Abstract
A founding premise of the human genome project was that knowledge of the spectrum of abnormalities that comprise cancers and other human diseases would lead to improved disease management by identifying molecular abnormalities that could guide disease detection and diagnosis, suggest new therapeutic strategies and be developed as markers to predict response to therapy. This project led to elucidation of a reference normal human genome sequence and normal polymorphisms therein against which sequences from diseased tissues can be compared to enable identification of causal abnormalities. It also stimulated development of an array of computational tools for genomic analysis and catalyzed public and private sector development of revolutionary tools for genome analysis that transformed analysis of whole genomes from an enterprise that required international teams and hundreds of millions of dollars to a process that can be carried out in core facilities for only a few thousand dollars per sample. Indeed, the $1000 genome is nearly upon us. Applications of these technologies to human cancers in international cancer genome projects are now revealing the spectra of abnormalities that comprise thousands of individual cancers. Analyses of these data are leading to the promised improvements in disease management. We review several aspects of cancer genomics with emphasis on aspects that are relevant to improving cancer therapy.
Keywords: Genomics, Targeted therapy, Personalized medicine
Highlights
Current status of genome‐wide analysis of a variety of tumor types.
How analyses of these above data are shaping our investigation of important therapeutic targets.
Methodology for development of therapeutically relevant modalities.
Progress of creating personalized medicine.
1. The cancer genome landscape
Somatic genomic and epigenomic abnormalities accumulate during life as a result of exposure to endogenous and exogenous reactive chemicals that modify the DNA sequence itself as well as the chromatin structure in which it is packaged. In most cases, elaborate protective mechanisms recognize these abnormalities and either repair them or eliminate and replace the cells carrying them. Considering the number of cells at potential risk of damage in an individual, estimated to be ∼1014, the efficacy of the protection/repair systems is remarkably high. Occasionally, however, a cell in a cancer‐prone tissue acquires an abnormality that initiates a chain of events that ultimately culminates in the development of a malignant lesion. These may be epigenomic events that enhance proliferation and/or events that damage aspects of the repair/protection systems that lead to increased rates of aberration accumulation. Still, this is not enough to enable accumulation of the full suite of abnormalities needed to generate the cancer hallmarks that comprise a complete metastatic phenotype. This likely requires reactivation of telomerase in order to sustain protective telomeres as needed for indefinite proliferation (Harley et al., 1990). This also happens rarely and typically requires coordinate inactivation of mechanisms such as p53‐mediated DNA damage surveillance (Hollstein et al., 1991) that eliminate cells with chromosomal damage typical of cells with short telomeres. Eventually, however, a cell may reactivate telomerase and accumulate a suite of abnormalities that enables development of most of the hallmarks of cancer – resistance to anoikis, reduced apoptosis, enhanced proliferation, angiogenesis, enhanced metabolic activity, etc. (Hanahan and Weinberg, 2000). Typically, several “driver” abnormalities accumulate to accomplish this deregulation. Since many aspects of aberration formation are stochastic – for example, mutagen exposure, variation in individual DNA repair capacity, random chromosome damage during growth with nonfunctional telomeres and the differentiation status of the cell in which abnormalities accumulate ‐ the spectrum of accumulated abnormalities can vary substantially between cancers that otherwise appear clinically similar (Kumar et al., 2011). These differences influence the detailed cancer phenotype and likely explain why cancers that otherwise appear clinically similar progress and respond differently to treatment. Robust technologies that allow detailed assessment of these abnormalities enable development of precision medicine – that is, use of the right treatment for the right patient at the right time. However, establishing the links between abnormality spectrum and optimal treatment is daunting because of the large number of abnormalities that can occur. Remarkably, the Catalogue of Somatic Mutations in Cancer (COSMIC) maintained by the Sanger Institute (http://www.sanger.ac.uk/genetics/CGP/Census) now lists over 100,000 different somatic mutations discovered during analysis of over 400,000 cancers. It is likely that only a small fraction of these are cancer “drivers” that are selected during cancer progression because they contribute to some aspect of cancer pathology. Separating drivers from bystander mutations that occur by chance during progression of a genomically unstable tumor is an ongoing international effort.
2. Cancer subtypes
Although individual tumors typically accumulate a unique spectrum of somatic genomic and epigenomic aberrations, driver mutations typically occur sufficiently frequently in multiple cancers such that they define cancer subsets (Alizadeh et al., 2000; Collisson et al., 2011; Sorlie et al., 2001). Identification of these subtypes is important since they define molecularly homogeneous cancer subsets that respond similarly to treatment. Importantly, cancer subsets depend so strongly on some driver mutations that compounds that inhibits the proteins or pathways activated by the drivers can have substantial therapeutic benefit (Druker et al., 2001; Lynch et al., 2004; Slamon et al., 2001). Particularly well‐established driver events include somatic and germline mutations involving KRAS, BRAF, RET, FLT3, PIK3CA, BRCA1/2, NF1/2, MLH1, MSH2, VHL, and TP53; amplification of MYC, MYCN, ERBB2, EGFR CCNE1, and ALK; deletion of RB1, CDKN2A, TP53 and PTEN and translocations involving ETS family transcription factors, ALK, ABL1, ABL2, PDGFRA/B (Vogelstein and Kinzler, 2004). Overall, COSMIC currently lists 474 candidate cancer gene aberrations that may contribute to the physiology of human cancers. Many of these are possible targets for novel therapies but only a few have been successfully targeted to date (see below).
Human cancer subtypes can be further defined using other omic endpoints including transcription profiles for coding and noncoding regulatory RNAs, promoter methylation patterns and protein profiles as illustrated in Figure 1. mRNA expression profiling is particularly well developed and is now widely used to define subtypes in most major human tumor types (The Cancer Genome Atlas, 2000, 2011, 2010, 2008, 2011). Although still not definitive, it appears that subtypes defined by these endpoints reflect the transcriptional and epigenomic status of the cell that became immortalized during cancer genesis. Transcriptional subtypes in some tumors are strongly associated with outcome so that the information can be used to identify patients at low risk of progressing for whom watchful waiting should be considered in lieu of chemotherapy or other aggressive treatment (van't Veer and Bernards, 2008).
Figure 1.
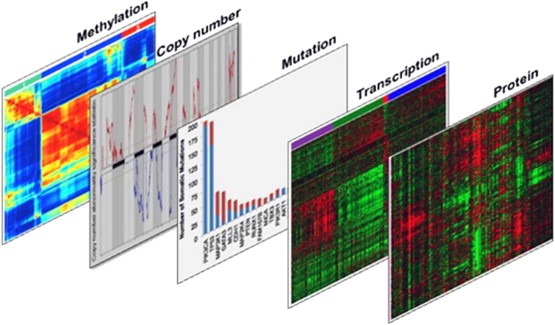
Omic endpoints to determine cancer subtypes. Tumors can be analyzed based on a variety of endpoints. The analyses can be used individually to stratify tumors into subtypes or in conjunction to further refine classification to determine appropriate targeted therapy in a highly specific manner.
2.1. International cancer genomics
These discoveries have motivated international efforts to survey cancers for additional actionable genomic and epigenomic events. These efforts have been accelerated by the development of genome‐wide analysis tools such as microarray‐based hybridization for analysis of cancer‐associated changes in mRNA and miRNA transcription, copy number and promoter methylation (Liu et al., 2004; Pinkel et al., 1998; Schena et al., 1995; Weber et al., 2005) as well as massively parallel nucleic acid sequencing for assessment of changes in transcription, copy number, genome structure and DNA sequence (Metzker, 2010). These analytical approaches are now being applied world‐wide to define cancer subtypes and identify new therapeutic approaches. Efforts by The Cancer Genome Atlas (TCGA) and the International Cancer Genome Consortium (ICGC) are particularly important in that effort. It seems likely that detailed genomic and epigenomic catalogs will be available for most of the major human tumor types by 2015 and that integrative analyses of these data will reveal therapy defining subtypes. Analyses of genomic and epigenomic profiles of several tumor types (e.g. breast, glioblastoma, colon, lung, pancreas, etc.) demonstrate the existence of recurrent subtypes defined by transcriptional patterns, genomic aberrations, promoter methylation patterns and protein expression patterns (The Cancer Genome Atlas, 2008, 2011). In most cases, aberration subtypes can be combined to refine subtypes. For example, estrogen receptor positive (luminal subtype) breast cancers can be further classified according to HER2 amplification status and further still according to the presence or absence of PIK3CA mutations (Loi et al., 2010; Prat and Perou, 2011). It is now becoming clear that responses to agents targeting specific aberrations and pathways can be influenced by the presence of other aberrations. In HER2‐positive breast cancer, for example, responses to trastuzumab and lapatinib appear to be diminished by the presence of PIK3CA mutations (Eichhorn et al., 2008; Junttila et al., 2009). Likewise, the efficacy of imatinib in CML patients whose tumors are driven by ABL1 translocations is diminished by the emergence of mutations that interfere with imatinib binding (Stein and Smith, 2010).
2.2. Pathways
The bewilderingly large number of different epigenomic and genomic abnormalities now being discovered in the course of international genomics efforts presents a significant challenge for therapy development since many cancers seem to be driven by a few recurrent abnormalities and an array of aberrations that occur at relatively low frequency and differ between tumors of the same type. This is particularly clear in the recently published analyses of glioblastoma (The Cancer Genome Atlas, 2008) and ovarian cancer (The Cancer Genome Atlas, 2011). Developing therapies to attack events deregulated by low prevalence abnormalities is not financially or logistically feasible. Fortunately, integrative analyses of these data make it clear that several key regulatory pathways including p53, RB, PI3K/RAS and NOTCH are recurrently deregulated by diverse genomic and epigenomic aberrations. Some initial attempts to organize these data suggest the existence of approximately a dozen core signaling pathways that are deregulated during progression to metastatic cancer (Jones et al., 2008). The specific aberrations and gene targets may vary between cancers as illustrated in Figure 2 for the RB and PI3K/RAS pathways in ovarian cancer. This suggests that it will be possible to move from targeting specific genomic aberrations to treatment of the deregulated pathways, regardless of the mechanism(s) by which they are deregulated.
