Abstract
Populations with two sexes are vulnerable to a pair of genetic conflicts: sexual antagonism that can arise when alleles have opposing fitness effects on females and males; and parental antagonism that arises when alleles have opposing fitness effects when maternally and paternally inherited. This paper extends previous theoretical work that found stable linkage disequilibrium (LD) between sexually antagonistic loci. We find that LD is also generated between parentally antagonistic loci, and between sexually and parentally antagonistic loci, without any requirement of epistasis. We contend that the LD in these models arises from the admixture of gene pools subject to different selective histories. We also find that polymorphism maintained by parental antagonism at one locus expands the opportunity for polymorphism at a linked locus experiencing parental or sexual antagonism. Taken together, our results predict the chromosomal clustering of loci that segregate for sexually and parentally antagonistic alleles. Thus, genetic conflict may play a role in the evolution of genomic architecture.
Keywords: sexual antagonism, parental antagonism, linkage disequilibrium
1. Introduction
In populations with two sexes, half the genes in the current population spent the previous generation in female bodies, whereas the other half resided in male bodies. This bisection of the gene pool is associated with two forms of antagonistic selection that can maintain allelic polymorphism in one-locus models.
(a). Sexual antagonism
An allele may confer above average fitness on one sex but below average fitness on the other sex [1–4]. This form of balancing selection results in a stable polymorphism when the strength of selection is roughly equal between males and females [1].
(b). Parental antagonism
An allele may be relatively beneficial when inherited from one sex but deleterious when inherited from the other. Under parental antagonism, reciprocal heterozygotes represent the extremes of fitness: one of these heterozygotes sees both alleles in their favoured parental-origin contexts; the other sees both alleles in their disfavoured parental-origin contexts. Just as a high-fitness genotype can become a low-fitness genotype when the sexual context of sexually antagonistic alleles is reversed, a high-fitness heterozygote at a parentally antagonistic locus becomes a low-fitness heterozygote when the parental origins of its alleles are reversed. Genes segregating for such parentally antagonistic effects on phenotype have been found [5], and such polymorphism is predicted from theory [6,7].
In previous papers, we considered sexually antagonistic selection at two loci [8,9]. We found that stable linkage disequilibrium (LD) is generated between sexually antagonistic loci in the absence of epistasis even when the loci are unlinked [8] and that sexually antagonistic polymorphism at one locus facilitates the invasion of sexually antagonistic alleles at linked loci [9]. This LD arises in each sexual generation because of admixture between genes that have been selected in male bodies and genes that have been selected in female bodies.
This paper extends our previous work by considering two models: one, a model of parentally antagonistic selection at two loci; and the other, a model of parentally antagonistic selection at one locus with sexually antagonistic selection at a second locus. We find that stable LD is present at polymorphic equilibria between a locus experiencing parental antagonism and a linked locus that experiences either parental or sexual antagonism although this is not the case when the loci are unlinked. Moreover, we find that the opportunity for parentally antagonistic polymorphism is expanded at loci linked to another locus segregating for either parentally or sexually antagonistic alleles. We argue that the LD in these models arises from subtle forms of admixture.
2. Methods
We present a standard two-locus, two-allele, diploid model [10]. We consider two loci, A and B, with two alleles each, A1, A2 and B1, B2, respectively. These loci recombine with frequency r. Let xi and yj be the frequencies of the ith and jth haplotypes in eggs and sperm such that x1, x2, x3, x4 and y1, y2, y3, y4 are the respective frequencies of A1B1, A1B2, A2B1, A2B2 haplotypes in eggs and in sperm. These haplotype frequencies are stored in vectors xT = [x1, x2, x3, x4] and yT = [y1, y2, y3, y4] (henceforth, lower-case bold letters represent vectors). Let pζ and qζ be the frequency of A1 and B1 in gametes, such that pζ = ζ1 + ζ2 and qζ = ζ1 + ζ3, with ζ taking a value of x for eggs and y for sperm.
Let u11, u12, u21, u22 be the fitness corresponding to genotypes A1A1, A1A2, A2A1, A2A2 at locus A. These fitnesses are stored in the matrix
Let v11f, v12f, v21f, v22f and v11m, v12m, v21m, v22m be the fitness corresponding to genotypes B1B1, B1B2, B2B1, B2B2 at locus B when expressed in females and males respectively. Unlike the A locus, the B locus is allowed to confer sex-specific fitness effects. These fitnesses are stored in the matrices
Let wijf and wijm be the respective fitnesses of female and male zygotes that develop from the union of the ith egg haplotype with the jth sperm haplotype. These zygotic fitnesses are stored in matrices Wf and Wm. For the purpose of eliminating epistasis within sexes [11], we assume that the fitness of a zygote results from the product of the one-locus genotypic fitnesses, that is
where ⊗ represents the Kronecker product of two matrices.
