Abstract
The microbial diversity and the community succession in the fermenting cover lees of Chinese Luzhou-flavor liquor were investigated by small-subunit rRNA (SSU rRNA) culture independent method. All sequences retrieved from the 1, 7 and 60 days fermented cover lees were respectively assigned into the genera of Streptococcus, Acetobacter, Arthrobacter, Bacillus, Staphylococcus, Serratia, Nocardia, Methanoculleus, Clostridium, Aneurinibacillus, Corynebacterium, Lactobacillus, Microbacterium, Trichosporon, Saccharomycopsis, Sagenomella, Talaromyces, Eurotium, Issatchenkia, Zygosaccharomyces, Saccharomyces and TM7 phylum. The fungal Issatchenkia, Saccharomycopsis and Talaromyces and the bacteria Staphylococcus and Lactobacillus were most abundant in the 1 day fermented cover lees, the fungal Issatchenkia, Saccharomyces and Talaromyces and the bacteria Bacillus and Streptococcus were dominant in the 7 days cover lees, the archaea Methanoculleus and the fungal Eurotium and Talaromyces were prevalent in the 60 days cover lees. When the microbial community profiles in three samples were compared at species level, the prokaryotic community similarity coefficient was from 0.4042 to 0.5703 and descended to 0.2222, and that of eukaryotic community was from 0.3000 to 0.6000 and followed to 0.5215. These results suggested that microbial diversity variability and community succession have happened in the cover lees associated with fermentation proceeding and such variability and succession respond for the appearance of some unique flavor of Luzhou-flavor liquor.
Keywords: Luzhou-flavor liquor, Microbial diversity, Community succession, Fermenting cover lees
Introduction
Chinese liquor is well known worldwide and very popular in China. In Chinese liquors, there are three main types: sauce fragrance liquor, intense fragrance liquor and light fragrance liquor [1]. Luzhou-flavor liquor is a typical representative of the Chinese intense fragrance liquor, and it is produced in southeast of Sichuan province in China. In 1996 and 2006, the traditional brewing technique of Luzhou-flavor liquor was listed in Chinese intangible cultural heritage list and in world cultural heritage tentative list, respectively [2].
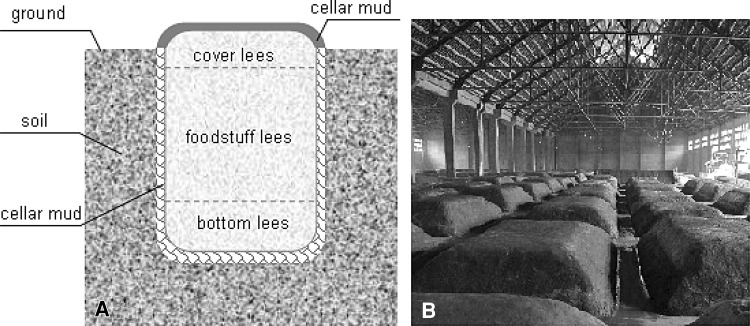
Luzhou-flavor liquor is distilled from the fermented liquor lees in the soil cellar. The liquor lees is the solid medium colonized on by many brewing microorganisms, and it is divided into the cover lees, the foodstuff lees and the bottom lees according to their material composition and position in the cellar (Fig. 1a). The foodstuff lees, located in the middle layer of soil cellar, is the main fermentation region and it consists of the mixture grains such as sorghum, corn, wheat, rice, cavings, the starter Chinese Qu and the distilled foodstuff lees from the last turn. The bottom lees in the bottom of soil cellar, usually named as mother lees, is foodstuff lees that has fermented for three times or more. The cover lees in the top layer contains the starter Chinese Qu and the distilled foodstuff lees from the last turn. After these lees are prepared as the rules defined by the process, they are placed into the soil cellar and covered by the cellar mud, then fermented at the natural condition for 60 days or more (Fig. 1b). During the fermenting, the brewing microorganisms, which come from the cellar mud, the starter Chinese Qu, the mother lees and the natural environment surround the soil cellar, are considered to play a key role [2]. And the unique sensorial appearance of Luzhou-flavor liquor is largely dependent on the microbial succession associated with fermenting [2].
