Abstract
Environmental awareness has led to a serious consideration for biological surfactants and hence non-edible vegetable oils may serve as a substitute carbon source for bio-surfactant production (rhamnolipid) which might be an alternative to complex synthetic surfactants. There are reports of rhamnolipid production from plant based oil giving higher production than that of glucose because of their hydrophobicity and high carbon content. Therefore the contribution of non-edible oil such as Mesua ferrea seed oil could serve as a good carbon source for rhamnolipid production. Moreover the use of rhamnolipid production from non-edible plant based seed oil has not been reported elsewhere. The present work focus on the optimal production of rhamnolipid by considering both micro and macro nutrients and culture conditions using response surface methodology. The study observes that micronutrients play a significant role in rhamnolipid production from Pseudomonas aeruginosa (MTCC 7815). The investigation results with the statistically optimize parameters able to produce a higher rhamnolipid production and this methodology could be used to optimize the nutrients requirements and culture conditions. The present findings would assist in bioremediation of crude oil contaminated ecosystems.
Electronic supplementary material
The online version of this article (doi:10.1007/s12088-013-0403-2) contains supplementary material, which is available to authorized users.
Keywords: Rhamnolipid, Non-edible oil, Response surface methodology, Carbon source
Introduction
Environmental awareness has led to a serious circumstance realizing biological surfactants as an alternative to synthetic surfactants [1]. Biosurfactants have higher specificity, low toxicity, higher biodegradability, stability at extreme pH, salinity and temperature compared to synthetic surfactants. Their unique structures provide unique properties that lacks in commercial surfactants [2]. Moreover, it has application in petroleum industry which includes enhanced oil recovery (EOR), bioremediation and waste treatment to remove hazardous materials, pharmaceutical-cosmetics formulas, agriculture, food processing industries, detergents, laundry supplies and paint industries [3–6]. Recently, biosurfactants have received much attention in nanobiotechnology too [7, 8].
Despite their potential applications, they are not employed extensively in industry because of its high production cost associated with inefficient methods for product recovery and expensive substrates that limit their commercialization. The use of cheap raw substrates is an important factor for the overall economic recovery as they account for 50 % of the total production cost [9, 10]. Also, the possible way for cost reduction is the use of an alternate substrates such as agro-industrial wastes which include molasses, whey milk, distillery waste, olive oil mill effluent, soap stock, cassava waste, potato substrates etc. [10, 11].
Alternate carbon substrates such as palm seed oil, olive oil, sunflower oil, safflower oil, canola oil, soybean oil and corn oil were evaluated for rhamnolipid production [12–18]. There are reports of rhamnolipid production from plant oil which is much higher than that of glucose because of their hydrophobicity and high carbon content per weight [10]. However, based on cost and global food supply, the use of food-grade oils for producing rhamnolipid is economically not remarkable. This may cause imbalance and depletion of food sources. It is both extravagant and unethical to use food source for rhamnolipid production. Besides, the recent food storage crisis, limited land availability for crop cultivation and food industry with increasing food demand have persuade the price of edible plant based oil to increase [19]. In this perspective, non-edible vegetable oils may be an alternate substrate for rhamnolipid production. Therefore, contributing non-edible oils such as Mesua ferrea seed oil could be a good carbon source for producing rhamnolipid. Mesua ferrea (Family: Calophyllaceae) is a slow-growing tree confined mainly to the wet and tropical parts of South-East Asia [20]. Mesua ferrea seed oil is typical triacylglycerols, consists of fatty acid and glycerol moieties. The major composition of fatty acids in its oil is myristic acid (2.13 %), palmitic acid (10.87 %) linoleic acid (13.68 %), oleic acid (55.93 %), stearic acid (14.19 %) and arachidic acid (2.92 %) [21].
Till date there are no reports for the use of non-edible Mesua ferrea seed oil for rhamnolipid production (Supplementary material Figs. S1, S2, S3, S4). It is easy to extract from the seed and available throughout the year. The present work aims to optimize the factors (nutrition and culture conditions) directly responsible for rhamnolipid production using statistical designs and response surface methodology (RSM) for higher yield. RSM is use in various experiments for optimization of products such as bacterial medium composition [22], enzymatic activity [23, 24] algal growth [25] etc.
A few works on synthesis and rhamnolipid production from Pseudomonas aeruginosa was reported earlier [26–29] however we are reporting for the first time the optimal production of rhamnolipid from Mesua ferrea seed oil. The present finding would assist in bioremediation of crude oil contaminated ecosystems and since rhamnolipid is known to be stable at extreme condition it will help in microbial enhanced oil recovery process (MEOR).
Materials and Method
Microorganism
Professor B. K. Konwar, Department of Molecular Biology and Biotechnology, Tezpur University, Assam, India provided the Pseudomonas aeruginosa (MTCC 7815) used in the present investigation. The strain was maintained in nutrient agar slants and plates with proper sub-culturing at an interval of 30 days (Supplementary material Fig. S5). The strain was preserve in 50 % glycerol and stored at −80 °C for future work.
Culture Conditions
The inoculum was prepared using bacterial cells transferred from the storage culture to a test tube containing 10 ml of the nutrient broth and incubated at 37 °C in an orbital incubator shaker at 180 rpm. The optical density of bacterial suspension was adjusted to 0.65 × 108 cfu/ml. 1.0 ml of the culture was inoculated into a 250 ml Erlenmeyer flask containing 100 ml of mineral salt medium (MSM).