Assuming all other evolutionary forces besides viability selection and recombination are absent, the frequency of each haplotype in eggs and sperm in the next generation is
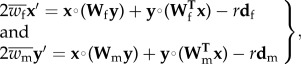 |
2.1 |
where 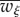 is the mean fitness, defined as
is the mean fitness, defined as 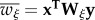 , and
, and 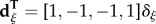 , with
, with 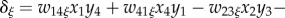
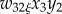 (ξ = f for females; ξ = m for males). Primes denote frequencies in the next generation, empty circles denote the Schur product, and superscript T denote the transpose.
(ξ = f for females; ξ = m for males). Primes denote frequencies in the next generation, empty circles denote the Schur product, and superscript T denote the transpose.
Non-random associations between loci in the haploid phase are measured by the coefficient of LD, D [12], with subscripts x for eggs and y for sperm:
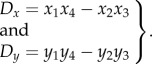 |
2.2 |
Non-random associations between loci in the diploid phase are measured by the difference in frequencies between coupling and repulsion double-heterozygotes:
| 2.3a |
which Úbeda et al. [8] have shown is equal to:
| 2.3b |
where the covariance is taken over the two sexes' gametes. It is worth noting that 2Covζ(p,q) = 1/2 (px – py)(qx – qy). We report both haploid and diploid measures of LD because these two measures can differ when there is sex-specific selection [8].
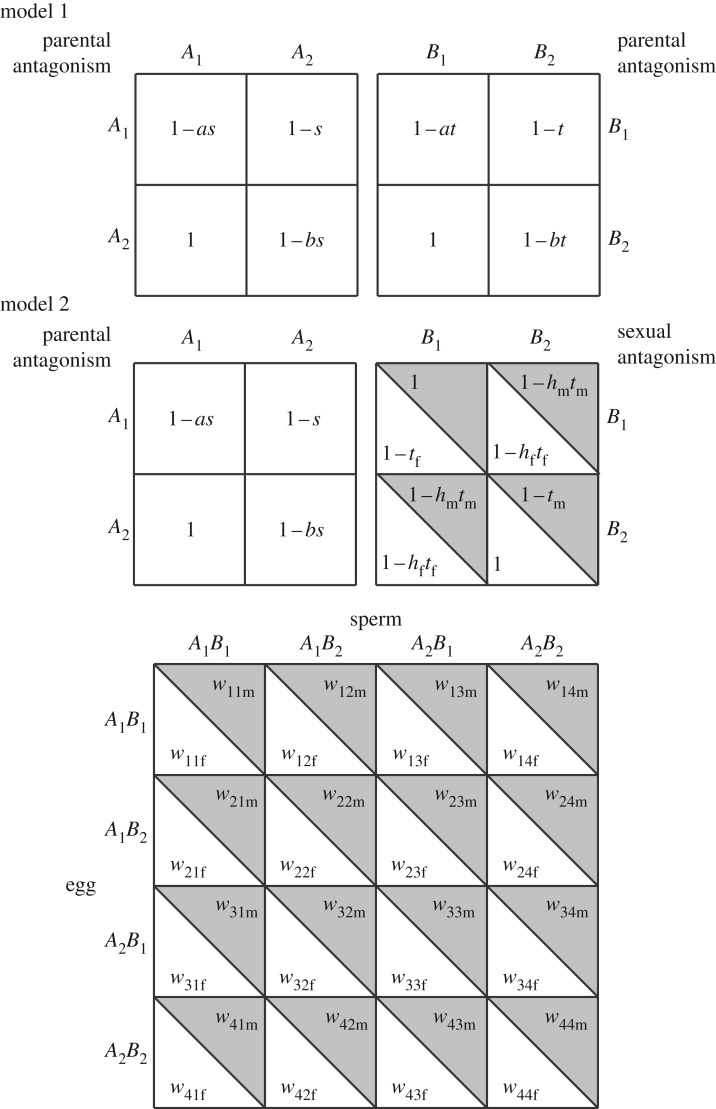
(a). Model 1: two parentally antagonistic loci
Let both the A and B loci experience parentally antagonistic selection (figure 1). A1 and B1 are assumed to confer high relative fitness when paternally derived and low relative fitness when maternally derived, and vice versa for A2 and B2. Thus, the reciprocal heterozygotes exhibit the extremes of fitness, with A2A1 and B2B1 the most fit genotypes and A1A2 and B1B2 the least fit genotypes.
Figure 1.
Model parametrizations. The A locus segregates for alleles with parentally antagonistic effects in both models. The B locus segregates for alleles with either parentally or sexually antagonistic effects, depending on the particular model. A two-locus genotype confers a fitness exactly equal to the product of the fitnesses specified by the genotypes at each locus.
Parental antagonism can be parametrized with selection coefficients t and s for locus A and B, 0 < t, s ≤ 1, and two scaling parameters a and b, 0 ≤ a, b ≤ 1, that are akin to dominance coefficients but which scale the homozygous fitnesses relative to the heterozygous fitness extremes. The fitness of genotypes A1A1, A1A2, A2A1, A2A2 is 1 – as, 1 – s, 1 and 1 – bs, respectively (figure 1). These fitnesses are stored in the matrix UP. The fitness of genotypes B1B1, B1B2, B2B1, B2B2 are 1 – at, 1 – t, 1 and 1 – bt, respectively (figure 1). These fitnesses are stored in the matrix VP.
