Abstract
Biosynthesis of 3-hydroxypropionic acid (3-HP) typically involves two sequential reactions catalyzed by glycerol dehydratase (DhaB) and aldehyde dehydrogenase (AldH). Although plasmid-dependent over-expression of the two enzymes is common, systematic investigation of gene arrangement in vector has not been reported. Here we show that gene arrangements have a noticeable influence on 3-HP production. Using Klebsiella pneumoniae as a host, three AldH-coding genes: ald4 from Saccharomyces cerevisiae, aldh from Escherichia coli, and puuC from host K. pneumoniae, were respectively ligated to dhaB. The recombinant Kp/pET-pk-ald4-dhaB (Kp refers to as K. pneumoniae, pk is a native promoter) produced the highest yield of 3-HP in comparison to both Kp/pET-pk-dhaB-ald4 and Kp/pET-pk-dhaB-pk-ald4, suggesting that the preferential expression of AldH can increase 3-HP production. Additionally, when different AldH-coding genes were respectively ligated downstream of dhaB, the recombinant Kp/pET-pk-dhaB-puuC produced more 3-HP than that by Kp/pET-pk-dhaB-aldh or Kp/pET-pk-dhaB-ald4, implying the intrinsic compatibility of native gene puuC with its host. These findings indicate the applicability of native AldH-coding gene and provide insights into strategies for metabolic engineering of multiple genes.
Electronic supplementary material
The online version of this article (doi:10.1007/s12088-013-0390-3) contains supplementary material, which is available to authorized users.
Keywords: Klebsiella pneumoniae, 3-Hydroxypropionic acid, Gene arrangement, Glycerol dehydratase, Aldehyde dehydrogenase
Introduction
The depletion of petro-resource and deterioration of the environment has made it imperative to look for alternative means of producing chemicals. Recently, microbial fermentation instead of chemical synthesis is widely used to generate chemicals. 3-Hydroxypropionic acid (3-HP) ranks the third among the 12 top-value platform chemicals proposed by United States Department of Energy [11, 21]. 3-HP can be readily converted into a panel of economically important compounds, such as 1,3-propanediol (1,3-PDO), acrylic acid, and acrylamide [11, 18]. In addition, 3-HP is a potential nematicide [16], or the monomer of poly-3-hydroxypropionic acid that has advantages over poly-β-hydroxybutyrate (PHB) in several physical properties such as tensile strength and elongation at break [1, 2]. To develop a low-cost 3-HP bioprocess, most research groups in the world use glycerol as carbon source because it is the main by-product of the flourishing biodiesel industries. However, the current 3-HP yield is too low to be commercialized.
Among the diverse groups of 3-HP-producing bacteria, Klebsiella pneumoniae is a competitive host mainly because of its powerful capacity to metabolize glycerol. One convincing evidence comes from a recent report showing that nearly 50 g/L of 1,3-PDO was achieved by scale-up fermentation [10]. Since 3-HP and 1,3-PDO are parallel metabolites derived from glycerol, 3-HP could be abundantly produced. In K. pneumoniae, glycerol flux is spilt into two branches: oxidation and reduction [19]. In oxidation pathway, glycerol dehydrogenase converts glycerol to dihydroxyacetone (DHA) (the activator of dha regulon), which is then converted to hydroxypyruvate phosphate by dihydroxyacetone kinase, and enters glycolytic pathway to provide energy for cell growth. In reductive pathway, glycerol dehydratase (GDHt, encoded by dhaB) catalyzes glycerol to 3-hydroxypropionaldehyde (3-HPA), which is subsequently converted into 1,3-PDO and 3-HP by 1,3-propanediol oxidoreductase (PDOR) and ALDH, respectively [4, 7, 8, 17].
The parallel oxidation/reduction pathways were designated as dha regulon. Under anaerobic or micro-aerobic conditions, K. pneumoniae can generate 1,3-PDO using glycerol as sole carbon source [8, 12]. Recent work reported a native AldH in K. pneumoniae named PuuC, can convert 3-HPA to 3-HP [3]. Because of the low activities of most AldHs identified so far, plasmid-dependent over-expression remains the predominant strategy for diverting carbon flux towards 3-HP. Park and his coworkers reported that 38.7 g/L of 3-HP was produced through over-expression of dhaB and aldh in E. coli [14]. Recently, another research group from China reported pronounced concentration of both 3-HP and 1,3-PDO generated by only expressing aldh [10].
