Abstract
Since its description from the Galapagos Rift in the mid-1980s, Archinome rosacea has been recorded at hydrothermal vents in the Pacific, Atlantic and Indian Oceans. Only recently was a second species described from the Pacific Antarctic Ridge. We inferred the identities and evolutionary relationships of Archinome representatives sampled from across the hydrothermal vent range of the genus, which is now extended to cold methane seeps. Species delimitation using mitochondrial cytochrome c oxidase subunit I (COI) recovered up to six lineages, whereas concatenated datasets (COI, 16S, 28S and ITS1) supported only four or five of these as clades. Morphological approaches alone were inconclusive to verify the identities of species owing to the lack of discrete diagnostic characters. We recognize five Archinome species, with three that are new to science. The new species, designated based on molecular evidence alone, include: Archinome levinae n. sp., which occurs at both vents and seeps in the east Pacific, Archinome tethyana n. sp., which inhabits Atlantic vents and Archinome jasoni n. sp., also present in the Atlantic, and whose distribution extends to the Indian and southwest Pacific Oceans. Biogeographic connections between vents and seeps are highlighted, as are potential evolutionary links among populations from vent fields located in the east Pacific and Atlantic Oceans, and Atlantic and Indian Oceans; the latter presented for the first time.
Keywords: deep sea, hydrothermal vents, cold methane seeps, cryptic species, polychaete
1. Introduction
It has been more than three decades since the discovery of deep ocean chemosynthetic communities. Over 600 animal species have been described from these habitats, mainly from hydrothermal vents near active tectonic plate boundaries, as well as from hydrocarbon seeps along continental margins [1–3]. Biodiversity patterns among deep-sea chemosynthetic fauna have been discussed at length in the context of taxonomic and environmental affinities leading to the designation of various biogeographic ‘provinces’ [1,3–6]. The few rigorous studies that have inferred these patterns in a phylogenetic context and on a broad scale [7–11] have focused on Pacific Ocean taxa [8,12–15]. Deep ocean currents, plate tectonics, seafloor spreading rates, oxygen levels, bathymetry, larval dispersal capabilities and sulfide or methane-rich communities, such as sunken wood and whale falls, as potential evolutionary ‘stepping stones’, are just some of the extrinsic factors that have been posited to drive species distributions in deep ocean chemosynthetic habitats [1,15–17].
Significant effort has been put forth in characterizing the faunal communities of these dynamic ecosystems. Traditional taxonomy, which emphasizes the characterization of morphological diversity, cannot always account for other biological attributes, such as developmental [18] and ecological adaptations [7,19,20], leading to over or underestimates of diversity [17,21]. Molecular systematics has been a useful tool to provide a testable framework to infer evolutionary relationships of genetic lineages, independent of phenotypic, ontogenetic and ecological variation. The integration of molecular data has greatly improved our knowledge of species delimitations and distributions, however with the caveat that taxonomic, genetic and geographical diversity estimates are all sensitive to sampling [22].
Annelids account for approximately 20% (approx. 111 species) of the named hydrothermal vent animal species [2]. The East Pacific Rise (EPR) has among the best-studied vent annelids [23–30] and the incorporation of molecular data has shed light on cryptic diversity found along this system [12,14,21,31,32]. The giant vestimentiferan tubeworm, Riftia pachyptila, is a dominant feature of hydrothermal vent sites along the EPR and was shown to be genetically homogeneous across a broad range (27° N–32° S), with a genetic break identified at the Easter microplate (approx. 26° S) [14]. The thermally tolerant Alvinella pompejana is known only from the EPR and although morphologically similar across a distance of approximately 5000 km (21° N–32° S), mitochondrial (mt) data revealed a north/south genetic break [14,33]. Species of Alvinella and Riftia are restricted to the east Pacific, whereas Paralvinella is amphi-Pacific, though so far not recorded outside of this ocean [2,34]. Major annelid clades are represented on a broad geographical scale throughout diverse chemosynthetic environments (e.g. Siboglinidae and Polynoidae), but among vent animals, only two ‘species’ have been recorded on a global scale: the ampharetid Amphisamytha galapagensis [8,35] and the amphinomid Archinome rosacea [36,37]; the latter being the focus of this study, while the former is now known to be a species complex [8].
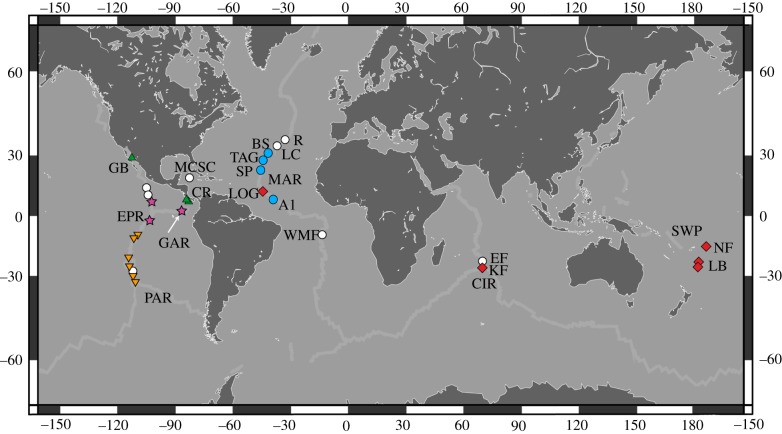
Amphinomids are best represented by the stinging fireworms (e.g. Eurythoe and Hermodice), which are common inhabitants of tropical reef environments [38,39]. Archinome rosacea was the first amphinomid described from chemosynthetic habitats from the original 1979 collections from Rose Garden, located at the Galapagos Rift (GAR; 0° N; 2400 m) in the eastern Pacific [36]. Since its description in 1985, Archinome has been recorded across major spreading centres in the Pacific, Atlantic and Indian Oceans (figure 1) [2,40]. Archinome specimens (figure 2 and electronic supplementary material, figure S1) are easily recognizable among vent fauna, with prominent calcareous, bifurcate (forked) chaetae, an elongate trilobed caruncle (figure 2b,c), a fusiform (spindle-like) body shape, prominent mid-ventral muscular scutes (figure 2g) and can range in size from just a few millimetres to several centimetres. In 2006, the distribution of A. rosacea was restricted to the GAR and the northeast Pacific Rise (NEPR) [2], in contrast to earlier accounts, which proposed a more widespread range including the Guaymas Basin (GB) sedimented vents, Mid-Atlantic Ridge (MAR) and Central Indian Ridge (CIR) vent systems [41,42]. Referencing unpublished data (J. Kudenov 2006), Desbruyères et al. [2] suggested the presence of at least three additional species, yet until recently A. rosacea remained the only named species. In 2009, Archinome storchi [40] was described from the Pacific Antarctic Ridge (PAR, 37° S). Also until recently, Archinome had only been recorded from hydrothermal vents. In 2009 and 2010, specimens were collected from cold methane seeps located at the Costa Rica margin (CRM) [43]. Archinome has been collected from a broad range of vent localities (figure 1) and depths (1000–3500 m) [40], however it is now known to occur at depths greater than 4000 m, including Ashadze-1 (A1; 12° N, MAR; 4080 m) [44].