Figure 2.
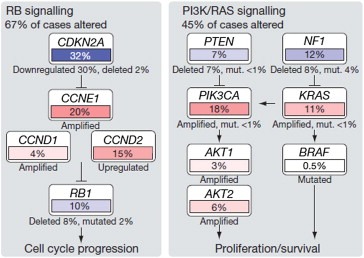
Altered pathways in high‐grade serous ovarian carcinoma. Multiple genes in the Rb and PI3K/Ras pathways are mutated at varying frequencies. First published: Nature 2006.
Development of pathway‐targeted therapeutic approaches is facilitated by integration of genomic information from multiple platforms in ways that reveal the deregulated pathways. This is challenging because relatively few of the low frequency aberrations have been functionally assessed. Nonetheless, several algorithms have been developed for this purpose. In most cases, these algorithms map genomic information on activating and inactivating aberrations onto regulatory pathway structures curated from the literature. Several web‐based resources provide curated pathway structures to enable such studies including KEGG (Kyoto Encyclopedia of Genes and Genomes; http://www.genome.jp/kegg/pathway.html), the Reactome (http://www.reactome.org/ReactomeGWT/entrypoint.html), and Pathway Commons (Cerami et al., 2011). Algorithms that interpret genomic information in the context of curated pathways include Ingenuity (http://www.ingenuity.com/), PARADIGM (PAthway Recognition Algorithm using Data Integration on Genomic Models (Vaske et al., 2010)), NetBox (Cerami et al., 2010), and ARACNE (Algorithm for the Reconstruction of Accurate Cellular Networks (Margolin et al., 2006)). One limitation of these approaches is that the curated pathways are heavily redundant. Heiser et al. addressed this problem in the context of PARADIGM by computationally eliminating redundant pathway elements to produce a “Superpathway” in which pathway activity differences between comparator populations are recognized as interconnected activity nodes (Heiser et al., 2011).
3. What makes a good therapeutic target?
Identifying aberrations and pathways that can be targeted for therapeutic benefit is challenging given the large number of both low and high prevalence genomic aberrations that are being discovered. However, a few criteria are emerging to prioritize therapeutically relevant targets for cancer therapy. First, a high priority target should be an aberrant protein or pathway on which cancers depend for survival when aberrantly up‐regulated so that inhibition leads to a cell death response. Some driver aberrations influence cancer hallmarks like proliferation or motility that may enhance cancer spread but may not lead to tumor cell death when inhibited. Second, an ideal target would be a pathway or protein that is uniquely up‐regulated in the target tumor and not in normal tissues. Aberrant pathways that result from genomic aberrations selected during tumor progression are attractive in this regard because these are present in a large percentage of the target tumor population. Examples include pathways deregulated by BCR‐Abl in chronic myelogenous leukemia, or associated with estrogen receptor (ER) expression or HER2 amplification in breast cancer. Unfortunately, some otherwise attractive targets will be present in only a small fraction of tumors of a particular type. For example, EGFR mutations occur only in about 15% of in non‐small cell lung cancers (Penzel et al., 2011) and some now being identified by international genomics efforts are present in only a few percent of tumors. These targets may be made more attractive by identifying multiple tumor types in which they are present. International genomics efforts will be particularly helpful in that regard. From a practical point of view, targets that are drivers in tumors or tumor subtypes that respond poorly to conventional therapies are easier to get into clinical trials and have a larger impact on outcome. Finally, the target should be druggable. MYC, KRAS and TP53 are among the most frequently aberrant driver genes in human cancers but have proven remarkably resistant to therapeutic intervention (Kessler et al., 2012; Luo et al., 2009). On the other hand, approaches to inhibiting kinases are well developed (Zhang et al., 2009).
4. Target validation
International omic analyses and meta‐analyses are identifying hundreds of genes and proteins as candidate targets. However, it is still a long journey from bedside to benchside and back to bedside – that is, gleaning a candidate from a list of clinically viable targets, validating that target in the laboratory, developing targeted therapies for it, and then returning to the clinical setting to test efficacy in patients. Once a target has been identified, the laboratory phase of the process begins and typically consists of using cell culture models, animal models, or both to validate the target and develop targeted therapies. Proper validation requires a judicious choice of cells that are being used to model mutations in vitro. While tumor cell lines accurately mirror some aspects of the genomic landscape of primary tumors (Chin et al., 2006; Neve et al., 2006), there are caveats to their use. Tumor lines selected to study one particular omic feature also bring along a variety of other genomic aberrations that may confound analyses (see following section ‘crosstalk between targets’). Additionally, tumor lines can be genetically unstable in vitro and can change phenotype when under a particular selective pressure. Researchers have used this phenomenon to their benefit to begin studying mechanisms of disease resistance, typically by chronically culturing tumor cells in the presence of therapeutic agent until only resistant cells exist (Abukhdeir et al., 2008; Kute et al., 2004). In this case, it is favorable to have a genetically unstable cell to study but other experimental questions are better answered with an established cell line that has a well‐defined, nearly diploid genotype that is stable in prolonged culture. For this, some tissue types have non‐tumorigenic cell lines with limited genomic aberrations that can be further manipulated – such as MCF‐10A cells for breast (Soule et al., 1990), HPDE cells for pancreas (Furukawa et al., 1996), RWPE‐1 cells for prostate (Bello et al., 1997), and others. These have been utilized with great success for in vitro target validation often in concert with available established tumor cell lines.
Both oncogenes and tumor suppressor genes are important in the initiation, growth and metastases of tumors and so both are viable targets for therapy. Greater success, by far, has been achieved in developing targeted therapies for oncogenes because it is more difficult to restore the function of a tumor suppressor gene than it is to blunt the activity of an overactive oncogene. It is possible, however, to develop inhibitors of proteins whose levels increase when a driver's tumor suppressor is lost. Identification of pathways that are activated by tumor suppressor gene loss is particularly attractive in this regard.
From a genomic perspective, identification of candidate driver cancer genes begins with identification of an aberration that occurs significantly more often that would be expected from chance. These identification efforts are informed by analysis of genome profiles from many tumors. In the case of copy number aberrations, the extents of the regions of abnormality also serve to narrow the region of interest. Cross‐species comparisons of genomic aberrations that arise spontaneously are also particularly helpful in identifying driver aberrations since the regions of genomic abnormality harboring driver cancer genes are often mirrored in syntenic regions in spontaneous mouse tumors of the same type (Hodgson et al., 2005; Maser et al., 2007).
Functional validation of oncogenes as cancer drivers typically proceeds by demonstrating increased malignant potential (increased growth, reduced cell death, increased motility, etc.) resulting from over expression of the candidate in cells in which it is expressed at normal levels and decreased malignant potential after treatment with interference RNA (RNAi) or pharmacological inhibitors in cancer cells in which it is over expressed. Validation of tumor suppressors is more difficult because by their nature loss of function of both alleles of the gene is required for the phenotype. Still, the converse experiments can be performed for tumor suppressor genes – either RNAi of the endogenous alleles resulting in increased malignant potential or restoring expression of nonfunctional alleles to demonstrate tumor suppressive effects. Genome‐wide knockdown and expression approaches are now being developed to facilitate these candidate cancer gene prioritization experiments (Dow et al., 2012; Prahallad et al., 2012; Root et al., 2006; Yang et al., 2011). Besides simple overexpression/knockdown studies, gene targeting of cells in vitro can be done to examine the role of particular mutations. While much more technically difficult, knock‐in of an activating mutation or knock‐out of a tumor suppressor, usually by viral‐mediated homologous recombination targeted to a particular genomic locus, provides a more physiologically relevant system and can unmask phenotypes that may be missed using simpler techniques (Gustin et al., 2009, 2007, 2011).
A higher level of validation is achieved by validating in vitro results in xenografts or genetically engineered mice or other in vivo systems. Animal models can be used for both validation of proteins as a therapeutically relevant target and, more commonly, for validation of a targeted therapy itself (van Miltenburg and Jonkers, 2012). Cancer research most commonly utilizes mouse models and, currently, xenograft or orthotopic transplantation of human tumor cells in mice is the most widely used method. However, more sophisticated techniques have allowed the development of other mouse models such as genetically engineered germline mutation models and chimeric animals which allow for spatial and/or temporal induction of tumorigenesis (for complete reviews of these models see (Heyer et al., 2010) and (van Miltenburg and Jonkers, 2012)). Use of these more sophisticated models is becoming more common because the models can be designed in immunocompetent mice, typically have high penetrance of the desired tumor type and are highly reproducible (Heyer et al., 2010). However, as with the use of cell lines, a well‐rounded understanding of strengths and limitations of each animal model is necessary for selection of an appropriate model based on the scientific question under investigation. As an example, genetically engineered mice with germline deletion of p53 are not exactly akin to the human counterpart of Li‐Fraumeni syndrome as they do not develop a similar range of epithelial cancers or at the same latency (Jacks et al., 1994), whereas mouse models with dominant‐negative gain‐of‐function mutations in p53 more accurately model the human disease (Olive et al., 2004). With the utilization of the appropriate model, in vivo experimentation remains the gold standard for target validation because the animal model more closely reflects the biology seen in human disease than any currently available in vitro system. The models can recapitulate tumor interaction with the microenvironment and, in the case of assessing a potential therapy, allow for evaluation of both the tolerability of therapy in other organ systems as well as the influence of metabolism on efficacy toward the target.
5. Successes in targeted therapy
To date, hundreds of candidate targets have been identified through biological and genomic studies and almost a thousand experimental anticancer therapies are now under development. However, only a few pathway‐targeted therapies have become standard of care (Ma and Adjei, 2009). Some of these are listed in Table 1. We describe a few of these in this section.
Table 1.
