Abstract
The use of mass spectrometry for characterization of small molecules, nucleotides, and proteins in model organisms as well as primary tissues and clinical samples continues to proliferate at a rapid pace. The complexity and dynamic range of target analytes in biological systems hinders comprehensive analysis and simultaneously drives improvements in instrument hardware and software. As a result, state-of-the-art commercial mass spectrometers are equipped with sophisticated embedded control systems that provide robust acquisition methods accessed through intuitive graphical interfaces. Although optimized for speed, these pre-configured scan functions are otherwise closed to end-user customization beyond simple, analytical-centric parameters supplied by the manufacturer. Here we present an open-source framework (mzAPI/Live) that enables users to generate arbitrarily complex LC-MSn acquisition methods via simple Python scripting. As a powerful proof-of-concept we demonstrate real-time assignment of tandem mass spectra through rapid query of NIST peptide libraries. This represents an unprecedented capability to make acquisition decisions based on knowledge of analyte structures determined during the run itself, thus providing a path toward biology-driven mass spectrometry data acquisition for the broader community.
Keywords: Quantitative proteomics, mass spectrometry, LC-MS, mass-informatics
While mass spectrometry continues to enjoy wider use in the analysis of biomolecules, the complexity and concentration dynamic range of these analytes in biological systems impedes efforts directed at comprehensive profiling. Despite significant improvements in mass spectrometry hardware and software, LC-MS/MS suffers from stochastic sampling of target molecules, which tends to compress the apparent dynamic range of detected analytes and leads to poor characterization of low-abundance species. To offset these limitations prioritization schemes have become commonplace in acquisition methods for tandem mass spectrometry. In practice, the most widely used algorithm relies on a real-time rank-ordering of analytes based on their signal intensities, with the expectation that the quality of fragment ion spectra will correlate with precursor abundance. Target m/z values interrogated by MS/MS are then put on a temporary exclusion list to minimize acquisition of redundant spectra. This general approach, termed “data-dependent analysis,” was first introduced by Finnigan-MAT (now ThermoFisher Scientific). Other vendors use similar prioritization schemes, including Waters’ data-directed analysis (DDA), Agilent’s data-directed acquisition and Ab Sciex’s information-dependent acquisition (IDA) [1]. This form of acquisition logic is fundamentally limited to rudimentary analytical metrics such as signal intensity or mass assignment. Unfortunately these measures cannot be mapped with meaningful specificity to any useful biological annotation.
Providing users with greater flexibility and autonomy in terms of mass spectrometry hardware, electronics, and instrument control is an important step towards community-wide experimentation with novel acquisition schemes. In fact there is a rich history in which close collaboration between manufacturers and customer laboratories resulted in new or improved hybrid instrument configurations [2-7], customized hardware to support novel ionization and fragmentation techniques [8-11], along with modifications that broadened the range of measurable molecular classes [12-14]. Although these projects yielded new scientific insight and in certain cases led to commercially viable instrument platforms, they were typically pursued through exclusive arrangements with specific laboratories. More recently, these efforts have been extended to refinements in data acquisition logic [15], including modification of instrument parameters in real-time (e.g., during data acquisition) [16, 17]. The latter studies leveraged the ThermoFisher COM object, an instrument control API that, in practice, requires a detailed understanding of Windows C++ programming as well as in-depth knowledge of the interfaces and workflows provided by the manufacturer (as is typically experienced through the GUI-based software). Herein we sought to improve the accessibility of COM object functionality and further democratize the ability of non-expert scientists to experiment with novel acquisition methods though mzAPI/Live, an open-source framework that enables customized mass spectrometry acquisition through simple Python scripts. Our approach mirrors similar proposals for post-acquisition data analysis [18, 19], whereby vendors distribute DLLs that provide direct access to mass spectrometer binary output files without disclosing proprietary information related to the underlying file formats.