Figure 1.
Distribution map of Archinome species. Symbols indicate all known records, with sites sampled for this study denoted by triangles (A. levinae n. sp.), stars (A. rosacea), inverted triangles (A. storchi), circles (A. tethyana n. sp.), diamonds (A. jasoni n. sp.) and open circles (unsampled records). A1, Ashadze-1; BS, Broken Spur; CIR, Central Indian Ridge; CRM, Costa Rica Margin; EF, Edmund Field; EPR, East Pacific Rise; GAR, Galapagos Rift; GB, Guaymas Basin; KF, Kairei Field; LOG, Logatchev; LB, Lau Basins (KLM and TML); LC, Lost City; MAR, Mid-Atlantic Ridge; MCSC, Mid-Cayman Spreading Center; PAR, Pacific Antarctic Ridge; R, Rainbow; SP, Snake Pit; SWP, southwest Pacific basins; TAG, TAG; WMF, Wideawake Mussel Field. (Online version in colour.)
Figure 2.
Archinome species. (a) (Live) whole body, dorsal view of A. levinae n. sp. (SIO-BIC A1316); (b) (live) dorsal view of anterior body segments of A. levinae n. sp. (SIO-BIC A1398; CRM, 9° N); (c) (live) dorsal view of anterior body segments of A. levinae n. sp. (SIO-BIC A1316); (d) (preserved) frontal view of A. jasoni n. sp. (SIO-BIC A2313; CIR); (e) (preserved) dorsal view of anterior body segments of A. jasoni n. sp. (SIO-BIC A2313); (f) (preserved) whole body, dorso-lateral view of A. jasoni n. sp. (SIO-BIC A2313); (g) (live) whole body, ventral view of A. jasoni n. sp. (KML); (h) (live) whole body, dorsal view of A. jasoni n. sp. (KML); (i) (live) dorsal view of A. storchi (PAR). Note within species variation in caruncle length and size for A. levinae n. sp. and A. jasoni n. sp. Scale bars, 1 mm. a, Anus; an, antennae; ac, accessory dorsal cirrus; br, branchia; c, caruncle; ch, chaetae; dc, dorsal cirrus; ma, median antenna; mvs, mid-ventral scutes; vc, ventral cirrus; numbers denote segments. (Online version in colour.)
Given Archinome's broad distribution and uncertainty as to the number of species within the genus, we used an integrative systematic approach to: (i) infer the identities of Archinome specimens from across the ‘cosmopolitan’ range among vent systems; (ii) infer the evolutionary relationships among vent and seep Archinome and (iii) explore the biogeographic links and diversification patterns across the Atlantic, Indian and Pacific Oceans.
2. Material and methods
(a). Sample collection
Archinome samples were collected using remotely operated vehicles including Woods Hole Oceanographic Institution's (WHOI) Jason I (R/V Knorr) and Jason II (R/V Melville), Monterey Bay Aquarium Research Institute's Tiburon (R/V Western Flyer) and Institut Français de Recherche pour l'Exploitation de la Mer's (IFREMER) Victor 6000 (R/V Pourquoi Pas?), and human occupied vehicles Alvin (WHOI) and Nautile (IFREMER) during deep-sea expeditions between 1990 through 2010. Figure 1 shows known records and sampling localities from vent and seep communities included in this study. Specimens were sampled from among larger vent fauna such as Vestimentifera and mytilid bivalves, as well as from upper sediment layer samples obtained from suction samplers and mesh scoops. Specimens were sorted aboard research vessels and when possible relaxed in a 50 : 50 (7% MgCl2: seawater) MgCl2 solution, followed by preservation in 10% formalin, then transferred to 70% ethanol for morphological evaluation and 80–95% ethanol or stored at −80°C for molecular work. Molecular samples were kept cold at 4°C or frozen at −80°C or −20°C. Collection and voucher information and details regarding evaluation of morphology can be found in the electronic supplementary material, text and tables S1, S4 and S5. Most specimens are lodged at the Scripps Institution of Oceanography Benthic Invertebrate Collection (SIO-BIC).
(b). Gene data collection, phylogenetic methods and genetic structure
Protocols for whole genomic DNA extraction, amplification and sequencing procedures are as reported by Borda et al. [45], unless stated otherwise. Electronic supplementary material, table S2 lists primers and annealing temperature profiles used for amplification of mt cytochrome c oxidase subunit I (COI), and mt 16S rDNA (16S). Amplification protocols for the nuclear internal transcribed spacer 1 (ITS1) and 28S rDNA (28S) followed Nygren & Pleijel [46] and Borda et al. [45], respectively. All data were analysed using maximum-likelihood (ML) and Bayesian inference (BI) procedures following methods described in [45], as was the choice of outgroup to root the analyses (i.e. Chloeia viridis). Notopygos ornata was included as an additional outgroup taxon based on hypothesized affinities associated with body shape and branchial morphology [37,45]. Phylogenetic trees (figure 3) are based on the BI topology, unless stated otherwise (see electronic supplementary material, figures S3 and S4), with support values (i.e. ML bootstrap (boot); posterior probabilities (pp)) indicated at nodes. Haplotype networks were generated for combined COI + 16S using TCS v. 1.21 [47], based on maximum parsimony and with a 95% probability (14-step connection limit) and fixed step connection limits ranging 10–50; gaps were treated as missing data. GenBank (16S, COI: JX027992–JX028115; 28S: JX028121–JX028141; ITS: KF288935–KF288959) and voucher accession numbers are provided in the electronic supplementary material, table S1. See also the electronic supplementary material, text for extended phylogenetic methods and sequence evaluation criteria.
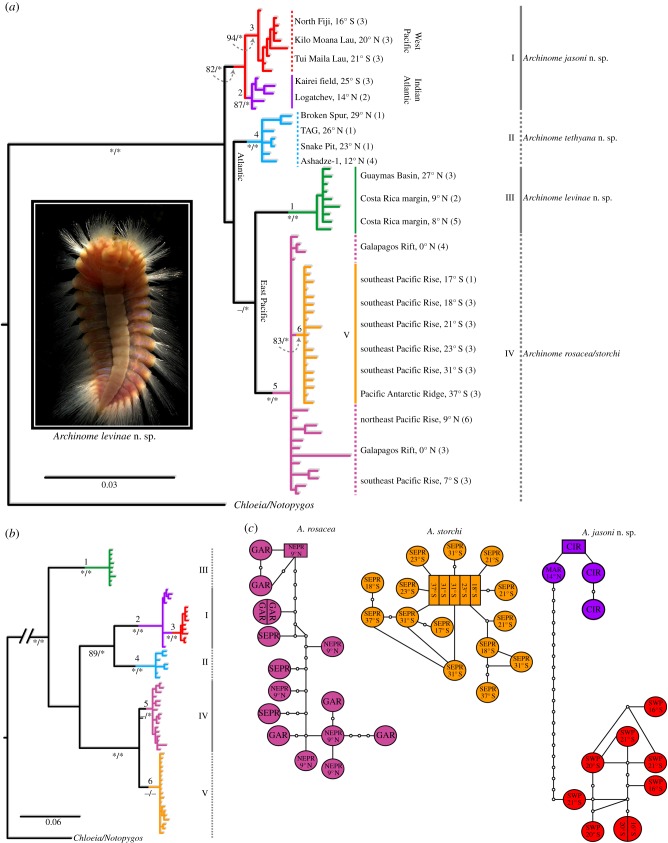
Figure 3.
