Abstract
The Helicobacter pylori outer membrane proteins play an important role in pathogenesis; the outer inflammatory protein A (OipA) is one of these proteins which play the main role in the development of inflammation. In this study, purification of recombinant H. pylori OipA was performed by Ni–NTA affinity chromatography. Gastric carcinoma epithelial cells (AGS cell) were treated by different concentrations of recombinant OipA for various lengths of time and cell viability was evaluated by the viability assay. Statistical analysis showed that OipA had toxic effects on AGS cells in a concentration of 500 ng/ml after 24 and 48 h, and this toxic dose was 256 ng/ml after 72 h. OipA had direct toxic effects on gastric epithelial cells and the toxicity was observed to depend on time and dose of H. pylori exposure. Attachment of H. pylori to gastric epithelial cells is a key part in the pathogenesis and enables H. pylori to damage the epithelial cells with OipA.
Keywords: Helicobacter pylori, Toxicity, Outer inflammatory protein A, Gastric epithelial cell
Introduction
The pathogenesis of Helicobacter pylori is complex and outer membrane proteins (OMPs) have an essential role in progress of H. pylori infection. The OMPs are necessary to attach to gastric epithelial cells [1–4]. Some of the OMPs cause inflammation and IL-8 secretion from gastric epithelial and immune system cells [5]. Outer inflammatory protein A (OipA) is an OMP and one of the most important inflammatory proteins in H. pylori. Some studies have shown several pathogenic effects of H. pylori OipA such as bacterial attachment to gastric epithelial cell, activation of focal adhesion kinase, re-organization of cytoskeleton and IL-8 secretion [5–7]. Almost all strains of H. pylori have alleles of this protein but the status of oipA is not always “on” and can change to “off” in some infections [8]. Expression of this protein is regulated by slipped-strand repair mechanism and oipA is expressed in most of East Asian strains [9]. OipA could potentially has an important role in the pathogenesis of H. pylori, and purification of this protein can aid to investigate pathogenic evaluation of this factor.
In this study we purified this outer membrane protein and evaluated toxicity effects of this protein on gastric epithelial cell.
Materials and Methods
DNA Genomics of Helicobacter pylori
DNA genomics of H. pylori 26695 was kindly provided by Dr Esmaeili (Tarbiat Modares University). Bacterial strain E. coli GM2163 and BL21 (DE3) were obtained from Fermentas (Lithonia) and Blood Transfusion Organization (Iran), respectively. pJET and pET-28a as cloning and expression vectors were purchased from Novagen. LB and 2XYT broth and agar (Merck, Germany) with and without 30 μg/ml kanamycin and ampicillin (Sigma, USA) were used for bacterial culture.
Genomic DNA was extracted by genomic DNA extraction kit (Fermentas, Lithonia). Specific primers were designed according to oipA completed sequences of H. pylori 26695 from NCBI. The sequence of forward primer containing a site for NcoI restriction enzyme was 5′AAT CCATGG TCCACGCTGAAAGGAATGGG-3′and reveres primer with a site for XhoI was 5′AGG CTCGAG CACTTTAACCCCTAATTCAACAC-3′. PCR products were ligated to pJET vector. This construct was transformed into E. coli GM2163 and was cultured on LB agar containing ampicillin to preserve sequence of oipA in prokaryotic system.
Plasmid extraction was performed by plasmid extraction kit (Bioneer, Korea) and pJET containing oipA and pET28a was extracted and two vectors were digested with NcoI and XhoI restriction enzymes to make cohesive ends. Then oipA gene and pET-28a were ligated with T4DNA ligase and were transformed into a competent E. coli BL21 (DE3) strain.
Induction was performed in 2XYT medium by 1 mmol/l IPTG and purification was applied by Ni–NTA affinity chromatography according to manufacturer’s protocol (Qiagen, Valencia, CA, USA) and was confirmed by polyclonal antibody against H. pylori and Western blot.
Human Gastric Cancer Cell Line (AGS Cells)
Human gastric cancer cell line (AGS cells) was provided from cell bank of the Pasteur Institute (Iran). AGS cells were cultured in RPMI 1640 that was supplemented with 100 μg/ml penicillin, streptomycin (GIPCO/Invitrogen) and 10 % FBS. The cells were incubated in 37 °C and 5 % CO2. RPMI 1640 and fetal bovine serum were obtained from GIPCO/Invitrogen.