The MSM (g/l) consists of: 5.0 g urea, 5.0 g (NH4)2SO4, 4.8 g Na2HPO4, 4.75 g KH2PO4, 1.0 g MgSO4·7H2O, 10 mg CaCl2·2H2O, 3.6 mg FeSO4·7H2O, 4.4 mg CuSO4·7H2O, 9.0 mg MnSO4·5H2O, 7.6 mg H3BO3, 3.6 mg ZnSO4·7H2O, 3.0 mg MnO3 and 30 ml of the test carbon source (sterilized by passing through 0.2 μm membrane filter) and the volume was adjusted up to 1,000 ml of distilled water.
The initial pH of the broth was adjusted to 6.8 using 6 N HCl and incubated at 37 °C for 9 days in an orbital incubator shaker at 180 rpm. Cell growth was monitored by measuring the protein content [30] and the increase in biomass was monitored by gravimetric method [31] continuously at an interval of 12 h for 9 days.
Oil Samples
Mesua ferrea seed oil (Supplementary material Fig. S4) was extracted using petroleum ether in a soxhlet apparatus. The solvent part was evaporated under vacuum at 40 °C in a rotary evaporator (Eyela, CCAS-1110, Tokyo, Rikakikai Co. Ltd.). The extracted oil was used in the culture medium as the sole source of carbon and energy. The physical and chemical characteristics of Mesua ferrea seed oil is shown in Table 1 as reported by Sayeed et al. [21].
Table 1.
Physical and chemical characteristics of Mesua ferrea seed oil as reported by Sayeed et al. [21]
| Physical characteristics | Characteristics value | Chemical characteristics | Characteristics value |
|---|---|---|---|
| Specific gravity at 31 °C | 0.929 | Iodine value | 91.17 |
| Refractive index at 30 °C | 1.474 | Saponification value | 200.82 |
| Solidification point (°C) | −4.0 | Saponification equivalent | 279.35 |
| Pour point (°C) | −1.3 | Acid value | 11.87 |
| Cloud point (°C) | 5.8 | Free fatty acids (%) as oleic | 5.96 |
| Flash point (°C) | 98 | Ester value | 188.95 |
| Fire point (°C) | 116 | Unsaponifiable matter (%) | 1.50 |
| Smoke point (°C) | 44 | Acetyl value | 2.70 |
| Peroxide value | 3.58 | ||
| Reichert-Meissl value | 5.961 | ||
| Polenske number | 0.7891 |
Extraction of Rhamnolipid
The bacterial culture was centrifuged at 12,000×rpm for 20 min at 4 °C and the bacterial cell mass was removed. Also, the unused vegetable oil remained in the supernatant was removed with the help of a separating funnel. The pH of the culture supernatant was adjusted to 2 using 6 N HCl and kept overnight at 4 °C to precipitate the rhamnolipid. The precipitate was harvested by centrifugation at 10,000×rpm for 20 min at the same temperature. The precipitate was further extracted thrice with ethyl acetate. The organic phase was collected and dried in a rotary evaporator (Eyela, CCAS-1110, Tokyo, Rikakikai Co. Ltd.) to remove the solvent (Supplementary material Fig. S6). Further, the viscous honey-coloured residue was washed twice with n-hexane and the final product was dissolved in ethyl acetate, filtered and concentrated using a rotary evaporator [15].
Statistical Designs
Plackett–Burman Design
A Plackett–Burman experimental design was used to screen the significant variables that influenced the rhamnolipid production in two levels: −1 for a low-level and +1 for a high-level. In the present investigation, 16 assigned variables were screened in 20 experimental designs. All experiments were carried out in triplicates and the average rhamnolipid production is taken as the response. Such design is practically useful when there are many factors and these prevails uncertainty on nearer setting to the optimum response [32]. Factors under investigation as well as levels of each factor used in the experimental design are shown in Table 2 and the design matrix was illustrated in Table 3. From the results obtained by the Plackett–Burman design, the fitted first order model is
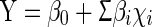 |
1 |
where Y is the predicted response, β0 and βi are constant coefficients, and χi are the coded independent variables or factors.
Table 2.
Media components, culture condition and test levels for Plackett–Burman experiment
| Variable | Variable code | Low-level (−1) | High-level (+1) |
|---|---|---|---|
| Culture volume (ml) | X1 | 50 | 25 |
| Culture pH | X2 | 5 | 8 |
| Aeration (rpm) | X3 | 150 | 200 |
| Carbon source (%, v/v) | X4 | 0.5 | 3 |
| Urea (%) | X5 | 0.05 | 0.5 |
| (NH4)2SO4 | X6 | 0.1 | 0.5 |
| KH2PO4 | X7 | 0.05 | 0.5 |
| Na2HPO4 | X8 | 0.05 | 0.5 |
| MgSO4·7H2O | X9 | 0.005 | 0.1 |
| CaCl2·2H2O | X10 | 0.001 | 0.01 |
| FeSO4·7H2O | X11 | 0 | 0.001 |
| CuSO4·7H2O | X12 | 0 | 0.001 |
| MnSO4·5H2O | X13 | 0 | 0.001 |
| H3BO3 (M) | X14 | 0 | 0.001 |
| ZnSO4·7H2O (M) | X15 | 0 | 0.001 |
| MnO3 (M) | X16 | 0 | 0.001 |
Table 3.