Phylogeny (BI topology shown) and genetic diversity of Archinome species. (a) COIno3rd + 16S + 28S + ITS1; (b) COIall + 16S + 28S + ITS1. Roman numerals specify species clades; numerals 1–6 (above nodes) correspond to clades recovered in (b); ML bootstrap and BI posterior probabilities (boot/pp) shown below nodes; asterisk (*) denotes boot > 90% and pp > 0.95; values below 80% denoted by a dash ‘–’. (c) COIall + 16S statistical parsimony haplotype networks (fixed 21-step connection limit) for A. rosacea, A. storchi and A. jasoni n. sp. Shaded circles and rectangles are scaled to size according to number of individuals per haplotype. Two or more names indicate shared haplotypes. Small open circles represent unsampled haplotypes. (Online version in colour.)
3. Results
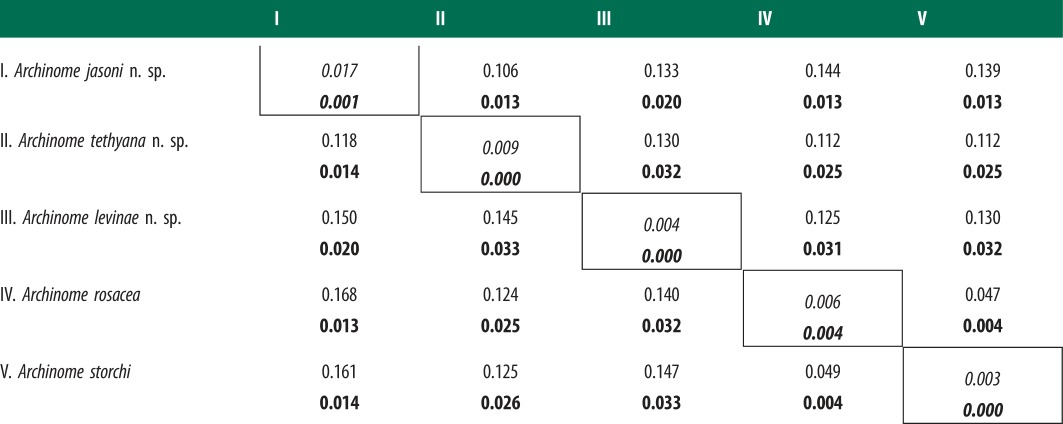
We inferred the phylogenetic relationships of Archinome specimens from COI (59 sequences; approx. 654 bp), 16S (65 sequences; approx. 472 bp), 28S (21 sequences; approx. 966 bp) and ITS1 (25 sequences; 572 bp). Table 1 provides mean intraclade and interclade TrN corrected and uncorrected pairwise distances for complete COI (dCOI) and ITS1 (dITS). COI exhibited the highest genetic divergences among clade terminals with the majority of synonymous changes occurring in third codon positions. COI saturation plots (see electronic supplementary material, figure S2) indicated that third position transitions reached saturation after approximately 13% sequence divergence. First and second codon position transitions and first through third codon position transversions were not saturated (results not shown). Interclade relationships and species identification were evaluated with the inclusion (COIall) and exclusion (COIno3rd) of COI third codon positions in combined analyses with 16S, 28S and ITS1 (figure 3). Results from individual and mt gene analyses can be found in the electronic supplementary material, figures S3 and S4. Mean COI interclade-corrected genetic distances were 12.5%, ranging 2.7–18.3%, and mean intraclade-corrected genetic distances was 0.5%, ranging 0–1.1%. ITS1 exhibited low divergences in comparison to COI. The highest corrected genetic pairwise distance was 3.6%. Mean ITS1 interclade-corrected genetic distance was 1.8%, ranging 1.0–3.6%, and mean intraclade-corrected genetic distance was 0.1%, ranging 0–1.0% (see table 1 and electronic supplementary material, table S3). Refer to the electronic supplementary material text for results regarding morphological evaluation.
Table 1.
Archinome pairwise distances. Mean Tamura Nei (TrN; below diagonal) and uncorrected (above diagonal) interclade and intraclade (TrN; italics along diagonal) pairwise distances for COI and ITS1 (bold).
 |
The phylogenetic relationships among Archinome species accepted here are based on COIno3rd + 16S + 28S + ITS (figure 3a). The data supported four Archinome clades, I–IV, of which three are regarded as new species and described in the electronic supplementary material, text. Numerical clades 1–6 above nodes correspond to those recovered in the analyses of concatenated COIall + 16S + 28S + ITS1 (figure 3b; see also the electronic supplementary material, figure S3A). Clade I (boot/pp = 82/0.94; dCOI = 1.7%), hereafter Archinome jasoni n. sp., included the southwest (SW) Pacific vent specimens (clade 3; boot/pp = 94/1.0; dCOI = 0.5%) from North Fiji (NF; 16° S; 1985 m), Kilo Moana Lau (KML; 20° S; 2650 m) and Tui Malila Lau (TML; 21° S; 1900 m) and clade 2 (boot/pp = 87/1.0; dCOI = 0.3%), which included specimens from Logatchev (LOG; 14° N, MAR, 3038 m) and Kairei field (25° S, CIR, 2432 m). Archinome jasoni n. sp. was supported as sister to the remaining Archinome species (boot/pp = 100/0.98). The highest A. jasoni n. sp. dCOI was 3.6% between specimens from NF/KML and LOG. The lowest interclade dCOI was 11.8% (CIR, clade 2) with Clade II (boot/pp = 100/1.0); hereafter, Archinome tethyana n. sp. The A. tethyana n. sp. clade included the northern MAR specimens (clade 4; boot/pp = 99/1.0). Sequence data for all four genes were available for A1 (MAR) specimens; only three representative 16S sequences (see electronic supplementary material, figure S3B) were available from Broken Spur (29° N; 3056 m), TAG (26° N; 3655 m) and Snake Pit (23° N; 3660 m). Clade III (clade 1; boot/pp = 98/1.0; mean dCOI = 0.4%), hereafter, Archinome levinae n. sp., included specimens from GB vents (27° N; approx. 2400 m) and CRM seeps (8–9° N; 1000–1800 m). The lowest interclade dCOI was 14.0% (with Clade IV). Archinome levinae n. sp. was sister to Clade IV (boot/pp = 98/1.0; dCOI = 2.7%), representing A. rosacea and A. storchi (Clade V) from the GAR, EPR and PAR (clades 5 and 6; figure 3b). Clade 5 (dCOI = 0.6%) included A. rosacea from GAR, as well as specimens from EPR 9° N (2500 m) and 7° S (2700 m). Clade 6 (dCOI = 0.3%; boot/pp = 83/1.0) comprised PAR specimens and those sampled northward along the southeast Pacific Rise (SEPR) from 31° S to 17° S (2200–2500 m). Clade 6 was a subclade nested among unresolved A. rosacea representatives (see also the electronic supplementary material, figures S3B and S4A). The highest dCOI was 5.0%, between representatives from the GAR (A. rosacea) and 17° S (A. storchi). The lowest interclade dCOI was 12.4%, between A. tethyana n. sp. and A. rosacea. The positions of A. tethyana n. sp. and A. levinae n. sp. received low (boot/pp = 52/0.78) to moderate support (boot/pp = 74/1.0), respectively.