Fig. 1.
The profile of the Luzhou-flavor liquor soil cellar (a) and the soil cellar filled with the fermenting lees (b)
The microbial succession is a universal phenomenon in natural fermentation process [3]. The microbial succession in several types of food, such as kimchi, traditional fermented mustard and Vietnamese alcohol starter has been examined by rRNA gene sequence analysis [2]. In recent, some researchers have already initiated the microbial succession analysis in Chinese traditional liquor fermentation by rRNA [1, 2, 4, 5]. The variability of bacteria and fungi present during the traditional fermentation process of Chinese light-fragrance Fen liquor lees was revealed in 2011 [5]. In 2012, Xiang et al. [2] firstly reported the microbial succession in foodstuff lees of traditional Chinese Luzhou-flavor liquor during the fermentation. In most cases, however, these investigations only focused on the microbes in the foodstuff lees but not in the cover lees. The cover lees is an important part of the Chinese traditional liquor cellar micro-ecosystem and it mainly contributes for the lees flavor of Chinese liquor. While, it is not yet known whether the microbial profiles in the cover lees are different from those in the foodstuff lees during the fermentation. In the present study, we have investigated the microbial diversity and their succession in the Luzhou-flavor liquor cover lees during its natural fermentation by 16S rRNA and 18S rRNA gene libraries. The information gathered may be useful to improve our understanding of the composition and succession of the microbes of Luzhou-flavor liquor cover lees during fermenting and to design the effective management of the fermentation process.
Materials and Methods
Liquor Lees Sampling
The experiments were carried out on an industrial scale traditional fermenting cover lees at the Luzhou Liquor Co. Ltd (Sichuan, China). The cover liquor lees fermented for 1 day (T1), 7 days (T7) and 60 days (T60) were respectively collected in sterile tubes and then frozen in liquid nitrogen until analysis.
DNA Extraction
The total DNAs were extracted and purified from microbial cells in the cover liquor lees according to the methods described by Xiang et al. [2].
Construction of SSU rRNA Library
Prokaryotic and eukaryotic SSU rRNA in the cover lees were respectively amplified by the 16S and 18S forward and reverse primers (Eu27F: 5′-AGAGTTTGATCCTGGCTCAG-3′ and 1490R: 5′-GGTTACCTTGTTACGACTT-3′ for bacterial 16S rRNA; Ar28F: 5′-TGGTTGATCCTGCCAGAGG-3′ and 1490R: 5′-GGTTACCTTGTTACGACTT-3′ for archaeal 16S rRNA; 18SF: 5′-AACCTGGTTGATCCTGCCAGT-3′ and 18SR: 5′-TGATCCTTCTGCAGGTTCACCTAC-3′ for fungal 18S rRNA) [2, 6]. The thermal cycling conditions for 16S and 18S primers were as previously described by Xiang et al. [2]. The amplified fragments were respectively cloned into a plasmid pGEM-T vector according to the manufacturer’s protocol. The recombined plasmids were then transformed into the chemically competent Escherichia coli DH 5α cells and transformants were identified on LB agar plates containing 100 μg/ml ampicilin, 40 μg/ml X-gal and 24 μg/ml IPTG. The white clones were used to construct a library of clones, which corresponded to a single SSU rRNA amplicon.
Sequencing and Phylogenetic Analysis of SSU rRNA
The recombined plasmids with SSU rRNA were used as templates for sequencing. The 16S rRNA and 18S rRNA sequences without chimeras were submitted to compare with sequences from GenBank and the percent similarity was then determined. The phylogenetic trees based on the 16S rRNA and 18S rRNA were respectively constructed with the MEGA 5.0 by the neighbor-joining method with the Kimura two-parameter model [7].
Statistical Analysis
BioEdit software was employed to carry out the pairwise comparison of all SSU rRNA sequences in this study. The rRNAs with 97 % or higher similarity were clustered into an operation taxonomic unit (OTU), while the others were grouped alone. The rarefaction analysis was performed by the Analytic Rarefaction 1.3 calculator based on the numbers of OTUs versus the numbers of clones [8]. More indices were calculated using Estimates Win 8.20 software for each library. These included Sobs, Schao1, SACE, Shannon diversity (H), Simpson and Coverage. Microbial community relationship of three samples was calculated using Baroni-Urbani and Buser similarity index by DPS 7.5.