Despite tremendous efforts to increase 3-HP production, merely 40 g/L concentration has been achieved so far. Therefore, it appears that uncovered bottlenecks exist, which highly limits 3-HP production [14]. For instance, the catalytic imbalance between GDHt and AldH may restrict 3-HP biosynthesis. To address this problem, vectors that express two enzyme genes should be used. In wild-type bacteria, function-related genes usually aggregate (so-called gene cluster) and are transcribed from a shared promoter, and prokaryotic expression of neighboring genes usually adopts head-to-tail tandem mode. By doing so, the gene adjacent to promoter is usually more expressed than that far away from it. Given the different activities of GDHt and AldH, gene arrangement in the vector may affect 3-HP yield.
Despite 3-HP biosynthesis has been fueled recently, much less is known about vector construction which may be really critical for 3-HP production. For this reason, we constructed a panel of vectors, where the two enzyme genes were arranged in different order to investigate their positional effect on 3-HP biosynthesis. Meanwhile, three AldH-coding genes from distinct microorganisms were ligated downstream of dhaB to determine which AldH is most appropriate for 3-HP production.
Materials and Methods
Strains, Plasmids and Cultivation Conditions
Escherichia coli DH5α, K. pneumoniae DSM 2026 and Saccharomyces cerevisiae (baker’s yeast) were purchased from DSMZ GmbH, Germany. The vector pET-28a (Novagen, Beijing) was used in this study with minor modification. The original T7 promoter was replaced by a native dhaB1 gene’s promoter named pk. The nucleotide sequence of pk promoter is from the termination codon of previous gene of dhaB1 to the initiation codon of dhaB1, the first subunit of dhaB gene cluster (GenBank U30903). The resulting vector is designated as pET-pk. E. coli was grown in Luria–Bertani (LB) medium. S. cerevisiae was grown in Yeast Extract Peptone Dextrose medium (g · L−1): yeast extract, 10; peptone, 20; glucose, 20. The medium (per liter) for producing 3-HP by recombinant K. pneumoniae contained the following components: K2HPO4·3H2O, 3.4 g; KH2PO4, 1.3 g; (NH4)2SO4, 4 g; MgSO4·7H2O, 0.5 g; CaCO3, 0.1 g; yeast extract, 3 g; glycerol, 40 g; and 1.25 mL of trace element solution. The trace element solution contained (per liter): ZnCl2·6H2O, 2.72 g; FeSO4, 32 g; MnCl2·4H2O, 0.68 g; CoCl2·6H2O, 1.88 g; H3BO3, 0.24 g; Na2MoO4, 0.02 g; CuCl2·2H2O, 1.88 g; and 40 mL of concentrated HCl. The recombinant was inoculated into 25 mL medium containing 50 μg/mL kanamycin in a 50 mL Erlenmeyer flask and microaerobically grown at 37 °C with shaking at 130 rpm. The microaerobic condition was maintained by using a foam stopper.
Reagents
Taq DNA polymerase and restriction enzymes were purchased from TaKaRa (Dalian, China). 3-HP was purchased from Tokyo Chemical Industry (TCI) Co. Ltd. (Tokyo, Japan). Other standard chemicals were products of Sigma. DNA synthesis and sequencing were performed by Beijing Sunbiotech Co. Ltd., China.
Construction of the Recombinants
Three representative AldHs were chosen to evaluate their activities for catalyzing 3-HPA to 3-HP. Tandem co-expression strategy was used to construct expression vector because spatial proximity may benefit continuous catalysis [5]. Three AldH-coding genes puuC, aldh(AAC74382) and ald4(NM_001183794) were PCR amplified from the genomic DNA of K. pneumoniae, E. coli, and S. cerevisiae (baker’s yeast), respectively. PCR parameters are below: 94 °C, 3 min; 94 °C, 50 s; 55 °C, 50 s; 72 °C, 1 min; 30 cycles; 72 °C, 5 min; 16 °C holding. All other molecular manipulations followed standard protocols [15].
Strains and plasmids were described in Table 1. Primers and restriction enzymes were listed in Table 2. The recombinant plasmids were transformed into K. pneumoniae, and the positive recombinants were screened by LB kanamycin plate and further identified by sequencing.
Table 1.