Evaluation of Toxicity Effects of OipA on AGS Cells
Evaluation of toxicity effects of OipA on AGS cells was performed by measuring MTT dye absorbance of viable cells. AGS cells were cultured in 96-well plates (10,000 cells/well) overnight, and treated by purified OipA for different concentrations and times. Serial dilutions of OipA from 1 ng/ml to 100 μg/ml were added on the cells and were incubated for 24, 48 and 72 h. BSA was used as control in the same concentrations. After incubation, 100 μl DMSO was added to every well, the formazan precipitate was dissolved in DMSO and the optical density of each well was read at 570 nm.
Statistical Analysis
Data were analyzed using one way ANOVA followed by Dunnett’s test (OriginPro v. 8.5.1.) and significant difference was set at p < 0.05.
Results
Cloning of OipA and Amplified by PCR
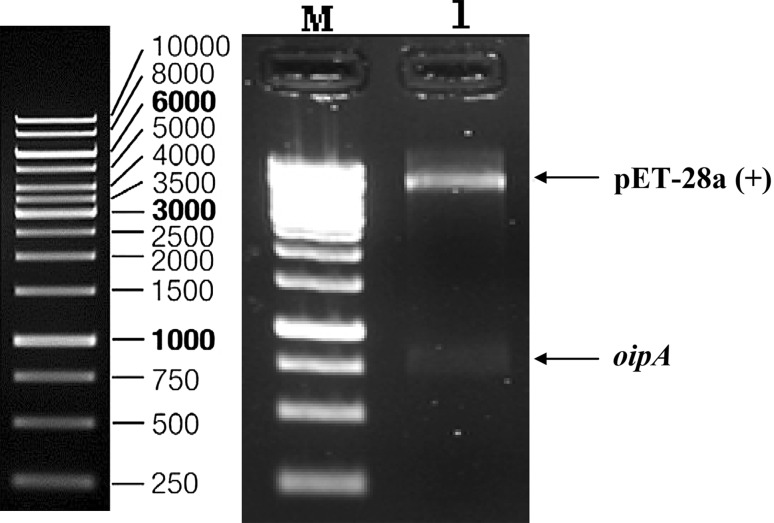
The gene encoding OipA protein was amplified by PCR with Primstar Taq DNA polymerase. The expected size, 783 bp was obtained. Cloning was confirmed by double digestion with NcoI and XhoI restriction enzymes (Fig. 1).
Fig. 1.
Confirmation of cloning with digestion. Lane M: 1 Kb DNA ladder, lane 1: oipA/pET28A double digestion bands
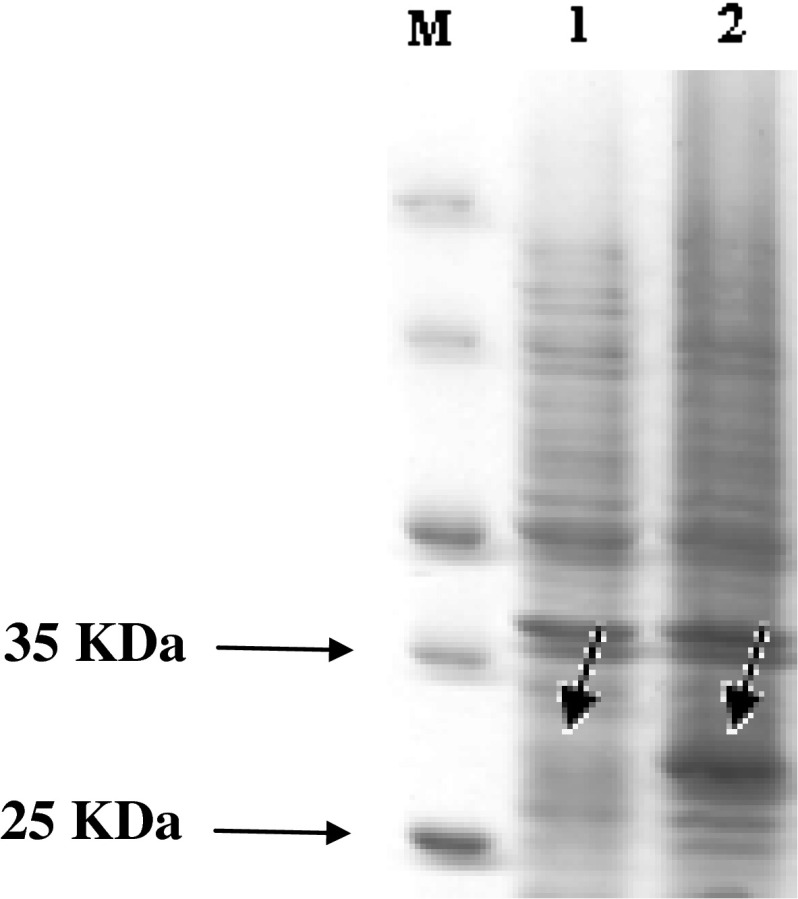
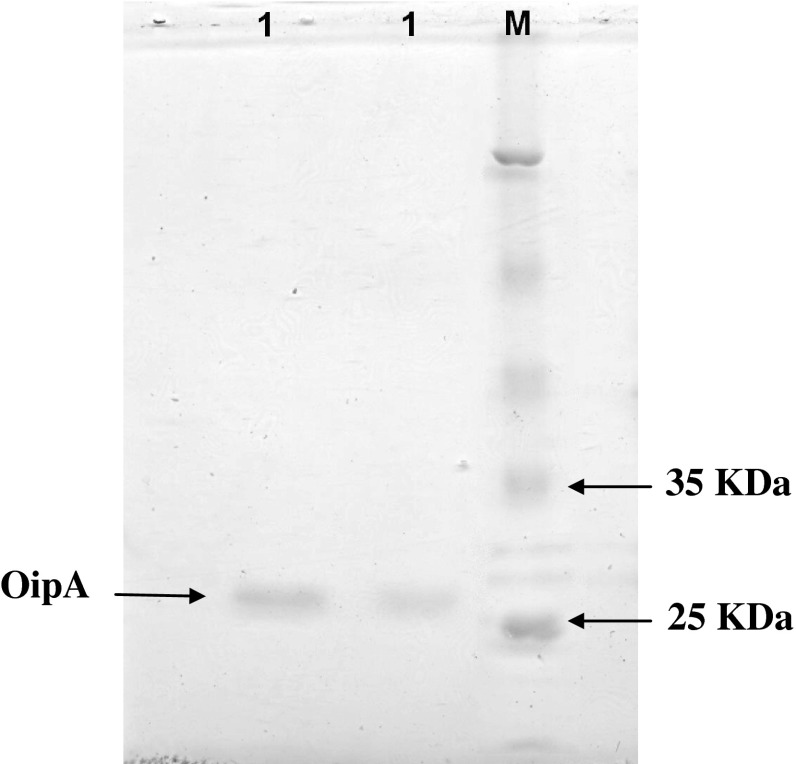
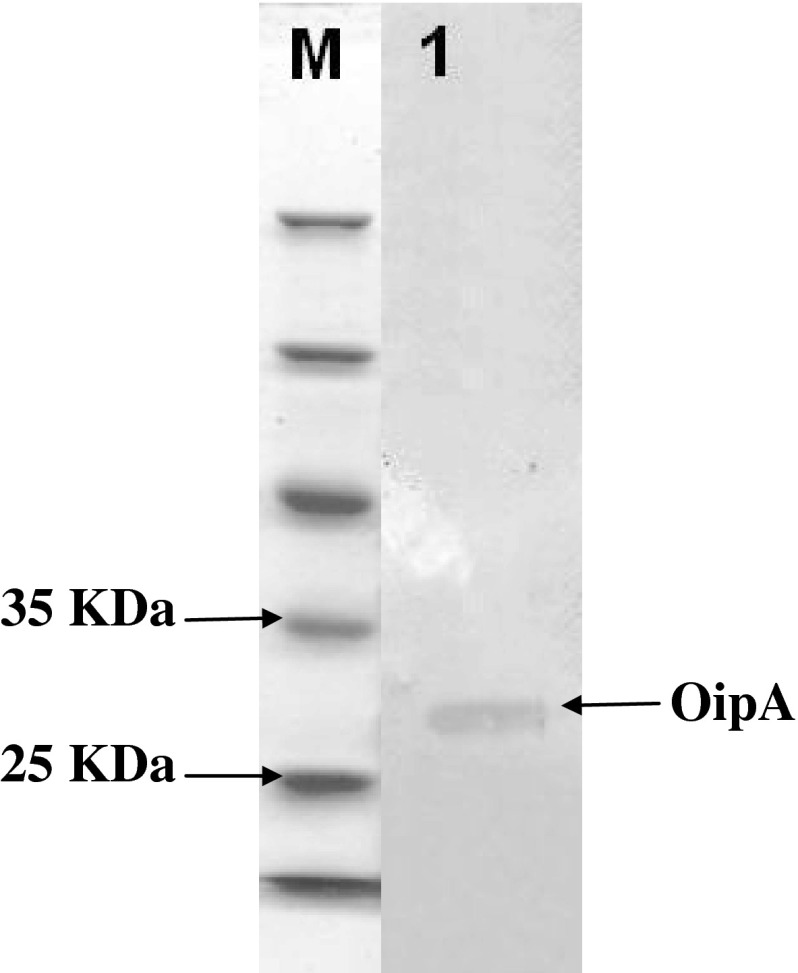