Randomized Plackett–Burman experimental design for evaluating factors influencing rhamnolipid production (g/l) from Pseudomonas aeruginosa (MTCC 7815)
| Runs | Physical variables | Carbon source | Nitrogen source | Energy source | Metal ions | Rhamnolipid yield (g/l)a | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Culture volume (ml) | Culture pH | Aeration (rpm) | Carbon source (%) | Urea (%) | (NH4)2SO4 | KH2PO4 | Na2HPO4 | MgSO4·7 H2O | CaCl2·2H2O | FeSO4·7H2O | CuSO4·7H2O | MnSO4·5H2O | H3BO3 (M) | ZnSO4·7H2O (M) | MnO3 (M) | ||
| 1 | 1 | 1 | −1 | −1 | −1 | −1 | 1 | −1 | 1 | −1 | 1 | 1 | 1 | 1 | −1 | −1 | 0.24 |
| 2 | −1 | 1 | −1 | 1 | −1 | 1 | 1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | 2.1 |
| 3 | −1 | 1 | 1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | −1 | −1 | 2.5 |
| 4 | 1 | −1 | 1 | 1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | −1 | 6.9 |
| 5 | −1 | −1 | −1 | 1 | −1 | 1 | −1 | 1 | 1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | 2.0 |
| 6 | 1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | −1 | −1 | 1 | −1 | 1 | −1 | 5.6 |
| 7 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | −1 | −1 | 1 | −1 | 1 | −1 | 1 | 4.1 |
| 8 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | 0.1 |
| 9 | −1 | −1 | −1 | −1 | 1 | −1 | 1 | −1 | 1 | 1 | 1 | 1 | −1 | −1 | 1 | 1 | 1.5 |
| 10 | 1 | −1 | 1 | −1 | 1 | 1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | 3.5 |
| 11 | −1 | −1 | 1 | −1 | 1 | −1 | 1 | 1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | 1 | 5.2 |
| 12 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | −1 | −1 | 1 | −1 | 1 | 1.4 |
| 13 | 1 | 1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 |
| 14 | −1 | 1 | 1 | −1 | −1 | −1 | −1 | 1 | −1 | 1 | −1 | 1 | 1 | 1 | 1 | −1 | 1.1 |
| 15 | 1 | −1 | 1 | 1 | −1 | −1 | −1 | −1 | 1 | −1 | 1 | −1 | 1 | 1 | 1 | 1 | 2 |
| 16 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | −1 | −1 | 1 | −1 | 1 | −1 | 1 | 1 | 1.1 |
| 17 | −1 | 1 | −1 | 1 | 1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | 6.1 |
| 18 | 1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | −1 | −1 | 1 | −1 | 3.7 |
| 19 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | −1 | −1 | 1 | −1 | 1 | −1 | 1 | 1 | 1 | 3 |
| 20 | 1 | −1 | −1 | −1 | −1 | 1 | −1 | 1 | −1 | 1 | 1 | 1 | 1 | −1 | −1 | 1 | 0.5 |
−1 low-level, +1 high-level
aRhamnolipid production (g/l) was determined after 9 days of incubation at 37 °C
Box–Behnken Design
Response surface method is an empirical modelling scheme used to assess the relationship between experimental variables and their corresponding responses [33]. In the present investigation, Box–Behnken design was used for four independent variables viz. oil (X1), urea (X2), NH4(SO4) (X3) and phosphates (X4) (Table 4) The ranges of the four variables studied in the present investigation are shown in Table 5. The factors of the highest confidence levels were represented in three levels, coded −1 for low, 0 for middle and +1 for high.
Table 4.
The levels of variables chosen for the Box–Behnken optimization experiment
| Variables | Variable code | −1 | 0 | +1 |
|---|---|---|---|---|
| Oil | X1 | 5.00 | 10.00 | 30 |
| Urea | X2 | 0.25 | 0.75 | 4 |
| NH4(SO4) | X3 | 0.50 | 1.00 | 5 |
| Phosphates | X4 | 1.00 | 3.00 | 8 |
Table 5.
Box–Behnken factorial experimental design, representing the response of rhamnolipid production (g/l) as influenced by oil, urea, NH4(SO4) and phosphates
| Runs | Oil | Urea | NH4(SO4) | Phosphates | Rhamnolipid production | Predicted value |
|---|---|---|---|---|---|---|
| 1 | −1 | −1 | 0 | 0 | 3.01 | 2.20125 |
| 2 | 1 | 0 | −1 | 0 | 2.16 | 1.64625 |
| 3 | 0 | 0 | 0 | 0 | 5.67 | 5.51000 |
| 4 | 1 | 0 | 0 | 1 | 3.86 | 3.98583 |
| 5 | 0 | 0 | −1 | 1 | 2.06 | 1.82625 |
| 6 | 0 | −1 | 1 | 0 | 3.14 | 3.35083 |
| 7 | 1 | −1 | 0 | 0 | 3.09 | 2.80458 |
| 8 | 0 | 0 | 1 | 1 | 6.95 | 5.94458 |
| 9 | 0 | 0 | 0 | 0 | 5.74 | 5.51000 |
| 10 | −1 | 0 | 1 | 0 | 3.35 | 4.52125 |
| 11 | −1 | 0 | 0 | −1 | 2.89 | 3.16917 |
| 12 | 1 | 1 | 0 | 0 | 4.37 | 4.11625 |
| 13 | 0 | 1 | 1 | 0 | 7.41 | 6.98250 |
| 14 | 0 | 0 | −1 | −1 | 2.31 | 2.25292 |
| 15 | 0 | 0 | 0 | 0 | 5.12 | 5.51000 |
| 16 | −1 | 0 | 0 | 1 | 2.89 | 3.24750 |
| 17 | 0 | 1 | 0 | −1 | 3.25 | 3.83292 |
| 18 | 0 | 0 | 1 | −1 | 5.04 | 4.21125 |
| 19 | −1 | 0 | −1 | 0 | 2.41 | 2.18792 |
| 20 | 0 | 1 | 0 | 1 | 4.24 | 4.92125 |
| 21 | −1 | 1 | 0 | 0 | 5.17 | 4.39292 |
| 22 | 0 | 1 | −1 | 0 | 1.87 | 2.06417 |
| 23 | 1 | 0 | 1 | 0 | 4.51 | 5.38958 |
| 24 | 0 | −1 | −1 | 0 | 1.36 | 2.19250 |
| 25 | 0 | −1 | 0 | −1 | 2.54 | 2.51625 |
| 26 | 0 | −1 | 0 | 1 | 2.66 | 2.73458 |
| 27 | 1 | 0 | 0 | −1 | 2.71 | 2.75750 |
The actual design matrix of 27 trials of experiments is given in Table 5. Experiments were performed in triplicates and the mean values were taken. The behaviour of the system is explained by the following quadratic equation
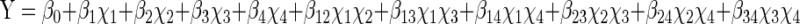 |
2 |
where Y is the predicted response, β0 model constant; χ1, χ2, χ3 and χ4 independent variables; β1, β2, β3 and β4 are linear coefficients; β12, β13, β14 and β23 are cross product coefficients and β 11, β22 and β33 are the quadratic coefficients.