Strains and plasmids used in this study
| Strains and plasmids | Description | Source |
|---|---|---|
| Strains | ||
| Klebsiella pneumoniae DSM 2026 | Source of gene dhaB and puuC | DSMZ GmbH, Germany |
| KP1ST | Wild-type K. pneumoniae DSM 2026 as the control | DSMZ GmbH, Germany |
| pET-pk | K. pneumoniae harboring vector pET-pk as the control | This work |
| E. coli Top 10 | Cloning host | Bio-med Beijing |
| E. coli BL21 | Source of gene aldh | Bio-med Beijing |
| 50-2-1 | Source of pk-ald4 and ald4 | Previous work |
| AB1-2-1 | K. pneumoniae harboring plasmid pET-pk-ald4-dhaB | Previous work |
| Pba | K. pneumoniae harboring plasmid pET-pk-dhaB-ald4 | This work |
| Pbpa | K. pneumoniae harboring plasmid pET-pk-dhaB-pk-ald4 | This work |
| Pb-aldh | K. pneumoniae harboring plasmid pET-pk-dhaB-aldh | This work |
| Pb-puuC | K. pneumoniae harboring plasmid pET-pk-dhaB-puuC | This work |
| Plasmids | ||
| pET-28a | Expression vector, kanr | Bio-med Beijing |
| pET-pk | Expression vector using promoter pk instead of T7, kanr | Previous work |
| pET-pk-ald4-dhaB | Ald4, dhaB in pET-pk vector, kanr | Previous work |
| pET-pk-b-aldh (‘b’ indicates dhaB) | dhaB, aldh in pET-pk vector, kanr | This work |
| pET-pk-b-puuC(‘b’ indicates dhaB) | dhaB, puuC in pET-pk vector, kanr | This work |
| pET-pk-b-ald4 (‘b’ indicates dhaB) | dhaB, ald4 in pET-pk vector, kanr | This work |
| pET-pk-b-pk-ald4 (‘b’ indicates dhaB) | dhaB, pk-ald4 in pET-pk vector, kanr | This work |
Table 2.
Primers used in this study (F forward; R reverse)
| Target gene | Primer | Sequence (5′–3′) | Restriction enzyme |
|---|---|---|---|
| dhaB | dhaB-F | 5′-CCGGAATTCATGAAAAGATCAAAACGATTTGCAGT-3′ | EcoR I |
| dhaB-R | 5′-TCCGAGCTCCCTTCTCTTAGCTTCCTTTACGCAGCTTAT-3′ | Sac I | |
| ald4 | ald4-F | 5′-TCCGAGCTCATGTTCAGTAGATCTACGCTCTGCTT-3′ | Sac I |
| ald4-R | 5′-CCCAAGCTTTTACTCGTCCAATTTGGCACGG-3′ | Hind III | |
| aldh | aldh-F | 5′-TCCGAGCTCATGAATTTTCATCATCTGGCTTACT-3′ | Sac I |
| aldh-R | 5′-CCCAAGCTTTCAGGCCTCCAGGCTTATCCAGAT-3′ | Hind III | |
| puuC | puuC-F | 5′-TCCGAGCTCATGAATTTTCAGCACCTGGCTTACT-3′ | Sac I |
| puuC-R | 5′-CCCAAGCTTTCAAGACTCCAGGGCAATCCAGAT-3′ | Hind III | |
| pk | pk-F | 5′-TCCGAGCTCCGTTATTTTGTCGCCCGCC-3′ | Sac I |
| clone PCR | pET-up | 5′-ATGCGTCCGGCGTAGA-3′ | |
| T7-down | 5′-TGCTAGTTATTGCTCAGCGG-3′ |
Flask Cultivation
The recombinants were grown in LB medium containing the following ingredients per liter: yeast extract 5 g, NaCl 10 g, peptone 10 g, and kanamycin 50 mg. 1 % of overnighted culture was inoculated to the medium containing the same concentration of antibiotics. Microaerobic environment was achieved by using 250 mL Erlenmeyer flask with 100 mL medium and shaking at 150 rpm, 37 °C.
Analytical Method
To examine gene expression, the recombinants were firstly grown in fermentation medium and then the cells were centrifuged at 10000 rpm for 10 min and incubated at 100 °C for 10 min. 10 μL samples were analyzed by 12 % (v/w) polyacrylamide gel electrophoresis (PAGE). Mini-Protein III Electrophoresis System (Bio-Rad, USA) was used to perform this experiment. Coomassie Brilliant Blue R-250 (0.2 %, w/v) was used to stain proteins on the gel and the concentration of proteins was measured by Bradford method with bovine serum albumin (BSA) as standard. Cell concentrations were measured by using Microplate reader at 600 nm with 200 μL fermentation medium added in the cuvette. The metabolite 3-HP was determined by a high performance liquid chromatography (HPLC) system (Shimazu, Kyoto, Japan) equipped with a C18 column and a SPD-20A UV detector. The residual glycerol concentration was monitored every 3 h by a titration method with NaIO4 (for control of glycerol). The mobile phase was 95 % H2O, 5 % methanol and 0.05 % phosphoric acid at the flow rate of 0.8 mL/min. All samples were filtered through 0.22-μm membrane filter.