Successful genomic aberration‐targeted therapeutic agents
| Aberration | Cancer | Therapeutic agent | Reference |
|---|---|---|---|
| ABL1 translocation | CML, ALL, T‐ALL | Imatinib, nilotinib, dasatinib | (Druker et al., 2006), (Kantarjian et al., 2006), (Steinberg, 2007) |
| ERBB2 amplification | Breast, ovarian, gastric, NSCLC | Trastuzumab, lapatinib | (Hudis, 2007), (Kopper, 2008) |
| EGFR mutation, amplification | Glioma, NSCLC, Lung | Erlotinib, cetuximab, panitumumab | (Iyer and Bharthuar, 2010), (Graham et al., 2004), (Saltz et al., 2006) |
| KIT mutation, amplification | GIST, mucosal melanoma, AML, TGCT | Imatinib | (Demetri et al., 2002), (Guo et al., 2011), (Kindler et al., 2004), (Cassier et al., 2011) |
| PDGFRA, FLT3 mutation | GIST, renal | Sorafenib, Sunitinib | (Wilhelm et al., 2006), (Motzer et al., 2006) |
| BRCA1/2 mutation | Breast, ovarian, pancreatic | Olaparib, iniparib | (Fong et al., 2009), (O'Shaughnessy et al., 2011) |
| ALK rearrangements | NSCLC, ALCL, neuroblastoma | Crizotinib | (Shaw et al., 2011), (Bresler et al., 2011) |
One classic example is the targeted treatment for chronic myeloid leukemia (CML). The characteristic chromosomal rearrangement (the BCR‐Abl fusion or Philadelphia chromosome) seen in the disease was first described in 1960s (Nowell and Hungerford, 1960). As a brief aside, we should note that the lengthy timeframe from discovery of pathogenic mutation to development of tractable clinical therapy may be exaggerated in this example as several research techniques which would play an important role in understanding the genesis of CML were developed after first report of the Philadelphia chromosome (e.g. improved cytogenetic analysis). Therefore it was nearly 25 years between discovery of the pathogenic translocation and identification of the genes involved (de Klein et al., 1982; Groffen et al., 1984). Many of the important criteria for a good therapeutic target, as outlined previously, were found with the BCR‐Abl fusion protein: It was a translocation specific to the leukemic cells in CML, it was expressed in a high percentage of cases, and it involved activation of a kinase making it easier to target than loss of a tumor suppressor (Druker and Lydon, 2000). Development of targeted therapy began with screening of a library of compounds, including some known to be tyrosine kinase inhibitors, for the ability to inhibit the activity of the mutant BCR‐Abl kinase in CML. A lead compound was identified as weakly inhibiting the kinase and subsequently modified to optimize its activity (Buchdunger et al., 1996). Subsequent validation of the optimized compound was carried out in cell culture models, animal models, and in vitro using blood and bone marrow samples collected from patients with CML (Druker et al., 1996; le Coutre et al., 1999). Finally in 1998 a clinical trial was initiated assessing the efficacy of the targeted tyrosine kinase inhibitor – now known as imatinib or Gleevec – in patients with CML (Druker et al., 2001). Since, it has outperformed the previously used therapies in side‐by‐side clinical trials and has become the standard of care for patients with newly diagnosed CML (Druker et al., 2006). However, as with most targeted therapies, the genetic instability of cancer eventually allows for the emergence of a population of cells that have the appropriate genetic aberrations to confer resistance to a selected therapy. So it has become necessary for researchers to develop second‐line therapies that can counteract recurrent disease that has lost sensitivity to initial therapy. In the case of imatinib, mutations in the BCR‐Abl protein alter its conformation making it difficult for the drug to bind (Burgess et al., 2005). For patients with CML that is imatinib‐resistant a second generation of tyrosine kinase inhibitors has been developed, including dasatinib and nilotinib. These drugs inhibit the BCR‐Abl kinase but via a different mechanism than imatinib and have shown promise therapeutically as second‐line treatment in CML (Stein and Smith, 2010).
In breast cancer, amplification of the HER2 gene defines a subset of disease that is typically highly aggressive. Once HER2 amplification and kinase activity was identified as a driving event in tumorigenesis (Hudziak et al., 1987; King et al., 1985) it became the focus as a target for therapy. Activity of the receptor requires dimerization which can be blocked by an antibody. A panel of mouse antibodies generated against HER2 were screened for their ability to recognize the receptor, inhibit dimerization and prevent growth of HER2‐positive breast cancer cells in vitro (Hudziak et al., 1989). Further pre‐clinical work was needed including humanization of the antibody so it could be tolerated by the immune system of a patient (Carter et al., 1992). After nearly a decade of development, trastuzumab was evaluated in the clinical setting as an effective targeted therapy for patients with HER2‐enriched breast cancer (Slamon et al., 2001). As with imatinib, trastuzumab demonstrated promising clinical responses in patients and has become the standard of care in HER2‐enriched breast cancer. However, also as seen with imatinib, resistance to trastuzumab occurs in a significant proportion of cancers through various mechanisms including compensatory signaling through parallel pathways such as the PI3‐kinase or Src pathways (Berns et al., 2007; Pohlmann et al., 2009; Zhang et al., 2011). However, unlike imatinib resistance which can be overcome with targeted therapy directed to the same molecule, resistance to HER2‐targeted therapy is more effectively overcome by using targeted therapies to multiple pathways (Tseng et al., 2006; Zhang et al., 2011).
Although difficult, there has also been success in developing targeted therapies for cancers that have loss of tumor suppressors as their driving feature such as breast and ovarian cancers arising from loss of function of the BRCA genes. It was stated earlier that, in general, it is difficult to pharmacologically target mutations in tumor suppressor genes and gene therapy is one of the few treatment modalities for tumors arising from loss of tumor suppressors. BRCA1 and BRCA2, however, are tumor suppressors whose loss of function can be treated pharmacological. The genes are involved in DNA damage repair therefore loss of function can result in a higher number of genomic aberrations accumulating in cells (even if only one copy of a BRCA gene is lost) (Konishi et al., 2011). As with resistance to targeted therapy, the cell uses other pathways to compensate for loss of DNA repair function mediated by BRCA1 and BRCA2. In this case, the cell relies heavily of the poly ADP‐ribose polymerase (PARP) pathway to repair damaged DNA, and subsequent pharmacological inhibition of the PARP pathway is synthetically lethal and causes the cell to undergo apoptosis (Farmer et al., 2005). Pharmacologic treatment is possible in this case because the therapy is targeted at a compensatory pathway. Strikingly this treatment has little adverse effects because non‐tumor cells in the patient still have a functional DNA repair pathway mediated by BRCA proteins and so are not sensitized to PARP inhibition (Fong et al., 2009). As denoted in Table 1, two PARP inhibitors – olaparib and iniparib – have passed pre‐clinical testing and are being used therapeutically for patients with BRCA‐driven breast cancer (Fong et al., 2009; O'Shaughnessy et al., 2011).
6. Interactions between pathways
A major difficulty in cancer treatment is the interconnectedness of intracellular pathways. Unlike in the stages of target validation using simplified cell or animal models, the development of new therapies directed at these targets must eventually encounter the convoluted domain of cellular signaling. To use breast cancer as an example, let us consider three major proteins used widely to determine clinical outcome and for targeted treatment: human epidermal growth factor receptor 2 (HER2) (Prat and Perou, 2011), estrogen receptor alpha (ER) (Prat and Perou, 2011), and the catalytic subunit of the PI3‐kinase family member of the gene encoded by PIK3CA (PIK3CA) (Kalinsky et al., 2009).
Breast cancers are generally classified into 5 subtypes: luminal A, luminal B, HER2‐enriched, claudin‐low and basal (Prat and Perou, 2011). Both luminal types are defined by expression of ER and luminal B tumors are additionally defined as being HER2‐enriched. Claudin‐low and basal subtypes are defined as negative for expression of ER and not HER2‐enriched (Prat and Perou, 2011) so for the purpose of this discussion will be omitted. PIK3CA status is not used as a criterion for stratification and it has been shown that mutations in PIK3CA significantly correlate with both luminal classes of breast cancer (Sorlie et al., 2001). Interestingly, in the luminal A subtype the presence of a PIK3CA mutation, perhaps paradoxically, is associated with better outcome to hormonal therapy such as tamoxifen (Loi et al., 2010). The exact mechanism for this is still unclear but it may be because although PIK3CA mutations are oncogenic they are only weakly activating and result in low levels of signaling through mTORC1 (a downstream effector of PI3K signaling) which permits the cell to remain sensitive to hormonal therapy. Indeed, others have reported that mutations in PIK3CA correlate with a favorable clinical outcome (Kalinsky et al., 2009). However, when in the context of amplification of the HER2 locus a different relationship prevails. In HER2‐enirched tumors, resistance to HER2‐targeted therapy is coupled with mutation in the PI3K pathway (Junttila et al., 2009) and progression‐free survival of patients after HER2‐targeted therapy is significantly worse in the presence of a PIK3CA mutation (Berns et al., 2007). It should be noted that in the latter study the mutational analysis was performed in patients that had already received HER2‐targeted therapy so it is impossible to know if the mutations in PIK3CA were harbored in the tumor prior to treatment and selected for with HER2‐targeted therapy or if they are de novo mutations that cause resistance to the therapy. Regardless, resistance to HER2‐targeted therapy can be reversed by pharmacologic inhibition of the PI3K pathway (Eichhorn et al., 2008). So the status of PIK3CA depends on the context in which it is present: In ER‐positive disease it is a good prognostic factor while in HER2‐enriched tumors it is associated with a therapy‐resistant phenotype.
