Abstract
Odontogenesis is the result of the reciprocal interactions between epithelial–mesenchymal cells leading to terminally differentiated odontoblasts. This process from dental papilla mesenchymal cells to odontoblasts is regulated by a complex signaling pathway. When isolated from the developing tooth germs, odontoblasts quickly lose their potential to maintain the odontoblast-specific phenotype. Therefore, generation of an odontoblast-like cell line would be a good surrogate model for studying the dental mesenchymal cell differentiation into odontoblasts and the molecular events of dentin formation. In this study, immortalized dental papilla mesenchymal cell lines were generated from the first mouse mandibular molars at postnatal day 3 using pSV40. These transformed cells were characterized by RT-PCR, immunohistochemistry, Western blot, and analyzed for alkaline phosphatase activity and mineralization nodule formation. One of these immortalized cell lines, iMDP-3, displayed a high proliferation rate, but retained the genotypic and phenotypic characteristics similar to primary cells as determined by expression of tooth-specific markers and demonstrated the ability to differentiate and form mineralized nodules. Furthermore, iMDP-3 cells had high transfection efficiency as well as were inducible and responded to BMP2 stimulation. We conclude that the establishment of the stable murine dental papilla mesenchymal cell line might be used for studying the mechanisms of dental cell differentiation and dentin formation.
Keywords: Cell differentiation, Dental papilla mesenchymal cells, Odontoblasts, Immortalization, SV-40 T antigen
Introduction
Tooth organogenesis is the result of the reciprocal interactions between epithelial–mesenchymal cells leading to the terminal differentiation of matrix producing cells (Zeichner-David et al. 1995; Thesleff and Nieminen 1996). Odontoblasts produce the dentin matrix and are responsible for the synthesis of dentin, which is the principal mineralized tissue of teeth (Linde and Goldberg 1993). Odontoblasts are original from dental papilla mesenchymal cells that have undergone a complex process of cell lineages. During dentinogenesis, differentiation of dental papilla mesenchymal cells into odontoblasts is regulated by many several growth factors, transcription factors, and others (Thesleff 2003; Chen et al. 2012). A consequence of odontoblast cytodifferentiation is the expression of specific gene products that form the dentin extracellular matrix (ECM). Dentin ECM is composed of collagenous and non-collagenous proteins (NCPs) to form a unique dentin ECM (MacDougall et al. 1997). Collagen type I accounts for about 85% of protein fractions in dentin and mutations of collagen type I are associated with bone/dentin genetic diseases called dentinogenesis imperfecta type I [also termed osteogenesis imperfecta (OI)] (Levin et al. 1988; Biria et al. 2012). Most of the NCPs are also expressed in dentin. However, expressional levels of numerous NCPs appear to be more specific in dentin even though they are also found in other tissues (D’Souza et al. 1997; Yuan et al. 2009). Among these NCPs, dentin sialoprotein (DSP) and dentin phosphoprotein (DPP) are highly expressed in teeth, especially in odontoblasts and predentin (D’Souza et al. 1997; Yuan et al. 2009). DSP mutations in humans are associated with dentinogenesis imperfecta type II (DGI-II) and dentin dysplasia type II (DD-II; Xiao et al. 2001; Rajpar et al. 2002; Kim and Simmer 2007) while DPP mutations cause dentinogenesis imperfecta type III (DGI-III; McKnight et al. 2008; Song et al. 2008). Various other NCPs synthesized by the odontoblasts such as dentin matrix protein 1 (DMP1) along with DSP and DPP also play important roles in calcifying organic matrix (George et al. 1993). Furthermore, many transcription factors and growth factors are essential for tooth-related gene expression and dentin formation (Thesleff 2003; Chen et al. 2012). Although studies have demonstrated the abundance of these collagenous and NCP proteins during odontogenesis, their precise roles in this process have still largely unresolved.
Deciphering the regulatory mechanisms involving the terminal cell differentiation and synthesis of odontoblast-specific ECM molecules require a homogenous cell population. Unlike bone and other tissues, it is relatively limited to collect enough amounts of cells from dental pulp. Also, it is laborious due to several obstacles: (a) difficulty in micro-dissection, (b) paucity of a pure population of cells, and (c) the fact that these primary odontoblastic cells cannot be maintained in cultures for a long time due to their finite life span (Sakamoto et al. 1985; Ruch et al. 1995). Several methodologies have been employed to develop cell lines from various mineralized tissues (Thonemann and Schmalz 2000; Galler et al. 2006). Therefore, generation of a dental papilla mesenchymal cell line would be a valuable tool for studying the molecular mechanisms of the process of ECM mediated odontoblast cytodifferentiation, biomineralization, and reparative or teritiary dentin formation.
As mouse is a comprehensive model for the study of tooth development and formation, in this study, we established an immortalized mouse dental papilla mesenchymal cell line using transfection with virus 40 T-antigen gene (SV40). We further observed these cell growth rates as well as their genotypic and phenotypic characteristics. Finally, we tested whether these immortalized cells were inducible by growth factors.
Materials and Methods
Primary mouse dental papilla mesenchymal cells.
Animal Welfare Committee at the University of Texas Health Science Center at San Antonio (UTHSCSA) approved the protocol of animal use. The dental pulps from the first molars of mandibles of 3-d-old C57BL/6 mice (The Jackson Laboratory, Bar Harbor, ME) were isolated using tying forceps under microscope and washed with phosphate buffered saline (PBS), digested for 1 h at 37°C in a solution of 3 mg/ml collagenase type I and 4 mg/ml of dispase (Worthington Biochem, Freehold, NJ). Then, the cells were grown with alpha minimum essential medium (α-MEM, Invitrogen, San Diego, CA) containing 10% fetal calf serum plus penicillin (100 unit/ml) and streptomycin (100 μg/ml), and cultured at 37°C in a humidified atmosphere of air containing 5% CO2. Medium was refreshed every 2 d and cells were spread after reaching confluence.
Gene transfer and selection of immortalized cells.
Primary dental papilla mesnechymal cells (70% confluence) in passage 3 were transfected with pSV3neo (ATCC, No. 37150, Manassas, VA, USA), a plasmid containing coding sequences of SV40 T-Ag and a neomycin (G418)-resistance gene by electroporation (Wu et al. 2009). One day after transfection, cells were trypsinized, divided at low densities and cultured in standard medium supplemented with 600 μg/ml of G418 (Sigma–Aldrich, St. Louis, MO). After G418 selection, several colonies were formed and treated with trypsin and re-plated at low densities to obtain the secondary selection. Several immortalized cell lines were established and confirmed by PCR and immunostaing using SV40 specific primers (Table 1) and anti-SV 40 antibody. One of the immortalized cell lines of passage 50, iMDP-3 (immortalized mouse dental papilla mesenchymal cells), was extensively characterized.
Table 1.