Result and Discussion
Characterization of Recombinant Strains
All recombinants were analyzed by restriction digestion and further confirmed by sequencing. The recombinants were added into fermentation medium and cultured for 12 h. The optical density at 600 nm (OD600) was monitored to ensure the same concentration of samples for SDS-PAGE analysis. The molecular weight of ALDH (from E. coli) and PuuC were ~55 kDa, while ALD4 was a slightly larger. Since sufficient glycerol (40 g/L) as a substrate was added into fermentation medium, all three subunits of dhaB in wild type K. pneumoniae (KP1ST) were highly expressed and visible on the PAGE gel. On the contrary, the PuuC bands in two control strains were too light to be observed.
Different Gene Arrangements on 3-HP Production
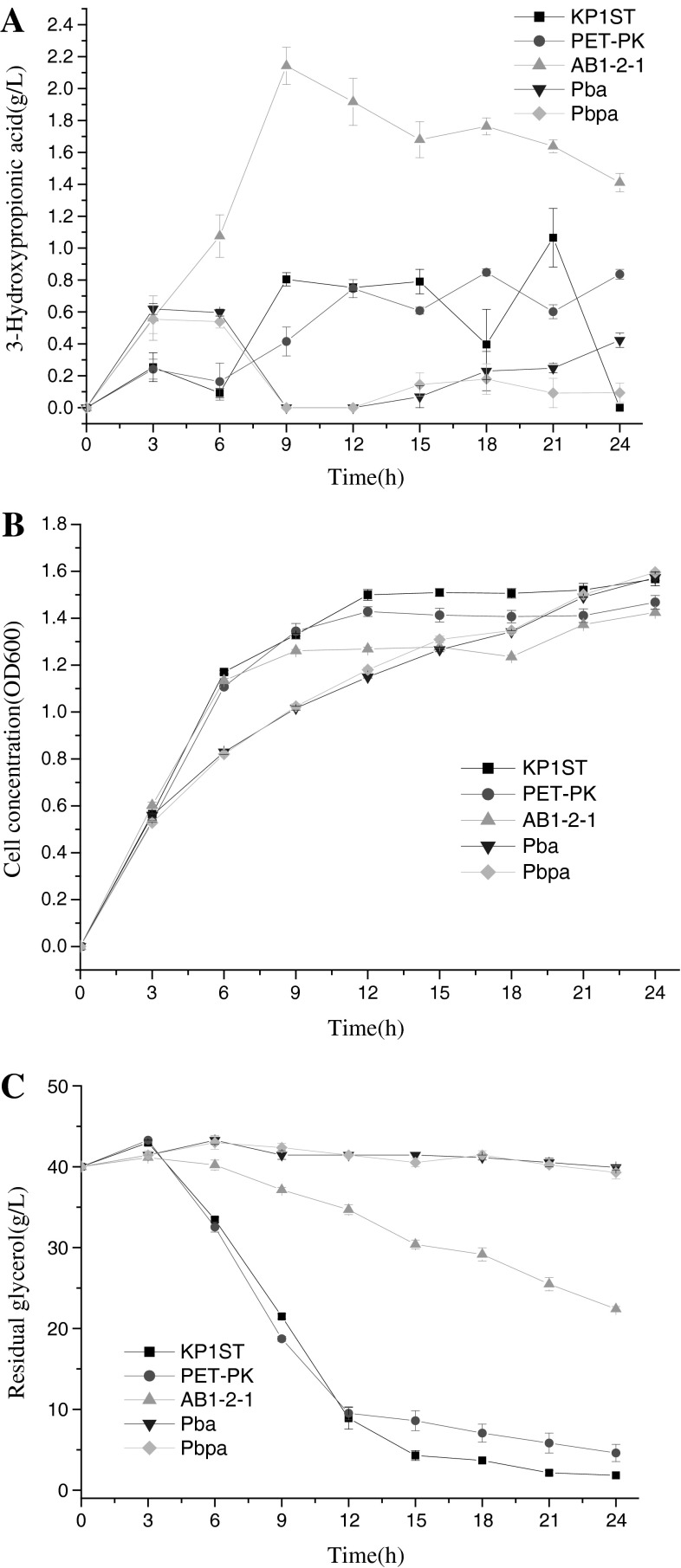
Even though pk is not as strong as T7 promoter, it is powerful enough to drive gene expression because it is the native promoter of dhaB and thereby compatible with the elements of the transcription machinery in K. pneumoniae. Thus we used native promoter pk to construct the coexpression vectors. From the engineered strains, AB1-2-1(Kp/pET-pk-ald4-dhaB) produced the highest 3-HP yield (Fig. 1a), whereas the 3-HP production in Pba (Kp/pET-pk-dhaB-ald4) and Pbpa (Kp/pET-pk-dhaB-pk-ald4) was lower than that produced by two control strains, indicating that different gene arrangements in the expression vector greatly affect 3-HP production. This result may be ascribed to 3-HPA, an intermediary metabolite toxic to host and therefore unfavorable to cell growth [17]. Compared with Kp/pET-pk-dhaB-ald4, Kp/pET-pk-ald4-dhaB produced more 3-HP because ald4 was preferentially expressed, and the enzyme ALD4 could convert 3-HPA to 3-HP immediately once 3-HPA was generated. In another words, such a tandem gene arrangement avoided the build-up of 3-HPA. Consistent with this deduction, Kp/pET-pk-ald4-dhaB in fact consumed more glycerol than Kp/pET-pk-dhaB-ald4 (Pba), because ald4 was preferentially expressed and no excessive 3-HPA accumulated, thereby facilitating cell growth (Fig. 1c). As predicted, this strain grew more vigorously than Pba (Kp/pET-pk-dhaB-ald4) and Pbpa (Kp/pET-pk-dhaB-pk-ald4) (Fig. 1b), which can also be explained by the less accumulation of 3-HPA. For strain Pbpa (Kp/pET-pk-dhaB-pk-ald4), the gene dhaB and aldh were respectively driven by promoter pk. The 3-HP, residual glycerol and biomass in this strain were nearly equal to that in Pba (Kp/pET-pk-dhaB-ald4), except little difference of 3-HP during late phase of fermentation. All together, the coordinated expression of two enzyme genes was revealed to be critical for 3-HP production and glycerol consumption.