Owing to the lack of sex-specificity, the frequencies of haplotypes in eggs and sperm are equal after one generation, i.e. x = y = z, and system (2.1) reduces to
| 2.4 |
where 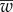 is the mean fitness, defined as
is the mean fitness, defined as 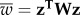 , and
, and 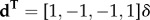 with
with 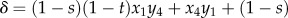
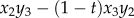 . LD in the haploid phase reduces to Dx = Dy = Dz = z1z4 – z2z3.
. LD in the haploid phase reduces to Dx = Dy = Dz = z1z4 – z2z3.
The lack of sex-specificity results in equal frequency of alleles in eggs and sperm, i.e. px = py and qx = qy, which yields Covζ(p,q) = 0. Substituting these into (2.3b), we find that LD is equal in the haploid and diploid phase, i.e. Dt = Dz.
(b). Model 2: one parentally antagonistic locus and one sexually antagonistic locus
Let the A locus experience parentally antagonistic selection and the B locus experience sexually antagonistic selection (figure 1). A1 is assumed to confer high relative fitness when paternally derived and low relative fitness when maternally derived, and vice versa for A2. Allele B1 is assumed to confer high relative fitness in males and low relative fitness in females, and vice versa for B2.
The fitness of genotypes A1A1, A1A2, A2A1, A2A2 is 1 – as, 1 – s, 1 and 1 – bs, respectively (figure 1). These fitnesses are stored in the matrix UP. Sexual antagonism in locus B is parametrized with selection coefficients tf and tm, 0 < tf, tm ≤ 1, and dominance parameters hf and hm, 0 ≤ hf, hm ≤ 1, for females and males, respectively. The fitness of genotypes B1B1, B1B2, B2B1, B2B2 is 1 – tf, 1 – hftf, 1 – hftf, and 1 when expressed in females and 1, 1 – hmtm, 1 – hmtm and 1 – tm when expressed in males. These fitnesses are stored in the matrices VSf and VSm. By limiting dominance and selection parameters to the range [0,1], we guarantee opposing directional selection in the two sexes. To avoid complexities that arise when dominance differs between the sexes, we assume throughout that hf = h and hm = 1 – h [13–15].
Substituting the fitness matrices into system (2.1) yields
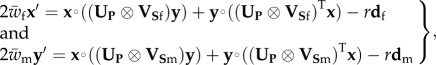 |
2.5 |
where 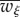 is the mean fitness, defined as
is the mean fitness, defined as 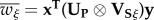 , and where
, and where 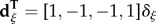 with
with 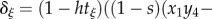
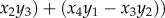 .
.
(c). Analysis
Owing to the complexity of these systems, we did not derive analytical solutions; instead, we used the recursions to carry out a numerical analysis of the equilibrium frequencies and the associated LD using code written for Matlab (R2012a). We initialize our populations near one of the four corners (corresponding to fixation of one of the four haplotypes) with all four haplotypes segregating, and we allow the population to evolve until an equilibrium state is reached. The equilibrium state of the population was found to be independent of the choice of corner.
3. Results
(a). Two parentally antagonistic loci
Results are summarized in figures 2 and 3. For the simplest presentation, we generate these figures with symmetric parametrizations (e.g. a = b = 0.75 in figure 2 and s = t in figure 3), but results are qualitatively similar when these symmetries are broken.
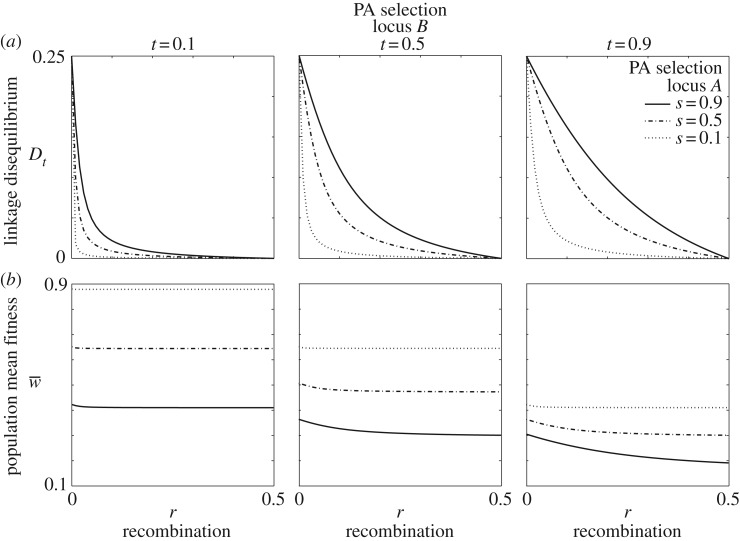
Figure 2.
(a–b) Linkage disequilibrium and mean fitness with parentally antagonistic selection at both loci. The magnitude of linkage disequilibrium (Dt) between pairs of parentally antagonistic loci declines with weaker selection (s,t) and greater recombination (r). The scaling parameters for both loci are a = b = 0.75.
