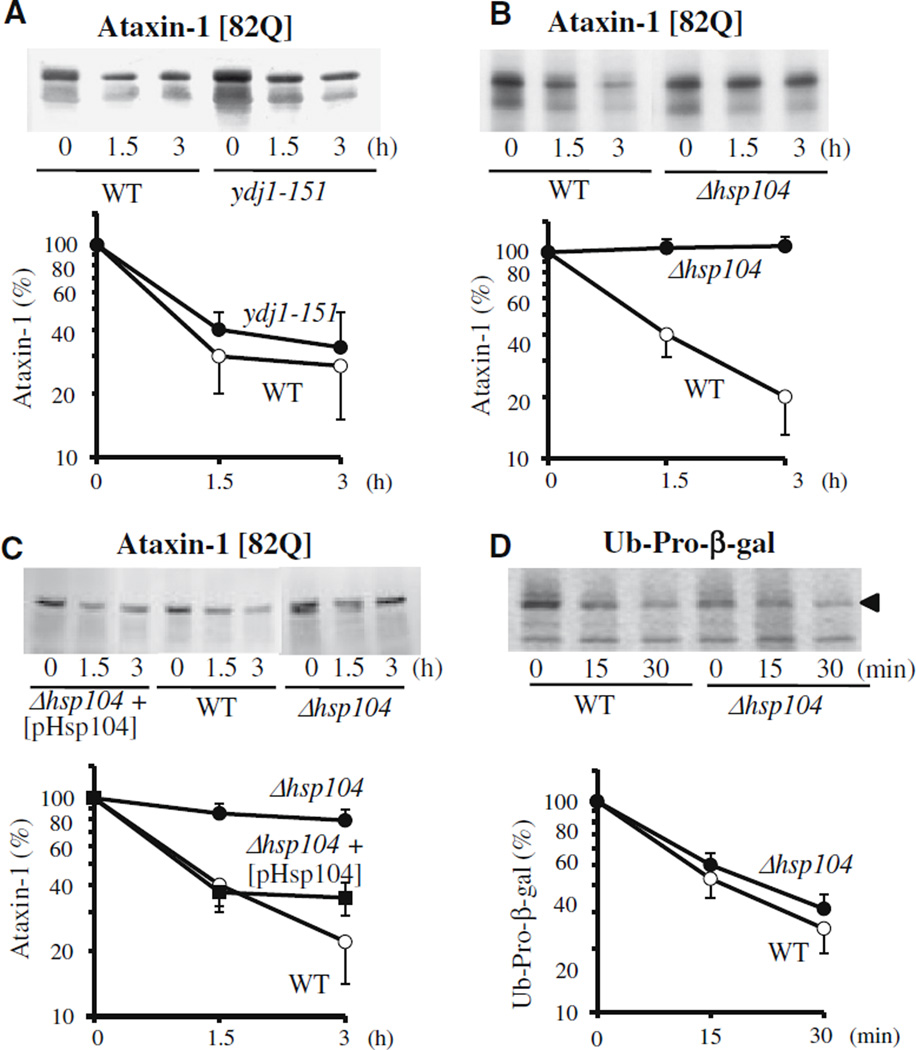
In yeast, the inactivation of Ssa proteins (Hsp70) and/or of the cofactor Ydj1p (Hsp40) causes a marked reduction in the rapid breakdown of certain misfolded proteins [3,14,15]. To examine whether the degradation of the mutant ataxin-1 [82Q] also requires these chaperones, we used the temperature-sensitive strain ydj1-151, in which Ydj1p is rapidly inactivated upon shift to 38 °C. Even though Ydj1p is essential for the rapid breakdown of a number of abnormal or short-lived proteins, it was not required for the degradation of mutant ataxin-1 (Fig. 2A).
Fig. 2.