Fig. 1.
Effects of gene arrangements on 3-HP production, cell growth and glycerol consumption. a 3-Hydroxypropionic acid, b cell concentration, c residual glycerol. KP1ST, wild-type K. pneumoniae; PET-PK, K. pneumoniae harboring blank vector pET-pk, pk is native promoter; AB1-2-1, K. pneumoniae harboring pET-pk-ald4-dhaB; Pba, K. pneumoniae harboring pET-pk-dhaB-ald4; Pbpa, K. pneumoniae harboring pET-pk-dhaB-pk-ald4
Production of 3-HP by Recombinants Harboring Distinct AldHs Downstream of GDHt
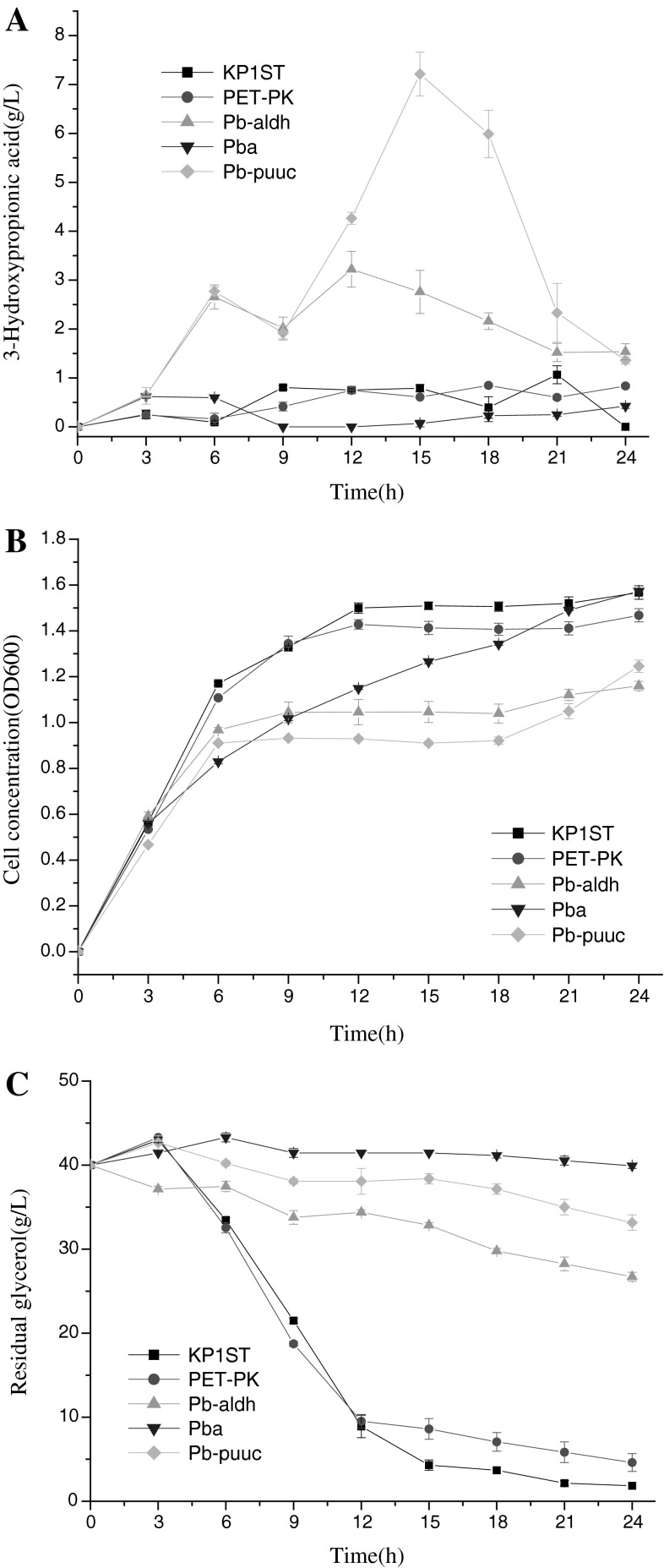
Three AldH-coding genes puuC, aldh, and ald4 were respectively ligated downstream of dhaB and transformed into K. pneumoniae. The recombinant Kp/pET-pk-dhaB-puuC produced the highest level of 3-HP, followed by Kp/pET-pk-dhaB-aldh and Kp/pET-pk-dhaB-ald4 (Fig. 2a). This result revealed the differential activities of three AldHs, which may be partially attributed to codon bias [9, 20]. As shown in Table 1, the native gene puuC was cloned from K. pneumoniae [3], whose nucleic acid sequence is similar to aldh from E. coli. Both puuC and aldh genes are from Gram negative bacteria. By contrast, aldh4 gene is from S. cerevisiae, a fungus genetically distant from bacteria. Thus, 3-HP yield may significantly depend upon the homogeneity between K. pneumoniae and the donor of AldH. For example, the recombinant Kp/pET-pk-dhaB-puuC generated the most 3-HP among three recombinants, because puuC is a native gene and should be compatible with the transcription machinery of K. pneumoniae.
Fig. 2.
Effects of expressing different aldehyde dehydrogenase genes downstream of dhaB on 3-HP production, cell growth, and glycerol consumption.