Primer sequences used for polymerase chain reactions
| Gene | Primer sequences | Product size |
|---|---|---|
| ALP | Forward: 5′-CGGGACTGGTACTCGGATAA-3′ | 209 |
| Reversed: 5′-TGAGATCCAGGCCATCTAGC-3′ | ||
| ATF4 | Forward: 5′-GAAACCTCATGGGTTCTCCA-3′ | 203 |
| Reversed: 5′-AGAGCTCATCTGGCATGGTT-3′ | ||
| Col1α1 | Forward: 5′-CCTGACGCATGGCCAAGAAGA-3′ | 145 |
| Reversed: 5′-GCATTGCACGTCATCGCACA-3′ | ||
| Dlx3 | Forward: 5′-GCGACACTCAGGAATCATTG-3′ | 108 |
| Reversed: 5′-CGGTCCATGCATTTGTTATC-3′ | ||
| Dmp1 | Forward: 5′-CAGTGAGGATGAGGCAGACA-3′ | 175 |
| Reversed: 5′-TCGATCGCTCCTGGTACTCT-3′ | ||
| Dspp | Forward: 5′-AACTCTGTGGCTGTGCCTCT-3′ | 171 |
| Reversed: 5′-TATTGACTCGGAGCCATTCC-3′ | ||
| Gapdh | Forward: 5′-CAAAGTTGTCATGGATGACC-3′ | 195 |
| Reversed: 5′-CCATGGAGAAGGCTGGGG-3′ | ||
| Lhx6 | Forward: 5′-AGAAGCTAGCGGACATGACG-3′ | 219 |
| Reversed: 5′-CTCTCAATGTAGCCGTGCAA-3′ | ||
| Lhx7 | Forward: 5′-TACTTCAGACGGTATGGGAC-3′ | 237 |
| Reversed: 5′-TCCCATTACCGTTCTCCACT-3′ | ||
| Mepe | Forward: 5′-CTGTGGATCCTTGTGAGAAT-3′ | 199 |
| Reversed: 5′-TAGAGGATTTTGGCTTTCTG-3′ | ||
| Oc | Forward: 5′-CTTGGTGCACACCTAGCAGA-3′ | 208 |
| Reversed: 5′-TTCTGTTTCCTCCCTGCTGT-3′ | ||
| Opn | Forward: 5′-TCTGATGAGACCGTCACTGC-3′ | 170 |
| Reversed: 5′-AGGTCCTCATCTGTGGCATC-3′ | ||
| Osx | Forward: 5′-ACTCATCCCTATGGCTCGTG-3′ | 238 |
| Reversed: 5′-GGTAGGGAGCTGGGTTAAGG-3′ | ||
| Runx2 | Forward: 5′-TACAAACCATACCCAGTCCCTGTTT-3′ | 197 |
| Reversed: 5′-AGTGCTCTAACCACAGTCCATGCA-3′ | ||
| SV40 | Forward: 5′-AGCAGACACTCTATGCCTGTGTGGAGTAAG-3′ | 751 |
| Reversed: 5′-GACTTTGGAGGCTTCTGGATGCAACTGAG-3′ |
ALP alkaline phosphatase, ATF4 activating transcription factor 4, Col1α1 alpha 1 collagen type, Dlx3 Distal-less 3, Dmp1 dentin matrix protein 1, Dspp dentin sialophosphoprotein, Gadph plyceraldhyde-3-phosphate dehydrogenase, Lhx LIM homeobox protein, Mepe matrix extracellular phosphglycoprotein, Oc osteocalcin, Opn osteopontin, Osx Osterix, SV40 SV40 large T-antigen
Cell morphology and proliferation assays.
Morphology of iMDP-3 cells was observed by a light inverted microscope. Cell proliferation assay was performed by direct cell counting and MTT methods. Briefly, cells were seeded into 6-well plates at 2.5 × 104 cells per well. The cells were trypsinized and counted daily using a hemocytometer for up to 9 d. For MTT assay, cells were seeded into 96-well plates with 1.5 × 103 cells per well and detected from days 1 to 9, respectively, using MTT cell proliferation assay kit (ATCC, No. 30-1010K, Manassas, VA).
Detection of transformation.
Simian virus 40 sequences were accessed in Genbank (Accession No. J02400) and specific primers were synthesized (Table 1). Genomic DNA was isolated from iMDP-3 cells. pSV3 neo plasmid was used as positive control. Two-hundred nanograms of DNA (for pSV3 neo plasmid DNA 10 ng) were diluted in a 25-μl polymerase chain reaction (PCR) mix (Sigma–Aldrich). The reactions were carried out at 95°C for 5 min for one cycle and then at 95°C for 30s, 55°C for 60s, and 72°C for 60s for 30 cycles, with a final 10 min extension at 72°C. Five microliters of PCR products was analyzed by agarose gel electrophoresis and visualized by ethidium bromide staining. For detection of SV40 protein expression, iMDP-3 cells were seeded on coverslips in 6-well plates and cultured for 48 h in standard α-MEM medium. The coverslips were rinsed with PBS and fixed with cold acetone and methanol (1:1). The cells were blocked with 10% goat serum and incubated with a primary anti-SV40 large T-antigen monoclonal antibody (1:100, Santa Cruz Biotechnology Inc., Santa Cruz, CA) for overnight at 4°C. Then the cells were washed with PBS containing 0.1% goat serum and incubated with the secondary antibody conjugated with Alexa Fluo® 488 green (Molecular Probes, Eugene, OR) for 1 h at room temperature. For negative control, the primary SV40 antibody was replaced by mouse IgG I (Dakocytomation, Carpinteria, CA). For cell nucleus staining, the cells were treated with DAPI (Sigma–Aldrich). Images of Alexa Fluo® 488 green staining of the SV40 protein were obtained at the Core Optical Imaging Facility at UTHSCSA under the same parameters in a Nikon inverted microscope.
Immunohistochemistry.
For detection of tooth-related proteins, the immortalized cells were fluorescently immunostained by antibodies directed against mouse Dmp1 (gifts from Dr. Larry Fisher, NIDCR, USA), Runx2, Osx (Sp7), Opn, Oc, Dsp, and Col1α1 (Santa Cruz Biotechnology Inc.) and Dlx3 (Abcam, Cambridge, MA). Negative control of mouse IgG I was purchased from Dakocytomation (Carpinteria, CA). Immunohistochemical assay was performed with corresponding secondary antibodies with Alexa Fluo® 488 green fluorescent labeling (Molecular Probes). Microphotographs were obtained under a Nikon microscope using a Nikon Cool pix 4500 digital camera.
Western blot analysis.
Primary and immortalized cells were washed with cold PBSand lysed with a RIPA buffer (Santa Cruz Biotechnology Inc.). The whole cell lysates were resolved by 7% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a membrane (Bio-Rad Laboratory, Hercules, CA). Western blot assay was performed as described earlier (Chen et al. 2008) using Dsp and Dmp1 antibodies, respectively. The anti-Dsp and anti-Dmp1 goat polyclonal antibodies were obtained as described above. Goat anti-mouse β-actin antibody (Santa Cruz Biotechnology Inc.) was used as an internal control.
RNA preparation and reverse transcription-polymerase chain reaction (RT-PCR).
Total RNA was extracted from the primary and immortalized dental papilla mesenchymal cells using RNA STAT-60 kit (Tel-Test, Inc., Friendswood, TX), treated with DNase I (Promega, Madison, WI), and purified with the RNeasy Mini Kit (Qiagen Inc., Valencia, CA). RNA concentration was determined at an optical density of OD260. The RNA was transcribed into cDNA by SuperScript II reverse transcriptase (Invitrogen). Specific primers for the PCR were synthesized in Table 1, and this included Alp, Atf4, Dlx3, Dmp1, Dspp, Lhx6, Lhx7, Mepe, Oc, Opn, Osx, Runx2, Gapdh, and collagen type I. The PCR reaction was first denatured at 95°C for 5 min, and then carried out at 95°C for 60 s, at 55–60°C for 60s and at 72°C for 60s for 30 cycles and with a final 10 min extension at 72°C. Five microliters of PCR products were analyzed by agarose gels with ethidium bromide staining. Corrective DNA was verified by DNA sequencing.
Alkaline phosphatase (ALP) and mineralization assays.
For detection of ALP activity, iMDP-3 cells were cultured in calcifying medium [α-MEM supplemented with 10% FBS, penicillin (100 unit/ml) and streptomycin (100 μg/ml), 50 μg/ml ascorbic acid, and 10 mM sodium β-glycerophosphate] at 37°C for 7 d. These cells were then fixed with 70% ethanol for 5 min and washed in the buffer (100 mM Tris–HCl, pH 9.5; 100 mM NaCl; 50 mM MgCl2). In situ ALP staining was performed according to the supplier’s instructions (Bio-Rad, Hercules, CA, USA). For mineralization assay, these cells were plated in a 6-well plate at a density of 4 × 105 per well and cultured in calcifying medium at 37°C for 4, 7, and 14 d, respectively. The cells were fixed in 10% formaldehyde neutral buffer solution and then stained with alizarin red S (Sigma-Aldrich).