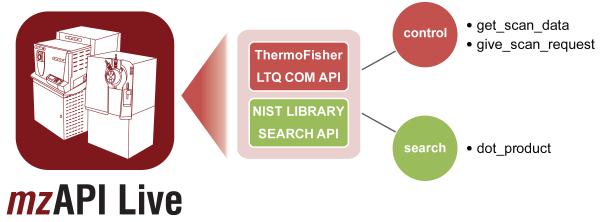
The mzAPI/Live framework consists of two components: a command-line wrapper for the LTQ COM object, and a Python script to control acquisition. The command-line tool was written in C++ using the Microsoft Foundation Class (MFC) libraries and Microsoft Visual C++. To dramatically reduce the learning curve for custom acquisition development we wrapped both the complexity of the C++ code and many details of the acquisition control in a simple Python-based API consisting of only two callbacks: get_scan_data and give_scan_request (Figure 1). To execute acquisition, the user simply specifies via command-line arguments the name of the Python module to use for acquisition control, and the name of the output data file. Following connection to the instrument, initialization of the Python code, and receipt of a start signal (e.g., an LC contact closure), scan data is passed to the Python script in a loop of request-and-answer, until a pre-set acquisition endpoint is reached. To facilitate analysis of multiple samples, a GUI was created using wxPython which allows users to create and launch sample lists of arbitrary length. This architecture provides a direct way for user-defined scripts to dynamically interact with the instrument embedded system control; this simple event-driven loop provides a message when data is made available (get_scan_data) and a request from the instrument for the definition of the next scan event (give_scan_request).
Figure 1. Instrument Control Through mzAPI/Live.
mzAPI/Live hides the complexity of both the LTQ COM library and the NIST search engine behind three easy to use Python callback/functions. Arbitrarily complex acquisition workflows can be programmed using the Python scripting language.
In general, mzAPI/Live is agnostic to the acquisition method itself - rather, it bridges the existing gap between the closed nature of current instrument control software and users’ desires to test and develop novel methods. The acquisition methods written using mzAPI/Live can be easily shared with other researchers though publications or other community resources. This approach also has the advantage of placing both the opportunity for innovation and the responsibility to justify changes upon the scientist. Over time, adoption of mzAPI/Live will facilitate distribution of novel acquisition methods and reduce the knowledge- and technology-based barriers to entry for scientists who utilize mass spectrometry within their research.
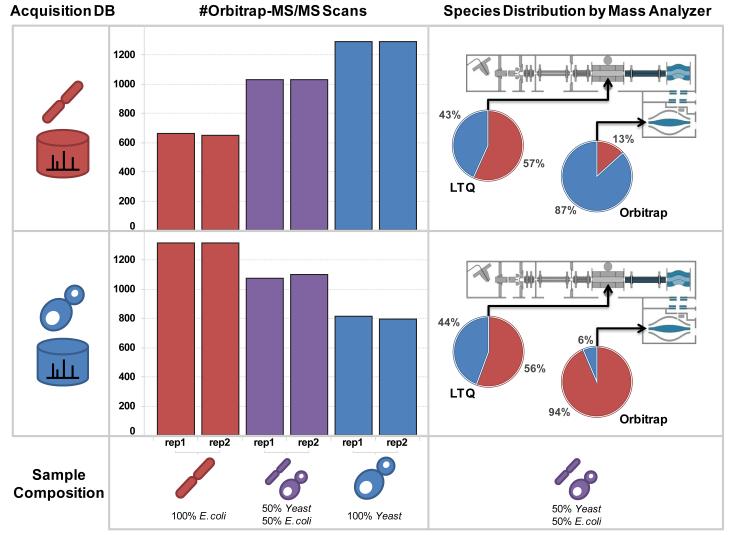
As a powerful example, we utilized mzAPI/Live to build acquisition logic based on biological annotation rather than simple analytical metrics. Towards this end we wrapped the NIST peptide library search engine (NIST MS DLL version 2.1.3 downloaded from http://peptide.nist.gov) in Python and implemented real-time sequence identification. Spectral libraries have long been proposed as an effective mechanism to capture and re-use knowledge acquired during previous experiments [20]. We modeled a scenario in which the instrument is provided with a library representing “prior” experiments and then is challenged with samples that contain increasingly large numbers of “novel” or “unexpected” chemical species. We did this by using a spectral library defined for one of two biological species (E. coli or