Effects of gene arrangements on 3-HP production, cell growth and glycerol consumption. a 3-Hydroxypropionic acid, b cell concentration, c residual glycerol.KP1ST, wild-type K. pneumoniae; PET-PK, K. pneumoniae harboring empty vector pET-pk, pk is native promoter; AB1-2-1, K. pneumoniae harboring pET-pk-ald4-dhaB; Pba, K. pneumoniae harboring pET-pk-dhaB-ald4; Pbpa, K. pneumoniae harboring pET-pk-dhaB-pk-ald4
Compared with two control strains, three recombinants grew poorly (Fig. 2b). There might be two reasons: (i) metabolic burden imposed on host due to plasmid replication; (ii) catalytic imbalance between two key enzymes which resulted in 3-HPA accumulation, cell death or slow growth. The glycerol consumption of three recombinants was roughly in accord with 3-HP titer (Fig. 2b, c) with the exception of two control strains, whereby the consumed glycerol was mainly converted into biomass.
The aldh gene from E. coli was verified to be more efficient than other homologous genes for producing 3-HP in K. pneumoniae [10]. Here we showed puuC gene from K. pneumoniae was more effective than other AldH-coding genes. This result does not contradict the former one because we focused on the coexpression of two genes instead of only expression of AldH. When ligated downstream of dhaB, AldH-coding gene may be partially influenced by dhaB.
From above results, the activity of AldH for 3-HP production may be more important than its expression level. For example, when puuC was overexpressed in E. coli based on high copy plasmid, only a trace of 3-HP could be detected (nearly equal to that in wild type strain, data not shown). Hence, the catalytic activity of PuuC may be significantly affected by codon bias and proper folding of the protein. Furthermore, this result pinpointed the drawback of high copy vector which usually results in the metabolic burden on cell growth due to plasmid replication, and the imbalance of the cofactors. Therefore, a mid- or low copy vector may be ideal for 3-HP production.