In addition to signaling through the PI3‐kinase pathway, the family of epidermal growth factor receptors – of which HER2 is a member – can signal through the mitogen‐activated protein kinase (MAPK) pathway. Additionally, the interaction between the MAP kinase pathway and nuclear hormone receptors in wild‐type cells has been shown (Kato et al., 1995). However within a tumor these pathways become more intimately involved as they can cooperate with or compensate for each other. As an example, resistance to tamoxifen is accompanied by increased levels of ERBB2 in the breast cancer cell line model MCF‐7 (Knowlden et al., 2003). In this case when the growth‐proliferative signals of ER are blunted with tamoxifen, the cells compensate by increasing expression of HER2 (and HER1). As expected, combining hormonal therapy with a therapy targeted to a member of the EGFR family effectively induces growth arrest in resistant cells (Chu et al., 2005; Gee et al., 2003; Johnston et al., 2009). A similar phenomenon is seen when focusing on the HER2 pathway for targeted therapy and ferreting out a mechanism of resistance. In a model of resistance to lapatinib (a small molecule inhibitor of HER1 and HER2 kinase activity), cells become dependent on ER signaling demonstrated by increased nuclear localization of ER and ER‐induced gene expression (Xia et al., 2006). Again, in this model combining lapatinib with an anti‐estrogenic compound is sufficient to overcome the resistance of the cells to the single treatment modality. So, it is clear when implementing combination therapy that the therapy must be omic context‐dependent (e.g. lapatinib with an anti‐estrogenic compound in a HER2‐enriched but ER‐negative tumor is likely useless). It is important, then, not just to understand an individual pathway in which a gene participates but how each of those pathways can interact inside the cell.
As discussed in the consideration of only three genes important for breast cancer (ER, HER2, and PIK3CA), the straightforward rationale of targeting each for therapy quickly becomes muddled when the interactions of respective pathways are taken into account. To reiterate, PIK3CA mutations are correlated with good response to hormonal therapy in ER‐positive breast cancer but associated with poor response and resistance to HER2‐targeted therapy in HER2‐enirched tumors. Even with the considerations enumerated here other genomic factors, such as microRNAs, or epigenetic alterations are not discussed. Clearly, when taken altogether the environment inside the cell is multi‐faceted and multi‐factorial, and researchers have the daunting task of trying to unravel its intricacies and understand how they are important in cancer.
7. Interactions with the microenvironment
The function of genomic aberrations in a cancer cell can be significantly conditioned by the microenvironment in which the cancer cell resides. A complete discussion of all components of the microenvironment and how they interact with a tumor is beyond the scope of this text (more thorough reviews can be found elsewhere, see ref. (Bissell and Hines, 2011)). However, we highlight a few important components of the microenvironment, explain how they interact with a tumor, and discuss the possibility of therapy directed toward microenvironmental factors.
One remarkable feature of the tumor microenvironment is its diversity. Cancer cells may reside in variable proximity to the stromal environment where they arise, immune cells may be recruited to the cancer, and soluble factors such as cytokines may be generated at sites distant from the tumor. These factors can influence the activity of the signaling pathways that are targeted by the therapeutic agents. Fibroblasts are the main cellular component of the stroma and are the cells responsible for depositing the extracellular matrix. There is dynamic signaling between the lesion and the stroma that aids in the progression to invasive disease. For example, the cytokine transforming growth factor‐beta (TGF‐β) is released by the cancer cells resulting in formation of the reactive stroma that remodels the architecture of the extracellular matrix in the proximity of the tumor (Ronnov‐Jessen et al., 1996). The newly remodeled extracellular matrix is more conducive to angiogenesis. An additional effect of TGF‐β release is the recruitment of immune cells such as macrophages to the region of the tumor (Sieweke et al., 1990). Recruited macrophages are of great interest because of the roles they play in angiogenesis, tumor progression, and metastasis (reviewed in Qian and Pollard, 2010). Macrophages have the ability to degrade the extracellular matrix of the stroma and the basement membrane encircling an in situ lesion which permits an invasive phenotype (Condeelis and Pollard, 2006; Wyckoff et al., 2004). Additionally, macrophages have been linked to the “angiogenic switch” – that is, an increase in vasculature to the tumor which is associated with tumor cell invasion and metastasis (Lin et al., 2006). This is a result of release of another important microenvironmental signaling factor, vascular endothelial growth factor (VEGF). So, by cooperating with its microenvironment, an in situ lesion poises itself to become not only an invasive cancer but potentially metastatic. Again, only a few players in the microenvironment milieu have been mentioned here and their role in carcinogenesis and tumor progression outlined. Multiple additional factors compose the microenvironment and are involved in the tumorigenic process; thus it is clear that the cancer cell does not act alone and in its environment is part of network of interactions as richly complex as those seen within the cell.
So it is possible that the microenvironment could be the target for therapy and indeed research in this area has seen success; each component of the microenvironment detailed in the brief discussion above can also be used as an example of a target for therapeutic intervention. Bevacizumab (Avastin) is an FDA‐approved drug that blocks the action of the soluble factor VEGF thereby inhibiting tumor angiogenesis (Presta et al., 1997). This is the best known example and probably the most successful as it has passed clinical trials and is used regularly in the treatment of certain cancers. In vivo studies also have shown that tumor‐associated macrophages can be targeted therapeutically and results in reduced tumor growth and metastasis (Luo et al., 2006). It has even been possible to pharmacologically target stromal tissue and in doing so increase efficacy of chemotherapy in a mouse model of pancreatic cancer (Olive et al., 2009). Many other examples exist but these give a small flavor of the potential utility of targeting the microenvironment in treating cancer.
8. Molecular markers
Clinical deployment of targeted therapeutic agents typically requires a molecular assay that defines the cancer subpopulation most likely to respond. Simple genomic markers that predict response include immunohistochemical and fluorescence in situ hybridization (FISH) based assays for HER2 amplification to identify potential responders to trastuzumab and lapatinib (Dybdal et al., 2005), FISH‐ and PCR‐based assays to detect the BCR‐Abl fusion gene to identify imatinib responders (Bhatia et al., 2003), and DNA sequence‐based assays to detect BRCA1/2 carriers that are likely to respond to olaparib or iniparib (Spurdle et al., 2012). More complex omic signatures are being explored to predict responses to a broad range of targeted and non‐targets therapies. Recent studies suggest that useful markers may be found to predict responses to a significant fraction of targeted therapies (Heiser et al., 2011; Spurdle et al., 2012; Wagle et al., 2012; Walsh et al., 2010). However, development of such markers is only beginning and careful assay standardization (e.g. development according to Clinical Laboratory Improvement Amendments (CLIA) guidelines) and clinical validation is needed before assays should be used to select patients for targeted therapies.
Altogether, the confluence of international efforts to identify mutations in human tumors, understand pathways that are deregulated, develop targeted therapies for those pathways and properly implement those therapies clinically is leading us to the desired horizon – an age of personalized medicine where the most efficacious treatment is dictated by and tailored to the tumor and the patient. While more work and likely many more years of effort are needed to reach this goal, this review provides insight into the methodology of developing targeted therapies and outlines successes that demonstrate the fight against cancer is not insurmountable.