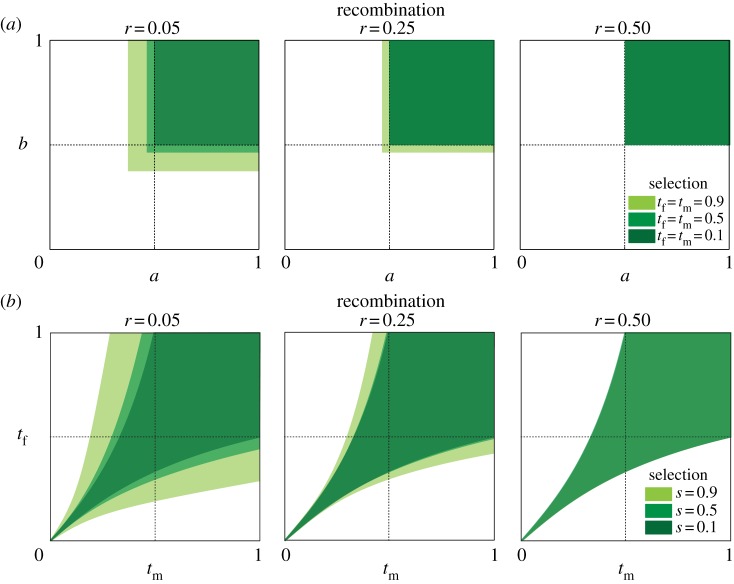
Figure 3.
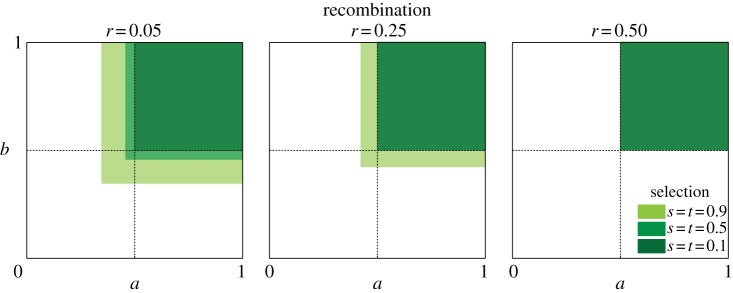
The opportunity for polymorphism with parentally antagonistic selection at both loci. The area of parameter space that allows polymorphism at linked parentally antagonistic loci expands with stronger selection and reduced recombination. The axes in each plot are the scaling parameters, a and b, which determine where between the heterozygous extremes the homozygote fitnesses fall. Low values of a and b correspond to relatively high fitness of the homozygotes. For each value of s and r, we simulated 10 000 combinations of a and b. The coloured space represents populations that achieved a polymorphic equilibrium with those values for a and b.
An excess of A1B1 and A2B2 haplotypes (positive LD) is present at the polymorphic equilibrium when r < 0.5 (figure 2). Greater recombination reduces the population mean fitness (figure 2). Stronger selection and weaker recombination are associated with greater LD in both haploid and diploid phases.
Parental antagonism at one locus changes the conditions for polymorphism under parental antagonism at a second locus. Stable polymorphism in a one-locus model of parental antagonism requires 1/2 < a,b ≤ 1 [6,7]. In our two-locus model, values of a and b outside this range permit polymorphism, provided recombination is sufficiently small and selection is sufficiently strong (figure 3). In the one-locus model, the strength of selection does not affect the opportunity for polymorphism, but polymorphism is more likely with larger values of s and t in the two-locus model for linked loci (figure 3).
(b). One sexually antagonistic locus and one parentally antagonistic locus
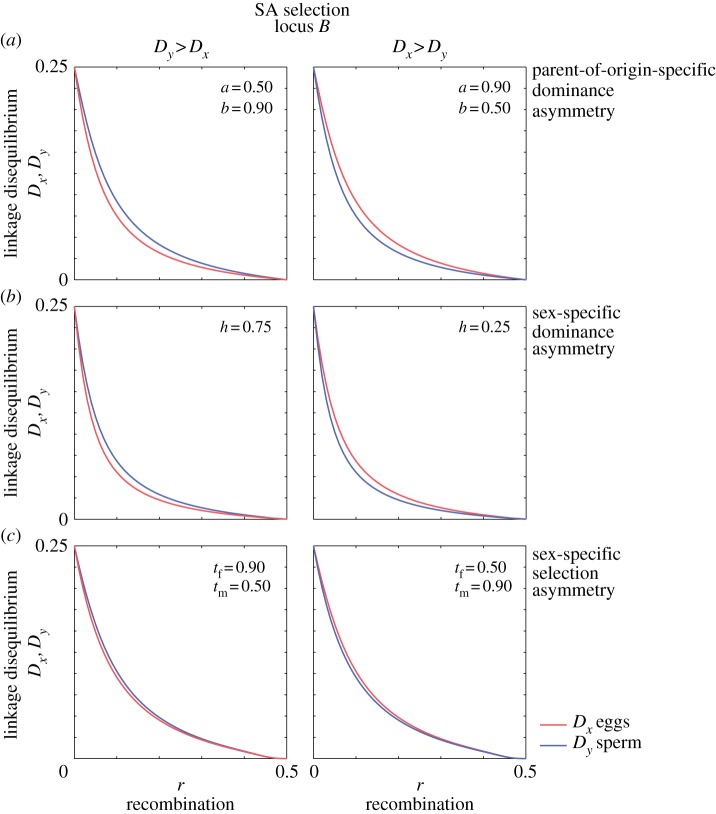
Results are summarized in figures 4 and 5. For the sake of simplicity, we elaborate these results for symmetric parametrizations (e.g. tf = tm, a = b = hf = hm = 1/2 in figure 4 and hf = hm = 1/2 in figure 5), but qualitatively similar results are obtained when these symmetries are broken. Cases in which asymmetric parametrizations produce a qualitatively novel result are summarized in figure 6.