Evaluation of concatenated COIall + 16S + 28S + ITS1 (figure 3b) supported that Archinome comprised five clades showing minimal geographical overlap. The resulting topology was similar to that of COIall (see electronic supplementary material, figures S3A and S2B), with the exception that A. jasoni n. sp. clade 3 was nested within clade 2, instead of showing reciprocal monophyly (figure 3a). The topology deviated from that observed in figure 3a, in that vent/seep A. levinae n. sp. was the sister group to the remaining Archinome species and reciprocally monophyletic (boot/pp = 95/1.0) A. rosacea (boot/pp = 77/0.66) and A. storchi (boot/pp = 75/1.0) clades were recovered; each clade with low support, however. Combined COIall + 16S data (n = 35) supported distinct networks (even with a fixed 50 step connection limit) for A. rosacea (n = 16) and A. storchi (n = 19), each containing 15 haplotypes. A single haplotype was shared between two A. rosacea individuals (GAR), while one haplotype was shared among five A. storchi individuals from the SEPR (figure 3c). No haplotypes were shared among A. rosacea (7° S) and A. storchi (17° S) individuals found approximately 1200 km apart. A single network (figure 3c; fixed 21-step limit connection), covering approximately 25 000 km distance, was recovered for A. jasoni (n = 13), with 12 haplotypes, of which one was shared between two individuals from SW Pacific basin (16° S, 20° S).
4. Discussion
(a). Delineation of cryptic species in the deep sea
Accounts of cryptic species in the marine realm are no longer new phenomena. Molecular phylogenies often deviate from those relying on traditional taxonomic tools and continue to reveal cryptic diversity [7,21,38,48]. In the deep sea, morphological stasis may not coincide with speciation events owing to stabilizing selection driven by extreme abiotic factors (e.g. low dissolved oxygen, low temperatures and darkness), in turn, introducing challenges in biodiversity estimates [21,49]. In recent years, mtDNA has been a primary tool for the detection of cryptic species [7,50], although the approach remains controversial [51–54], and can be sensitive to sampling [55]. As such, integrative taxonomic approaches (e.g. multi-locus datasets) are recommended [21,56,57]. Morphological taxonomic approaches (e.g. light microscopy, SEM) alone did not allow conclusive identification of new species, as sampling comprised individuals varying in size and exhibiting variable and/or overlapping morphologies, within and among clades (figure 2 and electronic supplementary material, table S5). Future work based on larger sample sizes and consideration of size-related variation, may reveal species-specific characters. Based on the currently available material, we designate new Archinome species on the basis of molecular evidence alone (see also [58]).
Our approach for estimating Archinome species diversity was to include broad geographical sampling and to use a multi-locus framework (figure 3). We recognize that our sampling exhibits large geographical gaps (figure 1) leaving an incomplete picture of species distributions. Our phylogenetic hypothesis for Archinome as a whole (figure 3a) required the exclusion of COI third codon position (owing to saturation), resulting in a conflicting topology when the third position was considered (figure 3b). The designation of A. levinae n. sp. and A. tethyana n. sp. was unambiguous, however, this was less so for the remaining species. In particular, A. rosacea appeared to be paraphyletic with respect to A. storchi (figure 3a). However, COI was not saturated at more restricted levels, and when the third codon position was included, it became clear that both species were reciprocally monophyletic (figure 3b). Furthermore, these two clades were disparate enough not to form a single haplotype network (figure 3c) and showed a nearly 5% COI divergence. Although we did not find clear morphological differences between A. rosacea and A. storchi in terms of the argued diagnostic features [40] (figure 2i; for further discussion, see the electronic supplementary material, table S5), we accept both as distinct species. On the same criteria, A. jasoni n. sp. was best left as a broadly distributed species (figure 3a–c), despite vast distances separating LOG, CIR and SW Pacific vent populations. COI sequence divergences were less than 4%, with no shared haplotypes. Given this low genetic divergence, the absence of clear morphological distinction and variable size classes among A. jasoni n. sp. populations (figure 2d–f), we do not have sufficient evidence to designate them as separate species at this time. We recognize the presence of two, possibly three lineages, as A. jasoni n. sp., which only further sampling will be able to resolve.
(b). Distribution and diversification of Archinome across chemosynthetic systems
The diversification of Archinome appears to align (in part) with Moalic et al.'s [5] hypothesis, which proposed west Pacific vent fauna as ‘ancestral’ and ‘central’ to those found elsewhere. Our phylogenetic hypothesis deviated with respect to identifying potential links between the Atlantic and eastern Pacific seep/vent communities. However, the biogeographic roles of cold seeps and the Mid-Cayman Spreading Center (MCSC) [59], for example, were not considered in their study. Archinome jasoni n. sp. was the sister taxon to the remaining species and included one clade that was exclusive to the SW Pacific basins. Although taxonomic affinities between the CIR and west Pacific have previously been reported [6,42], only a handful of phylogenetic studies have included CIR fauna, and none have evaluated annelids prior to this study. Archinome jasoni n. sp. also included a CIR–LOG clade. Van Dover et al. [42] proposed CIR as a mid-point for faunal exchange between the Atlantic and west Pacific along the southwest and southeast Indian Ridges, respectively. This scenario appears to be consistent with the presence of A. jasoni n. sp. in both regions.