DNA transfection.
For transfection experiment, iMDP-3, mouse immortalized odontoblast-like (MO6-G3; MacDougall et al. 1995) and mouse immortalized pre-odontoblast-like cells (MD10-F2; Chen et al. 2005) were seeded at 2 × 105 cells/35 mm diameter dishes and underwent transfection 18 h later with mammalian expression plasmid tagged with green fluorescent protein (GFP) gene (pEGFP-C2, Clontech Laboratories Inc., Mountain View, CA) using LipofectAMINE 2000 (Life Technologies, Grand Island, NY ) as specified by the manufacturer. Three hours after transfection, serum-free DNA containing medium was replaced by fresh growth medium with 10% serum. After 12-h transfection, cell transfection efficiency was observed using a Nikon inverted fluorescent microscope. Primary embryonic human kidney transformed cells with human adenovirus type 5 DNA (293; Cell Biolabs Inc., San Diego, CA) were used for positive control.
Induction of gene expression by recombinant BMP2 (rBMP2).
The immortalized murine dental papilla mesenchymal cells were maintained in α-MEM medium with 10% fetal calf serum plus penicillin (100 unit/ml) and streptomycin (100 μg/ml). The cells were then grown in α-MEM medium with 1% fetal calf serum plus penicillin (100 unit/ml) and streptomycin (100 μg/ml) and treated either with or without 300 ng/ml of recombinant human BMP2 (rBMP2; R&D Systems, Minneapolis, MN). After rBMP2 induction, the cells were rinsed with ice-cold PBS and fixed for 10 min on ice with methanol/acetone (1:1). Gene expression in the immortalized cells induced by rBMP2 was detected by immunohistochemistry as described above.
Statistical analysis.
All values were represented as the mean ± standard deviation (SD). Statistical significance was determined using the unpaired Student’s t test with a p value of <0.05 being statistical significant.
Results
Immortalization of mouse dental papilla mesenchymal cells.
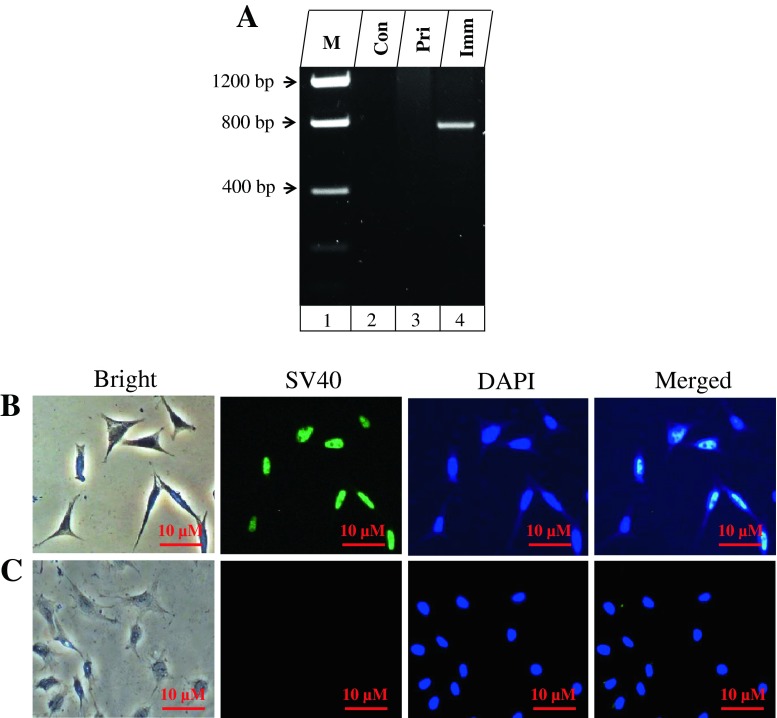
To generate immortalization of mouse dental papilla mesenchymal cell lines, primary dental papilla mesenchymal cells were isolated and transfected with SV3 neo plasmid containing SV40 T-Ag gene and then selected with G418. The colonies were formed after 2–3 wk selections and transfected cells from these colonies grew over 50 population doublings (PDLs) without significant growth retardation. One of the transfected cell lines, termed iMDP-3 (immortalized mouse dental papilla mesenchymal), was passaged at 50 generations and used for detailed characterization. SV40 T-Ag gene expression was seen in iMDP-3 cells by PCR analyses (Fig. 1A). Immunohistochemistry further confirmed that simian virus 40 T-Ag was expressed in all iMDP-3 cells (Fig. 1B).
Figure 1.
Identification of SV40 transformation. (A) Genomic DNA in the primary and immortalized dental papilla mesenchymal cells was isolated and amplified by the SV40 specific primer. PCR products were run on an agarose gel and stained with ethidium bromide. Lane 1, DNA marker (Low DNA mass ladder, Invitrogen); lane 2, control; lane 3, primary dental papilla mesenchymal cells; lane 4, immortalized cells. (B, C) Immunohistochemical staining with antibody against simian virus 40 large T-antigen (SV40 T-Ag) in iMDP-3 cells and immunolabeling of SV40 protein was mostly present in the nucleus (green color; B) whereas immunostaining was not seen in the primary cells (C). The cells were stained with DAPI for the nucleus. Con control, Pri primary, Imm immortalized.
Morphology and proliferation of immortalized dental papilla mesenchymal cells.
Cell morphology was observed using a light inverted microscope (Fig. 2A–D). The immortalized cell morphology displays a spindle shape like the primary dental papilla mesenchymal cells. There were no microscopical findings showing senescence or neoplastic nature in these cells. Proliferation of both the primary and immortalized cells appeared stable. However, the immortalized cells displayed a more rapid growth rate than the primary cells as revealed by cell counting and MTT assays (Fig. 2E, F).
Figure 2.
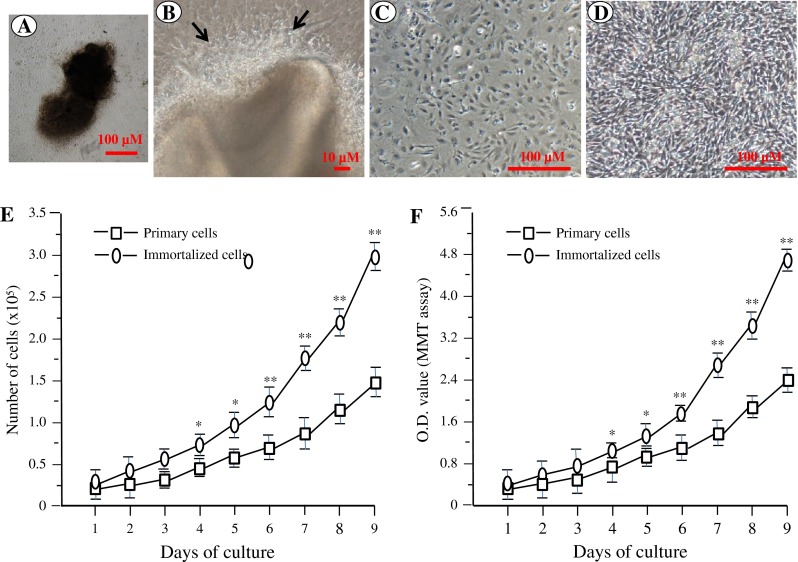
Cell morphology and proliferation of mouse dental papilla mesenchymal cells. (A) A microphotograph of an isolated dental pulp from a mouse first molar at postnatal day 3. (B). Primary cell growth was seen in the edge of the dental pulp (arrows). (C, D) The primary and immortalized dental papilla mesenchymal cells were photographed under a light microscope using a Nikon Coolpix 4500 digital camera. (E, F) Proliferation data of the primary and immortalized cells were observed by cell counting (E) and MTT (F) assays.