Garay Joseph P., Gray Joe W. (2012), Omics and therapy – A basis for precision medicine, Molecular Oncology, 6, doi: 10.1016/j.molonc.2012.02.009.
References
- Abukhdeir, A.M. , Vitolo, M.I. , Argani, P. , De Marzo, A.M. , Karakas, B. , Konishi, H. , Gustin, J.P. , Lauring, J. , Garay, J.P. , Pendleton, C. , Konishi, Y. , Blair, B.G. , Brenner, K. , Garrett-Mayer, E. , Carraway, H. , Bachman, K.E. , Park, B.H. , 2008. Tamoxifen-stimulated growth of breast cancer due to p21 loss. Proc. Natl. Acad. Sci. U S A. 105, 288–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alizadeh, A.A. , Eisen, M.B. , Davis, R.E. , Ma, C. , Lossos, I.S. , Rosenwald, A. , Boldrick, J.C. , Sabet, H. , Tran, T. , Yu, X. , Powell, J.I. , Yang, L. , Marti, G.E. , Moore, T. , Hudson, J. , Lu, L. , Lewis, D.B. , Tibshirani, R. , Sherlock, G. , Chan, W.C. , Greiner, T.C. , Weisenburger, D.D. , Armitage, J.O. , Warnke, R. , Levy, R. , Wilson, W. , Grever, M.R. , Byrd, J.C. , Botstein, D. , Brown, P.O. , Staudt, L.M. , 2000. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 403, 503–511. [DOI] [PubMed] [Google Scholar]
- Bello, D. , Webber, M.M. , Kleinman, H.K. , Wartinger, D.D. , Rhim, J.S. , 1997. Androgen responsive adult human prostatic epithelial cell lines immortalized by human papillomavirus 18. Carcinogenesis. 18, 1215–1223. [DOI] [PubMed] [Google Scholar]
- Berns, K. , Horlings, H.M. , Hennessy, B.T. , Madiredjo, M. , Hijmans, E.M. , Beelen, K. , Linn, S.C. , Gonzalez-Angulo, A.M. , Stemke-Hale, K. , Hauptmann, M. , Beijersbergen, R.L. , Mills, G.B. , van de Vijver, M.J. , Bernards, R. , 2007. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 12, 395–402. [DOI] [PubMed] [Google Scholar]
- Bhatia, R. , Holtz, M. , Niu, N. , Gray, R. , Snyder, D.S. , Sawyers, C.L. , Arber, D.A. , Slovak, M.L. , Forman, S.J. , 2003. Persistence of malignant hematopoietic progenitors in chronic myelogenous leukemia patients in complete cytogenetic remission following imatinib mesylate treatment. Blood. 101, 4701–4707. [DOI] [PubMed] [Google Scholar]
- Bissell, M.J. , Hines, W.C. , 2011. Why don't we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat. Med.. 17, 320–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresler, S.C. , Wood, A.C. , Haglund, E.A. , Courtright, J. , Belcastro, L.T. , Plegaria, J.S. , Cole, K. , Toporovskaya, Y. , Zhao, H. , Carpenter, E.L. , Christensen, J.G. , Maris, J.M. , Lemmon, M.A. , Mosse, Y.P. , 2011. Differential inhibitor sensitivity of anaplastic lymphoma kinase variants found in neuroblastoma. Sci. Transl Med.. 3, 108ra114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchdunger, E. , Zimmermann, J. , Mett, H. , Meyer, T. , Muller, M. , Druker, B.J. , Lydon, N.B. , 1996. Inhibition of the Abl protein-tyrosine kinase in vitro and in vivo by a 2-phenylaminopyrimidine derivative. Cancer Res.. 56, 100–104. [PubMed] [Google Scholar]
- Burgess, M.R. , Skaggs, B.J. , Shah, N.P. , Lee, F.Y. , Sawyers, C.L. , 2005. Comparative analysis of two clinically active BCR-ABL kinase inhibitors reveals the role of conformation-specific binding in resistance. Proc. Natl. Acad. Sci. U S A. 102, 3395–3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter, P. , Presta, L. , Gorman, C.M. , Ridgway, J.B. , Henner, D. , Wong, W.L. , Rowland, A.M. , Kotts, C. , Carver, M.E. , Shepard, H.M. , 1992. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc. Natl. Acad. Sci. U S A. 89, 4285–4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassier, P.A. , Gelderblom, H. , Stacchiotti, S. , Thomas, D. , Maki, R.G. , Kroep, J.R. , van der Graaf, W.T. , Italiano, A. , Seddon, B. , Domont, J. , Bompas, E. , Wagner, A.J. , Blay, J.Y. , 2011. Efficacy of imatinib mesylate for the treatment of locally advanced and/or metastatic tenosynovial giant cell tumor/pigmented villonodular synovitis. Cancer. [DOI] [PubMed] [Google Scholar]
- Cerami, E. , Demir, E. , Schultz, N. , Taylor, B.S. , Sander, C. , 2010. Automated network analysis identifies core pathways in glioblastoma. PLoS One. 5, e8918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami, E.G. , Gross, B.E. , Demir, E. , Rodchenkov, I. , Babur, O. , Anwar, N. , Schultz, N. , Bader, G.D. , Sander, C. , 2011. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res.. 39, D685–D690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin, K. , DeVries, S. , Fridlyand, J. , Spellman, P.T. , Roydasgupta, R. , Kuo, W.L. , Lapuk, A. , Neve, R.M. , Qian, Z. , Ryder, T. , Chen, F. , Feiler, H. , Tokuyasu, T. , Kingsley, C. , Dairkee, S. , Meng, Z. , Chew, K. , Pinkel, D. , Jain, A. , Ljung, B.M. , Esserman, L. , Albertson, D.G. , Waldman, F.M. , Gray, J.W. , 2006. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 10, 529–541. [DOI] [PubMed] [Google Scholar]
- Chu, I. , Blackwell, K. , Chen, S. , Slingerland, J. , 2005. The dual ErbB1/ErbB2 inhibitor, lapatinib (GW572016), cooperates with tamoxifen to inhibit both cell proliferation- and estrogen-dependent gene expression in antiestrogen-resistant breast cancer. Cancer Res.. 65, 18–25. [PubMed] [Google Scholar]
- Collisson, E.A. , Sadanandam, A. , Olson, P. , Gibb, W.J. , Truitt, M. , Gu, S. , Cooc, J. , Weinkle, J. , Kim, G.E. , Jakkula, L. , Feiler, H.S. , Ko, A.H. , Olshen, A.B. , Danenberg, K.L. , Tempero, M.A. , Spellman, P.T. , Hanahan, D. , Gray, J.W. , 2011. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med.. 17, 500–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condeelis, J. , Pollard, J.W. , 2006. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 124, 263–266. [DOI] [PubMed] [Google Scholar]
- le Coutre, P. , Mologni, L. , Cleris, L. , Marchesi, E. , Buchdunger, E. , Giardini, R. , Formelli, F. , Gambacorti-Passerini, C. , 1999. In vivo eradication of human BCR/Abl-positive leukemia cells with an Abl kinase inhibitor. J. Natl. Cancer Inst.. 91, 163–168. [DOI] [PubMed] [Google Scholar]
- Demetri, G.D. , von Mehren, M. , Blanke, C.D. , Van den Abbeele, A.D. , Eisenberg, B. , Roberts, P.J. , Heinrich, M.C. , Tuveson, D.A. , Singer, S. , Janicek, M. , Fletcher, J.A. , Silverman, S.G. , Silberman, S.L. , Capdeville, R. , Kiese, B. , Peng, B. , Dimitrijevic, S. , Druker, B.J. , Corless, C. , Fletcher, C.D. , Joensuu, H. , 2002. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N. Engl. J. Med.. 347, 472–480. [DOI] [PubMed] [Google Scholar]
- Dow, L.E. , Premsrirut, P.K. , Zuber, J. , Fellmann, C. , McJunkin, K. , Miething, C. , Park, Y. , Dickins, R.A. , Hannon, G.J. , Lowe, S.W. , 2012. A pipeline for the generation of shRNA transgenic mice. Nat. Protoc.. 7, 374–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druker, B.J. , Lydon, N.B. , 2000. Lessons learned from the development of an Abl tyrosine kinase inhibitor for chronic myelogenous leukemia. J. Clin. Invest.. 105, 3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druker, B.J. , Tamura, S. , Buchdunger, E. , Ohno, S. , Segal, G.M. , Fanning, S. , Zimmermann, J. , Lydon, N.B. , 1996. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat. Med.. 2, 561–566. [DOI] [PubMed] [Google Scholar]
- Druker, B.J. , Talpaz, M. , Resta, D.J. , Peng, B. , Buchdunger, E. , Ford, J.M. , Lydon, N.B. , Kantarjian, H. , Capdeville, R. , Ohno-Jones, S. , Sawyers, C.L. , 2001. Efficacy and safety of a specific inhibitor of the BCR-Abl tyrosine kinase in chronic myeloid leukemia. N. Engl. J. Med.. 344, 1031–1037. [DOI] [PubMed] [Google Scholar]
- Druker, B.J. , Guilhot, F. , O'Brien, S.G. , Gathmann, I. , Kantarjian, H. , Gattermann, N. , Deininger, M.W. , Silver, R.T. , Goldman, J.M. , Stone, R.M. , Cervantes, F. , Hochhaus, A. , Powell, B.L. , Gabrilove, J.L. , Rousselot, P. , Reiffers, J. , Cornelissen, J.J. , Hughes, T. , Agis, H. , Fischer, T. , Verhoef, G. , Shepherd, J. , Saglio, G. , Gratwohl, A. , Nielsen, J.L. , Radich, J.P. , Simonsson, B. , Taylor, K. , Baccarani, M. , So, C. , Letvak, L. , Larson, R.A. , 2006. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N. Engl. J. Med.. 355, 2408–2417. [DOI] [PubMed] [Google Scholar]
- Dybdal, N. , Leiberman, G. , Anderson, S. , McCune, B. , Bajamonde, A. , Cohen, R.L. , Mass, R.D. , Sanders, C. , Press, M.F. , 2005. Determination of HER2 gene amplification by fluorescence in situ hybridization and concordance with the clinical trials immunohistochemical assay in women with metastatic breast cancer evaluated for treatment with trastuzumab. Breast Cancer Res. Treat.. 93, 3–11. [DOI] [PubMed] [Google Scholar]