High rates of gene flow and low genetic variation have been reported for Rimicaris vent shrimp from 36° N to 4° S [60–64]. Zelnio & Hourdez [64] found west Pacific Chorocaris vandoverae as sister to Rimicaris exoculata + Chorocaris chacei (MAR); however, the phylogenetic placement of CIR Rimicaris kairei has not yet been inferred. The gastropod, Alviniconcha hessleri, reportedly occurs in the west Pacific and Indian Oceans [42], however A. aff. hessleri (CIR) was genetically distinct from its west Pacific counterpart, yet clustered among west Pacific Alviniconcha sp. Type 2 [65,66]. A CIR + SW Pacific clade has also been reported for Bathymodiolus mussels, showing little sequence divergences among them [10,11]. Low genetic divergences were also observed among CIR and SW Pacific A. jasoni n. sp., and the inclusion of MAR samples now corroborates previously reported affinities among Atlantic, Indian and western Pacific Ocean fauna [5,42]. Unlike widespread R. exoculata, we recovered two species in the MAR. However, our limited sampling could have missed the co-occurrence of A. jasoni n. sp. and A. tethyana n. sp. Alternatively, their colonizing routes leading to A1 and LOG might be significantly separate, and they may never be found in sympatry. Only more extensive sampling will be able to clarify this.
Biogeographic links between the Atlantic and east Pacific were proposed by Van Dover et al. [3] and were also observed here in the sister group relationship between the Atlantic A. tethyana n. sp. and the eastern Pacific species. Atlantic/east Pacific affinities have been shown for several annelid taxa [1,8,67] pointing towards a former connection between both oceans via a deep ocean passage [68] prior to the closure of the Isthmus of Panama. Recent discoveries of MCSC vent fauna suggest affinities with MAR fauna [59,69], including a new Rimicaris species [69] and Archinome spp. (A. Glover 2013, personal communication). Although A. tethyana n. sp. was sister to the east Pacific clades, its position was not highly supported. This could be attributed to missing data for northern MAR specimens and/or unsampled representatives from intermediate geographical regions (e.g. MCSC; to be evaluated elsewhere).
The diversification of A. rosacea, A. storchi and A. levinae n. sp. is likely attributed to vicariant events involving a formerly widespread ancestor that became isolated from the Atlantic; the latter possibly coincident with the rise of the Central American (CA) Isthmus (approx. 15 Ma; [68]) and subsequent tectonic shifts and subduction events of the Pacific, Cocos and Nazca Plates. The continental margin distribution of A. levinae n. sp. may be associated with vicariance coincident with the rise of the CA Isthmus and the formation of the Gulf of California in the Late Miocene (less than 8 Ma; [70,71]). Although records are few, species shared between GB and CRM have previously been reported [7,8], and now include A. levinae n. sp. Archinome samples from cold seeps at the GB (27°34′ N, 111°27′ W) were not available for this study, though we suspect A. levinae n. sp. may be found there given comparable depths (approx. 1700 m) and being located a mere 50 km north from the GB vent communities [72]. Hydrothermal vents at GB are particular with seeping fluids that circulate through thick sediment layers [73]. The presence of A. levinae n. sp. nearly 4000 km south at methane seeps of the CRM suggests either long distance dispersal capacity of larvae or perhaps the presence of overlooked chemosynthetic environments along the CA margin. Genetic isolation between A. levinae n. sp. and A. rosacea/A. storchi may have been caused by the formation of the deep Middle American Trench [70] having served as a dispersal barrier to vent populations at GAR (approx. 1000 km south) and the EPR. The genetic break between 7° S and 17° S (SEPR), as seen between A. rosacea and A. storchi, may be due to the sampling gap [22] or the result of vicariance associated with the formation and rotation of the Bauer microplate (between 10° and 15° S) in the Miocene [74]. This event has been proposed to have disrupted vent communities and flow of ocean currents along the SEPR, potentially restricting gene flow from more northerly populations (e.g. 7° S; [15]). Compared to other EPR taxa, Bathymodiolus, Lepetodrilus and Alvinella, appear to conform to this trend, whereas species distributions of Amphisamytha, Branchipolynoe, Hesiolyra, Riftia and Tevnia appear to be less constrained across this presumed dispersal barrier [8,14,15].
5. Conclusion
We evaluated the phylogeny of Archinome from chemosynthetic environments on a global scale to redefine the geographical distribution of A. rosacea and A. storchi, the former of which had been unclear, and revealed the presence of three previously undescribed cryptic species. Among these, A. levinae n. sp., inhabiting both vent and methane seep sites found 4000 km apart and A. jasoni n. sp., which for the first time potentially supports biogeographic links among Atlantic, Indian and Pacific Ocean vent systems. With the inclusion of representatives from poorly sampled chemosynthetic sites, in particular CIR and cold seep communities, we hope this study will provide a framework for continued elucidation of the diversification and evolution among deep-sea invertebrate species from chemosynthetic environments.
Acknowledgements
We thank the captains and crews of the R/V Atlantis, R/V Western Flyer, R/V Knorr, R/V Melville, R/V L'Atalante, R/V Pourquoi-Pas? and the pilots of ROVs Tiburon, ROVs Jason I and Jason II, and Victor 6000 and the pilots and crews of HOVs Alvin and Nautile for their technical support. Special thanks to Bob Vrijenhoek, chief scientist of the cruises to the SEPR (2005) and SWP (2005), for inviting G.W.R., F.P. and N.G.W. aboard. We are grateful to Didier Jollivet, chief scientist of the BioSpeedo 2004 cruise (R/V L'Atalante, HOV Nautile), Yves Fouquet, chief scientist of the Serpentine 2007 cruise (R/V Pourquoi Pas? ROV Victor 6000) and to Lisa Levin, chief scientist of the CRROCKS cruises (2009; 2010). Thanks also to Bob Vrijenhoek, Rich Lutz, Didier Jollivet and Cindy Van Dover for providing specimens from GB, MAR and CIR. We also thank Dieter Fiege and Gordon Bock for initial discussions on the morphology of Archinome. Harim Cha kindly accessioned the material into the SIO-BIC.
Funding statement
Financial support for this study was provided by NSF DBI-0706856, Census of Marine Life TAWNI, SSB Mini-PEET and EOL Rubenstein Fellowship (E.B.), with additional support from DBI-1036186 (A.S.) and OCE-1029160 (G.W.R.).