In our study designed the primers to make sure that the “on” phase of the oipA gene is quantified. Escherichia coli BL21 (DE3) was transformed with recombinant plasmid pET28a/oipA containing T7 promoter as expression host. Protein production was induced with IPTG (1 mM). SDS-PAGE 12/5 % gel detected proteins in approximately 30 kDa weight after staining with coomassie blue G-250 (Fig. 2). Purification was applied by Ni–NTA affinity chromatography according to manufacturer’s protocol (Fig. 3). In order to detect recombinant OipA proteins, Western blot analysis was performed. The major band observed in SDS-PAGE (30 kDa) was confirmed as recombinant OipA protein by Western blot analysis (Fig. 4). In our study, we used polyclonal whole cell H. pylori antibody.
Fig. 2.
Protein detection by SDS-PAGE 12/5 % (w/v). Lane M: standard protein size marker (kDa). Lane 1: non-induced OipA, lane 2: induced OipA with 1 mmol IPTG
Fig. 3.
Confirmation of purification of OipA with SDS-PAGE. Lane M: standard protein size marker (kDa). Lane 1: purified OipA
Fig. 4.
Confirmation of OipA with Western blot analysis. Lane M: standard protein size marker (kDa). Lane 1: OipA protein
Measurement the Viability of Gastric Epithelial Following Treatment with H. pylori OipA
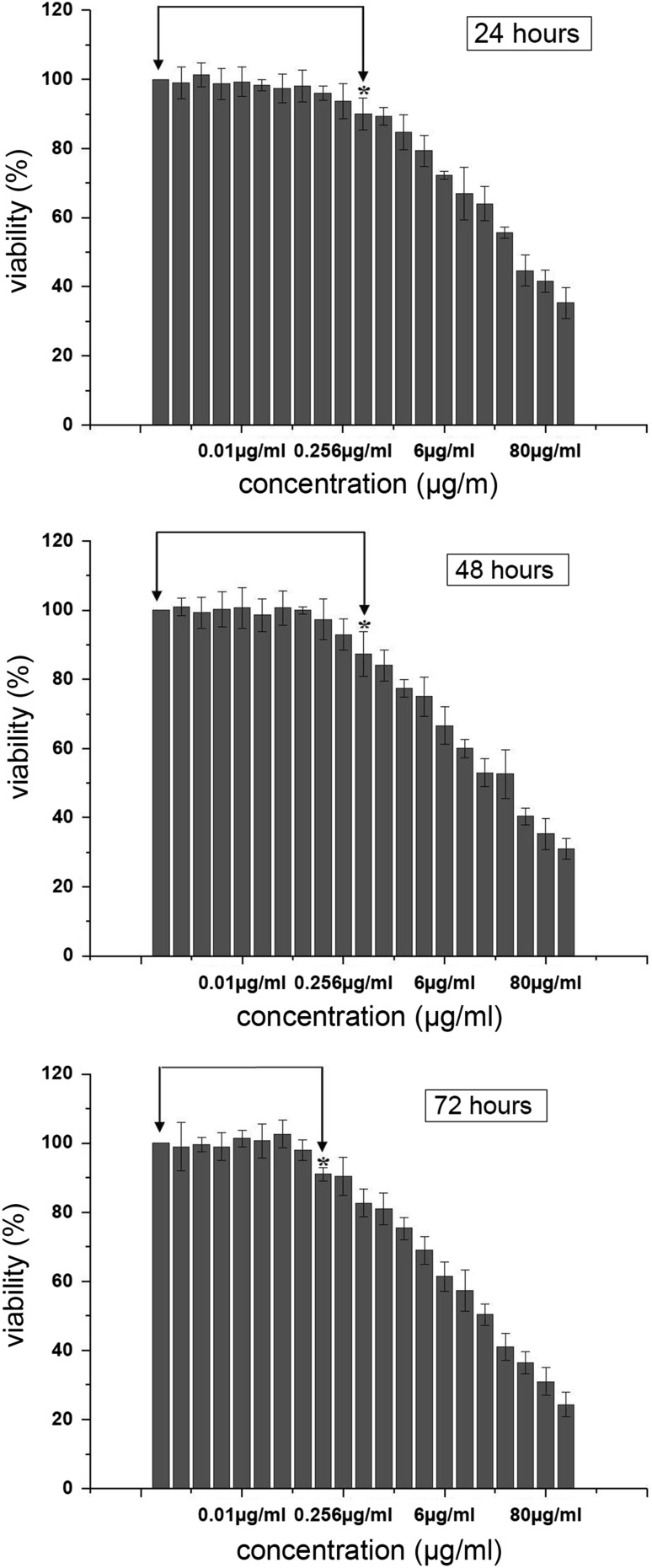
To evaluate the effect of OipA on the gastric epithelial cells, various concentrations of OipA treatment times were used. The results indicated that a concentration of 500 ng/ml we had a significant decrease in the viability of gastric epithelial cell at 24 and 48 h. With the increasing of time to 72 h we had a significant decrease in the viability of gastric epithelial cells in the concentration of 256 ng/ml. We had no change in viability of AGS cells that had treated with BSA as control. These results showed that with the increasing of OipA dose and time, the viability of gastric epithelial cells deceased (Fig. 5).