- Eichhorn, P.J. , Gili, M. , Scaltriti, M. , Serra, V. , Guzman, M. , Nijkamp, W. , Beijersbergen, R.L. , Valero, V. , Seoane, J. , Bernards, R. , Baselga, J. , 2008. Phosphatidylinositol 3-kinase hyperactivation results in lapatinib resistance that is reversed by the mTOR/phosphatidylinositol 3-kinase inhibitor NVP-BEZ235. Cancer Res.. 68, 9221–9230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer, H. , McCabe, N. , Lord, C.J. , Tutt, A.N. , Johnson, D.A. , Richardson, T.B. , Santarosa, M. , Dillon, K.J. , Hickson, I. , Knights, C. , Martin, N.M. , Jackson, S.P. , Smith, G.C. , Ashworth, A. , 2005. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 434, 917–921. [DOI] [PubMed] [Google Scholar]
- Fong, P.C. , Boss, D.S. , Yap, T.A. , Tutt, A. , Wu, P. , Mergui-Roelvink, M. , Mortimer, P. , Swaisland, H. , Lau, A. , O'Connor, M.J. , Ashworth, A. , Carmichael, J. , Kaye, S.B. , Schellens, J.H. , de Bono, J.S. , 2009. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med.. 361, 123–134. [DOI] [PubMed] [Google Scholar]
- Furukawa, T. , Duguid, W.P. , Rosenberg, L. , Viallet, J. , Galloway, D.A. , Tsao, M.S. , 1996. Long-term culture and immortalization of epithelial cells from normal adult human pancreatic ducts transfected by the E6E7 gene of human papilloma virus 16. Am. J. Pathol.. 148, 1763–1770. [PMC free article] [PubMed] [Google Scholar]
- Gee, J.M. , Harper, M.E. , Hutcheson, I.R. , Madden, T.A. , Barrow, D. , Knowlden, J.M. , McClelland, R.A. , Jordan, N. , Wakeling, A.E. , Nicholson, R.I. , 2003. The antiepidermal growth factor receptor agent gefitinib (ZD1839/Iressa) improves antihormone response and prevents development of resistance in breast cancer in vitro. Endocrinology. 144, 5105–5117. [DOI] [PubMed] [Google Scholar]
- Graham, J. , Muhsin, M. , Kirkpatrick, P. , 2004. Cetuximab. Nat. Rev. Drug Discov.. 3, 549–550. [DOI] [PubMed] [Google Scholar]
- Groffen, J. , Stephenson, J.R. , Heisterkamp, N. , de Klein, A. , Bartram, C.R. , Grosveld, G. , 1984. Philadelphia chromosomal breakpoints are clustered within a limited region, BCR, on chromosome 22. Cell. 36, 93–99. [DOI] [PubMed] [Google Scholar]
- Guo, J. , Si, L. , Kong, Y. , Flaherty, K.T. , Xu, X. , Zhu, Y. , Corless, C.L. , Li, L. , Li, H. , Sheng, X. , Cui, C. , Chi, Z. , Li, S. , Han, M. , Mao, L. , Lin, X. , Du, N. , Zhang, X. , Li, J. , Wang, B. , Qin, S. , 2011. Phase II, open-label, single-arm trial of imatinib mesylate in patients with metastatic melanoma harboring c-Kit mutation or amplification. J. Clin. Oncol.. 29, 2904–2909. [DOI] [PubMed] [Google Scholar]
- Gustin, J.P. , Karakas, B. , Weiss, M.B. , Abukhdeir, A.M. , Lauring, J. , Garay, J.P. , Cosgrove, D. , Tamaki, A. , Konishi, H. , Konishi, Y. , Mohseni, M. , Wang, G. , Rosen, D.M. , Denmeade, S.R. , Higgins, M.J. , Vitolo, M.I. , Bachman, K.E. , Park, B.H. , 2009. Knockin of mutant PIK3CA activates multiple oncogenic pathways. Proc. Natl. Acad. Sci. U S A. 106, 2835–2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan, D. , Weinberg, R.A. , 2000. The hallmarks of cancer. Cell. 100, 57–70. [DOI] [PubMed] [Google Scholar]
- Harley, C.B. , Futcher, A.B. , Greider, C.W. , 1990. Telomeres shorten during ageing of human fibroblasts. Nature. 345, 458–460. [DOI] [PubMed] [Google Scholar]
- Heiser, L.M. , Sadanandam, A. , Kuo, W.L. , Benz, S.C. , Goldstein, T.C. , Ng, S. , Gibb, W.J. , Wang, N.J. , Ziyad, S. , Tong, F. , Bayani, N. , Hu, Z. , Billig, J.I. , Dueregger, A. , Lewis, S. , Jakkula, L. , Korkola, J.E. , Durinck, S. , Pepin, F. , Guan, Y. , Purdom, E. , Neuvial, P. , Bengtsson, H. , Wood, K.W. , Smith, P.G. , Vassilev, L.T. , Hennessy, B.T. , Greshock, J. , Bachman, K.E. , Hardwicke, M.A. , Park, J.W. , Marton, L.J. , Wolf, D.M. , Collisson, E.A. , Neve, R.M. , Mills, G.B. , Speed, T.P. , Feiler, H.S. , Wooster, R.F. , Haussler, D. , Stuart, J.M. , Gray, J.W. , Spellman, P.T. , 2011. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc. Natl. Acad. Sci. U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer, J. , Kwong, L.N. , Lowe, S.W. , Chin, L. , 2010. Non-germline genetically engineered mouse models for translational cancer research. Nat. Rev. Cancer. 10, 470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson, J.G. , Malek, T. , Bornstein, S. , Hariono, S. , Ginzinger, D.G. , Muller, W.J. , Gray, J.W. , 2005. Copy number aberrations in mouse breast tumors reveal loci and genes important in tumorigenic receptor tyrosine kinase signaling. Cancer Res.. 65, 9695–9704. [DOI] [PubMed] [Google Scholar]
- Hollstein, M. , Sidransky, D. , Vogelstein, B. , Harris, C.C. , 1991. p53 mutations in human cancers. Science. 253, 49–53. [DOI] [PubMed] [Google Scholar]
- Hudis, C.A. , 2007. Trastuzumab – Mechanism of action and use in clinical practice. N. Engl. J. Med.. 357, 39–51. [DOI] [PubMed] [Google Scholar]
- Hudziak, R.M. , Schlessinger, J. , Ullrich, A. , 1987. Increased expression of the putative growth factor receptor p185HER2 causes transformation and tumorigenesis of NIH 3T3 cells. Proc. Natl. Acad. Sci. U S A. 84, 7159–7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudziak, R.M. , Lewis, G.D. , Winget, M. , Fendly, B.M. , Shepard, H.M. , Ullrich, A. , 1989. p185HER2 monoclonal antibody has antiproliferative effects in vitro and sensitizes human breast tumor cells to tumor necrosis factor. Mol. Cell Biol.. 9, 1165–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer, R. , Bharthuar, A. , 2010. A review of erlotinib – An oral, selective epidermal growth factor receptor tyrosine kinase inhibitor. Expert Opin. Pharmacother.. 11, 311–320. [DOI] [PubMed] [Google Scholar]
- Jacks, T. , Remington, L. , Williams, B.O. , Schmitt, E.M. , Halachmi, S. , Bronson, R.T. , Weinberg, R.A. , 1994. Tumor spectrum analysis in p53-mutant mice. Curr. Biol.. 4, 1–7. [DOI] [PubMed] [Google Scholar]
- Johnston, S. , Pippen, J. , Pivot, X. , Lichinitser, M. , Sadeghi, S. , Dieras, V. , Gomez, H.L. , Romieu, G. , Manikhas, A. , Kennedy, M.J. , Press, M.F. , Maltzman, J. , Florance, A. , O'Rourke, L. , Oliva, C. , Stein, S. , Pegram, M. , 2009. Lapatinib combined with letrozole versus letrozole and placebo as first-line therapy for postmenopausal hormone receptor-positive metastatic breast cancer. J. Clin. Oncol.. 27, 5538–5546. [DOI] [PubMed] [Google Scholar]
- Jones, S. , Zhang, X. , Parsons, D.W. , Lin, J.C. , Leary, R.J. , Angenendt, P. , Mankoo, P. , Carter, H. , Kamiyama, H. , Jimeno, A. , Hong, S.M. , Fu, B. , Lin, M.T. , Calhoun, E.S. , Kamiyama, M. , Walter, K. , Nikolskaya, T. , Nikolsky, Y. , Hartigan, J. , Smith, D.R. , Hidalgo, M. , Leach, S.D. , Klein, A.P. , Jaffee, E.M. , Goggins, M. , Maitra, A. , Iacobuzio-Donahue, C. , Eshleman, J.R. , Kern, S.E. , Hruban, R.H. , Karchin, R. , Papadopoulos, N. , Parmigiani, G. , Vogelstein, B. , Velculescu, V.E. , Kinzler, K.W. , 2008. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 321, 1801–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junttila, T.T. , Akita, R.W. , Parsons, K. , Fields, C. , Lewis Phillips, G.D. , Friedman, L.S. , Sampath, D. , Sliwkowski, M.X. , 2009. Ligand-independent HER2/HER3/PI3K complex is disrupted by trastuzumab and is effectively inhibited by the PI3K inhibitor GDC-0941. Cancer Cell. 15, 429–440. [DOI] [PubMed] [Google Scholar]
- Kalinsky, K. , Jacks, L.M. , Heguy, A. , Patil, S. , Drobnjak, M. , Bhanot, U.K. , Hedvat, C.V. , Traina, T.A. , Solit, D. , Gerald, W. , Moynahan, M.E. , 2009. PIK3CA mutation associates with improved outcome in breast cancer. Clin. Cancer Res.. 15, 5049–5059. [DOI] [PubMed] [Google Scholar]
- Kantarjian, H. , Giles, F. , Wunderle, L. , Bhalla, K. , O'Brien, S. , Wassmann, B. , Tanaka, C. , Manley, P. , Rae, P. , Mietlowski, W. , Bochinski, K. , Hochhaus, A. , Griffin, J.D. , Hoelzer, D. , Albitar, M. , Dugan, M. , Cortes, J. , Alland, L. , Ottmann, O.G. , 2006. Nilotinib in imatinib-resistant CML and Philadelphia chromosome-positive ALL. N. Engl. J. Med.. 354, 2542–2551. [DOI] [PubMed] [Google Scholar]
- Kato, S. , Endoh, H. , Masuhiro, Y. , Kitamoto, T. , Uchiyama, S. , Sasaki, H. , Masushige, S. , Gotoh, Y. , Nishida, E. , Kawashima, H. , Metzger, D. , Chambon, P. , 1995. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science. 270, 1491–1494. [DOI] [PubMed] [Google Scholar]
- Kessler, J.D. , Kahle, K.T. , Sun, T. , Meerbrey, K.L. , Schlabach, M.R. , Schmitt, E.M. , Skinner, S.O. , Xu, Q. , Li, M.Z. , Hartman, Z.C. , Rao, M. , Yu, P. , Dominguez-Vidana, R. , Liang, A.C. , Solimini, N.L. , Bernardi, R.J. , Yu, B. , Hsu, T. , Golding, I. , Luo, J. , Osborne, C.K. , Creighton, C.J. , Hilsenbeck, S.G. , Schiff, R. , Shaw, C.A. , Elledge, S.J. , Westbrook, T.F. , 2012. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science. 335, 348–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler, T. , Breitenbuecher, F. , Marx, A. , Beck, J. , Hess, G. , Weinkauf, B. , Duyster, J. , Peschel, C. , Kirkpatrick, C.J. , Theobald, M. , Gschaidmeier, H. , Huber, C. , Fischer, T. , 2004. Efficacy and safety of imatinib in adult patients with c-kit-positive acute myeloid leukemia. Blood. 103, 3644–3654. [DOI] [PubMed] [Google Scholar]