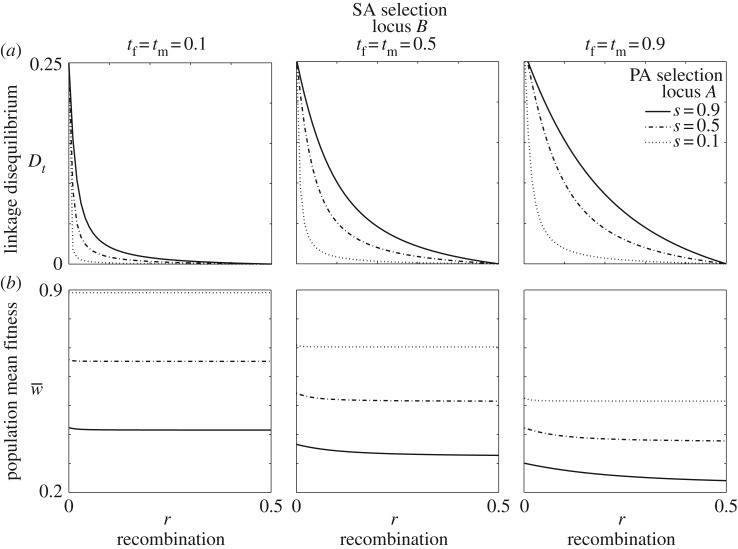
Figure 4.
(a–b) Linkage disequilibrium (Dt) and mean fitness with parentally and sexually antagonistic selection. The magnitude of Dt in the diploid phase between a sexually antagonistic locus and a parentally antagonistic locus declines with weaker selection and greater recombination. In the data shown, the scaling parameters for the parentally antagonistic loci are a = b = 0.75, tm = tf = t, and the allelic effects for the sexually antagonistic locus are additive (hm = hf = 1/2).
Figure 5.
The opportunity for polymorphism with parentally and sexually antagonistic selection. (a) Polymorphism for parentally antagonistic alleles when linked to a sexually antagonistic locus. Shown is the opportunity for polymorphism for the A locus, which experiences parental antagonism, while it is linked to a sexually antagonistic locus, B, at different recombination distances. The opportunity for polymorphism is sensitive to the strength of sexually antagonistic selection (tf, tm). The selection coefficient for the parentally antagonistic locus was s = 0.5 for all runs. The allelic effects for the sexually antagonistic locus are additive (hm = hf = 1/2). For each value of tf (tm) and r, we simulated 10 000 combinations of a and b, the scaling parameters that dictate the fitness of the homozygotes for the A locus. The coloured space represents populations that achieved a polymorphic equilibrium with those values for a and b. (b) Polymorphism for sexually antagonistic alleles when linked to a parentally antagonistic locus. Shown is the opportunity for polymorphism for the B locus, which segregates for sexually antagonistic alleles, whereas it is linked to a parentally antagonistic locus, A, at different recombination distances. The opportunity for polymorphism is sensitive to the strength of parentally antagonistic selection (s) and the recombination rate between loci (r). In the data shown, a = b = 0.75 for the parentally antagonistic locus and the allelic effects for the sexually antagonistic locus are additive (hm = hf = 1/2). For each value of s and r, we simulated 10 000 combinations of tf and tm. The coloured space represents populations that achieved a polymorphic equilibrium with those values for tf and tm.
Figure 6.
(a–c) Linkage disequilibrium under asymmetric parametrization and linkage disequilibrium. Linkage disequilibrium arises between a sexually and a parentally antagonistic locus. Dx and Dy, the linkage disequilibrium in eggs and sperm, differ when asymmetries are introduced to the fully symmetric parametrization. Unless noted, parameter values were a = b = h = 1/2; s = tm = tf = 0.5).
Positive LD accompanies all polymorphic equilibria with r < 0.5 (figure 4). This means that the allele favoured in females is coupled with the allele favoured when maternally derived, and the allele favoured in males is coupled with the allele favoured when paternally derived. Stronger selection and weaker recombination produce greater LD at equilibrium in both haploid and diploid phases. Population mean fitness decreases as the recombination rate increases (figure 4).