Statistical Analysis of Data
The experimental data obtained were studied and interpreted using Minitab 15 statistical software [34] to estimate the response of the dependent variable. Each experiment was performed in triplicates and the mean values were taken. The quality of fitness of the polynomial model equation was expressed by the coefficient of determination (R2), and its statistical significance was checked by an F test and the significance of the regression coefficient was tested by a t test.
Results and Discussion
Rhamnolipid Production
The performance of rhamnolipid production with the carbon sources used in the present investigation depicted in Table 5, clearly explained an alternate potential carbon substrate for rhamnolipid production. There are certain reports that mark the type and concentration of carbon substrates which affect the production of rhamnolipid yield [18, 35]. Wei et al. (2005) reported using major types of carbon sources such as carbohydrate, hydrocarbon and vegetable oils for rhamnolipid production by P. aeruginosa J4 strain.
Additionally, there are reports of vegetable oils depicting as more effective substrate in rhamnolipid production from P. aeruginosa compared to glucose, glycerol and hydrocarbons [15, 35–37]. However, in contrast to these findings Pseudomonas EMI strain used by Benincasa et al. [14] showed that carbon sources such as glucose and glycerol were superior over the vegetable oils like olive and soya bean oil when tested with rhamnolipid yield and productivity. This report clearly suggests that the carbon source preference for rhamnolipid production mainly depends on the behaviour of P. aeruginosa strain.
Additionally, the authors would like to highlight the point that the use of two nitrogen sources enhanced the rhamnolipid production and yield. In the present investigation a combination of urea and (NH4)2SO4 (both nitrogen sources) was found to be more productive. This is because P. aeruginosa (MTCC 7815) is able to use these nitrogen sources. However, in order to obtain high concentrations of rhamnolipid, it is necessary to have restrained conditions of this macro-nutrient [38]. Also, there are several reports where two different nitrogen sources were used for the rhamnolipid production. Bordoloi and Konwar [39] used (NH4)2SO4 as inorganic and urea as organic nitrogen sources. While Xia et al. [40] used (NH4)2SO4 as inorganic and yeast extract as an organic nitrogen sources.
Evaluation of the Factors that Influence Features on Producing Rhamnolipid Biosurfactant
For explanation of various fermentation factors affecting the rhamnolipid production from P. aeruginosa (MTCC 7815), the independent variables examined in the Plackett–Burman experiments and their settings are shown in Table 2. Sixteen different variables including medium constituents and fermentation conditions were selected to perform the optimization process. The mean of rhamnolipid production for the different trials are listed in Table 3.
The main effect of each variable was calculated according to the difference between averages of measurements made on high-level (+1) and low-level (−1) of that variable which is shown in Table 3. The variation suggests the need of the importance for optimized medium to reach for a higher productivity. The data in Table 3 showed a wide variation of rhamnolipid yield ranging from 0.1 to 6.9 g/l out of the 20 experimental run conditions.
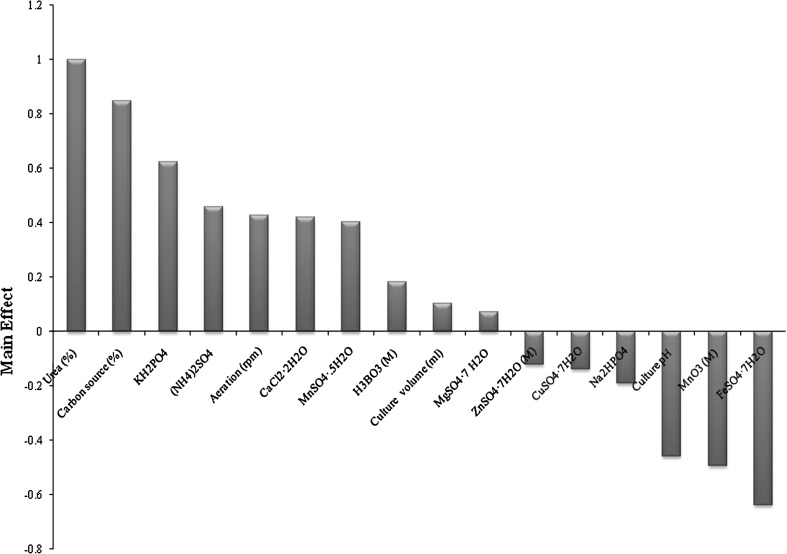
Further, the Placket–Burman experimental design used in the present investigation which is based on the first order model for the analysis of the data (main effects) is graphically represented in Fig. 1. From Fig. 1 and Table 6, i.e. the regression analysis coefficient data suggest that the culture volume (ml), aeration (rpm), carbon source (Mesua ferrea seed oil), urea, (NH4)2SO4, KH2PO4, MgSO4·7H2O, CaCl2·2H2O, MnSO4·5H2O and H3BO3 affect the rhamnolipid production positively. While, culture pH, Na2HPO4, FeSO4·7H2O, CuSO4·7H2O, ZnSO4·7H2O and MnO3 contributed negatively. However the confidence level of the culture volume (ml), MgSO4·7H2O, CuSO4·7H2O, ZnSO4·7H2O (M) is comparatively low (shown in Fig. 1; Table 6) compared to other factors. Additionally, the complementary factors i.e. aeration (rpm) and culture volume (ml) showed positive affect for rhamnolipid production which might be a major factor for the optimal production. Also from the Pareto chart from Fig. 1, for these physical parameters, the effect of aeration (rpm) contribute to a much higher level compared to culture volume (ml).