Hsp104 is required for the selective degradation of mutant human ataxin-1. (A) Selective degradation of mutant ataxin-1 [82Q] does not require Ydj1p, an Hsp40 homolog, (B) but does require Hsp104. Plasmids carrying the mutant [82Q] ataxin-1 were transfected into (A) the wild-type (W303-1b) and temperature-sensitive mutant strain of Ydj1p (ACY 17b: ydj1–151) or (B) the wild-type (W303a) and Hsp104-deletion mutant strain (YS 483: Δhsp104). Degradation of ataxin-1 was then measured as described in Fig. 1. (C) Transfection of Hsp104 into the Hsp104-deletion strain restores the degradation of mutant ataxin-1 back to the control level. A CEN plasmid carrying the wild-type Hsp104 (under the GAL1 promoter) was introduced into Δhsp104 cells expressing the mutant ataxin-1, and its degradation was measured. (D) Hsp104 deletion does not affect the degradation of the model substrate ubiquitin-Pro-β-galactosidase (Ub-Pro-β-gal).
Hsp104, another major chaperone in yeast, plays a role in cellular thermotolerance and catalyzes the disaggregation and refolding of aggregated proteins with an aid of Hsp70/Hsp40 system [16,17]. Over-expression of Hsp104 can suppress the toxicity of huntingtin in C. elegans and mammalian cells [9,10]. Interestingly, the deletion of Hsp104 in yeast blocked the aggregation of mutant huntingtin [8]. In addition, aggregation of the chimeric protein containing polyQdomain of huntingtin fused to GFP was dependent on the presence of prion proteins, which requires the function of Hsp104 [11]. Such observations may imply that Hsp104 plays a more direct role in the degradation of polyQ proteins. In fact, we observed that the breakdown of mutant ataxin-1 [82Q] was completely blocked in Δhsp104 mutant (Fig. 2B). To verify that such a defect in ataxin-1 degradation is due to the lack of Hsp104, we re-introduced Hsp104 into this mutant and measured ataxin-1 degradation.When the level of Hsp104 was restored to that in wild-type cells, the mutant ataxin-1 was again rapidly degraded (Fig. 2C). By contrast Δhsp104 mutant showed no reduction in the rapid breakdownof a model substrate for UFD pathway, Ub-Pro-β-gal, which requires Ydj1p and Ssa proteins (Fig. 2D), or in the degradation of short-lived cell proteins in general (data not shown).
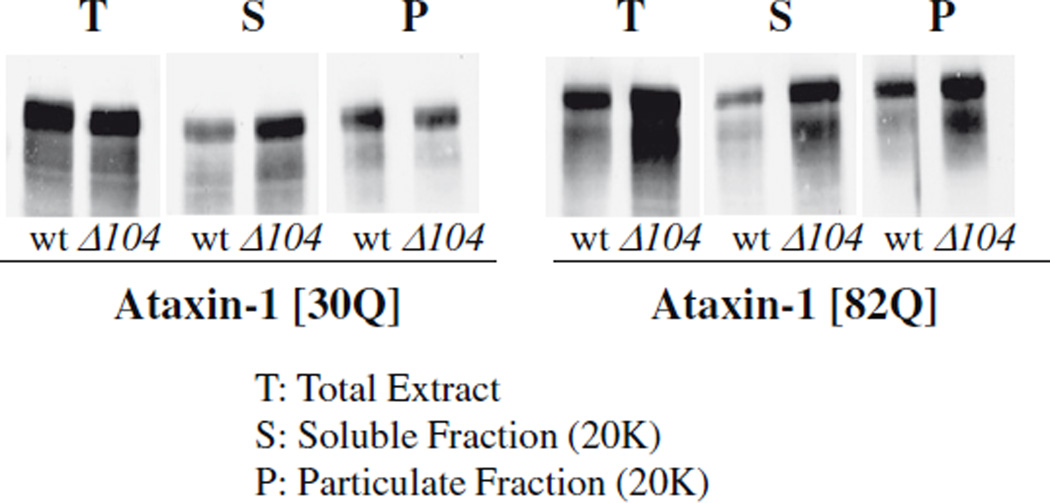
Molecular chaperones may promote degradation of abnormal proteins by simply maintaining them in a soluble form that is easily attacked by the cells’ degradative machinery [18]. However there are also a number of examples where chaperones (especially, co-chaperones such as CHIP) play more direct role in the degradation of polyglutamine proteins, perhaps as recognition elements [4]. To test if Hsp104 promotes the degradation of mutant ataxin- 1 by maintaining them in a soluble form, we compared the subcellular distribution of ataxin-1 in the wild-type and Δhsp104 mutant. After cells were lysed, the extract was centrifuged at 20,000g for 30 min and the relative amounts of ataxin-1 in the soluble (supernatant) and particulate (pellet) fractions were then assayed by immunoblotting. At steady state, the wild-type [30Q] and mutant [82Q] ataxin-1 showed similar subcellular distributions; both were soluble although a significant fraction of each was also found in the particulate fraction. Surprisingly, there was no difference in their distribution in the wild-type and Δhsp104 strains (Fig. 3). The ataxin-1 in the supernatant also remained soluble upon further centrifugation at 100,000g for an hour (data not shown). Thus these proteins do not appear to be present in aggregates even in Δhsp104 mutant. Therefore, the failure to degrade the mutant ataxin-1 in the chaperone-deficient cells is not due to substrate accumulation in a particulate form.