The opportunity for polymorphism at a locus experiencing parental or sexual antagonism is increased when it is linked to a polymorphic locus experiencing the other form of antagonism (figure 5). Consider first the opportunity for polymorphism at a locus experiencing parental antagonism linked to a locus experiencing sexual antagonism (figure 5a). There are parametrizations in which neither a nor b are greater than a half but both parentally antagonistic alleles persist (figure 5a). Therefore, barriers to invasion of a parentally antagonistic allele are lowered at loci linked to a sexually antagonistic locus. Consider now the opportunity for polymorphism at a locus experiencing sexual antagonism linked to a locus experiencing parental antagonism (figure 5b). There are regions of the parameter space where polymorphism is maintained at a sexually antagonistic locus that is linked to a parentally antagonistic locus but not for a sexually antagonistic locus that is unlinked (figure 5b). Note that the condition for polymorphism under sexual antagonism at an unlinked locus is tf/(1 + tf) < tm < tf/(1 – tf) [14]. Stronger selection expands opportunities for polymorphism in both cases (figure 5).
With a symmetric parametrization (i.e. tf = tm, a = b, hf = hm = 1/2), LD does not differ between eggs and sperm (figure 4) and population mean fitness does not differ between females and males. Not surprisingly, these symmetries are lost with asymmetric parametrizations. Considering parental-origin-specific asymmetries, LD at equilibrium is greater in eggs than sperm (Dx > Dy) when the fitness of A1A1 is greater than the fitness of A2A2, i.e. when a > b (figure 6a). Considering sex-specific asymmetries, LD at equilibrium is greater in eggs than sperm (Dx > Dy) when the male-beneficial allele B1 is dominant (h > 1/2; figure 6b) and when the selective disadvantage of B1 in males is greater than the selective disadvantage of B2 in females (when tm > tf; figure 6c). LD at equilibrium is greater in sperm than eggs (Dy > Dx) when the direction of any of these asymmetries is reversed (figure 6).
4. Discussion
Two significant findings emerge from our models. First, stable LD is present at equilibrium between a locus segregating for parentally antagonistic alleles and a linked locus that segregates for either parentally or sexually antagonistic alleles. Second, the opportunity for parentally or sexually antagonistic polymorphism is expanded at loci linked to another locus that remains polymorphic owing to parental antagonism.
Prout [16] has noted that one-locus models show antagonistic selection to be a rather poor explanation for variation in nature, as the opportunity for polymorphism is appreciable only for rather strong selection. Correlated selection on linked loci favours polymorphism because it increases the ‘strength’ of selection relative to a single-locus model. In the limiting case of zero recombination, the two-locus model reduces to a single-locus model with two haplotypes (equivalent to alleles) maintained at a polymorphic equilibrium. The selection coefficients for these haplotypes combine the weaker selection coefficients of the two constituent loci and are therefore more conducive to the maintenance of polymorphism. As the recombination fraction grows, this ‘single-locus’ model with stronger selection grades into a model of weaker selection on two increasingly independent loci.
Our results can be understood intuitively. Both LD and the expanded opportunity for polymorphism arise because alleles at closely linked loci have correlated recent histories. Consider the pair of alleles at a parentally antagonistic locus. The paternally beneficial allele will have spent more of its history as a patrigene (i.e. a paternally derived gene) and the maternally beneficial allele more as a matrigene (i.e. a maternally derived gene). As a necessary corollary, the paternally beneficial allele will have spent more of its history in male bodies, and the maternally beneficial allele will have spent more of its history in female bodies. Paternally beneficial alleles will therefore develop statistical associations with alleles at closely linked loci that are also paternally beneficial or male beneficial (and maternally beneficial alleles with alleles that are maternally beneficial or female beneficial). Conditions for sexually antagonistic and parentally antagonistic polymorphism are relaxed at closely linked loci because each allele spends more of its history in the environment in which it is favoured than it would have spent if the loci were unlinked but subject to the same selective forces. Therefore, our findings can be understood because alleles at closely linked loci have correlated recent histories. Recombination diminishes the correlation in recent selective history of alleles at linked loci.
The ultimate source of the association between sexually and parentally antagonistic loci is a subtle form of admixture [8,17,18]. Sperm are enriched for male-beneficial alleles. Males collectively are more likely to obtain male-beneficial alleles from their fathers than from their mothers because of sexual antagonism in the parental generation. Therefore, male-beneficial alleles preferentially occur on paternally derived haplotypes that will be enriched for paternally beneficial alleles at parentally antagonistic loci. Conversely, eggs are enriched for female-beneficial alleles that preferentially occur on maternally derived haplotypes that will be enriched for maternally beneficial alleles. These correlations arise because individuals are more likely to inherit beneficial sexually antagonistic alleles from their paternal grandfather and maternal grandmother than from their maternal grandfather and paternal grandmother [19]. Úbeda et al. [8] showed that LD between sexually antagonistic loci is formally equivalent to that arising from population admixture where the two populations are male and female bodies. This admixture is recurrent because eggs and sperm unite in each sexual generation. Similarly, in this paper, LD arises between parentally antagonistic loci because matrigenes and patrigenes of mothers are mixed together as matrigenes of offspring, and matrigenes and patrigenes of fathers are mixed together as patrigenes of offspring.