Fig. 1.
Effect of nutrients and culture condition factors on the production of rhamnolipid by Pseudomonas aeruginosa (MTCC 7815)
Table 6.
Statistical analysis of Plackett–Burman design showing coefficient values, t and p values for each variable on rhamnolipid production (g/l)
| Variables | Coefficient | t statistics | p value | Confidence level (%) |
|---|---|---|---|---|
| Intercept | 2.682 | – | – | (1−p value) × 100 |
| Culture volume (ml) | 0.102 | 0.640 | 0.567 | 43.3 |
| Culture pH | −0.458 | −2.875 | 0.064 | 93.6 |
| Aeration (rpm) | 0.428 | 2.687 | 0.075 | 92.5 |
| Carbon source (%) | 0.848 | 5.324 | 0.013 | 98.7 |
| Urea (%) | 0.998 | 6.266 | 0.008 | 99.2 |
| (NH4)2SO4 | 0.458 | 2.875 | 0.064 | 93.6 |
| KH2PO4 | 0.622 | 3.905 | 0.030 | 97 |
| Na2HPO4 | −0.192 | −1.205 | 0.314 | 68.6 |
| MgSO4·7 H2O | 0.072 | 0.452 | 0.682 | 31.8 |
| CaCl2·2H2O | 0.418 | 2.624 | 0.079 | 92.1 |
| FeSO4·7H2O | −0.638 | −4.005 | 0.028 | 97.2 |
| CuSO4·7H2O | −0.138 | −0.866 | 0.450 | 55 |
| MnSO4·.5H2O | 0.402 | 2.524 | 0.086 | 91.4 |
| H3BO3 (M) | 0.182 | 1.143 | 0.336 | 66.4 |
| ZnSO4·7H2O (M) | −0.122 | −0.766 | 0.499 | 50.1 |
| MnO3 (M) | −0.492 | −3.089 | 0.054 | 94.6 |
The correlation between the sixteen variables and the rhamnolipid production based on the calculated t values and confidence level (%) which is shown in Table 6 can be described by the polynomial model:
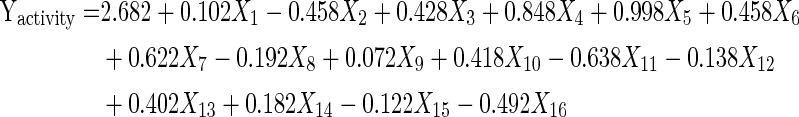 |
From Table 3, it is depicted that the presence of high-level of urea (0.5 g/l), Mesua ferrea seed oil (30 ml/l), KH2PO4 (0.5 g/l), (NH4)2SO4 (0.5 g/l), aeration (200 rpm), CaCl2·2H2O (0.01 g/l), MnSO4·5H2O (0.001 g/l), H3BO3 (0.001 g/l), culture volume (25 ml), MgSO4·7H2O (0.1 g/l) were found to be the most significant factors affecting the rhamnolipid production positively. Whereas low-level of Na2HPO4 (0.05 g/l), incubation at low pH (5) and without adding ZnSO4·7H2O, CuSO4·7H2O, MnO3, FeSO4·7H2O will enhance the rhamnolipid production. To evaluate the accuracy of the Placket–Burman design, a verification experiment was conducted in triplicate. By applying the Placket–Burman design, the experiment was carried out with the following composition (g/l): urea, 0.5; KH2PO4, 0.5; (NH4)2SO4, 0.5; CaCl2·2H2O, 0.01; MnSO4·5H2O, 0.001; H3BO3, 0.001; MgSO4·7H2O, 0.1; Na2HPO4, 0.05; Mesua ferrea seed oil, 30 ml (v/v), while pH is 5 incubated at a constant shaking of 200 rpm and the culture volume of 25 ml. The production of rhamnolipid increases about 6.95 g/l which is nearly equals to 2.0 times higher than that produced from basal medium (3.54 g/l). Based on t-statistic value thus obtained (Table 6), concentration of urea in the culture medium was considered to be the highly considerable variable for rhamnolipid production by the bacterial strain.