Fig. 3.
Mutant ataxin-1 does not accumulate in the particulate fraction in the Hsp104-deletion mutant. The wild-type and Δhsp104 cells expressing ataxin-1 were grown until OD600 = 1.5. The cells were collected, disrupted by vortexing with an equal volume of glass-beads and the unbroken cells and debris were removed by gentle centrifugation at 500g for 10 min. The resulting extracts were further divided into the soluble (supernatant) and particulate (pellet) fractions by centrifugation at 20,000g for 30 min. The presence of ataxin-1 in these fractions was detected by immunoblotting with anti-ataxin-1 antibody 11750.
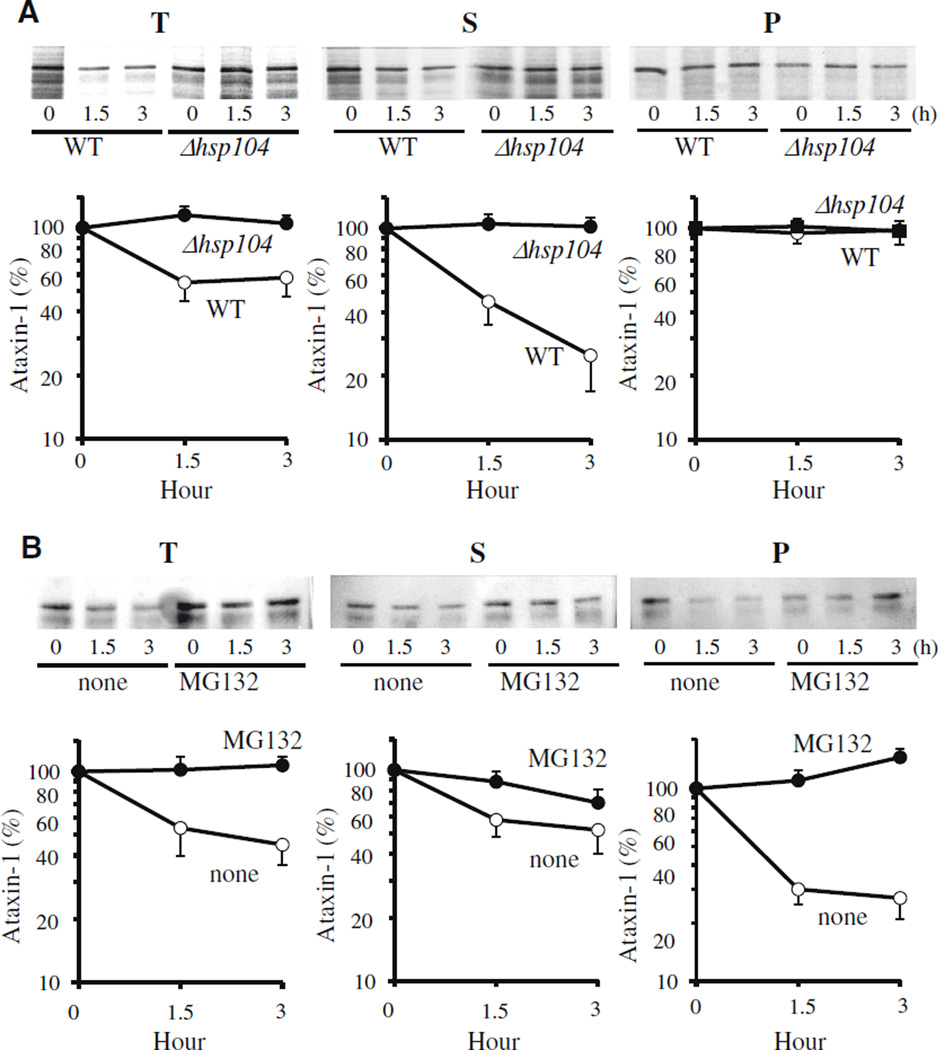
This finding was confirmed by pulse-chase analyses of mutant ataxin-1 [82Q] in the soluble and particulate fractions. The newly synthesized molecules in the soluble fraction were lost rapidly during the first hour and accounted for the great majority of ataxin-1 degradation (Fig. 4). When degradation was prevented in the Δhsp104 mutant, the non-degraded ataxin-1 remained largely soluble. The small amount of ataxin-1 in the 20,000g pellet did not change in the absence of Hsp104 (Fig. 4A). When ataxin-1 degradation was blocked by MG132 treatment, there was a reproducible tendency of the non-degraded ataxin-1 to shift with time from the soluble to the particulate fraction (evident after 90 min) (Fig. 4B). A similar accumulation of non-degraded proteins generally in inclusions or “ aggresomes” has been observed in many studies with proteasome inhibitors in mammalian cells [19,20]. Since ataxin-1 accumulated in the pellets in wild-type strain but not in Δhsp104 mutant, Hsp104 may play a role in the aggregation process. In any case, Hsp104 clearly does not promote ataxin-1 degradation by altering its solubility but instead somehow enhances the susceptibility of soluble molecules to the ubiquitin– proteasome system. Because these results cannot be simply reconciled with Hsp104’s ability to disassemble aggregates and to promote refolding [16,17], our observation may indicate a new molecular action of Hsp104 facilitating proteasomal degradation of certain proteins.