References
- 1.Tunnicliffe V, McArthur AG, McHugh D. 1998. A biogeographical perspective of the deep-sea hydrothermal vent fauna. Adv. Mar. Biol. 34, 353–442 (doi:10.1016/S0065-2881(08)60213-8) [Google Scholar]
- 2.Desbruyères D, Segonzac M, Bright M. 2006. Handbook of deep-sea hydrothermal vent fauna. Linz, Austria: Denisia 18 [Google Scholar]
- 3.Van Dover CL, German CR, Speer KG, Parson LM, Vrijenhoek RC. 2002. Evolution and biogeography of deep-sea vent and seep invertebrates. Science 295, 1253–1257 (doi:10.1126/science.1067361) [DOI] [PubMed] [Google Scholar]
- 4.Rogers AD, et al. 2012. The discovery of new deep-sea hydrothermal vent communities in the southern ocean and implications for biogeography. PLoS Biol. 10, e1001234 (doi:10.1371/journal.pbio.1001234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moalic Y, Desbruyères D, Duarte CM, Rozenfeld AF, Bachraty C, Arnaud-Haond S. 2011. Biogeography revisited with network theory: retracing the history of hydrothermal vent communities. Syst. Biol. 61, 127–137 (doi:10.1093/sysbio/syr088) [DOI] [PubMed] [Google Scholar]
- 6.Bachraty C, Legendre P, Desbruyères D. 2008. Biogeographic relationships among deep-sea hydrothermal vent faunas at global scale. Deep-Sea Res. 56, 1371–1378 (doi:10.1016/j.dsr.2009.01.009) [Google Scholar]
- 7.Johnson SB, Waren A, Vrijenhoek RC. 2008. DNA barcoding of Lepetodrilus limpets reveals cryptic species. J. Shellfish Res. 27, 43–51 (doi:10.2983/0730-8000) [Google Scholar]
- 8.Stiller J, Rousset V, Pleijel F, Chevaldonné P, Vrijenhoek RC, Rouse GW. 2013. Phylogeny, biogeography and systematics of hydrothermal vent and methane seep Amphsamytha (Ampharetidae, Annelida), with descriptions of three new species. Syst. Biodivers. 11, 35–65 (doi:10.1080/14772000.2013.772925) [Google Scholar]
- 9.Gollner S, Fontaneto D, Martinez Arbizu P. 2011. Molecular taxonomy confirms morphological classification of deep-sea hydrothermal vent copepods (Dirivultidae) and suggests broad physiological tolerance of species and frequent dispersal along ridges. Mar. Biol. 158, 221–231 (doi:10.1007/s00227-010-1553-y) [Google Scholar]
- 10.Kyuno A, Shintaku M, Fujita Y, Matsumoto H, Utsumi M, Watanabe H, Fujiwara Y, Miyazaki J. 2009. Dispersal and differentiation of deep-sea mussels of the genus Bathymodiolus (Mytilidae, Bathymodiolinae). J. Mar. Biol. 2009, 625672 (doi:10.1155/2009/625672) [Google Scholar]
- 11.Miyazaki J, de Oliveira ML, Fujita Y, Matsumoto H, Fujiwara Y. 2010. Evolutionary process of deep-sea Bathymodiolus mussels. PLoS ONE 5, e10363 (doi:10.1371/journal.pone.0010363) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chevaldonné P, Jollivet D, Desbruyères D, Lutz RA, Vrijenhoek RC. 2002. Sister-species of eastern Pacific hydrothermal vent worms (Ampharetidae, Alvinellidae, Vestimentifera) provide new mitochondrial COI clock calibration. Cah. Biol. Mar. 43, 367–370 [Google Scholar]
- 13.Tyler PA, German CR, Ramirez-Llodra E, Van Dover CL. 2002. Understanding the biogeography of chemosynthetic ecosystems. Oceanol. Acta 25, 227–241 (doi:10.1016/S0399-1784(02)01202-1) [Google Scholar]
- 14.Hurtado LA, Lutz RA, Vrijenhoek RC. 2004. Distinct patterns of genetic differentiation among annelids of eastern Pacific hydrothermal vents. Mol. Ecol. 13, 2603–2615 (doi:10.1111/j.1365-294X.2004.02287.x) [DOI] [PubMed] [Google Scholar]
- 15.Plouviez S, Shank TM, Faure B, Daguin-Thiebaut C, Viard F, Lallier FH, Jollivet D. 2009. Comparative phylogeography among hydrothermal vent species along the East Pacific Rise reveals vicariant processes and population expansion in the South. Mol. Ecol. 18, 3903–3917 (doi:10.1111/j.1365-294X.2009.04325.x) [DOI] [PubMed] [Google Scholar]
- 16.Tyler PA, Young CM. 1999. Reproduction and dispersal at vents and cold seeps. J. Mar. Biol. Assoc. UK 79, 193–208 (doi:10.1017/S0025315499000235) [Google Scholar]
- 17.Vrijenhoek RC. 2010. Genetic diversity and connectivity of deep-sea hydrothermal vent metapopulations. Mol. Ecol. 19, 4391–4411 (doi:10.1111/j.1365-294X.2010.04789.x) [DOI] [PubMed] [Google Scholar]
- 18.Shank TM, Lutz RA, Vrijenhoek RC. 1998. Molecular systematics of shrimp (Decapoda: Bresiliidae) from deep-sea hydrothermal vents, I: enigmatic ‘small orange’ shrimp from the Mid-Atlantic Ridge are juvenile Rimicaris exoculata. Mol. Mar. Biol. Biotechnol. 7, 88–96 [PubMed] [Google Scholar]
- 19.Goffredi SK, Hurtado LA, Hallam S, Vrijenhoek RC. 2003. Evolutionary relationships of deep-sea vent and cold seep clams (Mollusca: Vesicomyidae) of the ‘pacifica/lepta‘ species complex. Mar. Biol. 142, 311–320 (doi:10.1007/s00227-002-0941-3) [Google Scholar]
- 20.Johnson SB, Young CR, Jones WJ, Waren A, Vrijenhoek RC. 2006. Migration, isolation, and speciation of hydrothermal vent limpets (Gastropoda; Lepetodrilidae) across the Blanco Transform Fault. Biol. Bull. 210, 140–157 (doi:10.2307/4134603) [DOI] [PubMed] [Google Scholar]
- 21.Vrijenhoek RC. 2009. Cryptic species, phenotypic plasticity, and complex life history: assessing deep-sea fauna diversity with molecular markers. Deep-Sea Res. 56, 1713–1723 (doi:10.1016/j.dsr2.2009.05.016) [Google Scholar]
- 22.Audzijonyte A, Vrijenhoek RC. 2010. When gaps really are gaps: statistical phylogeography of hydrothermal vent invertebrates. Evolution 64, 2369–2384 (doi:10.1111/j.1558-5646.2010.00987.x) [DOI] [PubMed] [Google Scholar]
- 23.Cavanaugh CM, Gardiner SL, Jones ML, Jannasch HW, Waterbury JB. 1981. Prokaryotic cells in the hydrothermal vent tube worm Riftia pachyptila Jones: possible chemoautotrophic symbionts. Science 213, 340–342 (doi:10.1126/science.213.4505.340) [DOI] [PubMed] [Google Scholar]
- 24.Marsh AG, Mullineaux LS, Young CM, Manahan DT. 2001. Larval dispersal potential of the tubeworm Riftia pachyptila at deep-sea hydrothermal vents. Nature 411, 77–80 (doi:10.1038/35075063) [DOI] [PubMed] [Google Scholar]