Nucleotide Sequence Accession Numbers
The represent SSU rRNA sequences of OTUs in this study were deposited in the GenBank databases at the NCBI. Prokaryotic 16S rRNA sequences GenBank ID: KC337085-KC337102; Eukaryotic 18S rRNA sequences GenBank ID: KC337075-KC337084.
Results
SSU rRNA Libraries of Luzhou Flavor Cover Lees
Each sample was used to produce a library of SSU rRNA clones. A total of 32 archaea clones, 143 bacterial clones and 150 eukaryotic clones were recovered from three fermented cover lees, 27 bacterial clones and 33 eukaryotic clones from T1 sample, 10 archaea clone, 99 bacterial clones and 75 eukaryotic clones from T7 sample, 22 archaea clone, 17 bacterial clones and 42 eukaryotic clones from T60 sample. All clone sequences above without chimeras were sorted into OTUs. It was determined that 6 bacterial OTUs and 6 eukaryotic OTUs were in T1 lees; 1 archaea OTU, 12 bacterial OTUs and 7 eukaryotic were in T7 lees; 2 archaea OTU, 7 bacterial OTUs and 4 eukaryotic OTUs were in T60 lees; and that most OTUs contained multiple clones, while only few OTUs were unique, represented by only a single clone.
Estimated Microbial Diversity of Cover Lees
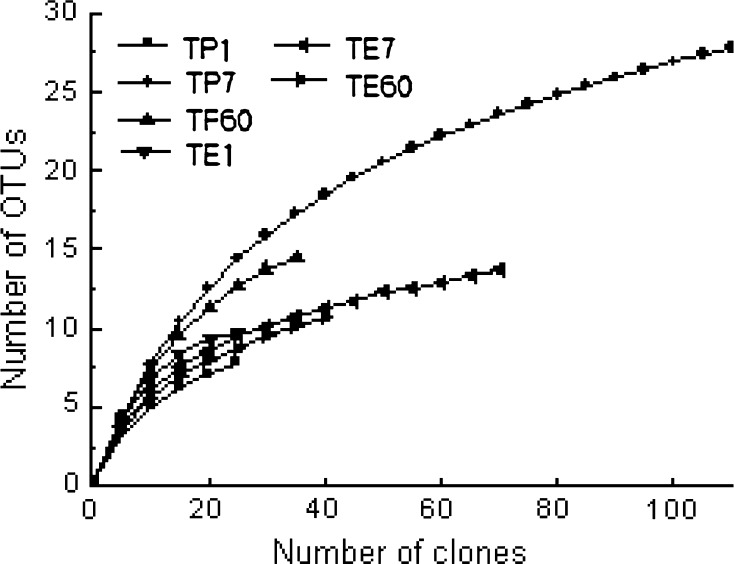
The rarefaction curves based on the 16S rRNA and 18S rRNA sequences in three samples were respectively obtained by plotting the number of OTUs observed against the number of clones (Fig. 2). It was calculated that some curves did not reach clear saturation. This result indicates that some microbes might escape from detection. However, the obvious decrease in the rate of OTUs detection on the curve indicated that the major part of the diversity of prokayotic and eukayotic micorbes in current study had already been detected (Fig. 2). This conclusion was further supported by calculating the coverage of six libraries, which was from 55.7 % to 100 % (Table 1). Biodiversity were calculated for prokaryotic and eukaryotic populations using the Shannon-wiener (H) diversity index and the Simpson dominancy index. The H index and the Simpson index respectively varied from 1.93 to 2.83 and from 6.11 to 13.72 for six gene libraries (prokaryotic libraries: TP1, TP7 and TP60; eukayotic libraries: TE1, TE7 and TE60) (Table 1). Maximum OTU richness was respectively estimated with SChao1 and SACE, and it revealed that SChao1 12 and SACE 10 for TP1, SChao1 35.66 and SACE 32 for TP7, SChao1 16.33 and SACE 15.85 for TP60, SChao1 10 and SACE 10 for TE1, SChao1 24 and SACE 24 for TE7 and SChao1 23.5 and SACE 16 for TE60 (Table 1). Based on these estimations, 55.7 %–100 % of the diversities were covered by the applied sampling survey.