Expression of tooth-related genes in dental papilla mesenchymal cells.
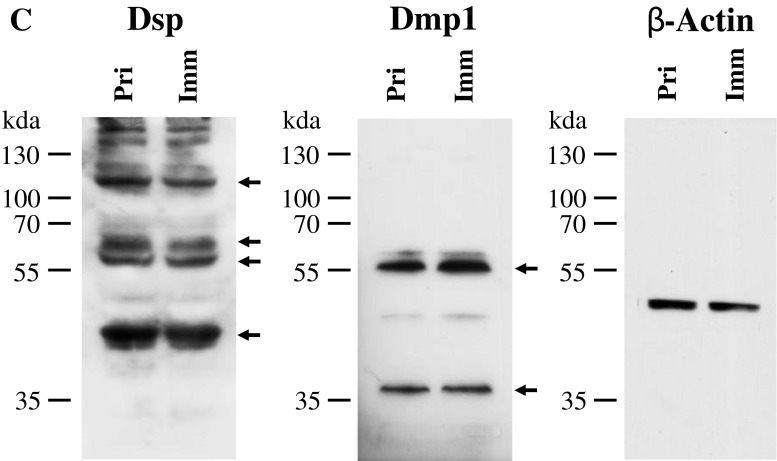
To determine whether the immortalized cells express tooth-related genes, we tested expression of various tooth-related genes using RT-PCR assay. Figure 3A showed that the transformed cells similar to the primary cells synthesized collagenous and non-collagenous protein genes such as collagen type I, ALP, Dmp1, Dspp, Mepe, Oc, and Opn. Also, these cells expressed Atf4, Dlx3, Lhx6, Lhx7, Osx, and Runx2 that are involved in the odontoblast differentiation and tooth mineralization (Fig. 3A). Immunohistochemistry studies further confirmed that these primary and immortalized cells express these proteins as well (Fig. 3B). Furthermore, expression of Dsp and Dmp1 proteins was detected in the primary and immortalized cells by Western blot assay (Fig. 3C). These results indicate that the immortalized cells, with a long term culture, retain their genotypic and phenotypic characteristics. Furthermore, when these cells were treated with recombinant bone morphogenetic protein 2 (rBMP2), BMP2 was able to induce translocation of Dlx3 and Osx transcription factors into the nucleus, conversely, both the proteins were mostly distributed within the cytoplasm without BMP2 stimulation (Fig. 4). It indicates that BMP2 activates Dlx3 and Osx in the immortalized cells.
Figure 3.
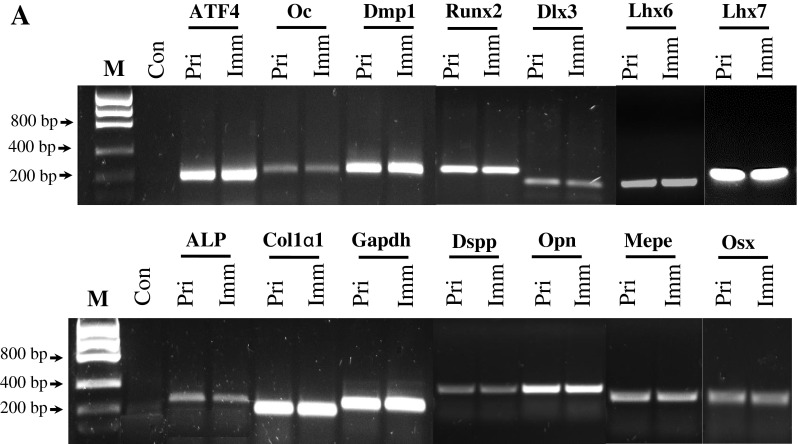
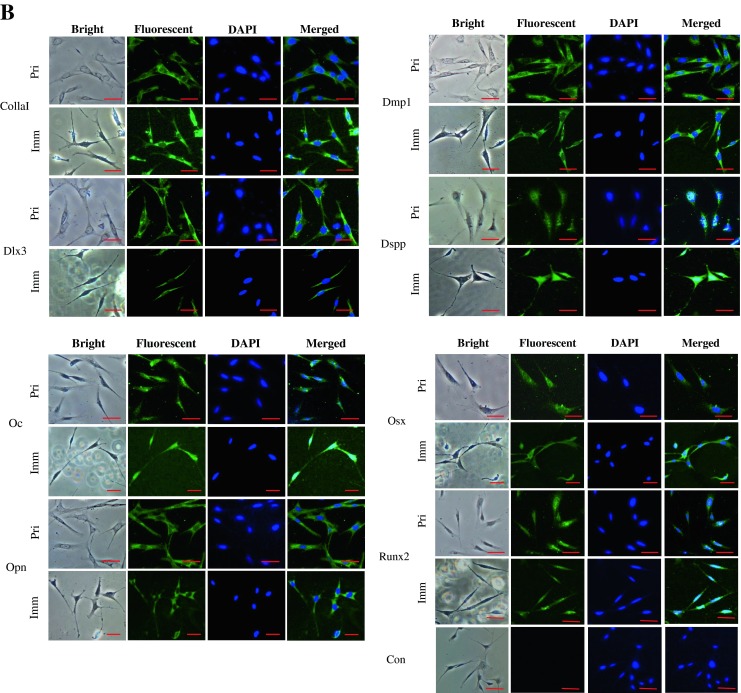
Expression of tooth-related genes in the primary and immortalized dental papilla mesenchymal cells. (A) Total RNAs from the primary and immortalized cells were extracted and reversely transcribed. The cDNAs were amplified by PCR using specific primers shown in Table 1. The PCR products were run on 1% agarose gels and stained with ethidium bromide. M DNA marker (Low DNA mass ladder, Invitrogen), Con negative control. (B) Immunohistochemistry staining with antibodies against Col1a1, Dlx3, Dmp1, Dsp, Oc, Opn, Osx, and Runx2 in the primary and immortalized murine dental papilla mesenchymal cells. Scale bar = 10 μM. (C). Expression of Dsp and Dmp1 proteins in the primary and immortalized mouse dental papilla mesenchymal cells was analyzed by Western blot assay using anti-Dsp and anti-Dmp1 antibodies. Two major fragments of Dmp1 were observed as 57- and 37-kDa, respectively, in a SDS-PAGE gele using anti-Dmp1 antibody. Multiple fragments of Dsp polypeptides were detected in MD10-F2 cells using anti-Dsp antibodiy. Major bands are approximately 120, 65, 60, and 40 kDa as indicated by arrows. β-actin was used as an internal control. Pri and Imm represent the primary and immoratalized dental papilla mesenchymal cells.
Figure 4.