Productivity of Strains and Feedback Inhibition
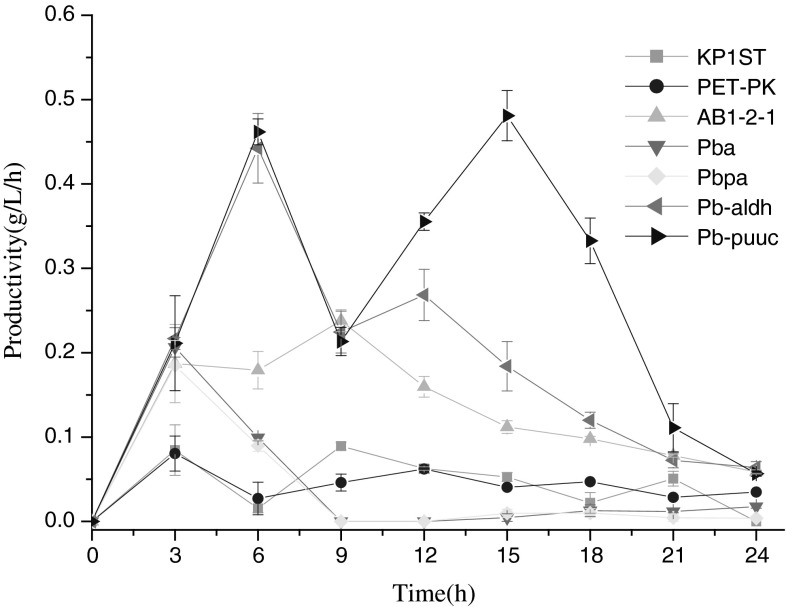
To determine the efficiency of strains for producing 3-HP, we calculated the productivity (g/L/h). As shown in Fig. 3, productivity varied among strains. Despite the fluctuation during fermentation, the strain Kp/pET-pk-dhaB-puuc exhibited higher productivity than Kp/pET-pk-dhaB-aldh and Kp/pET-pk-dhaB-ald4, implying that the AldH-coding gene when ligated downstream of dhaB diversifies 3-HP yield. For strains harboring enzyme genes in different order, Kp/pET-pk-ald4-dhaB (AB1-2-1) showed higher efficiency than Kp/pET-pk-dhaB-ald4 (Pba) and Kp/pET-pk-dhaB-pk-aldh4 (Pbpa), indicating that the different gene arrangements affect 3-HP formation.
Fig. 3.
3-HP productivities of the recombinants. KP1ST, wild-type K. pneumoniae; PET-PK, K. pneumoniae harboring blank vector pET-pk; Pb-aldh, K. pneumoniae harboring pET-pk-dhaB-aldh; Pb-puuc, K. pneumoniae harboring pET-pk-dhaB-puuC; Pba, K. pneumoniae harboring pET-pk-dhaB-ald4; AB1-2-1, K. pneumoniae harboring pET-pk-ald4-dhaB; Pbpa, K. pneumoniae harboring pET-pk-dhaB-pk-aldh4
One noticeable phenomenon was feedback inhibition during late phase of fermentation, which may be ascribed to the rigidity and plasticity of dha regulon. On one hand, GDHt is a multi-subunit enzyme tailor-made for conversion of glycerol to 3-HPA, an irreversible step to guarantee the rigidity of dha regulon. On the other hand, 3-HPA is catalyzed into 3-HP or 1,3-PDO by AldH and PDOR respectively, the two reversible reactions which are recognized as a buffer mechanism evolved to cope with stimuli.
Conclusion
Collectively, we show here the prominent influence of gene arrangements on 3-HP production. Coordinated expression of two key enzymes is revealed to be critical for production of 3-HP. Owing to the lower activity of AldH (compared with GDHt) and the toxicity of intermediary metabolite 3-HPA to the host, preferential expression of AldH-coding genes (ald4, aldh or puuC) would divert more carbon flux towards 3-HP. In addition, we compared three AldH-coding genes for their capacity to produce 3-HP when ligated downstream of dhaB. The native gene puuC was shown to be most efficient in conversion of 3-HPA to 3-HP. Apart from enzymatic activity, the intrinsic compatibility of puuC to K. pneumoniae is also a key factor for 3-HP biosynthesis. Given the evolving nature of ALDH family [10, 13], upcoming work may be the determination of efficient enzymes and engineering of their spatial organization [5, 6]. Collectively, this study has provided keys for better understanding of metabolic engineering which involves multiple enzyme genes.
Electronic supplementary material
Schematic diagram of vector construction (DOC 191 kb)
Acknowledgments
This work was supported by National Natural Science Foundation of China (No. 20876009, 21076013, 21276014) and National Basic Research Program of China (973 Program) (2012CB725200).