Fig. 2.
Rarefaction analysis of fermenting cover lees prokaryotic 16S rRNA and eukaryotic 18S rRNA libraries, displaying the number of OTUs detected versus the number of sequences analyzed. TP1, TP7 and TP60 respectively represented 16S rRNA libraries from 1 day, 7 days and 60 days fermented cover lees. TE1, TE7 and TE60 respectively represented 18S rRNA libraries from 1 day, 7 days and 60 days fermented cover lees
Table 1.
Richness and diversity estimations of OTUs derived from 1 day, 7 days and 60 days fermented cover lees prokaryotic 16S rRNA and eukaryotic 18S rRNA gene libraries
| Sample | Num of clone | Sobs | Schao1 | SACE | Coveragea | H | Simpson |
|---|---|---|---|---|---|---|---|
| 16S rRNA analysis | |||||||
| TP1 | 27 | 8 | 12 | 10 | 72.7 | 1.93 | 9.55 |
| TP7 | 112 | 25 | 35.66 | 32 | 73.9 | 2.83 | 13.72 |
| TP60 | 39 | 15 | 16.33 | 15.85 | 93.2 | 2.49 | 13 |
| 18S rRNA analysis | |||||||
| TE1 | 33 | 10 | 10 | 10 | 100 | 2.23 | 11.53 |
| TE7 | 75 | 14 | 24 | 24 | 58.3 | 2.17 | 7.06 |
| TE60 | 42 | 11 | 23.5 | 16 | 55.7 | 1.96 | 6.11 |
aPercentage of coverage: Sobs/mean (SChao1, SACE) × 100
Microbial Distribution and Phylogenesis Based on SSU rRNA
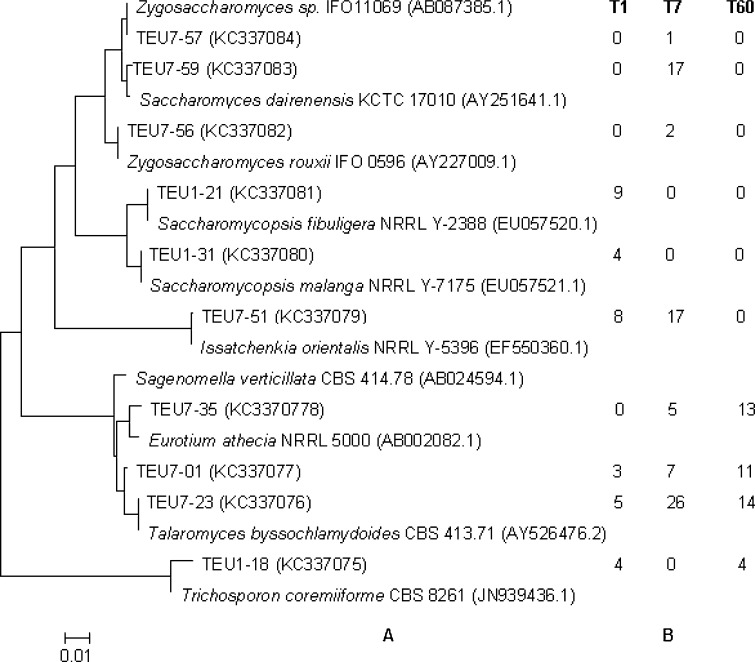
The prokaryotic 16S rRNA without chimeras were rapidly and accurately classified to genus level by program RDP MultiClassifier. The 18S rRNA represent sequences were compared with those in GenBank and were also assigned to a same phylotype. All prokaryotic sequences retrieved in this study respectively fell into the genera of Lactobacillus, Streptococcus, Bacillus, Staphylococcus, Clostridium, Acetobacter, Serratia, Nocardia, Aneurinibacillus, Corynebacterium, Arthrobacter, Microbacterium and TM7 phylum, a lineage of bacteria domain, and acheae domain Methanoculleus and Methanospirillum genera (Fig. 3a), and those eukaryotic sequences were assigned into Saccharomyces, Zygosaccharomyces, Saccharomycopsis, Issatchenkia, Eurotium, Talaromyces and Trichosporon, Sagenomella (Fig. 4a). Among these genera, the fungal Issatchenkia (8/33), Saccharomycopsis (13/33) and Talaromyces (8/33) and bacteria Staphylococcus (9/27), Lactobacillus (13/27) were most abundant in the T1 lees, the fungal Issatchenkia (17/75), Saccharomyces (17/75) and Talaromyces (33/75) and the bacteria Bacillus (19/109) and Streptococcus (20/109) were dominant in the T7 lees, the archaea Methanoculleus (18/39) and the fungal Eurotium (13/42) and Talaromyces (25/42) were prevalent in the T60 lees (Figs. 3b, 4b).