- 25.Fisher CR, et al. 1988. Physiology, morphology, and biochemical composition of Riftia pachyptila at Rose Garden in 1985. Deep-Sea Res. 35, 1745–1758 (doi:10.1016/0198-0149(88)90047-7) [Google Scholar]
- 26.Fisher CR, Childress JJ, Sanders NK. 1988. The role of vestimentiferan hemoglobin in providing an environment suitable for chemoautotrophic sulfide-oxidizing endosymbionts. Symbiosis 5, 229–246 [Google Scholar]
- 27.Grzymski JJ, et al. 2008. Metagenome analysis of an extreme microbial symbiosis reveals eurythermal adaptation and metabolic flexibility. Proc. Natl Acad. Sci. USA 105, 17 516–17 521 (doi:10.1073/pnas.0802782105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Bris N. 2007. How does the annelid Alvinella pompejana deal with an extreme hydrothermal environment? Rev. Environ. Sci. Biotech. 6, 197–221 (doi:10.1007/s11157-006-9112-1) [Google Scholar]
- 29.Pradillon F, Gaill F. 2007. Adaptation to deep-sea hydrothermal vents: some molecular and developmental aspects. J. Mar. Sci. Technol. 15, 37–53 [Google Scholar]
- 30.Gagnière N, et al. 2010. Insights into metazoan evolution from Alvinella pompejana cDNAs. BMC Genomics 11, 634 (doi:10.1186/1471-2164-11-634) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jollivet D, Desbruyeres D, Bonhomme F, Moraga D. 1995. Genetic differentiation of deep-sea hydrothermal vent alvinellid populations (Annelida: Polychaeta) along the East Pacific Rise. Heredity 74, 376–391 (doi:10.1038/hdy.1995.56) [Google Scholar]
- 32.Young CR, Fujio S, Vrijenhoek RC. 2008. Directional dispersal between mid-ocean ridges: deep-ocean circulation and gene flow in Ridgeia piscesae. Mol. Ecol. 17, 1718–1731 (doi:10.1111/j.1365-294X.2008.03609.x) [DOI] [PubMed] [Google Scholar]
- 33.Plouviez S, Le Guen D, Lecompte O, Lallier FH, Jollivet D. 2010. Determining gene flow and the influence of selection across the equatorial barrier of the East Pacific Rise in the tube-dwelling polychaete Alvinella pompejana. BMC Evol. Biol. 10, 220 (doi:10.1186/1471-2148-10-220) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rousset V, Rouse GW, Feral J-P, Desbruyeres D, Pleijel F. 2003. Molecular and morphological evidence of Alvinellidae relationships (Terebelliformia, Polychaeta, Annelida). Zool. Scr. 32, 185–197 (doi:10.1046/j.1463-6409.2003.00110.x) [Google Scholar]
- 35.Zottoli RA. 1983. Amphisamytha galapagensis, a new species of ampharetid polychaete from the vicinity of abyssal hydrothermal vents in the Galapagos Rift, and the role of this species in rift ecosystems. Proc. Biol. Soc. Wash. 96, 379–391 [Google Scholar]
- 36.Blake JA. 1985. Polychaeta from the vicinity of deep-sea geothermal vents in the eastern Pacific. 1. Euphrosinidae, Phyllodocidae, Hesionidae, Nereidae, Glyceridae, Dorvilleidae, Orbiniidae, and Maldanidae. Bull. Biol. Soc. Wash. 6, 67–101 [Google Scholar]
- 37.Kudenov JD. 1991. A new family and genus of the order Amphinomida (Polychaeta) from the Galapagos hydrothermal vents. In Proceedings of the 2nd International Polychaeta Conference (eds Petersen ME, Kirkegaard JB.), Copenhagen, 1986 Systematics, Biology and Morphology of World Polychaeta. Ophelia (Supplement) 5, 111–120 [Google Scholar]
- 38.Barroso R, Klautau M, Sole-Cava AM, Paiva PC. 2010. Eurythoe complanata (Polychaeta: Amphinomidae), the ‘cosmopolitan’ fireworm, consists of at least three cryptic species. Mar. Biol. 157, 69–80 (doi:10.1007/s00227-009-1296-9) [Google Scholar]
- 39.Arhens J, Borda E, Barroso R, Campbell AM, Wolf A, Nugues M, Paiva P, Rouse GW, Schulze A. 2013. The curious case of Hermodice carunculata (Annelida: Amphinomidae): genetic homogeneity throughout the Atlantic Ocean and adjacent basins. Mol. Ecol. 22, 2280–2291 (doi:10.1111/mec.12263) [DOI] [PubMed] [Google Scholar]
- 40.Fiege D, Bock G. 2009. A new species of Archinome (Polychaeta: Archinomidae) from hydrothermal vents on the Pacific Antarctic Ridge 37° S. J. Mar. Biol. Assoc. UK 89, 689–696 (doi:10.1017/S0025315409000174) [Google Scholar]
- 41.Desbruyères D, Segonzac M. (eds) 1997. Handbook of deep-sea hydrothermal vent fauna. Brest, France: IFREMER [Google Scholar]
- 42.Van Dover CL, et al. 2001. Biogeography and ecological setting of Indian Ocean hydrothermal vents. Science 294, 818–823 (doi:10.1126/science.1064574) [DOI] [PubMed] [Google Scholar]
- 43.Levin LA, et al. 2012. A hydrothermal seep on the Costa Rica margin: middle ground in a continuum of reducing ecosystems. Proc. R. Soc. B 279, 2580–2588 (doi:10.1098/rspb.2012.0205) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fabri MC, Bargain A, Briand P, Gebruk A, Fouquet Y, Morineaux M, Desbruyères D. 2011. Hydrothermal vent community of a new deep-sea field Ashadze-1, 12° 58′ N on the Mid-Atlantic Ridge and a comparison of all northern Atlantic chemosynthetic ecosystems. J. Mar. Biol. Assoc. UK 91, 1–13 (doi:10.1017/S0025315410000731) [Google Scholar]
- 45.Borda E, Kudenov JD, Beinhold C, Rouse GW. 2012. Towards a revised Amphinomidae (Annelida: Amphinomida): description and affinities of a new genus and species from the Nile Deep-sea Fan, Mediterranean Sea. Zool. Scr. 41, 307–325 (doi:10.1111/j.1463-6409.2012.00529.x) [Google Scholar]
- 46.Nygren A, Pleijel F. 2011. From one to ten in a single stroke: resolving the European Eumida sanguinea (Phyllodocidae, Annelida) species complex. Mol. Phylogenet. Evol. 58, 132–141 (doi:10.1016/j.ympev.2010.10.010) [DOI] [PubMed] [Google Scholar]
- 47.Clement M, Posada D, Crandall KA. 2000. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 4, 331–346 [DOI] [PubMed] [Google Scholar]
- 48.Knowlton N. 2000. Molecular genetic analyses of species boundaries in the sea. Hydrobiologia 420, 73–90 (doi:10.1023/A:1003933603879) [Google Scholar]
