Abstract
Taxonomic identification of pollen and spores uses inherently qualitative descriptions of morphology. Consequently, identifications are restricted to categories that can be reliably classified by multiple analysts, resulting in the coarse taxonomic resolution of the pollen and spore record. Grass pollen represents an archetypal example; it is not routinely identified below family level. To address this issue, we developed quantitative morphometric methods to characterize surface ornamentation and classify grass pollen grains. This produces a means of quantifying morphological features that are traditionally described qualitatively. We used scanning electron microscopy to image 240 specimens of pollen from 12 species within the grass family (Poaceae). We classified these species by developing algorithmic features that quantify the size and density of sculptural elements on the pollen surface, and measure the complexity of the ornamentation they form. These features yielded a classification accuracy of 77.5%. In comparison, a texture descriptor based on modelling the statistical distribution of brightness values in image patches yielded a classification accuracy of 85.8%, and seven human subjects achieved accuracies between 68.33 and 81.67%. The algorithmic features we developed directly relate to biologically meaningful features of grass pollen morphology, and could facilitate direct interpretation of unsupervised classification results from fossil material.
Keywords: palynology, Poaceae, microscopy, pattern analysis, computational image analysis
1. Introduction
The taxonomic identification of pollen and spores, in common with many other biological sciences that rely on morphological comparison, uses inherently qualitative descriptors of shape and ornamentation. As a result, identifications are restricted to taxonomic groupings that can be reliably classified by multiple analysts and subtle morphological differences are often ignored. A consequence of this conservative approach is the coarse taxonomic resolution of the pollen and spore record [1]. The development of intuitive, numerical measures of shape and ornamentation would directly address these limitations of pollen and spore identification, and allow researchers to translate morphological differences observed under the microscope into quantitative, repeatable measurements. Treating morphology as measurement, rather than description, allows a broader range of observations to be incorporated into the analysis and identification of pollen and spores.
Grass pollen presents a classic demonstration of the taxonomic limits of current analytical approaches. The grass family (Poaceae) is an exceptionally successful group of plants, and can be found in a wide range of habitats from the tropics to the arctic. However, although Poaceae contains more than 11 000 recognized species [2] and its genetic diversity is visible in the diverse morphology of grass flowers [3], the gross morphology of grass pollen is remarkably similar throughout the family. The pollen is generally spheroidal with a single pore surrounded by an annulus [4,5]. This simple morphology has led researchers to suggest that pollen morphology is ‘uniform’ within the family [6] and has little to contribute to the reconstruction of grass evolution and diversification. Instead, direct evidence for the palaeoecology and evolutionary history of grasses has been provided mostly by other fossil groups such as phytoliths (microscopic silica bodies formed in plant tissues) [6,7].
Yet, owing to their high abundance in terrestrial and marine sediments, and standardized protocols that allow relative abundances of different plant groups to be directly compared through time, pollen grains provide a potentially rich source of information on the evolutionary history of grasses. As a result, palynologists have attempted to increase the taxonomic precision of grass pollen by measuring characters such as pollen grain length, grain width, pore diameter and annulus width [8–11], and by noting differences in the organization of the grass pollen exine [5,9]. High-resolution scanning electron microscopy (SEM) studies have revealed a diversity of surface ornamentation patterns that may have taxonomic significance [5,12–16] but are not visible when viewed using traditional light microscopy. However, exine ornamentation has not been widely used to classify grass pollen because of the difficulty in comparing the relatively small differences in surface patterning [5,13,14].
In this paper, we classify 240 specimens of grass pollen from 12 species in three subfamilies within Poaceae (table 1) by quantifying the size and density of sculptural elements and the complexity of the surface ornamentation that they form. In doing so, we develop a means of quantifying morphological features that have traditionally been described solely in qualitative terms. Our results provide a potential solution to the problem of classifying grass pollen, and demonstrate how computational image analysis, combined with high-resolution microscopy, holds the potential to dramatically increase the taxonomic resolution of pollen and spore records of Earth's vegetation [18,19].
Table 1.