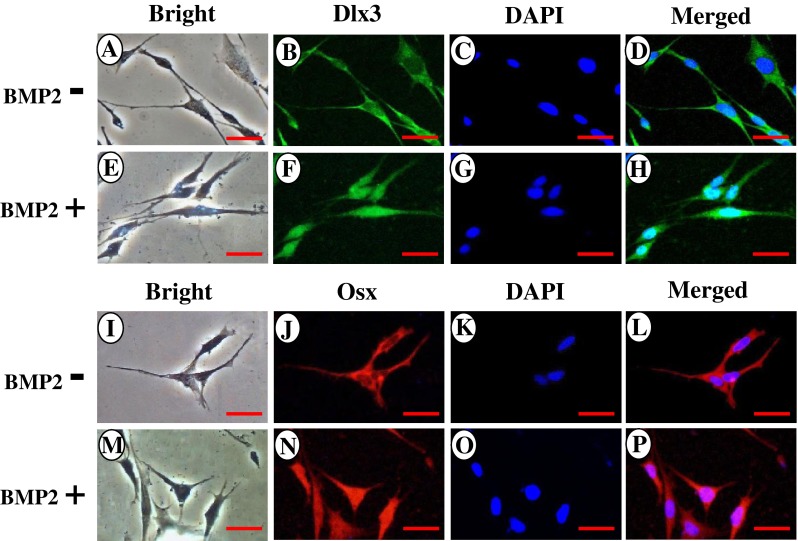
BMP2 induced Dlx3 and Osx protein translocation into the nucleus. Immortalized mouse dental papilla mesenchymal cells were treated with or without 300 ng/ml of recombinant BMP2 for 20 min in DMEM medium containing 1% serum. The cells were rinsed with ice-cold PBS and fixed for 10 min on ice with methanol/acetone (1:1). The cells were incubated with first antibodies against Dlx3 and Osx, respectively, followed by secondary antibodies with Alexa Fluo® 488 green fluorescent and Alexa Fluo® 568 red fluorescent labeling. The results show that without BMP2 treatment, Dlx3 and Osx proteins are mostly localized in the cytoplasm (B, J) whereas BMP2 induces both protein translocation into the nucleus (F, N). C, G, K, O The cells were stained with DAPI for the nucleus. D, H, L, P Images were merged. Scale bar = 10 μM.
Cell differentiation and mineralization.
To determine the differentiation and mineralization activities of these immortalized cells, we examined ALP activity by in situ ALP histochemistry as ALP is a marker of dental cell differentiation (Katagiri et al. 1990; Yamaguchi et al. 1991). At 4-d induction in calcifying medium, ALP expression in the immortalized cells was seen, but mineralized nodules were not detected (data not shown). However, at 7 and 14-d induction, both ALP expression and mineralized nodules were visualized by using ALP assay and alizarin red S staining, respectively (Fig. 5). ALP expression levels and mineralized nodule densities and sizes were increased with a long cell induction.
Figure 5.
Alkaline phosphatase activity and mineralized nodule formation in the immortalized dental papilla mesenchymal cells. (A, B) In situ histochemistry of ALP activity in the immortalized cells was visualized after a culture of 7 and 14 d in calcifying media. (C, D) The immortalized cells were treated with calcifying medium for 7 and 14 d. Nodule formation in the immortalized cells was seen with alizarin red S staining. Scale bars show 100 μM.
Cell transfection.
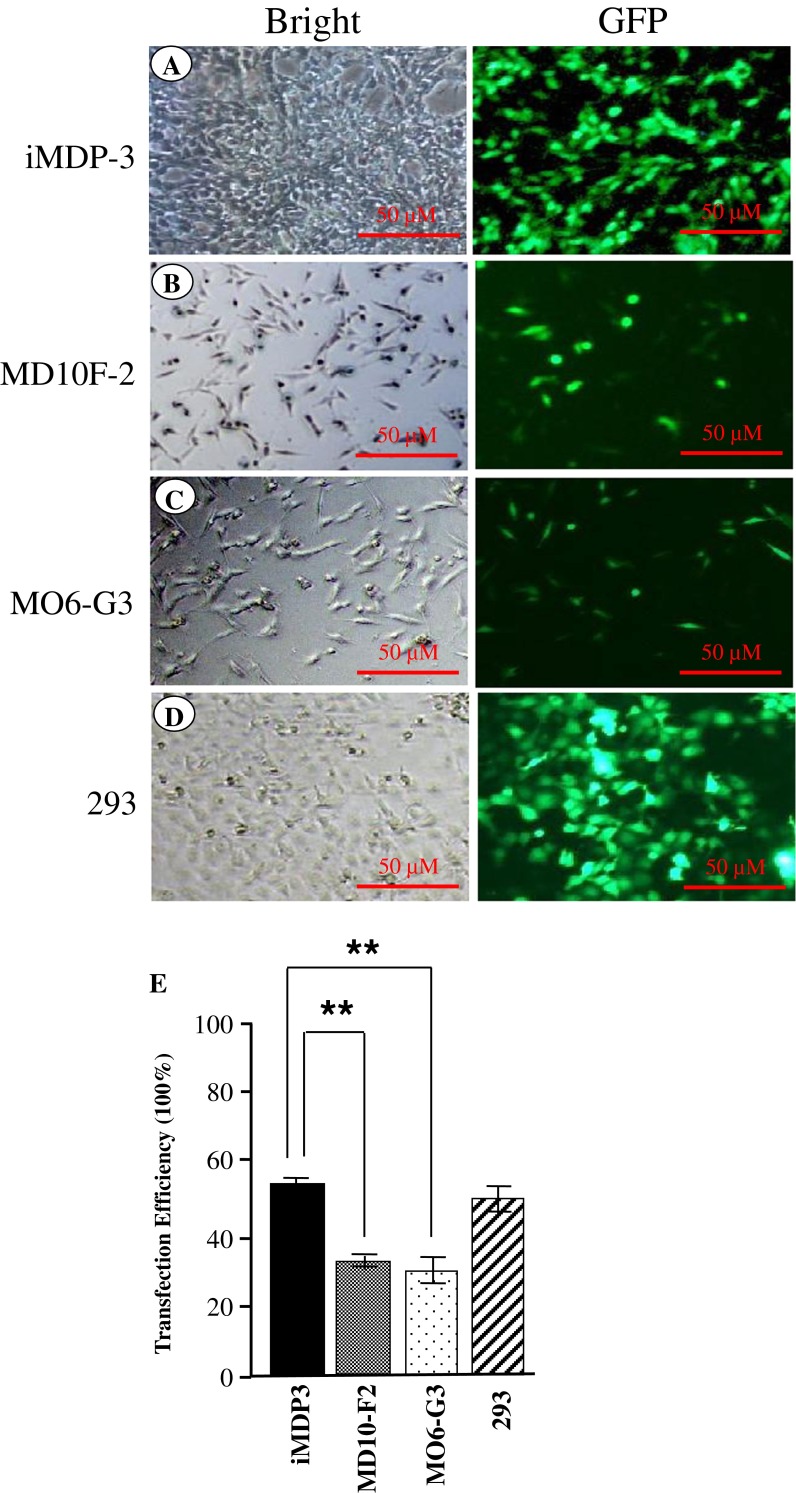
To determine cell transfection efficiency, the iMDP-3 cells were transiently transfected with GFP expression vector. After 24-h transfection, GFP expression in the immortalized cells was observed using the reverted fluorescent microscope (Fig. 6A). Fifty-four percent of GFP expression was seen in the transfected iMDP-3 cells and GFP expression was also observed in the immortalized cells after 72-h transfection (data not shown). Compared to mouse immortalized odontoblast-like (MO6-G3; 35%) and pre-odontoblast-like (MD10-F2; 37%) cell lines, the iMDP-3 cells had high transfection efficient activity (Fig. 6E).
Figure 6.
Cell transfection. pEGFP-C2 plasmid was transiently transfected into iMDP-3, MO6-G3, MD10-F2, and 293 cells, respectively. Twenty-four hours after transfection, GFP expression was observed in four cell lines using the inverted fluorescent microscope. iMDP-3 had a high transfection activity (A) compared to mouse immortalized odontoblast-like (MO6-G3; B) and pre-odontoblast-like (MD10-F2) cells (C). Two hundred ninety-three cells (D) were used for positive control of DNA transfection efficiency. (E) The data are the means ± SD from three independent observations. Significant differences are shown as follows: **p < 0.01 by student’s test. *p <0.05 is significant.