- 49.Bickford D, Lohman DJ, Sodhi NS, Ng PKL, Meier R, Winker K, Ingram KK, Das I. 2007. Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 22, 148–155 (doi:10.1016/j.tree.2006.11.004) [DOI] [PubMed] [Google Scholar]
- 50.Hebert PD, Penton EH, Burns JM, Janzen DH, Hallwachs W. 2004. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl Acad. Sci. USA 101, 14 812–14 817 (doi:10.1073/pnas.0406166101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meyer CP, Paulay G. 2005. DNA barcoding: error rates based on comprehensive sampling. PLoS Biol. 3, e422 (doi:10.1371/journal.pbio.0030422) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cameron S, Rubinoff D, Will K. 2006. Who will actually use DNA barcoding and what will it cost? Syst. Biol. 55, 844–847 (doi:10.1080/10635150600960079) [DOI] [PubMed] [Google Scholar]
- 53.Rubinoff D, Cameron S, Will K. 2006. A genomic perspective on the shortcomings of mitochondrial DNA for ‘barcoding’ identification. J. Hered. 97, 581–594 (doi:10.1093/jhered/esl036) [DOI] [PubMed] [Google Scholar]
- 54.Moritz C, Cicero C. 2004. DNA barcoding: promise and pitfalls. PLoS Biol. 2, e354 (doi:10.1371/journal.pbio.0020354) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wiemers M, Fiedler K. 2007. Does the DNA barcoding gap exist? A case study in blue butterflies (Lepidoptera: Lycaenidae). Front. Zool. 4, 8 (doi:10.1186/1742-9994-4-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Will KW, Mishler BD, Wheeler QD. 2005. The perils of DNA barcoding and the need for integrative taxonomy. Syst. Biol. 54, 844–851 (doi:10.1080/10635150500354878) [DOI] [PubMed] [Google Scholar]
- 57.Dasmahapatra KK, Elias M, Hill RI, Hoffman JI, Mallet J. 2010. Mitochondrial DNA barcoding detects some species that are real, and some that are not. Mol. Ecol. Resour. 10, 264–273 (doi:10.1111/j.1755-0998.2009.02763.x) [DOI] [PubMed] [Google Scholar]
- 58.Cook LG, Edwards RD, Crisp MD, Hardy NB. 2010. Need morphology always be required for new species descriptions? Invert. Syst. 24, 322–326 (doi:10.1071/IS10011) [Google Scholar]
- 59.Connelly DP, et al. 2012. Hydrothermal vent fields and chemosynthetic biota on the world's deepest seafloor spreading centre. Nat. Commun. 3, 620 (doi:10.1038/ncomms1636) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williams AB, Rona PA. 1986. Two new caridean shrimps (Bresiliidae) from a hydrothermal field on the Mid-Atlantic Ridge. J. Crus. Biol. 6, 446–462 (doi:10.2307/1548184) [Google Scholar]
- 61.Shank TM, Black MB, Halanych KM, Lutz RA, Vrijenhoek RC. 1999. Miocene radiation of deep-sea hydrothermal vent shrimp (Caridea: Bresiliidae): evidence from mitochondrial cytochrome oxidase subunit I. Mol. Phylogenet. Evol. 13, 244–254 (doi:10.1006/mpev.1999.0642) [DOI] [PubMed] [Google Scholar]
- 62.Teixeira S, Serrão EA, Arnaud-Haond S. 2012. Panmixia in a fragmented and unstable environment: the hydrothermal shrimp Rimicaris exoculata disperses extensively along the Mid-Atlantic Ridge. PLoS ONE 7, e38521 (doi:10.1371/journal.pone.0038521) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Teixeira S. 2010. Recent population expansion and connectivity in the hydrothermal shrimp Rimicaris exoculata along the Mid-Atlantic Ridge. J. Biogeogr. 38, 564–574 (doi:10.1111/j.1365-2699.2010.02408.x) [Google Scholar]
- 64.Zelnio KA, Hourdez S. 2009. A new species of Alvinocaris (Crustacea: Decapoda: Caridea: Alvinocarididae) from hydrothermal vents at the Lau Basin, southwest Pacific, and a key to the species of Alvinocarididae. Proc. Biol. Soc. Wash. 122, 52–71 (doi:10.2988/07-28.1) [Google Scholar]
- 65.Kojima S, Fujikura K, Okutani T, Hashimoto J. 2004. Phylogenetic relationship of Alviniconcha gastropods from the Indian Ocean to those from the Pacific Ocean (Mollusca: Provannidae) revealed by nucleotide sequences of mitochondrial DNA. Venus 63, 65–68 [Google Scholar]
- 66.Suzuki Y, et al. 2006. Host–symbiont relationships in hydrothermal vent gastropods of the genus Alviniconcha from the southwest Pacific. Appl. Environ. Microb. 72, 1388–1393 (doi:10.1128/AEM.72.2.1388-1393.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Black MB, Halanych KM, Maas PAY, Hoeh WR, Hashimoto J, Desbruyères D, Lutz RA, Vrijenhoek RC. 1997. Molecular systematics of vestimentiferan tubeworms from hydrothermal vents and cold-water seeps. Mar. Biol. 130, 141–149 (doi:10.1007/s002270050233) [Google Scholar]
- 68.Burton KW, Ling H-F, O'Nions RK. 1997. Closure of the Central American Isthmus and its effect on deep-water formation in the North Atlantic. Nature 386, 382–385 (doi:10.1038/386382a0) [Google Scholar]
- 69.Nye V, Copley J, Plouviez S. 2011. A new species of Rimicaris (Crustacea: Decapoda: Caridea: Alvinocarididae) from hydrothermal vent fields on the Mid-Cayman Spreading Centre, Caribbean. J. Mar. Biol. Assoc. UK 92, 1057–1072 (doi:10.1017/S0025315411002001) [Google Scholar]
- 70.Helenes J, Carreno AL, Carrillo RM. 2009. Middle to Late Miocene chronostratigraphy and development of the northern Gulf of California. Mar. Micropaleontol. 72, 10–25 (doi:10.1016/j.marmicro.2009.02.003) [Google Scholar]
- 71.Fisher RL. 1961. Middle America trench; topography and structure. Geol. Soc. Am. Bull. 72, 703–719 (doi:10.1130/0016-7606(1961)72[703:MATTAS]2.0.CO;2) [Google Scholar]
- 72.Simoneit BRT, Lonsdale PF, Edmond JM, Shanks WC., III 1990. Deep-water hydrocarbon seeps in Guaymas Basin, Gulf of California. Appl. Geochem. 5, 41–49 (doi:10.1016/0883-2927(90)90034-3) [Google Scholar]
- 73.Lonsdale P, Becker K. 1985. Hydrothermal plumes, hot springs, and conductive heat flow in the Southern Trough of Guaymas basin. Earth Planet. Sci. Lett. 73, 211–225 (doi:10.1016/0012-821X(85)90070-6) [Google Scholar]
- 74.Eakins BW, Lonsdale PF. 2003. Structural patterns and tectonic history of the Bauer microplate, eastern tropical Pacific. Mar. Geophys. Res. 24, 171–205 (doi:10.1007/s11001-004-5882-4) [Google Scholar]