Fig. 3.
The phylogenetic tree based on the complete16S rRNA sequences of representative clones from prokaryotic OTUs by using the neighbor-joining method (a) and the representative clone distributions in three samples (b). The scale bar corresponds to 0.02-estimated nucleotide substitution per sequence position. Bootstrap values from 1000 replicates are included
Fig. 4.
The phylogenetic tree based on the complete 18S rRNA sequences of representative clones from eukaryotic OTUs by using the neighbor-joining method (a) and the representative clone distributions in three samples (b). The scale bar corresponds to 0.01-estimated nucleotide substitution per sequence position. Bootstrap values from 1000 replicates are included
To further disclose the microbial taxonomic position and relationship in the Luzhou-flavor liqour cover lees, all represent SSU rRNA sequences were submitted to the similarity searches in a public database to infer a possible phylogenetic classification. The BLAST searches revealed that most represent sequences shared more than 99 % similarity with strains in public database at the SSU rRNA level. But the microbes represented by clone TP7-111 showed less than 99 % to the strains available, and it was assigned into TM7 phylum in bacteria domain. The phylogenetic trees based on 16S rRNA and 18S rRNA of the representative clones and their relational strains in GenBank were respectively constructed and shown in Figs. 3a and 4a. The trees revealed the phylogenetic affiliation of microbes inhabited in fermentation cover lees during the fermentation.
Microbial Community Succession
In general, the microbial succession is a universal phenomenon observed in natural fermentation process and is also a reflection of microbial interaction, competition for intrinsic growth factors [2]. With fermentation proceeding, the prokaryotic community similarity coefficient in three samples was from 0.4042 to 0.5703, and descended to 0.2222 when compared at the species level. And that of eukaryotic community was from 0.3000 to 0.6000 and followed to 0.5215 (Table 2). This result suggested that the microbial community succession has happened in the cover lees associated with fermentation proceeding. Furthermore, the same result was also proclaimed by the variations of H index and simpson index.
Table 2.
The similarity coefficient of Baroni-Urbani and Buser based on prokaryotic and eukaryotic microbial community pairwise comparisons in 1 day, 7 days and 60 days fermented cover lees
| Similarity coefficient | T1 | T7 | T60 |
|---|---|---|---|
| Prokaryotic libraries | |||
| T1 | 1.0000 | 0.4042 | 0.5703 |
| T7 | 1.0000 | 0.2222 | |
| T60 | 1.0000 | ||
| Eukaryotic libraries | |||
| T1 | 1.0000 | 0.3000 | 0.6000 |
| T7 | 1.0000 | 0.5215 | |
| T60 | 1.0000 | ||
Discussion
Compared with microbes in the foodstuff lees, the tendency of microbial diversity in the cover lees was also from low to high and then followed to low during the fermentation process (Table 2). While, microbial distribution in the cover lees is different from those in the foodstuff lees. In the beginning of fermentation process, the microbes inhabited in the cover lees mainly came from the starter. The fungal Issatchenkia, Saccharomycopsis and Talaromyces and bacteria Staphylococcus and Lactobacillus were most abundant in the T1 sample. In the fermentation industry, some mycelial moulds such as Aspergillus and Rhizopus are often employed for the degradation and saccharification of grains [9]. While, the strains Saccharomycopsis fibuligera, Issatchenkia orientalis and Talaromyces byssochlamydoides are detected as dominant fungi in T1 sample, but not Aspergillus and Rhizopus. In previous work, we found that S. fibuligera, I. orientalis and T. byssochlamydoides were also prevalent at the initial fermentation of Luzhou-falvor liquor foodstuff lees [2]. The S. fibuligera, I. orientalis and T. byssochlamydoides have strong amylolytic activity and their roles on hydrolysis of grain starch to sugar have been agreed by many authors [4, 10, 11]. In addition, the strain S. fibuligera has a very efficient conversion ability of glucose, starchy biomass and waste starchy substrates into the ethanol [10, 12]. Therefore, it was suggested that the degradation and saccharification of grains is mediated by S. fibuligera, I. orientalis and T. byssochlamydoides in the cove lees during the initial fermentation.