S. cerevisiae) while analyzing a series of samples with an increasing proportion of tryptic peptides derived from lysates of the other “novel” biological species. Spectral library searches were executed on the fly by the acquisition Python script through calls to the dot-product function, which passed MS/MS spectra (isolation width=2.8 Da, collision\ energy=35%, q=0.25, electron multiplier detection) for the 10 most abundant precursors in each Orbitrap MS scan (m/z 300-2000, resolution=60,000) to the underlying NIST MS DLL [21] and returned NIST similarity scores for the best matches. We programmed the instrument to acquire a high resolution MS/MS spectrum in the Orbitrap (isolation width=2.8 Da, collision energy=35%, q=0.25, resolution=7500) for any precursor yielding a fragment ion spectrum in the linear ion trap that exhibited a particularly poor NIST dot-product score (score < 200). Note also that we easily re-created much of the standard data-dependent logic, such as charge state determination, precursor selection based on signal intensity, and exclusion lists. Using LTQ Instrument Control version 2.6 in conjunction with LTQ Tune 2.1 on an Orbitrap Velos, we observed an overhead of ~1.3s per FTMS scan, and 60 ms per ion trap CAD scan, both of which were independent of the embedded Python interpreter. Encouragingly, dot-product matches against the NIST spectral library were executed in an average time of 10 ms, suggesting that improvements in the underlying COM object code would enable creation of novel acquisition schemes that may rival the speed of pre-configured methods provided by the manufacturer. Figure 2 shows that the number and overall proportion of Orbitrap MS/MS scans increased with a shift in concentration from E.coli, the “known” species as defined by the acquisition database, to yeast (the “novel” species); the opposite trend was observed when the acquisition database was replaced with a yeast spectral library. We used multiplierz [19, 22] to collate peptides derived from a Mascot search (1% FDR) of each LC-MS/MS acquisition and observed a striking degree of species enrichment based on real-time decisions to acquire Orbitrap fragment ion spectra for unexpected chemical species (see Supplemental Tables S1-S4).
Figure 2. LDA Enables Species-dependent Acquisition of MS/MS Spectra in an Orbitrap Mass Analyzer.
The acquisition database (left column) is set to either S. cerevisiae (top row) or E. coli (bottom row). Mixtures of tryptic peptides from each species (middle column) are analyzed by LC-MS/MS in replicate for each acquisition database. Fragment ion MS/MS spectra acquired in the linear ion trap are interrogated in real time by dot-product search against NIST spectral libraries; those peptides not assigned with high confidence are subjected to high resolution MS/MS in the Orbitrap mass analyzer. All MS/MS spectra were searched post-acquisition using Mascot. For a 1:1 mixture of S. cerevisiae and E. coli peptides, the linear ion trap faithfully reproduces the underlying species composition (right column, LTQ piechart) while the majority of MS/MS scans in the Orbitrap are devoted to the “novel” species (right column, Orbitrap piechart). The species-specific trend is well represented in the relative proportions of Orbitrap MS/MS scans acquired across a wide range of species mixtures (middle column).