Discussion
Odontoblasts are derived from dental papilla mesenchymal cells and this process of these cell lineage commitments is regulated by a variety of growth factors, transcription factors and NCPs (Thesleff 2003; Chen et al. 2012). Mutations of these genes such as BMP2, DSPP and DMP1 cause dentin genetic disorders (Xiao et al. 2001; Ye et al. 2004; Kim and Simmer 2007; Feng et al. 2011). The understanding of signaling pathways of these factors involved in odontoblast differentiation and dentin formation remains still unknown as odontoblast differentiation in cultured system has not been well characterized due to two major limitations: it is hard to collect enough amounts of dental cells from a single tooth and limited life span of primary cells. We anticipate that the development and characterization of a dental papilla mesenchymal cell line will not only be useful for studying signaling pathways of odontoblast differentiation and identifying novel targets, but also help in the field of gene therapy for dentin regeneration.
In the present study, we established and characterized an immortalized dental papilla mesenchymal cell line from primary mouse dental papilla mesenchymal cells at postnatal day 3 by transfection with SV40 T-Ag. The immortalized cells stained SV40 positive and retained high ALP activity and mineralization ability. Also, these cells expressed all of the identification markers of tooth-related genes. These data suggest that the cell line, iMDP-3, is functionally active and displays all of genotypic and phenotypic characteristics similar to the primary dental papilla mesenchymal cells. The immortalized cells were inducible and BMP2 stimulated activity of two bone/tooth-relate transcription factors, Dlx3 and Osx, in these cells. Moreover, advantage of the iMDP-3 cell line has a high transfection efficiency compared to mouse MO6-G3 and MD10-F2 cells that have been used for biological function studies (Chen et al. 2005).
Various tissue-specific cell lines have been immortalized using SV40-Tg, a viral oncogene (Heath et al. 1989; MacDougall et al. 1995; Hanks et al. 1998; Thonemann and Schmalz 2000; Galler et al. 2006; Iwata et al. 2007; Suguro et al. 2008; Wu et al. 2009; Wu et al. 2010; Lin et al. 2013). These cell lines have been used well in many applications, including the study of normal and pathological cell differentiation and mineralization, the dental tissue repair and regeneration, toxicity and biocompatibility testing, gene regulation and signaling pathway. We observed that the transformed cells had the necessary biochemical program for cell differentiation and were capable of producing mineralizing extracellular matrices.
In particular, we analyzed several transcription factors known to be required for tooth development including Runx2, Osx, Lhx 6, Lhx 7, Dlx3, and ATF4. Both Runx2 and Osx are important factors for not only osteogenesis and chondrogenesis, but also dentinogenesis (Ducy et al. 1997; Komori et al. 1997; Mundlos et al. 1997; Otto et al. 1997; Nakashima et al. 2002). Mutations of Runx2 and Osx genes result in abnormal bone and tooth development and formation (Lee et al. 1997; D’Souza et al. 1999; Nakashima et al. 2002). Lhx 6 and 7 are two LIM-domain genes encoding homeodomain proteins produced by dental mesenchyme during tooth development (Priam et al. 2005; Arany et al. 2009). Mutations of both Lhx 6 and Lhx 7 resulted in lack of molar teeth in mice (Denaxa et al. 2009). Dlx3 is a homeodomain transcriptional factor necessary for tooth development. Dlx 3 gene mutations in humans are associated with an autosomal dominant genetic disorder called tricho–dento–osseous syndrome (TDO) characterized by abnormalities in hair, tooth and bone development (Price et al. 1998; Dong et al. 2005; Duverger et al. 2008). ATF4 is essential to osteoblast differentiation and skeletal development (Yang et al. 2004; Xiao et al. 2005; Ameri and Harris 2008). Furthermore, it has been found that these transcriptional factors are regulated by BMP2 and other growth factors (Park and Morasso 2002; Javed et al. 2008; Matsubara et al. 2008; Ulsamer et al. 2008; Fei et al. 2010). We found that BMP2 was able to stimulate Dlx3 and Osx activity in the immortalized cells. Therefore, having a cell line will be a valuable asset for pinpointing transcription factors that control the differentiation and biomineralization of the dental papilla mesenchymal cells into odontoblasts.
The immortalized cells are capable of synthesizing principal extracellular matrices that are necessary for the assembly of dentin matrix, namely collagen type I, which forms the scaffold for mineral deposition, and NCPs, which are responsible for nucleating and regulating the hydroxyapatite crystal size. In particular, DSPP and DMP1 define important markers during odontogenesis (George et al. 1993; Linde and Goldberg 1993; Ritchie et al. 1994; Zeichner-David et al. 1995; D’Souza et al. 1997; MacDougall et al. 1997). Mutations of DSPP gene are associated with dentinogenesis imperfect (Xiao et al. 2001; Rajpar et al. 2002; Kim and Simmer 2007; McKnight et al. 2008; Song et al. 2008). Also, DMP1 gene mutations impair odontoblast differentiation and dentin formation (Ye et al. 2004). We observed that the immortalized cells are able to differentiate and form mineralized nodules as detected by ALP and alizarin red S assays when cultured in calcifying medium as well as retain the genotypic and phenotypic characteristics similar to odontoblast-like cells described by our and other laboratories (MacDougall et al. 1995; Iwata et al. 2007; Wu et al. 2010).
Compared to other cell lines, only few mouse dental mesenchymal cell lines were generated (MacDougall et al. 1995; Hanks et al. 1998; Wu et al. 2010; Lin et al. 2013). These cells have been used for the study of gene regulation and signaling pathway transduction, but these cell lines have low transfection efficiency (Chen et al. 2005). In this study, we generated the iMDP-3 cell line that has a high transfection efficiency compared to two mouse odontoblast-like (MO6-G3) and pre-odontoblast-like (MD10-F2) cells (Fig. 6). It suggests that the cell line can be used for gene regulation and signal transduction pathway studies.
Conclusions
In the summary, the present study demonstrates that the immortalized mouse dental papilla mesenchymal cells have a unique odontogenic potential along with expression of odontoblast-specific markers and the capability of forming mineralized matrices. At first, advantage in the establishment of such a stable dental papilla mesenchymal cell is to provide a large consistent source of dental papilla mesenchymal cell products like dentin extracellular matrix suitable for biochemical analysis. Secondly, it provides excellent resource suitable for studying the mechanisms of dental papilla mesenchyaml cell proliferation, differentiation and mineralization as well as potential application of these cells for reparative formation and regeneration of dentin. Thirdly, the cell line is well responsible to growth factor stimulation and has high transfection efficiency for studying gene regulation and signaling transduction pathways.
Acknowledgment
This work was supported in part by National Institute of Health Grant DE019892 and San Antonio Area Foundation.
Conflict of interests
The authors declare that there is no conflict of interests.
Footnotes
Feng Wang and Li-An Wu contributed equally to this work.
Contributor Information
Wei Wang, Phone: +86-135-50412009, FAX: +86-591-22862454, Email: Wangwei0976@163.com.
Shuo Chen, Phone: +1-210-5673511, FAX: +1-210-5676603, Email: chens0@uthscsa.edu.