The Saccharomycescerevisiae mostly responded for alcoholic fermentation of the Luzhou-flavor liquor foodstuff lees fermented for 7 days [2]. However, S.cerevisiae was not detected in the T7 lees although it was predominant in the 7 days foodstuff lees (Fig. 4). And it was replaced by Saccharomyces dairenensis. The S. dairenensis, an unconventional yeast, can very efficiently selective ferment raw and untouched sugar from many biomass [13]. Furthermore, it is reported that I. orientalis has ability to produce ethanol from lignocellulose hydrolysates and cellobiose under different stress conditions [14, 15]. This hinted that the alcoholic fermentation in the T7 lees is mainly employed by S. dairenensis and I. orientalis. The genus Eurotium is an important microbe in the degradation of grains in Chinese Luzhou-flavor liquor lees [4]. And it was abundant in the foodstuff lees when the lees fermented for 7 days [2]. However, in the cover lees, it was not prevalent in the T7 lees. Interestingly, it was the dominant in T60 lees. In the Luzhou-flavor cover lees, the Eurotium athecia assigned into the genus Eurotium was firstly reported and might also play a primal role in degrading grains. The genera Methanoculleus and Methanospirillum were dominant in the T60 lees. This result was accordant with many previous studies [1, 2]. Indeed, it is generally accepted that the Methanoculleus and Methanospirillum can provide material basis for microbial propagating such as caproic acid bacteria, bulyric acid bacteria etc. and contribute to produce some characteristic flavor factor of Luzhou-liquor in the process [1].
Usually, only several types of bacteria and fungi are the major microbes contributing to the liquor fermentation process [5, 16]. Our results showed that the predominant microbial community distribution in the cover lees is different from that in the foodstuff lees, and that the collective bio-actions of different dominant archaea, bacteria and fungi living might play important roles in the alcoholic fermentation and producing flavor factor of Luzhou-flavor liquor. However, whether the inferior other microbes did also tamper with the alcoholic fermentation and the formation of characteristic flavor factor has not yet been known so far. The more detailed information about their activities in the process should receive more attention and further studies.
Acknowledgments
This work was financially supported by the grant from the Chunhui Program of ministry of education of the people’s republic of China (Z2010101) and the Key Laboratory of Food Biotechnology of Sichuan, Xihua University, Sichuan, China (SZJJ2009-014).