Fig. 4.
(A) Mutant ataxin-1 is primarily degraded in the soluble fraction. The radio-labeled cell extract from the wild-type and Δhsp104 mutant was further centrifuged at 20,000g for 30 min and divided into soluble (supernatant) and particulate (pellet) fractions. The pellets were re-suspended in IP buffer containing 0.05% SDS and briefly sonicated before the immunoprecipitation. (B) Proteasome inhibition leads to the some accumulation of mutant ataxin-1 in the particulate fraction from the wild-type yeast cells (JN284).
The ubiquitin–proteasome system in yeast closely resembles that in higher eukaryotes and therefore this capacity to selectively degrade the mutant ataxin-1 is not likely restricted to yeast. In cultured mammalian cells, polyQ-expanded androgen receptor is degraded by proteasomes which is enhanced by Hsp70/Hsp40 [21]. In brains from HD patients, proteasomes and ubiquitin are co-localized with polyglutamine aggregates and proteasome inhibitors prevent the clearance of mutant huntingtin in HD mouse models [22]. Surprisingly, purified eukaryotic proteasomes only digest polyQ sequences very poorly and during degradation release these highly aggregation-prone sequences for further hydrolysis by cytosolic peptidases [23]. The degradation of various polyglutamine proteins is therefore likely to differ in different cell types since inclusion formation and pathology caused by the mutant huntingtin and ataxin-1 occur in different neurons. In addition, in extracts of HeLa cells, the mutant ataxin-1 appears more resistant to proteasomal degradation than the wild-type protein, despite similar rates of ubiquitin conjugation [24]. Perhaps critical chaperones necessary for hydrolysis of the mutant ataxin-1 are present in yeast, but are lacking in the HeLa extract. It is noteworthy in this context that no homolog of Hsp104 has yet been found in higher eukaryotic cells. The loss of this chaperone during evolution is surprising since if hsp104 is expressed in mammalian cells, it can inhibit aggregate formation and cell death caused by expanded polyglutamine repeats proteins [10].