Classification and specimen information for the 12 species of grass investigated here. Subfamily classification of grasses follows [17]. ‘Group’ refers to the four morphological clusters created by sculptural element size and density (figure 2).
| subfamily | genus and species | species code | group |
|---|---|---|---|
| Pooideae | Anthoxanthum odoratum | 1 | 1 |
| Dactylis glomerata | 2 | 2 | |
| Phalaris arundinacea | 3 | 1 | |
| Poa australis | 4 | 1 | |
| Stipa tenuifolia | 5 | 4 | |
| Chloridoideae | Cynodon dactylon | 6 | 3 |
| Eragrostis mexicana | 7 | 3 | |
| Sporobolus pyramidalis | 8 | 2 | |
| Triodia basedowii | 9 | 1 | |
| Panicoideae | Bothriochloa intermedia | 10 | 1 |
| Digitaria insularis | 11 | 3 | |
| Oplismenus hirtellus | 12 | 2 |
2. Scanning electron microscopy image acquisition
Grass pollen from the 12 species was prepared for SEM imaging using standard palynological methods (see the electronic supplementary information). Twenty grains of each species were imaged at ×2000, ×6000 and ×12 000 magnification (see the electronic supplementary information, dataset S1). Analyses were undertaken on 400 × 400 pixel windows that were manually cropped from the ×6000 images (see the electronic supplementary information, dataset S2). In these images, 1 pixel measures 16.(6) nm. SEM images of all grass pollen analysed in this paper can be found at https://www.ideals.illinois.edu/handle/2142/43358.
3. Grouping morphologically similar species
(a). Quantifying sculptural element size and density
SEM revealed that the surface ornamentation of the 12 species is considerably diverse. Using terminology from descriptive palynology [20], some species are characterized by scabrate ornamentation consisting of granula, whereas others have more complex areolate ornamentation, with polygonal areas separated by grooves that form a negative reticulum. We first clustered the species into groups with similar patterns of surface ornamentation. To do this, we developed a scalar feature ι that represents the size of the sculptural elements on the surface of each specimen, and a feature ν that is related to the density of the granula on the surface of each specimen (figure 1).
Figure 1.
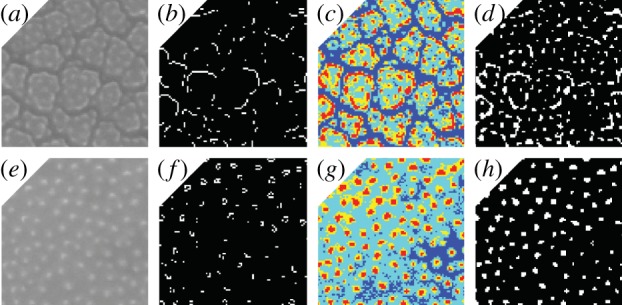
Thumbnails showing the image processing steps taken during the construction of features ι and ν. (a–d) show a species with areolae (Oplismenus hirtellus). (e–h) show a species with scabrate ornamentation consisting of granlua (Anthoxanthum odoratum). (a,e) show original ×6000 SEM images of each specimen cropped to 100 × 100 pixels. (b,f) show binary images with white pixels representing edges delineated by a Sobel edge detector (used to generate feature ι). (c,g) show quantized images with pixels clustered into four groups from low (dark blue) to high (red) intensity. (d,h) show binary images with pixels in the cluster of highest intensity shown as white pixels (used to generate feature ν).
The size of sculptural elements on the surface of each pollen grain was quantified from five windows measuring 100 × 100 pixels randomly chosen and cropped from each ×6000 SEM image of each specimen. Working with smaller windows decreased the influence of variations in the brightness and contrast of each image. A Sobel edge detector was applied to each 100 × 100 window, which turned each of these into a binary image with the white pixels representing the detected edges (figure 1b,f). The white pixels were then clustered by forming a graph with the white pixels as nodes and two nodes connected with an edge if one node fell within the 3 × 3 pixel neighbourhood of the other. The connected components of this graph were identified using a standard Dulmage–Mendelsohn decomposition of the adjacency matrix [21]. We selected the connected component C with the largest number of pixels and applied principal component analysis to the coordinates of the centres of the pixels in C. We quantified the size of C using the largest eigenvalue ιC > 0 of the covariance matrix, which may be interpreted as the variance along the first principal direction. We employed the average value of ιC over the five 100 × 100 pixel windows as a scalar feature ι that represents the size of the sculptural elements on the surface of each specimen.
The density of the sculptural elements was quantified using the same 100 × 100 pixel windows. Minimum variance quantization [22] was used to cluster the pixels in each window into four groups. We interpreted the pixels in the cluster H of highest intensity as the pixels that form the granula of the scabrate ornamentation on each pollen grain (figure 1c,g). We constructed a graph with the pixels in H as nodes, and used the same 3 × 3 pixel neighbouring relations employed in the analysis of sculptural element size to identify the connected components of H. We used the number of connected components νH to estimate the number of granula in each 100 × 100 pixel window, and used the average value ν over the five 100 × 100 windows as a feature related to the density of the granula on the surface of each specimen. To balance out the scales of ι and ν that are related to sculptural element size and density, respectively, we centred both features by subtracting their mean values and scaling them to have unit variance.