While the above example is purely demonstrative, it is clear that the capability to perform LDA through mzAPI/Live provides significant incentive for construction of peptide spectral libraries, which have heretofore been touted primarily for their offline search benefits, namely an increase in search speed and sensitivity [23, 24]. By contrast, real-time structural determination provides a higher level of decision logic as compared to existing manufacturers’ acquisition methods. Labs which populate and curate spectral libraries tailored to their specific sample types (e.g., biochemical enrichment class, clinical cohort, etc.) will likely realize an increase in effective instrument time during acquisition, conserving relatively slow scan types (ETD, HCD-FT or CID-FT) only for precursors that are verified on the fly as being novel or of particular interest. Unlike methods based on inclusion/exclusion lists, which are typically defined relative to LC elution time, LDA is resilient to run-to-run variation in upstream fractionation performance. Finally, and in contrast to similar recent studies [16, 17], our approach leverages NIST spectral libraries directly, meaning that mzAPI/Live is immediately extensible to other analyte classes simply by substituting the appropriate NIST library. The code for mzAPI/Live is freely available for download from SourceForge (upon publication) and we welcome user feedback.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Dr. Dmitrii Tchechovskoi at the National Institute for Standards and Technology for providing the NIST DLL and associated documentation. In addition the authors thank Prof. John Yates and Dr. Aleksey Nakorchevsky at the Scripps Research Institute for technical assistance with the Xcalibur COM object. Finally the authors thank Brijesh Garg, as well as Drs. Guillaume Adelmant, Tim Sikorski, and Feng Zhou in the Marto lab for their assistance with preparation of E. coli and S. cerevisiae samples. Generous support for this work was provided the Dana-Farber Cancer Institute, and the National Institutes of Health, NINDS (P01NS047572).
ABBREVIATIONS
- LDA
Library Dependent Acquisition
Footnotes
AUTHOR CONTRIBUTIONS J.T.W and M.A. designed mzAPI/Live
J.T.W. implemented mzAPI/Live
S.B.F. and M.A.I. wrote Python scripts and other code for deisotoping, standard data dependent logic, and LC synchronization.
M.A. designed and implemented LDA
M.A. and S.B.F. planned and performed the experiments
J.A.M. conceived of the study and wrote the paper with assistance from J.T.W., M.A., and S.B.F.
COMPETING FINANCIAL INTERESTS The authors declare that they have no competing financial interests.
REFERENCES
- [1].Watson JT, Sparkman OD. Introduction to mass spectrometry : instrumentation, applications and strategies for data interpretation. John Wiley & Sons; Chichester, England ; Hoboken, NJ: 2007. [Google Scholar]
- [2].Cody RB, Jr., Amster IJ, McLafferty FW. Peptide mixture sequencing by tandem Fourier-transform mass spectrometry. Proc Natl Acad Sci U S A. 1985;82:6367–6370. doi: 10.1073/pnas.82.19.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hu Q, Noll RJ, Li H, Makarov A, et al. The orbitrap: a new mass spectrometer. Journal of Mass Spectrometry. 2005;40:430–443. doi: 10.1002/jms.856. [DOI] [PubMed] [Google Scholar]
- [4].Medzihradszky KF, Campbell JM, Baldwin MA, Falick AM, et al. The characteristics of peptide collision-induced dissociation using a high-performance MALDI-TOF/TOF tandem mass spectrometer. Anal Chem. 2000;72:552–558. doi: 10.1021/ac990809y. [DOI] [PubMed] [Google Scholar]
- [5].Michalski A, Damoc E, Hauschild JP, Lange O, et al. Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol Cell Proteomics. 2011;10:M111–011015. doi: 10.1074/mcp.M111.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Morris HR, Paxton T, Dell A, Langhorne J, et al. High sensitivity collisionally-activated decomposition tandem mass spectrometry on a novel quadrupole/orthogonal-acceleration time-of-flight mass spectrometer. Rapid Commun Mass Spectrom. 1996;10:889–896. doi: 10.1002/(SICI)1097-0231(19960610)10:8<889::AID-RCM615>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- [7].Syka JEP, Marto JA, Bai DL, Horning S, et al. Novel Linear Quadrupole Ion Trap/FT Mass Spectrometer: Performance Characterization and Use in the Comparative Analysis of Histone H3 Post-translational Modifications. Journal of Proteome Research. 2004;3:621–626. doi: 10.1021/pr0499794. [DOI] [PubMed] [Google Scholar]