References
- Ameri K, Harris AL. Activating transcription factor 4. Int. J. Biochem. Cell Biol. 2008;40:14–21. doi: 10.1016/j.biocel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- Arany S, Kawagoe M, Sugiyama T. Application of spontaneously immortalized odontoblast cells in tooth regeneration. Biochem. Biophys. Res. Commun. 2009;381:84–927. doi: 10.1016/j.bbrc.2009.02.025. [DOI] [PubMed] [Google Scholar]
- Biria M, Abbas FM, Mozaffar S, Ahmadi R. Dentinogenesis imperfecta associated with osteogenesis imperfect. Den. Res. J. (Isfahan) 2012;9:489–494. [PMC free article] [PubMed] [Google Scholar]
- Chen S.; Chen L.; Jahangiri A.; Chen B.; Wu Y.; Chuang H.H.; Qin C.; MacDougall M. Expression and processing of small integrin-binding ligand N-linked glycoproteins in mouse odontoblastic cells. Arch. Oral Biol. 53: 879–89; 2008. [DOI] [PMC free article] [PubMed]
- Chen S, McDaniel JS, Feng J, Rakian A, MacDougall M. DSPP Gene Transcription and Cellular Signaling Pathways. In: Goldberg M, editor. Frontiers between Science and Clinic in Odontology. France: Bentham Science Publishers; 2012. pp. 186–202. [Google Scholar]
- Chen S, Rani S, Wu Y, Unterbrink A, Gu TT, Gluhak-Heinrich J, Chuang HH, Macdougall M. Differential regulation of dentin sialophosphoprotein expression by Runx2 during odontoblast cytodifferentiation. J. Biol. Chem. 2005;280:29717–29727. doi: 10.1074/jbc.M502929200. [DOI] [PubMed] [Google Scholar]
- Denaxa M, Sharpe PT, Pachnis V. The LIM homeodomain transcription factors Lhx6 and Lhx7 are key regulators of mammalian dentition. Dev. Biol. 2009;333:324–336. doi: 10.1016/j.ydbio.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Amor D, Aldred MJ, Gu T, Escamilla M, MacDougall M. DLX3 mutation associated with autosomal dominant amelogenesis imperfecta with taurodontism. Am. J. Med. Genet. A. 2005;133A:138–141. doi: 10.1002/ajmg.a.30521. [DOI] [PubMed] [Google Scholar]
- D’Souza RN, Aberg T, Gaikwad J, Cavender A, Owen M, Karsenty G, Thesleff I. Cbfa1 is required for epithelial-mesenchymal interactions regulating tooth development in mice. Development. 1999;126:2911–2920. doi: 10.1242/dev.126.13.2911. [DOI] [PubMed] [Google Scholar]
- D’Souza RN, Cavender A, Sunavala G, Alvarez J, Ohshima T, Kulkarni AB, MacDougall M. Gene expression patterns of murine dentin matrix protein 1 (Dmp1) and dentin sialophosphoprotein (DSPP) suggest distinct developmental functions in vivo. J. Bone Miner. Res. 1997;12:2040–2049. doi: 10.1359/jbmr.1997.12.12.2040. [DOI] [PubMed] [Google Scholar]
- Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/S0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- Duverger O, Lee D, Hassan MQ, Chen SX, Jaisser F, Lian JB, Morasso MI. Molecular consequences of a frameshifted DLX3 mutant leading to Tricho-Dento-Osseous syndrome. J. Biol. Chem. 2008;283:20198–20208. doi: 10.1074/jbc.M709562200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Y, Xiao L, Hurley MM. Fibroblast growth factor 2 positively regulates expression of activating transcription factor 4 in osteoblasts. Biochem. Biophys. Res. Commun. 2010;391:335–339. doi: 10.1016/j.bbrc.2009.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Yang G, Yuan G, Gluhak-Heinrich J, Yang W, Wang L, Chen Z, McDaniel SJ, Donly KJ, Harris SE, Macdougall M, Chen S. Abnormalities in the Enamel in Bmp2-Deficient Mice. Cells Tissues Organs. 2011;194:216–221. doi: 10.1159/000324644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galler KM, Schweikl H, Thonemann B, D’Souza RN, Schmalz G. Human pulp-derived cells immortalized with Simian Virus 40 T-antigen. Eur. J. Oral Sci. 2006;114:138–146. doi: 10.1111/j.1600-0722.2006.00327.x. [DOI] [PubMed] [Google Scholar]
- George A, Sabsay B, Simonian PA, Veis A. Characterization of a novel dentin matrix acidic phosphoprotein. Implications for induction of biomineralization. J. Biol. Chem. 1993;268:12624–12630. [PubMed] [Google Scholar]
- Hanks CT, Sun ZL, Fang DN, Edwards CA, Wataha JC, Ritchie HH, Butler WT. Cloned 3T6 cell line from CD-1 mouse fetal molar dental papillae. Connect Tissue Res. 1998;37:233–249. doi: 10.3109/03008209809002442. [DOI] [PubMed] [Google Scholar]
- Heath JK, Rodan SB, Yoon K, Rodan G. Rat calvarial cell lines immortalized with SV-40 large T antigen: constitutive and retinoic acid-inducible expression of osteoblastic features. Endocrinology. 1989;124:3060–3068. doi: 10.1210/endo-124-6-3060. [DOI] [PubMed] [Google Scholar]
- Iwata T, Yamakoshi Y, Simmer JP, Ishikawa I, Hu JC. Establishment of porcine pulp derived cell lines and expression of recombinant dentin sialoprotein and recombinant dentin matrix protein-1. Eur. J. Oral Sci. 2007;115:48–56. doi: 10.1111/j.1600-0722.2007.00426.x. [DOI] [PubMed] [Google Scholar]
- Javed A, Bae JS, Afzal F, Gutierrez S, Pratap J, Zaidi SK, Lou Y, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Structural coupling of Smad and Runx2 for execution of the BMP2 osteogenic signal. J. Biol. Chem. 2008;283:8412–8422. doi: 10.1074/jbc.M705578200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri T, Yamaguchi A, Ikeda T, Yoshiki S, Wozney JM, Rosen V, Wang EA, Tanaka H, Omura S, Suda T. The non-osteogenic mouse pluripotent cell line, C3H10T1/2, is induced to differentiate into osteoblastic cells by recombinant human bone morphogenetic protein-2. Biochem. Biophys. Res. Commun. 1990;172:295–299. doi: 10.1016/S0006-291X(05)80208-6. [DOI] [PubMed] [Google Scholar]
- Kim JW, Simmer JP. Hereditary dentin defects. J. Dent. Res. 2007;86:392–399. doi: 10.1177/154405910708600502. [DOI] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/S0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- Lee B, Thirunavukkarasu K, Zhou L, Pastore L, Baldini A, Hecht J, Geoffroy V, Ducy P, Karsenty G. Missense mutations abolishing DNA binding of the osteoblast-specific transcription factor OSF2/CBFA1 in cleidocranial dysplasia. Nat. Genet. 1997;16:307–310. doi: 10.1038/ng0797-307. [DOI] [PubMed] [Google Scholar]
- Levin LS, Young RJ, Pyeritz RE. Osteogenesis imperfecta type I with unusual dental abnormalities. Am. J. Med. Genet. 1988;31:921–932. doi: 10.1002/ajmg.1320310427. [DOI] [PubMed] [Google Scholar]
- Lin H, Liu H, Sun Q, Yuan G, Zhang L, Chen Z. Establishment and characterization of a tamoxifen-mediated reversible immortalized mouse dental papilla cell line. In Vitro Cell. Dev. Biol. Anim. 2013;49:114–121. doi: 10.1007/s11626-012-9576-y. [DOI] [PubMed] [Google Scholar]
- Linde A, Goldberg M. Dentiogenesis. Crit. Rev. Oral Biol. Med. 1993;4:679–728. doi: 10.1177/10454411930040050301. [DOI] [PubMed] [Google Scholar]
- MacDougall M, Simmons D, Luan X, Nydegger J, Feng J, Gu TT. Dentin phosphoprotein and dentin sialoprotein are cleavage products expressed from a single transcript coded by a gene on human chromosome 4. Dentin phosphoprotein DNA sequence determination. J. Biol. Chem. 1997;272:835–842. doi: 10.1074/jbc.272.2.835. [DOI] [PubMed] [Google Scholar]