- King, C.R. , Kraus, M.H. , Aaronson, S.A. , 1985. Amplification of a novel v-erbB-related gene in a human mammary carcinoma. Science. 229, 974–976. [DOI] [PubMed] [Google Scholar]
- de Klein, A. , van Kessel, A.G. , Grosveld, G. , Bartram, C.R. , Hagemeijer, A. , Bootsma, D. , Spurr, N.K. , Heisterkamp, N. , Groffen, J. , Stephenson, J.R. , 1982. A cellular oncogene is translocated to the Philadelphia chromosome in chronic myelocytic leukaemia. Nature. 300, 765–767. [DOI] [PubMed] [Google Scholar]
- Knowlden, J.M. , Hutcheson, I.R. , Jones, H.E. , Madden, T. , Gee, J.M. , Harper, M.E. , Barrow, D. , Wakeling, A.E. , Nicholson, R.I. , 2003. Elevated levels of epidermal growth factor receptor/c-erbB2 heterodimers mediate an autocrine growth regulatory pathway in tamoxifen-resistant MCF-7 cells. Endocrinology. 144, 1032–1044. [DOI] [PubMed] [Google Scholar]
- Konishi, H. , Karakas, B. , Abukhdeir, A.M. , Lauring, J. , Gustin, J.P. , Garay, J.P. , Konishi, Y. , Gallmeier, E. , Bachman, K.E. , Park, B.H. , 2007. Knock-in of mutant K-ras in nontumorigenic human epithelial cells as a new model for studying K-ras mediated transformation. Cancer Res.. 67, 8460–8467. [DOI] [PubMed] [Google Scholar]
- Konishi, H. , Mohseni, M. , Tamaki, A. , Garay, J.P. , Croessmann, S. , Karnan, S. , Ota, A. , Wong, H.Y. , Konishi, Y. , Karakas, B. , Tahir, K. , Abukhdeir, A.M. , Gustin, J.P. , Cidado, J. , Wang, G.M. , Cosgrove, D. , Cochran, R. , Jelovac, D. , Higgins, M.J. , Arena, S. , Hawkins, L. , Lauring, J. , Gross, A.L. , Heaphy, C.M. , Hosokawa, Y. , Gabrielson, E. , Meeker, A.K. , Visvanathan, K. , Argani, P. , Bachman, K.E. , Park, B.H. , 2011. Mutation of a single allele of the cancer susceptibility gene BRCA1 leads to genomic instability in human breast epithelial cells. Proc. Natl. Acad. Sci. U S A. 108, 17773–17778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopper, L. , 2008. Lapatinib: a sword with two edges. Pathol. Oncol. Res.. 14, 1–8. [DOI] [PubMed] [Google Scholar]
- Kumar, A. , White, T.A. , MacKenzie, A.P. , Clegg, N. , Lee, C. , Dumpit, R.F. , Coleman, I. , Ng, S.B. , Salipante, S.J. , Rieder, M.J. , Nickerson, D.A. , Corey, E. , Lange, P.H. , Morrissey, C. , Vessella, R.L. , Nelson, P.S. , Shendure, J. , 2011. Exome sequencing identifies a spectrum of mutation frequencies in advanced and lethal prostate cancers. Proc. Natl. Acad. Sci. U S A. 108, 17087–17092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kute, T. , Lack, C.M. , Willingham, M. , Bishwokama, B. , Williams, H. , Barrett, K. , Mitchell, T. , Vaughn, J.P. , 2004. Development of Herceptin resistance in breast cancer cells. Cytometry A. 57, 86–93. [DOI] [PubMed] [Google Scholar]
- Lin, E.Y. , Li, J.F. , Gnatovskiy, L. , Deng, Y. , Zhu, L. , Grzesik, D.A. , Qian, H. , Xue, X.N. , Pollard, J.W. , 2006. Macrophages regulate the angiogenic switch in a mouse model of breast cancer. Cancer Res.. 66, 11238–11246. [DOI] [PubMed] [Google Scholar]
- Liu, C.G. , Calin, G.A. , Meloon, B. , Gamliel, N. , Sevignani, C. , Ferracin, M. , Dumitru, C.D. , Shimizu, M. , Zupo, S. , Dono, M. , Alder, H. , Bullrich, F. , Negrini, M. , Croce, C.M. , 2004. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc. Natl. Acad. Sci. U S A. 101, 9740–9744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loi, S. , Haibe-Kains, B. , Majjaj, S. , Lallemand, F. , Durbecq, V. , Larsimont, D. , Gonzalez-Angulo, A.M. , Pusztai, L. , Symmans, W.F. , Bardelli, A. , Ellis, P. , Tutt, A.N. , Gillett, C.E. , Hennessy, B.T. , Mills, G.B. , Phillips, W.A. , Piccart, M.J. , Speed, T.P. , McArthur, G.A. , Sotiriou, C. , 2010. PIK3CA mutations associated with gene signature of low mTORC1 signaling and better outcomes in estrogen receptor-positive breast cancer. Proc. Natl. Acad. Sci. U S A. 107, 10208–10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, Y. , Zhou, H. , Krueger, J. , Kaplan, C. , Lee, S.H. , Dolman, C. , Markowitz, D. , Wu, W. , Liu, C. , Reisfeld, R.A. , Xiang, R. , 2006. Targeting tumor-associated macrophages as a novel strategy against breast cancer. J. Clin. Invest.. 116, 2132–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, J. , Emanuele, M.J. , Li, D. , Creighton, C.J. , Schlabach, M.R. , Westbrook, T.F. , Wong, K.K. , Elledge, S.J. , 2009. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 137, 835–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, T.J. , Bell, D.W. , Sordella, R. , Gurubhagavatula, S. , Okimoto, R.A. , Brannigan, B.W. , Harris, P.L. , Haserlat, S.M. , Supko, J.G. , Haluska, F.G. , Louis, D.N. , Christiani, D.C. , Settleman, J. , Haber, D.A. , 2004. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med.. 350, 2129–2139. [DOI] [PubMed] [Google Scholar]
- Ma, W.W. , Adjei, A.A. , 2009. Novel agents on the horizon for cancer therapy. CA Cancer J. Clin.. 59, 111–137. [DOI] [PubMed] [Google Scholar]
- Margolin, A.A. , Nemenman, I. , Basso, K. , Wiggins, C. , Stolovitzky, G. , Dalla Favera, R. , Califano, A. , 2006. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinform.. 7, (Suppl. 1) S7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser, R.S. , Choudhury, B. , Campbell, P.J. , Feng, B. , Wong, K.K. , Protopopov, A. , O'Neil, J. , Gutierrez, A. , Ivanova, E. , Perna, I. , Lin, E. , Mani, V. , Jiang, S. , McNamara, K. , Zaghlul, S. , Edkins, S. , Stevens, C. , Brennan, C. , Martin, E.S. , Wiedemeyer, R. , Kabbarah, O. , Nogueira, C. , Histen, G. , Aster, J. , Mansour, M. , Duke, V. , Foroni, L. , Fielding, A.K. , Goldstone, A.H. , Rowe, J.M. , Wang, Y.A. , Look, A.T. , Stratton, M.R. , Chin, L. , Futreal, P.A. , DePinho, R.A. , 2007. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 447, 966–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzker, M.L. , 2010. Sequencing technologies – The next generation. Nat. Rev. Genet.. 11, 31–46. [DOI] [PubMed] [Google Scholar]
- van Miltenburg, M.H. , Jonkers, J. , 2012. Using genetically engineered mouse models to validate candidate cancer genes and test new therapeutic approaches. Curr. Opin. Genet. Dev.. [DOI] [PubMed] [Google Scholar]
- Motzer, R.J. , Hoosen, S. , Bello, C.L. , Christensen, J.G. , 2006. Sunitinib malate for the treatment of solid tumours: a review of current clinical data. Expert Opin. Investig. Drugs. 15, 553–561. [DOI] [PubMed] [Google Scholar]
- Neve, R.M. , Chin, K. , Fridlyand, J. , Yeh, J. , Baehner, F.L. , Fevr, T. , Clark, L. , Bayani, N. , Coppe, J.P. , Tong, F. , Speed, T. , Spellman, P.T. , DeVries, S. , Lapuk, A. , Wang, N.J. , Kuo, W.L. , Stilwell, J.L. , Pinkel, D. , Albertson, D.G. , Waldman, F.M. , McCormick, F. , Dickson, R.B. , Johnson, M.D. , Lippman, M. , Ethier, S. , Gazdar, A. , Gray, J.W. , 2006. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 10, 515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell, P.C. , Hungerford, D.A. , 1960. A minute chromosome in human chronic granulocytic leukemia. Science. 132, 1497 [DOI] [PubMed] [Google Scholar]
- Olive, K.P. , Tuveson, D.A. , Ruhe, Z.C. , Yin, B. , Willis, N.A. , Bronson, R.T. , Crowley, D. , Jacks, T. , 2004. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 119, 847–860. [DOI] [PubMed] [Google Scholar]
- Olive, K.P. , Jacobetz, M.A. , Davidson, C.J. , Gopinathan, A. , McIntyre, D. , Honess, D. , Madhu, B. , Goldgraben, M.A. , Caldwell, M.E. , Allard, D. , Frese, K.K. , Denicola, G. , Feig, C. , Combs, C. , Winter, S.P. , Ireland-Zecchini, H. , Reichelt, S. , Howat, W.J. , Chang, A. , Dhara, M. , Wang, L. , Ruckert, F. , Grutzmann, R. , Pilarsky, C. , Izeradjene, K. , Hingorani, S.R. , Huang, P. , Davies, S.E. , Plunkett, W. , Egorin, M. , Hruban, R.H. , Whitebread, N. , McGovern, K. , Adams, J. , Iacobuzio-Donahue, C. , Griffiths, J. , Tuveson, D.A. , 2009. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 324, 1457–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shaughnessy, J. , Osborne, C. , Pippen, J.E. , Yoffe, M. , Patt, D. , Rocha, C. , Koo, I.C. , Sherman, B.M. , Bradley, C. , 2011. Iniparib plus chemotherapy in metastatic triple-negative breast cancer. N. Engl. J. Med.. 364, 205–214. [DOI] [PubMed] [Google Scholar]
- Penzel, R. , Sers, C. , Chen, Y. , Lehmann-Muhlenhoff, U. , Merkelbach-Bruse, S. , Jung, A. , Kirchner, T. , Buttner, R. , Kreipe, H.H. , Petersen, I. , Dietel, M. , Schirmacher, P. , 2011. EGFR mutation detection in NSCLC–assessment of diagnostic application and recommendations of the German Panel for Mutation Testing in NSCLC. Virchows Arch.. 458, 95–98. [DOI] [PubMed] [Google Scholar]