It is intriguing that the degradation of mutant ataxin-1 does not require Hsp40 (and Hsp70), which are important for hydrolysis of misfolded proteins generally and enhances the degradation of androgen receptor [21], but instead involves Hsp104, which seems to function here without Hsp70/Hsp40 as essential cofactors. A number of groups have reported that high amount of all these chaperones can suppress the toxic effect of polyQ-repeat proteins and reduce the appearance of intracellular inclusions [5,7]. The present findings of Hsp104-mediated degradation may account for the ability of this chaperone to protect against both the intracellular accumulation and neurodegeneration induced by mutant huntingtin [8,9]. It remains unclear if the formation of these inclusions contributes to the neurodegeneration or if the formation of these inclusions represents a cellular defense mechanism that sequesters the potentially toxic polypeptides and prevents their interfering with the other cellular processes or triggering neuronal cell death [4,7]. In the present experimental system, the mutant ataxin-1 did not affect growth or viability of yeast (data not shown).
Hsp104 has also been found to be required specifically for the ER-associated degradation of a misfolded GFP-fusion protein [13]. Probably some unusual structural features of the mutant ataxin-1 and this model substrate lead to specific interactions with Hsp104. It is noteworthy that cells lacking this chaperone showed no defect in the degradation of short-lived proteins generally or of the model substrate Ub-Pro-β-gal. This fusion protein is known to be degraded by the UFD (ubiquitin fusion degradation) pathway [25] and its degradation requires Hsp70 (data not shown) and Hsp40 (Ydj1p) [14]. Thus, this effect of Hsp104 on protein degradation is specific to certain substrates. It will be important to determine how Hsp104 promotes either the recognition of the mutant ataxin-1 or its subsequent hydrolysis by proteasomes, especially since such information may help us understand not only the turnover of polyglutamine proteins in normal cells and their abnormal accumulation in affected neurons of patients but also an important new role of Hsp104 in elimination of certain types of abnormal potentially toxic polypeptides.
Acknowledgments
We are grateful to Drs. Huda Zoghbi and Susan Lindquist for providing the yeast strains and plasmids, and our coworkers for their assistance. This research was supported by the grants from Hereditary Disease and High Q Foundations and the NIGMS to A.L.G.
Contributor Information
Do Hee Lee, Email: do_lee@swu.ac.kr, dhleedo@yahoo.co.kr.
Alfred L. Goldberg, Email: alfred_goldberg@hms.harvard.edu.
References
- 1.Rubinsztein D. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Nijbroek G, Sullivan ML, McCracken AA, Watkins SC, Michaelis S, Brodsky JL. Hsp70 molecular chaperone facilitates endoplasmic reticulum-associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol. Biol. Cell. 2001;12:1303–1314. doi: 10.1091/mbc.12.5.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park SH, Bolender N, Eisele F, Kostova Z, Takeuchi J, Coffino P, Wolf DH. The cytoplasmic Hsp70 chaperone machinery subjects misfolded and endoplasmic reticulum import-incompetent proteins to degradation via the ubiquitin-proteasome system. Mol. Biol. Cell. 2007;18:153–165. doi: 10.1091/mbc.E06-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams AJ, Paulson HL. Polyglutamine neurodegeneration: protein misfolding revisited. Trends Neurosci. 2008;31:521–528. doi: 10.1016/j.tins.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakahira H, Breuer P, Hayer-Hartl MK, Hartl FU. Molecular chaperones as modulators of polyglutamine protein aggregation and toxicity. Proc. Natl. Acad. Sci. USA. 2002;99:16412–16418. doi: 10.1073/pnas.182426899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muchowski PJ, Wacker JL. Modulation of neurodegeneration by molecular chaperones. Nat. Rev. Neurosci. 2005;6:11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- 7.Shao J, Diamond MI. Polyglutamine diseases; emerging concepts in pathogenesis and therapy. Hum. Mol. Genet. 2007;16:R115–R123. doi: 10.1093/hmg/ddm213. [DOI] [PubMed] [Google Scholar]