- MacDougall M, Thiemann F, Ta H, Hsu P, Chen LS, Snead ML. Temperature sensitive simian virus 40 large T antigen immortalization of murine odontoblast cell cultures: establishment of clonal odontoblast cell line. Connect Tissue Res. 1995;33:97–103. doi: 10.3109/03008209509016988. [DOI] [PubMed] [Google Scholar]
- Matsubara T, Kida K, Yamaguchi A, Hata K, Ichida F, Meguro H, Aburatani H, Nishimura R, Yoneda T. BMP2 regulates Osterix through Msx2 and Runx2 during osteoblast differentiation. J. Biol. Chem. 2008;283:29119–29125. doi: 10.1074/jbc.M801774200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight DA, Hart SP, Hart TC, Hartsfield JK, Wilson A, Wright JT, Fisher LW. A comprehensive analysis of normal variation and disease-causing mutations in the human DSPP gene. Hum. Mutat. 2008;29:1392–1404. doi: 10.1002/humu.20783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, Albright S, Lindhout D, Cole WG, Henn W, Knoll JH, Owen MJ, Mertelsmann R, Zabel BU, Olsen BR. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89:773–779. doi: 10.1016/S0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/S0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/S0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- Park GT, Morasso MI. Bone morphogenetic protein-2 (BMP-2) transactivates Dlx3 through Smad1 and Smad4: alternative mode for Dlx3 induction in mouse keratinocytes. Nucleic Acids Res. 2002;30:515–522. doi: 10.1093/nar/30.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priam F, Ronco V, Locker M, Bourd K, Bonnefoix M, Duchêne T, Bitard J, Wurtz T, Kellermann O, Goldberg M, Poliard A. New cellular models for tracking the odontoblast phenotype. Arch. Oral Biol. 2005;50:271–277. doi: 10.1016/j.archoralbio.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Price JA, Bowden DW, Wright JT, Pettenati MJ, Hart TC. Identification of a mutation in DLX3 associated with tricho-dento-osseous (TDO) syndrome. Hum. Mol. Genet. 1998;7:563–569. doi: 10.1093/hmg/7.3.563. [DOI] [PubMed] [Google Scholar]
- Rajpar MH, Koch ML, Davies RM, Mellody KT, Kielty CM, Dixon MJ. Mutation of the signal peptide region of the bicistronic gene DSPP affects translocation to the endoplasmic reticulum and results in defective dentine biomineralization. Hum. Mol. Genet. 2002;11:2559–2565. doi: 10.1093/hmg/11.21.2559. [DOI] [PubMed] [Google Scholar]
- Ritchie HH, Hou H, Veis A, Butler WT. Cloning and sequence determination of rat dentin sialoprotein, a novel dentin protein. J. Biol. Chem. 1994;269:98–702. [PubMed] [Google Scholar]
- Ruch JV, Lesot H, Bègue-Kirn C. Odontoblast differentiation. Int. J. Dev. Biol. 1995;39:51–68. [PubMed] [Google Scholar]
- Sakamoto S, Baird SA, Sakamoto M. Collagenase synthesis in clonal rabbit odontoblast-like cells (RP cells) Biochem. Biophys. Res. Commun. 1985;133:709–716. doi: 10.1016/0006-291X(85)90962-3. [DOI] [PubMed] [Google Scholar]
- Song YL, Wang CN, Fan MW, Su B, Bian Z. Dentin phosphoprotein frameshift mutations in hereditary dentin disorders and their variation patterns in normal human population. J. Med. Genet. 2008;45:457–464. doi: 10.1136/jmg.2007.056911. [DOI] [PubMed] [Google Scholar]
- Suguro H, Asano M, Kaneko Y, Omagari D, Ogiso B, Moro I, Komiyama K. Characterization of human dental pulp-derived cell lines. Int. Endod. J. 2008;41:609–616. doi: 10.1111/j.1365-2591.2008.01409.x. [DOI] [PubMed] [Google Scholar]
- Thesleff I. Epithelial-mesenchymal signalling regulating tooth morphogenesis. J. Cell Sci. 2003;116:1647–1648. doi: 10.1242/jcs.00410. [DOI] [PubMed] [Google Scholar]
- Thesleff I, Nieminen P. Tooth morphogenesis and cell differentiation. Curr. Opin. Cell Biol. 1996;8:44–850. doi: 10.1016/S0955-0674(96)80086-X. [DOI] [PubMed] [Google Scholar]
- Thonemann B, Schmalz G. Immortalization of bovine dental papilla cells with simian virus 40 large t antigen. Arch. Oral Biol. 2000;45:857–869. doi: 10.1016/S0003-9969(00)00056-X. [DOI] [PubMed] [Google Scholar]
- Ulsamer A, Ortuño MJ, Ruiz S, Susperregui AR, Osses N, Rosa JL, Ventura F. BMP-2 induces Osterix expression through up-regulation of Dlx5 and its phosphorylation by p38. Biol. Chem. 2008;283:3816–3826. doi: 10.1074/jbc.M704724200. [DOI] [PubMed] [Google Scholar]
- Wu LA, Feng J, Wang L, Mu YD, Baker A, Donly KJ, Gluhak-Heinrich J, Harris SE, MacDougall M, Chen S. Immortalized mouse floxed Bmp2 dental papilla mesenchymal cell lines preserve odontoblastic phenotype and respond to BMP2. J. Cell. Physiol. 2010;225:132–139. doi: 10.1002/jcp.22204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LA, Yuan G, Yang G, Ortiz-Gonzalez I, Yang W, Cui Y, MacDougall M, Donly KJ, Harris S, Chen S. Immortalization and characterization of mouse floxed Bmp2/4 osteoblasts. Biochem. Biophys. Res. Commun. 2009;386:89–95. doi: 10.1016/j.bbrc.2009.05.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao G, Jiang D, Ge C, Zhao Z, Lai Y, Boules H, Phimphilai M, Yang X, Karsenty G, Franceschi RT. Cooperative interactions between activating transcription factor 4 and Runx2/Cbfa1 stimulate osteoblast-specific osteocalcin gene expression. J. Biol. Chem. 2005;280:30689–30696. doi: 10.1074/jbc.M500750200. [DOI] [PubMed] [Google Scholar]
- Xiao S, Yu C, Chou X, Yuan W, Wang Y, Bu L, Fu G, Qian M, Yang J, Shi Y, Hu L, Han B, Wang Z, Huang W, Liu J, Chen Z, Zhao G, Kong X. Dentinogenesis imperfecta 1 with or without progressive hearing loss is associated with distinct mutations in DSPP. Nat. Genet. 2001;27:201–204. doi: 10.1038/84848. [DOI] [PubMed] [Google Scholar]
- Yamaguchi A, Katagiri T, Ikeda T, Wozney JM, Rosen V, Wang EA, Kahn AJ, Suda T, Yoshiki S. Recombinant human bone morphogenetic protein-2 stimulates osteoblastic maturation and inhibits myogenic differentiation in vitro. J. Cell Biol. 1991;113:681–687. doi: 10.1083/jcb.113.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Matsuda K, Bialek P, Jacquot S, Masuoka HC, Schinke T, Li L, Brancorsini S, Sassone-Corsi P, Townes TM, Hanauer A, Karsenty G. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell. 2004;117:387–398. doi: 10.1016/S0092-8674(04)00344-7. [DOI] [PubMed] [Google Scholar]
- Ye L, MacDougall M, Zhang S, Xie Y, Zhang J, Li Z, Lu Y, Mishina Y, Feng JQ. Deletion of dentin matrix protein-1 leads to a partial failure of maturation of predentin into dentin, hypomineralization, and expanded cavities of pulp and root canal during postnatal tooth development. J. Biol. Chem. 2004;279:19141–19148. doi: 10.1074/jbc.M400490200. [DOI] [PubMed] [Google Scholar]
- Yuan G, Wang Y, Gluhak-Heinrich J, Yang G, Chen L, Li T, Wu LA, Chen Z, MacDougall M, Chen S. Tissue-specific expression of dentin sialophosphoprotein (DSPP) and its polymorphisms in mouse tissues. Cell Biol. Int. 2009;33:816–829. doi: 10.1016/j.cellbi.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeichner-David M, Diekwisch T, Fincham A, Lau E, MacDougall M, Moradian-Oldak J, Simmer J, Snead M, Slavkin HC. Control of ameloblast differentiation. Int. J. Dev. Biol. 1995;39:69–92. [PubMed] [Google Scholar]