Optimization of the Rhamnolipid Production by Box–Behnken Design
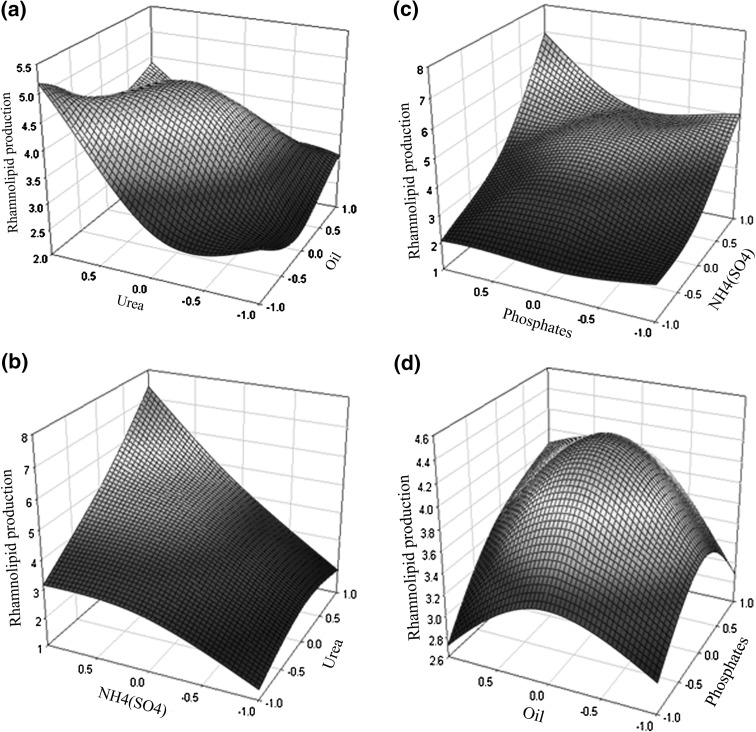
Placket–Burman design is based on the first order model and it doesn’t show the interaction within the variables and used for screening and evaluation of important variables that influence the response [41]. Therefore, Box–Behnken design was used as complementary step [42]. In this step of optimization, the level of four independent variables viz. oil (X1), urea (X2), NH4(SO4)2, (X3) and phosphates (X4) were further investigated at three different level which is listed in Table 4. While the design matrix of the variables with their experimental results for rhamnolipid production is shown in Table 5. All experiments were carried out in triplicates and the mean values were taken for observation. The experimental results were displayed in the form of three-dimensional response surface plots. Figure 2a showed that gradual increase in urea concentration with the high-level of oil (g/l) that will promote the rhamnolipid production, while further increase in urea concentration beyond the optimum level will decrease rhamnolipid production. According to Fig. 2b, the NH4SO4 concentration affects the rhamnolipid production more significantly than urea. Figure 2b also suggest that with the increase in NH4(SO4) the increase in rhamnolipid production and with the increase in urea concentration the increase in rhamnolipid production.
Fig. 2.
Rhamnolipid production (g/l) response surface from Pseudomonas aeruginosa (MTCC 7815) as affected by (a) urea and oil (b) NH4(SO4) and urea (c) phosphates and NH4(SO4) and (d) oil and phosphates
The interaction between phosphates and NH4SO4 concentration (g/l), where a maximum peak of rhamnolipid production was appeared which is shown in Fig. 2c. While Fig. 2c also suggest with the increase in NH4SO4, the higher yield of rhamnolipid while the phosphate concentration remains the same. Lastly Fig. 2d showed the concentration of oil (g/l) and phosphates (g/l) that affects the rhamnolipid production. It also suggests that the rhamnolipid production increases with the optimal amount of oil concentration and phosphate concentration. Such experimental design is a very useful tool for media optimization for enhanced rhamnolipid production.
For predicting the optimal point, a second order polynomial function was fitted to the experimental results of rhamnolipid production:
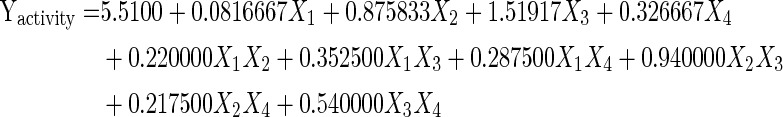 |
3 |
where, X1, X2, X3 and X4 are the oil, urea, NH4(SO4)2 and phosphates respectively.
The regression model Eq. (3) fits the experimental data well with a high R2 (coefficient of determination) value of 0.87. The value of R2 thus obtained suggests a high degree of correlation between the experimental and the predicted values.
Verification of the Model
To find out the optimum condition and to confirm the accuracy of the model, an experiment was carried out under basal and predicted optimal conditions where rhamnolipid production was monitored compared to the calculated data from the model. The estimated rhamnolipid production was 5.012 g/l compared to 7.22 g/l of the predicted value from the polynomial model. Thus, it points out the accuracy of the model with more than 69.42 %, which is an evidence for the model validation. However this variance of rhamnolipid production might be because of slight variation in the experimental conditions.
Conclusion
Statistical experiment design was applied to find out the optimal concentration of each media components for the better production of rhamnolipid. The experimental design was found to be a valuable tool for improving medium composition that leads to a higher degree of rhamnolipid production. Hence pointing out statistical design as a tool for media optimization for enhanced productivity and yield. The results also suggest the optimized conditions improve the rhamnolipid production rate. As a concluding remark, the authors would like to point that, using the method of experimental factorial design and RSM analysis, it was possible to find out the optimal handling conditions which would obtain a higher growth, thus a higher rhamnolipid production. Additionally, the validity of the model was proven by fitting the values of the variables in the model equation and by actually carrying out the experiment with those values of the variables resulting with a good accuracy of the statistical design. The present findings would aid in higher rhamnolipid yield which would assist in bioremediation of crude oil contaminated ecosystems such as soil and water bodies.
Electronic supplementary material
Acknowledgments
The authors would like to thank Yasser R. Abdel-Fattah, Bioprocess Development Department, Genetic Engineering and Biotechnology Research Institute, Alexandria, Egypt for his valuable suggestions in carrying out the statistical analysis part.
Conflict of interest
The authors have declared that no competing interests exist.