References
- 1.Andreessen B, Lange AB, Robenek H, Steinbuchel A. Conversion of glycerol to poly(3-hydroxypropionate) in recombinant Escherichia coli. Appl Environ Microbiol. 2010;76(2):622–626. doi: 10.1128/AEM.02097-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andreessen B, Steinbuchel A. Biosynthesis and biodegradation of 3-hydroxypropionate-containing polyesters. Appl Envir Microbiol. 2010;76(15):4919–4925. doi: 10.1128/AEM.01015-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashok S, Raj S, Rathnasingh C, Park S. Development of recombinant Klebsiella pneumonia ΔdhaT strain for the co-production of 3-hydroxypropionic acid and 1,3-propanediol from glycerol. Appl Microbiol Biotechnol. 2011;90:1253–1265. doi: 10.1007/s00253-011-3148-z. [DOI] [PubMed] [Google Scholar]
- 4.Celinska E. Debottlenecking the 1,3-propanediol pathway by metabolic engineering. Biotechnol Adv. 2010;28(4):519–530. doi: 10.1016/j.biotechadv.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Conrado RJ, Varner JD, DeLisa MP. Engineering the spatial organization of metabolic enzymes: mimicking nature’s synergy. Curr Opin Biotechnol. 2008;19:492–499. doi: 10.1016/j.copbio.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Dueber JE, Wu GC, Malmirchegini GR, Moon TS, Petzold CJ, Ullal AV, Prather KL, Keasling JD. Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotechnol. 2009;27(8):753–759. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- 7.Forage RG, Foster MA. Glycerol fermentation in Klebsiella pneumoniae: functions of the coenzyme B12-dependent glycerol and diol dehydratases. J Bacteriol. 1982;149:413–419. doi: 10.1128/jb.149.2.413-419.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forage RG, Lin EC. DHA system mediating aerobic and anaerobic dissimilation of glycerol in Klebsiella pneumoniae NCIB 418. J Bacteriol. 1982;151:591–599. doi: 10.1128/jb.151.2.591-599.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hense W, Anderson N, Hutter S, Stephan W, Parsch J, Carlini DB. Experimentally increased codon bias in the Drosophila Adh gene leads to an increase in larval, but not adult, alcohol dehydrogenase activity. Genetics. 2010;184(2):547–555. doi: 10.1534/genetics.109.111294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Y, Li Z, Shimizu K, Ye Q. Simultaneous production of 3-hydroxypropionic acid and 1,3-propanediol from glycerol by a recombinant strain of Klebsiella pneumoniae. Bioresour Technol. 2012;103(1):351–359. doi: 10.1016/j.biortech.2011.10.022. [DOI] [PubMed] [Google Scholar]
- 11.Jiang X, Meng X, Xian M. Biosynthetic pathways for 3-hydroxypropionic acid production. Appl Microbiol Biotechnol. 2009;82(6):995–1003. doi: 10.1007/s00253-009-1898-7. [DOI] [PubMed] [Google Scholar]
- 12.Johnson EA, Lin EC. Klebsiella pneumoniae 1,3-propanediol:NAD+ oxidoreductase. J Bacteriol. 1987;169(5):2050–2054. doi: 10.1128/jb.169.5.2050-2054.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo LH, Kim CH, Heo SY, Oh BR, Hong WK, Kim S, Kim DH, Seo JW. Production of 3-hydroxypropionic acid through propionaldehyde dehydrogenase PduP mediated biosynthetic pathway in Klebsiella pneumoniae. Bioresour Technol. 2012;103(1):1–6. doi: 10.1016/j.biortech.2011.09.099. [DOI] [PubMed] [Google Scholar]
- 14.Rathnasingh C, Raj SM, Jo JE, Park S. Development and evaluation of efficient recombinant Escherichia coli strains for the production of 3-hydroxypropionic acid from glycerol. Biotechnol Bioeng. 2009;104(4):729–739. doi: 10.1002/bit.22429. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3. New York: Cold Spring Harbor Laboratory; 2001. [Google Scholar]
- 16.Schwarz M, Kopcke B, Weber RW, Sterner O, Anke H. 3-Hydroxypropionic acid as a nematicidal principle in endophytic fungi. Phytochemistry. 2004;65(15):2239–2245. doi: 10.1016/j.phytochem.2004.06.035. [DOI] [PubMed] [Google Scholar]
- 17.Slininger PJ, Bothast RJ. Optimizing aerobic conversion of glycerol to 3-hydroxypropionaldehyde. Appl Envir Microbiol. 1985;50:1444–1450. doi: 10.1128/aem.50.6.1444-1450.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suthers PF, Cameron DC (2001) Production of 3-hydroxypropionic acid in recombinant organisms. WO Patent No. 01-16346
- 19.Tobimatsu T, Azuma M, Matsubara H, Takatori H, Niida T, Nishimoto K, Satoh H, Hayashi R, Toraya T. Cloning, sequencing, and high level expression of the genes encoding adenosylcobalamin-dependent glycerol dehydrase of Klebsiella pneumoniae. J Biol Chem. 1996;271:22352–22357. doi: 10.1074/jbc.271.37.22352. [DOI] [PubMed] [Google Scholar]
- 20.Tuller T, Waldman YY, Kupiec M, Ruppin E. Translation efficiency is determined by both codon bias and folding energy. Proc Natl Acad Sci USA. 2010;107(8):3645–3650. doi: 10.1073/pnas.0909910107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Werpy T, Petersen G. Top value added chemicals from biomass. Washington, DC: U.S. DOE; 2004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic diagram of vector construction (DOC 191 kb)