Unlike the result that two sexually antagonistic loci exhibit stable LD even when there is free recombination [8], here we find that free recombination eliminates LD in models with one or two parentally antagonistic loci. The reason for this difference is that the sources of LD in two sexually antagonistic loci are (i) their historic association in haplotypes, measured by Dx and Dy, and (ii) their association in the immediately preceding generation, measured by Covζ(p,q) = 1/2(px – py)(qx – qy) (equation (2.3b)) [8]. With free recombination between two sexually antagonistic loci, the LD in the haploid phase is eliminated (Dx = Dy = 0), but not the LD in the diploid phase, which remains positive, whereas selection is sex-specific and px ≠ py and qx ≠ qy. In the absence of sex-specific selection at both loci or at one locus (our model 1 and model 2, respectively), the allelic frequency is the same in both gametes for at least one of the loci. The covariance term of equation (2.3b) thus becomes zero with free recombination and LD in the diploid phase vanishes.
Stable LD in our models resembles stable LD observed in single-sex models of two heterotic loci [20,21]. However, we did not find LD between a heterotic locus and a locus experiencing either sexual or parental antagonism (not shown). For this reason, we favour the interpretation that the LD in our model results from the correlation of recent selective histories and admixture rather than from heterosis.
The work presented here is theoretically reminiscent of the work of others. Immler et al. [22] modelled the interaction between ploidally antagonistic selection and sex-specific selection at a single locus in both haploid and diploid stages of the life cycle. They showed that the simultaneous strengthening of selection in favour of an allele in haploid males and against the same allele in diploid females (or vice versa, with the sexes reversed) was conducive for the maintenance of polymorphism. Immler et al. called this a negative ploidy-by-sex interaction. Another way to say the same thing is that polymorphism is favoured when the direction of selective forces aligns in haploid and diploid individuals of the same sex. The similarity to our model becomes clear: the haploid-sex-specific selection in Immler et al.'s study is analogous to our parental-origin-specific selection [23]. Therefore, their one-locus model agrees with our two-locus model in predicting that associations between diploid male-beneficial effects and (haploid) patriline-beneficial effects and diploid female-beneficial effects and (haploid) matriline-beneficial effects will enhance the opportunity for polymorphism.
Our models predict the formation of haplotypes that accumulate paternally beneficial effects and haplotypes that accumulate maternally beneficial effects. This finding may be relevant to understanding the evolution of the unexplained arrangement of imprinted genes in clusters [24]. Similarly, our previous models [8,9] predict the formation of haplotypes that accumulate male-beneficial effects and haplotypes that accumulate female-beneficial effects. This finding may be relevant to understanding the evolution of sex chromosomes [25]. Our models show that more sexually antagonistic and parentally antagonistic variation in fitness can be maintained on these haplotypes than would be possible if the loci were unlinked. Care should be taken not to overstate the scope of these results, however. For all of them, their magnitude depends on the strength of selection and the tightness of linkage. One imagines that in certain edges and corners of our parameter space the influence of stochastic forces will swamp the more subtle effects of natural selection that we focused on.
In addition to their relevance to an understanding of gene clusters, our results might also contribute to an enhanced understanding of the evolution of sex-specific recombination rates [26,27]. Previous models to explain sex-specific recombination rates require certain of the properties of our model and results, namely haploid (or pseudo-haploid) selection pressure and sex-specific LD.
The models of this paper, and of our previous papers on sexually antagonistic loci [8,9], predict stable polymorphisms of multi-locus haplotypes. At such equilibria, there are high-fitness haplotypes that combine paternally beneficial with male-beneficial alleles and maternally beneficial with female-beneficial alleles. The other haplotypes represent a genetic load generated by recombination between these high-fitness haplotypes. There is substantial genetic variation in fitness at equilibrium because of (i) this recombinational load, and (ii) both high-fitness haplotypes are equally likely to be transmitted to male or female offspring and thus half of the time experience the ‘wrong’ environment.
Acknowledgements
We thank Richard Harrison, Jeremy Van Cleve and an anonymous reviewer for helpful comments on the work presented here.