References
- 1.Nitschke M, Costa SG, Haddad R, Gonçalves LA, Eberlin MN, Contiero J. Oil wastes as unconventional substrates for rhamnolipid biosurfactant production by Pseudomonas aeruginosa LBI. Biotechnol Prog. 2005;21(5):1562–1566. doi: 10.1021/bp050198x. [DOI] [PubMed] [Google Scholar]
- 2.Banat IM, Makkar RS, Cameotra SS. Potential commercial applications of microbial surfactants. Appl Microbiol Biotechnol. 2000;53(5):495–508. doi: 10.1007/s002530051648. [DOI] [PubMed] [Google Scholar]
- 3.Wei YH, Cheng CL, Chien CC, Wan HM. Enhanced di-rhamnolipid production with an indigenous isolate Pseudomonas aeruginosa J16. Process Biochem. 2008;43:769–774. doi: 10.1016/j.procbio.2008.03.009. [DOI] [Google Scholar]
- 4.Rahman KS, Rahman TJ, Kourkoutas Y, Petsas I, Marchant R, Banat IM. Enhanced bioremediation of n-alkane in petroleum sludge using bacterial consortium amended with rhamnolipid and micronutrients. Bioresour Technol. 2003;90(2):159–168. doi: 10.1016/S0960-8524(03)00114-7. [DOI] [PubMed] [Google Scholar]
- 5.Kitamoto D, Isoda H, Nakahara T. Functions and potential applications of glycolipid biosurfactants–from energy-saving materials to gene delivery carriers. J Biosci Bioeng. 2002;94(3):187–201. doi: 10.1263/jbb.94.187. [DOI] [PubMed] [Google Scholar]
- 6.Maier RM, Soberón-Chávez G. Pseudomonas aeruginosa rhamnolipids: biosynthesis and potential applications. Appl Microbiol Biotechnol. 2000;54(5):625–633. doi: 10.1007/s002530000443. [DOI] [PubMed] [Google Scholar]
- 7.del Pozo-Rodríguez A, Delgado D, Solinís MA, Pedraz JL, Echevarría E, Rodríguez JM, Gascón AR. Solid lipid nanoparticles as potential tools for gene therapy: in vivo protein expression after intravenous administration. Int J Pharm. 2010;385:157–162. doi: 10.1016/j.ijpharm.2009.10.020. [DOI] [PubMed] [Google Scholar]
- 8.Cevc G, Vierl U. Nanotechnology and transdermal rout: a state of the art review and critical appraisal. J Control Release. 2010;141:277–299. doi: 10.1016/j.jconrel.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 9.Rodrigues L, Moldes A, Teixeira J, Oliveira R. Kinetic study of fermentative biosurfactant production by Lactobacillus strains. Biochem Eng J. 2006;28(2):109–116. doi: 10.1016/j.bej.2005.06.001. [DOI] [Google Scholar]
- 10.Makkar RS, Cameotra SS. Biosurfactant production by microorganisms on unconventional carbon sources—a review. J Surf Det. 1999;2:237–241. doi: 10.1007/s11743-999-0078-3. [DOI] [Google Scholar]
- 11.Mercade ME, Manresa MA, Robert M, Espuny MJ, Andres C, Guinea J. Olive oil mill effluent (OOME). New substrate for biosurfactant production. Bioresour Technol. 1993;43:1–6. doi: 10.1016/0960-8524(93)90074-L. [DOI] [Google Scholar]
- 12.Oliveira FJS, Vazquez L, de Campos NP, de França FP. Production of rhamnolipids by a Pseudomonas alcaligenes strain. Process Biochem. 2009;44:383–389. doi: 10.1016/j.procbio.2008.11.014. [DOI] [Google Scholar]
- 13.Manresa MA, Bastida J, Mercade ME, Robert M, de Andres C, Espung MJ, et al. Kinetic studies on surfactant production by Pseudomonas aeruginosa 4471. J Ind Microbiol. 1991;8:133–136. doi: 10.1007/BF01578765. [DOI] [Google Scholar]
- 14.Benincasa M, Contiero J, Manresa MA, Moraes IO. Rhamnolipid production by Pseudomonas aeruginosa LBI growing on soapstock as the sole carbon source. J Food Eng. 2002;54:283–288. doi: 10.1016/S0260-8774(01)00214-X. [DOI] [Google Scholar]
- 15.Wu JY, Yeh KL, Lu WB, Lin CL, Chang JS. Rhamnolipid production with indigenous Pseudomonas aeruginosa EM1 isolated from oil-contaminated site. Bioresour Technol. 2008;99(5):1157–1164. doi: 10.1016/j.biortech.2007.02.026. [DOI] [PubMed] [Google Scholar]
- 16.Rahman KS, Rahman TJ, McClean S, Marchant R, Banat IM. Rhamnolipid biosurfactants production by strains of Pseudomonas aeruginosa using low cost raw materials. Biotechnol Prog. 2002;18:277–281. doi: 10.1021/bp020071x. [DOI] [PubMed] [Google Scholar]
- 17.Sim L, Ward OP, Li ZY. Production and characterisation of a biosurfactant isolated from Pseudomonas aeruginosa UW-1. J Ind Microbiol Biotechnol. 1997;19:232–238. doi: 10.1038/sj.jim.2900450. [DOI] [PubMed] [Google Scholar]
- 18.Mata-Sandoval JC, Karns J, Torrents A. High-performance liquid chromatography method for the characterization of rhamnolipid mixtures produced by Pseudomonas aeruginosa UG2 on corn oil. J Chromatogr A. 1999;864:211–220. doi: 10.1016/S0021-9673(99)00979-6. [DOI] [PubMed] [Google Scholar]
- 19.Ko-Sin N, Wei-Yang O, Lay-Koon G, Rajaiah S, Kumar S. Evaluation of jatropha oil to produce poly(3-hydroxybutyrate) by Cupriavidus necator H16. Polym Degrad Stab. 2010;95:1365–1369. doi: 10.1016/j.polymdegradstab.2010.01.021. [DOI] [Google Scholar]
- 20.Kostermans AJGH. Clusiaceae (Guttiferae) In: Dassanayaka MD, Fosberg FR, editors. A revised handbook to the flora of Ceylon. New Delhi: Amerind; 1980. pp. 107–110. [Google Scholar]
- 21.Sayeed MA, Ali MA, Sohel FI, Khan AM, Yeasmin MS. Physico-chemical characteristics of Mesua ferrea seed oil and nutritional composition of its seed and leaves. Bull Chem Soc Ethiop. 2004;18(2):157–166. [Google Scholar]