Fig. 5.
Viability of gastric epithelial cells (AGS) after 24, 48 and 72 h treatment with H. pylori OipA. The results are express as the mean ± SD, n = 3. *, p < 0.05 as compare with the control group
Discussion
Yamaoka was the first to report the discovery of a novel 33 kDa protein as potent virulence factor. He observed that this protein can induce inflammatory processes and was present in 80 % of H. pylori samples that were isolated from peptic ulcers [5, 10]. OipA protein is a very important virulence factor that has a synergic effect with cagA [6]. This protein does not express in all of the strains and expression of oipA is under control of “ct” repeat in signal sequence of oipA gene [5]. In this study we have evaluated toxicity effects of purified recombinant OipA on Gastric epithelial cells (AGS cells) and our research revealed that this protein is toxic on epithelial cells of the stomach (Fig. 5).
We had previously reported the N-terminal sequence of OipA to be important in attachment and signal transduction. Bioinformatics studies revealed that OipA, same as VacA, is an auto transporter protein [11]. Signal sequence of oipA was deleted by primer designing, because signal sequence is eliminated before expressing on surface of H. pylori.
Attachment of H. pylori to gastric epithelial cells is very important for pathogenesis of the bacteria [12] and OipA has direct toxic effects on gastric epithelial cells when exposed to H. pylori. As we have shown in this study, this toxicity depends on the duration of exposure and dose of the bacteria which have expressed this outer membrane protein.
Cell death or cell proliferation determines the pathogenic pathway of H. pylori. Gastric and duodenal ulcers result from gastric cell death and cell proliferation result in metaplasia and cancer. Imbalance between these two phenomena almost causes stomach disorder by H. pylori [13].
Some statistical investigations have shown that the presence of OipA is associated with high frequency of peptic and duodenal ulcers [10, 14, 15] and that the presence of this factor is not related to gastric atrophy and intestinal metaplasia. Other factors such as VacA and CagA are believed to be associated with gastric adenocarcinoma [10]. Our findings revealed that OipA causes direct death of gastric epithelial cells and has a role in inducing peptic and duodenal ulcers. These results confirm previous statistical studies which on the role of OipA in stomach ulcer.
Acknowledgments
This work was supported financially by Tarbiat Modares University (Tehran, Iran).
Contributor Information
Ashraf Mohabati Mobarez, Phone: +98-21-82-88-38-62, FAX: +98-21-82-88-38-62, Email: mmmobarez@modares.ac.ir.
Zuhair Mohammad Hassan, Phone: +98-21-8288-3565, FAX: +98-21-8800 6544, Email: hasan_zm@modares.ac.ir.