References
- 1.Owen ARG. 1953. A genetical system admitting of two distinct stable equilibria under natural selection. Heredity 7, 97–102 (doi:10.1038/hdy.1953.9) [Google Scholar]
- 2.Mande SPH. 1971. Owen's model of a genetical system with differential viability between sexes. Heredity 26, 49–63 (doi:10.1038/hdy.1971.5) [DOI] [PubMed] [Google Scholar]
- 3.Bonduriansky R, Chenoweth SF. 2009. Intralocus sexual conflict. Trends Ecol. Evol. 24, 280–288 (doi:10.1016/j.tree.2008.12.005) [DOI] [PubMed] [Google Scholar]
- 4.van Doorn GS. 2009. Intralocus sexual conflict. Ann. NY Acad. Sci. 1168, 52–71 (doi:10.1111/j.1749-6632.2009.04573.x) [DOI] [PubMed] [Google Scholar]
- 5.Kong A, et al. 2009. Parental origin of sequence variants associated with complex diseases. Nature 462, 868–874 (doi:10.1038/nature08625) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pearce GP, Spencer HG. 1992. Population genetic models of genomic imprinting. Genetics 130, 899–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Úbeda F, Haig D. 2004. Sex-specific meiotic drive and selection at an imprinted locus. Genetics 167, 2083–2095 (doi:10.1534/genetics.103.021303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Úbeda F, Haig D, Patten MM. 2011. Stable linkage disequilibrium owing to sexual antagonism. Proc. R. Soc. B 278, 855–862 (doi:10.1098/rspb.2010.1201) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patten MM, Haig D, Úbeda F. 2010. Fitness variation due to sexual antagonism and linkage disequilibrium. Evolution 64, 3638–3642 (doi:10.1111/j.1558-5646.2010.01100.x) [DOI] [PubMed] [Google Scholar]
- 10.Karlin S. 1975. General two-locus selection models: some objectives, results, and interpretations. Theor. Popul. Biol. 7, 364–398 (doi:10.1016/0040-5809(75)90025-8) [DOI] [PubMed] [Google Scholar]
- 11.Wade MJ, Winther RG, Agrawal AF, Goodnight CJ. 2001. Alternative definitions of epistasis: dependence and interaction. Trends Ecol. Evol. 16, 498–504 (doi:10.1016/S0169-5347(01)02213-3) [Google Scholar]
- 12.Lewontin RC, Kojima K. 1960. The evolutionary dynamics of complex polymorphisms. Evolution 14, 458–472 (doi:10.2307/2405995) [Google Scholar]
- 13.Fry JD. 2010. The genomic location of sexually antagonistic variation: some cautionary comments. Evolution 64, 1510–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kidwell JF, Clegg MT, Stewart FM, Prout T. 1977. Regions of stable equilibria for models of differential selection in two sexes under random mating. Genetics 85, 171–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Connallon T, Clark AG. 2012. A general population genetic framework for antagonistic selection that accounts for demography and recurrent mutation. Genetics 190, 1477–1489 (doi:10.1534/genetics.111.137117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prout T. 2000. How well does opposing selection maintain variation? In Evolutionary genetics: from molecules to morphology (eds Singh RS, Krimbas CB.), pp. 157–181 Cambridge, UK: Cambridge University Press [Google Scholar]
- 17.Nei M, Li WH. 1973. Linkage disequilibrium in subdivided populations. Genetics 75, 213–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li WH, Nei M. 1974. Linkage disequilibrium without epistasis in a subdivided population. Theor. Popul. Biol. 6, 173–183 (doi:10.1016/0040-5809(74)90022-7) [DOI] [PubMed] [Google Scholar]
- 19.Day T, Bonduriansky R. 2004. Intralocus sexual conflict can drive the evolution of genomic imprinting. Genetics 167, 1537–1546 (doi:10.1534/genetics.103.026211) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bodmer WF, Felsenstein J. 1967. Linkage and selection: theoretical analysis of the deterministic two locus random mating model. Genetics 57, 237–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karlin S, Feldman MW. 1978. Simultaneous stability of D = 0 and D ≠ 0 for multiplicative viabilities at two loci. Genetics 90, 813–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Immler S, Arnqvist G, Otto SP. 2012. Ploidally antagonistic selection maintains stable genetic polymorphism. Evolution 66, 55–65 (doi:10.1111/j.1558-5646.2011.01399.x) [DOI] [PubMed] [Google Scholar]
- 23.Haig D, Wilczek A. 2006. Sexual conflict and the alternation of haploid and diploid generations. Phil. Trans. R. Soc. B 361, 335–343 (doi:10.1098/rstb.2005.1794) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edwards CA, Ferguson Smith AC. 2007. Mechanisms regulating imprinted genes in clusters. Curr. Opin. Cell. Biol. 19, 281–289 (doi:10.1016/j.ceb.2007.04.013) [DOI] [PubMed] [Google Scholar]
- 25.Qiu S, Bergero R, Charlesworth D. 2013. Testing for the footprint of sexually antagonistic polymorphisms in the pseudo-autosomal region of a plant sex chromosome pair. Genetics 194, 663–672 (doi:10.1534/genetics.113.152397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lenormand T, Dutheil J. 2005. Recombination difference between the sexes: a role for haploid selection. PLoS Biol. 3, e63 (doi:10.1371/journal.pbio.0030063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brandvain Y, Coop G. 2012. Scrambling eggs: meiotic drive and the evolution of female recombination rates. Genetics 190, 709–723 (doi:10.1534/genetics.111.136721) [DOI] [PMC free article] [PubMed] [Google Scholar]