- 22.Rao KJ, Kim CH, Rhee SK. Statistical optimization of medium for the production of recombinant hirudin from Saccharomyces cerevisiae using response surface methodology. Process Biochem. 2000;35:639–647. doi: 10.1016/S0032-9592(99)00129-6. [DOI] [Google Scholar]
- 23.Kunamneni A, Singh S. Response surface optimization of enzymatic hydrolysis of maize starch for higher glucose production. Biochem Eng J. 2005;27:179–190. doi: 10.1016/j.bej.2005.08.027. [DOI] [Google Scholar]
- 24.Saeed HM, Abdel-Fattah YR, Gohar YM, Elbaz MA. Improved production of Pseudomonas aeruginosa uricase by optimization of process parameters through statistical experimental designs. Process Biochem. 2005;40:1707–1714. doi: 10.1016/j.procbio.2004.06.048. [DOI] [Google Scholar]
- 25.Song L, Qin JG, Su S, Xu J, Clarke S, et al. Micronutrient requirements for growth and hydrocarbon production in the oil producing green alga Botryococcus braunii (Chlorophyta) PLoS ONE. 2012;7(7):e41459. doi: 10.1371/journal.pone.0041459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer-Hoffert U, Zimmermann A, Czapp M, Bartels J, Koblyakova Y, et al. Flagellin delivery by Pseudomonas aeruginosa rhamnolipids induces the antimicrobial protein psoriasin in human skin. PLoS ONE. 2011;6(1):e16433. doi: 10.1371/journal.pone.0016433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamatkar NG, Shrout JD. Surface hardness impairment of quorum sensing and swarming for Pseudomonas aeruginosa. PLoS ONE. 2011;6(6):e20888. doi: 10.1371/journal.pone.0020888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bjarnsholt T, Jensen PØ, Jakobsen TH, Phipps R, Nielsen AK, et al. Quorum sensing and virulence of Pseudomonas aeruginosa during lung infection of cystic fibrosis patients. PLoS ONE. 2010;5(4):e10115. doi: 10.1371/journal.pone.0010115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macé C, Seyer D, Chemani C, Cosette P, Di-Martino P, et al. Identification of biofilm-associated cluster (bac) in Pseudomonas aeruginosa involved in biofilm formation and virulence. PLoS ONE. 2008;3(12):e3897. doi: 10.1371/journal.pone.0003897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 31.Bharali P, Konwar BK. Production and physico-chemical characterization of a biosurfactant produced by Pseudomonas aeruginosa OBP1 isolated from petroleum sludge. Appl Biochem Biotechnol. 2011;164(8):1444–1460. doi: 10.1007/s12010-011-9225-z. [DOI] [PubMed] [Google Scholar]
- 32.Strobel RJ, Sullivan GR. Experimental design for improvement of fermentations. In: Demain AL, Davies GE, editors. Manual of industrial microbiology and biotechnology. Washington, DC: ASM; 1999. pp. 80–93. [Google Scholar]
- 33.Box GEP, Behnken DW. Some new three level designs for the study of quantitative variables. Technometrics. 1960;2:455–475. doi: 10.1080/00401706.1960.10489912. [DOI] [Google Scholar]
- 34.Minitab 15 Statistical Software, Minitab Inc., PA, USA
- 35.Maier RM, Soberon-Chavez G. Pseudomonas aeruginosa rhamnolipids: biosynthesis and potential applications. Appl Microbiol Biotechnol. 2000;54:625–633. doi: 10.1007/s002530000443. [DOI] [PubMed] [Google Scholar]
- 36.Sandoval JCM, Karns J, Torrents A. Effect of nutritional and environmental conditions on the production and composition of rhamnolipids by Pseudomonas aeruginosa UG2. Microbiol Res. 2001;155:249–256. doi: 10.1016/S0944-5013(01)80001-X. [DOI] [PubMed] [Google Scholar]
- 37.Costa SGVAO, Nitschke M, Haddad R, Eberlin MN, Contiero J. Production of Pseudomonas aeruginosa LBI rhamnolipids following growth on Brazilian native oils. Proc Biochem. 2006;41:483–488. doi: 10.1016/j.procbio.2005.07.002. [DOI] [Google Scholar]
- 38.Rashedi H, Mazaheri AM, Jamshidi E, Bonakdarpour B. Optimization of the production of biosurfactant by Psuedomonas aeruginosa HR Isolated from an Iranian southern oil well. Iran J Chem Chem Eng. 2006;25:25–30. [Google Scholar]
- 39.Bordoloi NK, Konwar BK. Microbial surfactant-enhanced mineral oil recovery under laboratory conditions. Colloids Surf B. 2008;63:73–82. doi: 10.1016/j.colsurfb.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 40.Xia WJ, Luo ZB, Dong HP, Yu L, Cui QF, Bi YQ. Synthesis, characterization, and oil recovery application of biosurfactant produced by indigenous pseudomonas aeruginosa WJ-1 using waste vegetable oils. Appl Biochem Biotechnol. 2012;166(5):1148–1166. doi: 10.1007/s12010-011-9501-y. [DOI] [PubMed] [Google Scholar]
- 41.El-Sersy NA, Ibrahim HAH, Abou-Elela GM. Response surface methodology as a tool for optimizing the production of antimicrobial agents from Bacillus licheniformis SN2. Curr Res Bacteriol. 2010;3:1–14. doi: 10.3923/crb.2010.1.14. [DOI] [Google Scholar]
- 42.Banik RM, Pandey SK. Selection of metal salts for alkaline phosphatase production using response surface methodology. Bioprocess Food Ind. 2009;42:470–475. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.