- [8].Hunt DF, Stafford GC, Crow FW, Russell JW. Pulsed positive negative ion chemical ionization mass spectrometry. Analytical Chemistry. 1976;48:2098–2104. [Google Scholar]
- [9].Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Xia Y, Chrisman PA, Erickson DE, Liu J, et al. Implementation of Ion/Ion Reactions in a Quadrupole/Time-of-Flight Tandem Mass Spectrometer. Analytical Chemistry. 2006;78:4146–4154. doi: 10.1021/ac0606296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Xia Y, Chrisman PA, Erickson DE, Liu J, et al. Implementation of ion/ion reactions in a quadrupole/time-of-flight tandem mass spectrometer. Anal Chem. 2006;78:4146–4154. doi: 10.1021/ac0606296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Amster IJ, McLafferty FW, Castro ME, Russell DH, et al. Detection of mass 16 241 ions by Fourier-transform mass spectrometry. Anal Chem. 1986;58:483–485. doi: 10.1021/ac00293a049. [DOI] [PubMed] [Google Scholar]
- [13].Hunt DF, Shabanowitz J, McIver RT, Jr., Hunter RL, Syka JE. Ionization and mass analysis of nonvolatile compounds by particle bombardment tandem-quadrupole Fourier transform mass spectrometry. Anal Chem. 1985;57:765–768. doi: 10.1021/ac00280a043. [DOI] [PubMed] [Google Scholar]
- [14].Ruotolo BT, Giles K, Campuzano I, Sandercock AM, et al. Evidence for Macromolecular Protein Rings in the Absence of Bulk Water. Science. 2005;310:1658–1661. doi: 10.1126/science.1120177. [DOI] [PubMed] [Google Scholar]
- [15].Olsen JV, Schwartz JC, Griep-Raming J, Nielsen ML, et al. A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol Cell Proteomics. 2009;8:2759–2769. doi: 10.1074/mcp.M900375-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bailey DJ, Rose CM, McAlister GC, Brumbaugh J, et al. Instant spectral assignment for advanced decision tree-driven mass spectrometry. Proceedings of the National Academy of Sciences. 2012;109:8411–8416. doi: 10.1073/pnas.1205292109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Graumann J, Scheltema RA, Zhang Y, Cox J, Mann M. A Framework for Intelligent Data Acquisition and Real-Time Database Searching for Shotgun Proteomics. Molecular & Cellular Proteomics. 2012;11 doi: 10.1074/mcp.M111.013185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24:2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Askenazi M, Parikh JR, Marto JA. mzAPI: a new strategy for efficiently sharing mass spectrometry data. Nat Methods. 2009;6:240–241. doi: 10.1038/nmeth0409-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ausloos P, Clifton CL, Lias SG, Mikaya AI, et al. The critical evaluation of a comprehensive mass spectral library. Journal of the American Society for Mass Spectrometry. 1999;10:287–299. doi: 10.1016/S1044-0305(98)00159-7. [DOI] [PubMed] [Google Scholar]
- [21].Stein SE, Rudnick PA, editors. NIST Peptide Tandem Mass Spectral Libraries. Yeast and E. coli Peptide Mass Spectral Reference Data, S. cerevisiae and E. coli, ion trap, Official Build Dates: May. 24, 2011. National Institute of Standards and Technology; Gaithersburg, MD: Sep, 2011. 20899. Downloaded from http://peptide.nist.gov. [Google Scholar]
- [22].Parikh JR, Askenazi M, Ficarro SB, Cashorali T, et al. multiplierz: an extensible API based desktop environment for proteomics data analysis. BMC bioinformatics. 2009;10:364. doi: 10.1186/1471-2105-10-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lam H, Deutsch EW, Eddes JS, Eng JK, et al. Building consensus spectral libraries for peptide identification in proteomics. Nat Methods. 2008;5:873–875. doi: 10.1038/nmeth.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Craig R, Cortens JC, Fenyo D, Beavis RC. Using annotated peptide mass spectrum libraries for protein identification. Journal of Proteome Research. 2006;5:1843–1849. doi: 10.1021/pr0602085. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.