- 8.Krobitsch S, Lindquist S. Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc. Natl. Acad. Sci. USA. 2000;97:1589–1594. doi: 10.1073/pnas.97.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Satyal SH, Schmidt E, Kitagawa K, Sondheimer N, Lindquist S, Kramer JM, Morimoto RI. Polyglutamine aggregates alter protein folding homeostasis in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2000;97:5750–5755. doi: 10.1073/pnas.100107297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vacher C, Garcia-Oroz L, Rubinsztein DC. Overexpression of yeast hsp104 reduces polyglutamine aggregation and prolongs survival of a transgenic mouse model of Huntington’s disease. Hum. Mol. Genet. 2005;14:3425–3433. doi: 10.1093/hmg/ddi372. [DOI] [PubMed] [Google Scholar]
- 11.Meriin AB, Zhang X, He X, Newnam GP, Chernoff YO, Sherman MY. Huntingtin toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J. Cell Biol. 2002;157:997–1004. doi: 10.1083/jcb.200112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cashikar AG, Duennwald M, Lindquist SL. A chaperone pathway in protein disaggregation. J. Biol Chem. 2005;289:23869–23875. doi: 10.1074/jbc.M502854200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taxis C, Hitt R, Park S-H, Deak PM, Kostova Z, Wolf DH. Use of modular substrates demonstrates mechanistic diversity and reveals differences in chaperone requirement of ERAD. J. Biol Chem. 2003;278:35903–35913. doi: 10.1074/jbc.M301080200. [DOI] [PubMed] [Google Scholar]
- 14.Lee DH, Sherman MY, Goldberg AL. Involvement of the molecular chaperone Ydj1 in the ubiquitin-dependent degradation of short-lived and abnormal protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:4773–4781. doi: 10.1128/mcb.16.9.4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill K, Cooper AA. Degradation of unassembled Vph1p reveals novel aspects of the yeast ER quality control system. EMBO J. 2000;19:550–561. doi: 10.1093/emboj/19.4.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doyle SM, Wickner S. Hsp104 and ClpB: protein disaggregating machines. Trends Biochem. Sci. 2009;34:40–48. doi: 10.1016/j.tibs.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 17.Bösl B, Grimminger V, Walter S. The molecular chaperone Hsp104 -a molecular machine for protein disaggregation. J. Struct Biol. 2006;156:139–148. doi: 10.1016/j.jsb.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 18.Wickner S, Maurizi MR, Gottesman S. Posttranslational quality control: folding, refolding, and degrading proteins. Science. 1999;286:1888–1893. doi: 10.1126/science.286.5446.1888. [DOI] [PubMed] [Google Scholar]
- 19.Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 20.Zaarur N, Meriin AB, Gabai VL, Sherman MY. Triggering aggresome formation. Dissecting aggresome-targeting and aggregation signals in synphilin 1. J. Biol Chem. 2008;283:27575–27584. doi: 10.1074/jbc.M802216200. [DOI] [PubMed] [Google Scholar]
- 21.Bailey CK, Andriola IF, Kampinga HH, Merry DE. Molecular chaperones enhance the degradation of expanded polyglutamine repeat androgen receptor in a cellular model of spinal and bulbar muscular atrophy. Hum. Mol. Genet. 2002;11:515–523. doi: 10.1093/hmg/11.5.515. [DOI] [PubMed] [Google Scholar]
- 22.Davies JE, Sarkar S, Rubinsztein DC. The ubiquitin proteasome system in Huntington’s disease and the spinocerebellar ataxias. BMC Biochem. 2007;8(Suppl. 1):S2. doi: 10.1186/1471-2091-8-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhutani N, Venkatraman P, Goldberg AL. Puromycin-sensitive aminopeptidase is the major peptidase responsible for digesting polyglutamine sequences released by proteasomes during protein degradation. EMBO J. 2007;26:1385–1396. doi: 10.1038/sj.emboj.7601592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cummings CJ, Reinstein E, Sun Y, Antalffy B, Jiang Y, Ciechanover A, Orr HT, Beaudet AL, Zoghbi HY. Mutation of the E6-AP ubiquitin ligase reduces nuclear inclusion frequency while accelerating polyglutamine-induced pathology in SCA1 mice. Neuron. 1999;24:879–892. doi: 10.1016/s0896-6273(00)81035-1. [DOI] [PubMed] [Google Scholar]
- 25.Isano E, Saeki Y, Yokosawa H, Toh-e A. Rpn7 is required for the structural integrity of the 26S proteasome of Saccharomyces cerevisiae. J. Biol Chem. 2004;279:27168–27176. doi: 10.1074/jbc.M314231200. [DOI] [PubMed] [Google Scholar]