- Pinkel, D. , Segraves, R. , Sudar, D. , Clark, S. , Poole, I. , Kowbel, D. , Collins, C. , Kuo, W.L. , Chen, C. , Zhai, Y. , Dairkee, S.H. , Ljung, B.M. , Gray, J.W. , Albertson, D.G. , 1998. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat. Genet.. 20, 207–211. [DOI] [PubMed] [Google Scholar]
- Pohlmann, P.R. , Mayer, I.A. , Mernaugh, R. , 2009. Resistance to trastuzumab in breast cancer. Clin. Cancer Res.. 15, 7479–7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahallad, A. , Sun, C. , Huang, S. , Di Nicolantonio, F. , Salazar, R. , Zecchin, D. , Beijersbergen, R.L. , Bardelli, A. , Bernards, R. , 2012. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. [DOI] [PubMed] [Google Scholar]
- Prat, A. , Perou, C.M. , 2011. Deconstructing the molecular portraits of breast cancer. Mol. Oncol.. 5, 5–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presta, L.G. , Chen, H. , O'Connor, S.J. , Chisholm, V. , Meng, Y.G. , Krummen, L. , Winkler, M. , Ferrara, N. , 1997. Humanization of an anti-vascular endothelial growth factor monoclonal antibody for the therapy of solid tumors and other disorders. Cancer Res.. 57, 4593–4599. [PubMed] [Google Scholar]
- Qian, B.Z. , Pollard, J.W. , 2010. Macrophage diversity enhances tumor progression and metastasis. Cell. 141, 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronnov-Jessen, L. , Petersen, O.W. , Bissell, M.J. , 1996. Cellular changes involved in conversion of normal to malignant breast: importance of the stromal reaction. Physiol. Rev.. 76, 69–125. [DOI] [PubMed] [Google Scholar]
- Root, D.E. , Hacohen, N. , Hahn, W.C. , Lander, E.S. , Sabatini, D.M. , 2006. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat. Methods. 3, 715–719. [DOI] [PubMed] [Google Scholar]
- Saltz, L. , Easley, C. , Kirkpatrick, P. , 2006. Panitumumab. Nat. Rev. Drug Discov.. 5, 987–988. [DOI] [PubMed] [Google Scholar]
- Schena, M. , Shalon, D. , Davis, R.W. , Brown, P.O. , 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 270, 467–470. [DOI] [PubMed] [Google Scholar]
- Shaw, A.T. , Yasothan, U. , Kirkpatrick, P. , 2011. Crizotinib. Nat. Rev. Drug Discov.. 10, 897–898. [DOI] [PubMed] [Google Scholar]
- Sieweke, M.H. , Thompson, N.L. , Sporn, M.B. , Bissell, M.J. , 1990. Mediation of wound-related Rous sarcoma virus tumorigenesis by TGF-beta. Science. 248, 1656–1660. [DOI] [PubMed] [Google Scholar]
- Slamon, D.J. , Leyland-Jones, B. , Shak, S. , Fuchs, H. , Paton, V. , Bajamonde, A. , Fleming, T. , Eiermann, W. , Wolter, J. , Pegram, M. , Baselga, J. , Norton, L. , 2001. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N. Engl. J. Med.. 344, 783–792. [DOI] [PubMed] [Google Scholar]
- Sorlie, T. , Perou, C.M. , Tibshirani, R. , Aas, T. , Geisler, S. , Johnsen, H. , Hastie, T. , Eisen, M.B. , van de Rijn, M. , Jeffrey, S.S. , Thorsen, T. , Quist, H. , Matese, J.C. , Brown, P.O. , Botstein, D. , Eystein Lonning, P. , Borresen-Dale, A.L. , 2001. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. U S A. 98, 10869–10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soule, H.D. , Maloney, T.M. , Wolman, S.R. , Peterson, W.D. , Brenz, R. , McGrath, C.M. , Russo, J. , Pauley, R.J. , Jones, R.F. , Brooks, S.C. , 1990. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res.. 50, 6075–6086. [PubMed] [Google Scholar]
- Spurdle, A.B. , Healey, S. , Devereau, A. , Hogervorst, F.B. , Monteiro, A.N. , Nathanson, K.L. , Radice, P. , Stoppa-Lyonnet, D. , Tavtigian, S. , Wappenschmidt, B. , Couch, F.J. , Goldgar, D.E. , 2012. ENIGMA-evidence-based network for the interpretation of germline mutant alleles: an international initiative to evaluate risk and clinical significance associated with sequence variation in BRCA1 and BRCA2 genes. Hum. Mutat.. 33, 2–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, B. , Smith, B.D. , 2010. Treatment options for patients with chronic myeloid leukemia who are resistant to or unable to tolerate imatinib. Clin. Ther.. 32, 804–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg, M. , 2007. Dasatinib: a tyrosine kinase inhibitor for the treatment of chronic myelogenous leukemia and Philadelphia chromosome-positive acute lymphoblastic leukemia. Clin. Ther.. 29, 2289–2308. [DOI] [PubMed] [Google Scholar]
- Taylor, B.S. , Schultz, N. , Hieronymus, H. , Gopalan, A. , Xiao, Y. , Carver, B.S. , Arora, V.K. , Kaushik, P. , Cerami, E. , Reva, B. , Antipin, Y. , Mitsiades, N. , Landers, T. , Dolgalev, I. , Major, J.E. , Wilson, M. , Socci, N.D. , Lash, A.E. , Heguy, A. , Eastham, J.A. , Scher, H.I. , Reuter, V.E. , Scardino, P.T. , Sander, C. , Sawyers, C.L. , Gerald, W.L. , 2010. Integrative genomic profiling of human prostate cancer. Cancer Cell. 18, 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas, 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 455, 1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas, 2011. Integrated genomic analyses of ovarian carcinoma. Nature. 474, 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng, P.H. , Wang, Y.C. , Weng, S.C. , Weng, J.R. , Chen, C.S. , Brueggemeier, R.W. , Shapiro, C.L. , Chen, C.Y. , Dunn, S.E. , Pollak, M. , Chen, C.S. , 2006. Overcoming trastuzumab resistance in HER2-overexpressing breast cancer cells by using a novel celecoxib-derived phosphoinositide-dependent kinase-1 inhibitor. Mol. Pharmacol.. 70, 1534–1541. [DOI] [PubMed] [Google Scholar]
- Vaske, C.J. , Benz, S.C. , Sanborn, J.Z. , Earl, D. , Szeto, C. , Zhu, J. , Haussler, D. , Stuart, J.M. , 2010. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics. 26, i237–i245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van't Veer, L.J. , Bernards, R. , 2008. Enabling personalized cancer medicine through analysis of gene-expression patterns. Nature. 452, 564–570. [DOI] [PubMed] [Google Scholar]
- Vogelstein, B. , Kinzler, K.W. , 2004. Cancer genes and the pathways they control. Nat. Med.. 10, 789–799. [DOI] [PubMed] [Google Scholar]
- Wagle, N. , Berger, M.F. , Davis, M.J. , Blumenstiel, B. , DeFelice, M. , Pochanard, P. , Ducar, M. , Van Hummelen, P. , MacConaill, L.E. , Hahn, W.C. , Meyerson, M. , Gabriel, S.B. , Garraway, L.A. , 2012. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov.. 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, T. , Lee, M.K. , Casadei, S. , Thornton, A.M. , Stray, S.M. , Pennil, C. , Nord, A.S. , Mandell, J.B. , Swisher, E.M. , King, M.C. , 2010. Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing. Proc. Natl. Acad. Sci. U S A. 107, 12629–12633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, M. , Davies, J.J. , Wittig, D. , Oakeley, E.J. , Haase, M. , Lam, W.L. , Schubeler, D. , 2005. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet.. 37, 853–862. [DOI] [PubMed] [Google Scholar]
- Wilhelm, S. , Carter, C. , Lynch, M. , Lowinger, T. , Dumas, J. , Smith, R.A. , Schwartz, B. , Simantov, R. , Kelley, S. , 2006. Discovery and development of sorafenib: a multikinase inhibitor for treating cancer. Nat. Rev. Drug Discov.. 5, 835–844. [DOI] [PubMed] [Google Scholar]
- Wyckoff, J. , Wang, W. , Lin, E.Y. , Wang, Y. , Pixley, F. , Stanley, E.R. , Graf, T. , Pollard, J.W. , Segall, J. , Condeelis, J. , 2004. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res.. 64, 7022–7029. [DOI] [PubMed] [Google Scholar]
- Xia, W. , Bacus, S. , Hegde, P. , Husain, I. , Strum, J. , Liu, L. , Paulazzo, G. , Lyass, L. , Trusk, P. , Hill, J. , Harris, J. , Spector, N.L. , 2006. A model of acquired autoresistance to a potent ErbB2 tyrosine kinase inhibitor and a therapeutic strategy to prevent its onset in breast cancer. Proc. Natl. Acad. Sci. U S A. 103, 7795–7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. , Boehm, J.S. , Yang, X. , Salehi-Ashtiani, K. , Hao, T. , Shen, Y. , Lubonja, R. , Thomas, S.R. , Alkan, O. , Bhimdi, T. , Green, T.M. , Johannessen, C.M. , Silver, S.J. , Nguyen, C. , Murray, R.R. , Hieronymus, H. , Balcha, D. , Fan, C. , Lin, C. , Ghamsari, L. , Vidal, M. , Hahn, W.C. , Hill, D.E. , Root, D.E. , 2011. A public genome-scale lentiviral expression library of human ORFs. Nat. Methods. 8, 659–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Yang, P.L. , Gray, N.S. , 2009. Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer. 9, 28–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S. , Huang, W.C. , Li, P. , Guo, H. , Poh, S.B. , Brady, S.W. , Xiong, Y. , Tseng, L.M. , Li, S.H. , Ding, Z. , Sahin, A.A. , Esteva, F.J. , Hortobagyi, G.N. , Yu, D. , 2011. Combating trastuzumab resistance by targeting SRC, a common node downstream of multiple resistance pathways. Nat. Med.. 17, 461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]