References
- 1.Abdallah FB, Lagha R, Ellafi A, Namane A, Rousselle JC, Lenormand P, Kallel H. Identification of outer membrane proteins altered in response to UVC-radiation in Vibrio parahaemolyticus and Vibrio alginolyticus. Indian J Microbiol. 2012;52:660–665. doi: 10.1007/s12088-012-0299-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamaoka Y, Ojo O, Fujimoto S, Odenbreit S, Haas R, Gutierrez O, El-Zimaity HM, Reddy R, Arnqvist A, Graham DY. Helicobacter pylori outer membrane proteins and gastroduodenal disease. Gut. 2006;55:775–781. doi: 10.1136/gut.2005.083014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamaoka Y. Virulence factors of Helicobacter pylori: up-to-date. Nippon Shokakibyo Gakkai Zasshi. 2010;107:1262–1272. [PubMed] [Google Scholar]
- 4.Kalia VC. Quorum sensing inhibitors: an overview. Biotechnol Adv. 2013;31:224–245. doi: 10.1016/j.biotechadv.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Yamaoka Y, Kwon DH, Graham DY. A M(r) 34,000 proinflammatory outer membrane protein (oipA) of Helicobacter pylori. Proc Natl Acad Sci USA. 2000;97:7533–7538. doi: 10.1073/pnas.130079797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franco AT, Johnston E, Krishna U, Yamaoka Y, Israel DA, Nagy TA, Wroblewski LE, Piazuelo MB, Correa P, Peek RM., Jr Regulation of gastric carcinogenesis by Helicobacter pylori virulence factors. Cancer Res. 2008;68:379–387. doi: 10.1158/0008-5472.CAN-07-0824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dabiri H, Maleknejad P, Yamaoka Y, Feizabadi MM, Jafari F, Rezadehbashi M, Nakhjavani FA, Mirsalehian A, Zali MR. Distribution of Helicobacter pylori cagA, cagE, oipA and vacA in different major ethnic groups in Tehran, Iran. J Gastroenterol Hepatol. 2009;24:1380–1386. doi: 10.1111/j.1440-1746.2009.05876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matteo MJ, Armitano RI, Granados G, Wonaga AD, Sanches C, Olmos M, Catalano M. Helicobacter pylori oipA, vacA and dupA genetic diversity in individual hosts. J Med Microbiol. 2010;59:89–95. doi: 10.1099/jmm.0.011684-0. [DOI] [PubMed] [Google Scholar]
- 9.Kawai M, Furuta Y, Yahara K, Tsuru T, Oshima K, Handa N, Takahashi N, Yoshida M, Azuma T, Hattori M, Uchiyama I, Kobayashi I. Evolution in an oncogenic bacterial species with extreme genome plasticity: Helicobacter pylori East Asian genomes. BMC Microbiol. 2011;11:104. doi: 10.1186/1471-2180-11-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamaoka Y, Kikuchi S, el-Zimaity HM, Gutierrez O, Osato MS, Graham DY. Importance of Helicobacter pylori oipA in clinical presentation, gastric inflammation, and mucosal interleukin 8 production. Gastroenterology. 2002;123:414–424. doi: 10.1053/gast.2002.34781. [DOI] [PubMed] [Google Scholar]
- 11.Teymournejad O, Mobarez AM, Hassan ZM, Moazzeni SM, Yakhchali B, Eskandari V. In silico prediction of exposure amino acid sequences of outer inflammatory protein A of Helicobacter pylori for surface display on Escherichia coli. Indian J Hum Genet. 2012;18:83–86. doi: 10.4103/0971-6866.96659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanaka H, Yoshida M, Azuma T. The role of cagA in H. pylori infection. Nihon Rinsho. 2009;67:2245–2249. [PubMed] [Google Scholar]
- 13.Ge Z, Schauer DB, Fox JG. In vivo virulence properties of bacterial cytolethal-distending toxin. Cell Microbiol. 2008;10:1599–1607. doi: 10.1111/j.1462-5822.2008.01173.x. [DOI] [PubMed] [Google Scholar]
- 14.Mansour KB, Fendri C, Zribi M, Masmoudi A, Labbene M, Fillali A, Mami NB, Najjar T, Meherzi A, Sfar T, Burucoa C. Prevalence of Helicobacter pylori vacA, cagA, iceA and oipA genotypes in Tunisian patients. Ann Clin Microbiol Antimicrob. 2010;9:10. doi: 10.1186/1476-0711-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salih BA, Abasiyanik MF, Ahmed N. A preliminary study on the genetic profile of cag pathogenicity-island and other virulent gene loci of Helicobacter pylori strains from Turkey. Infect Genet Evol. 2007;7:509–512. doi: 10.1016/j.meegid.2007.03.002. [DOI] [PubMed] [Google Scholar]