References
- 1.Zhang WX, Qia ZW, Shigematsu T, Tang YQ, Hu C, Morimura S, Kida K. Analysis of the bacterial community in Zaopei during production of Chinese Luzhou-flavor liquor. J Inst Brew. 2005;111:215–222. doi: 10.1002/j.2050-0416.2005.tb00669.x. [DOI] [Google Scholar]
- 2.Xiang WL, Li K, Liu S, Xing YG, Li MY, Che ZM. Microbial succession in the traditional Chinese Luzhou-flavor liquor fermentation process as evaluated by SSU rRNA profiles. World J Microbiol Biot. 2013;29:559–567. doi: 10.1007/s11274-012-1210-3. [DOI] [PubMed] [Google Scholar]
- 3.Singh AK, Ramesh A. Succession of dominant and antagonistic lactic acid bacteria in fermented cucumber: insights from a PCR-based approach. Food Microbiol. 2008;25:278–287. doi: 10.1016/j.fm.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 4.Zhang WX, Qiao ZW, Tang YQ, Hu C, Sun Q, Morimura S, Kida K. Analysis of the fungal community in Zaopei during the production of Chinese Luzhou-flavour liquor. J Inst Brew. 2007;113:21–27. doi: 10.1002/j.2050-0416.2007.tb00251.x. [DOI] [Google Scholar]
- 5.Li XR, Ma EB, Yan LZ, Meng H, Du XW, Zhang SW, Quan ZX. Bacterial and fungal diversity in the traditional Chinese liquor fermentation process. Int J Food Microbiol. 2011;146:31–37. doi: 10.1016/j.ijfoodmicro.2011.01.030. [DOI] [PubMed] [Google Scholar]
- 6.Marshall MM, Amos RN, Henrich VC, Rublee PA. Developing SSU rDNA metagenomic profiles of aquatic microbial communities for environmental assessments. Ecol Indic. 2008;8:442–453. doi: 10.1016/j.ecolind.2007.04.007. [DOI] [Google Scholar]
- 7.Korpole S, Sharma R, Verma D. Characterization and phylogenetic diversity of carboxymethyl cellulase producing Bacillus species from a landfill ecosystem. Indian J Microbiol. 2011;51:531–535. doi: 10.1007/s12088-011-0153-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sundarakrishnan B, Pushpanathan M, Jayashree S, Rajendhran J, Sakthivel N, Jayachandran S, Gunasekaran P. Assessment of microbial richness in pelagic sediment of Andaman sea by bacterial tag encoded FLX titanium amplicon pyrosequencing (bTEFAP) Indian J Microbiol. 2012;52:544–550. doi: 10.1007/s12088-012-0310-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang HW, Kim KH, Nam YD, Roh SW, Kim MS, Jeon CO, Oh HM, Bae JW. Analysis of yeast and archaeal population dynamics in kimchi using denaturing gradient gel electrophoresis. Int J Food Microbiol. 2008;126:159–166. doi: 10.1016/j.ijfoodmicro.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 10.Hostinová E. Amylolytic enzymes produced by the yeast Saccharomycopsis fibuligera. Biol Bratislava. 2002;57(Suppl):247–251. [Google Scholar]
- 11.Dhaliwal SS, Oberoi HS, Sandhu SK, Nanda D, Kumar D, Uppal SK. Enhanced ethanol production from sugarcane juice by galactose adaptation of a newly isolated thermotolerant strain of Pichia kudriavzevii. Bioresour Technol. 2011;102:5968–5975. doi: 10.1016/j.biortech.2011.02.015. [DOI] [PubMed] [Google Scholar]
- 12.Chi ZM, Chi Z, Liu GL, Wang F, Ju L, Zhang T. Saccharomycopsis fibuligera and its applications in biotechnology. Biotechnol Adv. 2009;27:423–431. doi: 10.1016/j.biotechadv.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Ohara S, Fukushima Y, Sugimoto A, Terajima Y, Ishida T, Sakoda A. Rethinking the cane sugar mill by using selective fermentation of reducing sugars by Saccharomyces dairenensis, prior to sugar crystallization. Biomass Bioenergy. 2012;42:78–85. doi: 10.1016/j.biombioe.2012.03.024. [DOI] [Google Scholar]
- 14.Isono N, Hayakawa H, Usami A, Mishima T, Hisamatsu M. A comparative study of ethanol production by Issatchenkia orientalis strains under stress conditions. J Biosci Bioeng. 2012;113:76–78. doi: 10.1016/j.jbiosc.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 15.Kwon YJ, Wang F, Liu CZ. Deep-bed solid state fermentation of sweet sorghum stalk to ethanol by thermotolerant Issatchenkia orientalis IPE 100. Bioresour Technol. 2011;102:11262–11265. doi: 10.1016/j.biortech.2011.09.103. [DOI] [PubMed] [Google Scholar]
- 16.Fleet GH. Wine yeasts for the future. FEMS Yeast Res. 2008;8:979–995. doi: 10.1111/j.1567-1364.2008.00427.x. [DOI] [PubMed] [Google Scholar]