(b). Identification and validation of morphological groups
Using a scatterplot of features ι and ν as a guide, we clustered the 12 species into four groups (figure 2 and table 1). These four morphological groups were validated by a leave-one-out experiment with a k-nearest-neighbour classifier in which 228 of the 240 specimens were classified correctly (95% accuracy; we selected k = 9 based on classification performance). Group one contains five species that are characterized by granula spread relatively densely over the surface of the pollen grain and poorly defined areolae (figure 2). Group two contains three species that are characterized by areolae and granula spread relatively densely over the surface of the pollen grain (figure 2). Group three contains three species that are characterized by areolae and granula distributed relatively sparsely over the pollen surface (figure 2). The simple scabrate surface ornamentation of Stipa tenuifolia lacks areolae and has sparsely distributed granula. This surface ornamentation is sufficiently different to any of the other species that it clusters alone (figure 2).
Figure 2.
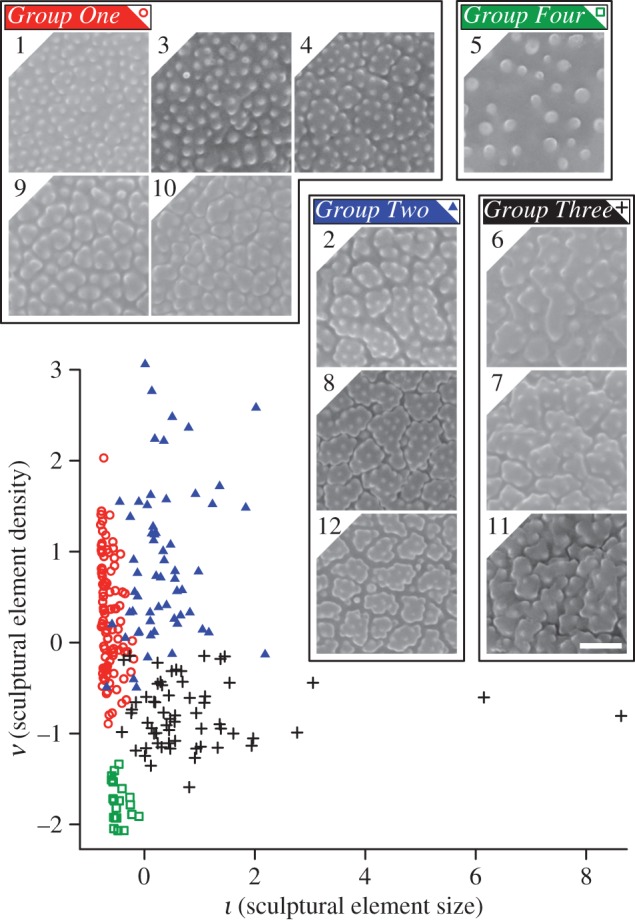
Pollen from the 12 species of grass investigated here clustered into four groups at 95% accuracy using features ι and ν. Thumbnails show the surface ornamentation of species within each group. Group one (red open circles) contains five species: A. odoratum (1), Phalaris arundinacea (3), Poa australis (4), Triodia basedowii (9) and Bothriochloa intermedia (10). Group two (blue solid triangles) contains three species: Dactylis glomerata (2), Sporobolus pyramidalis (8) and O. hirtellus (12). Group three (black plus signs) contains three species: Cynodon dactylon (6), Eragrostis mexicana (7) and Digitaria insularis (11). The two outliers are specimens 13 and 20 of E. mexicana. Group four (green open squares) contains a single species: S. tenuifolia (5). Images taken using SEM and shown at ×12 000 magnification. Scale bar measures 120 pixels and represents 1 µm. In these images, 1 pixel measures 8.(3) nm. Numeric species codes from table 1.
4. Classification of species within each morphological group
Next, we classified the species of grass contained within each of the four morphological groups. To do this, we approached the pixels in each image of a pollen grain as nodes in a network, with two nodes connected with an edge using a neighbouring relation. We treated the sculptural elements on the surface of a pollen grain as foreground objects (white pixels in figure 3c,f,i), and used the notion of network centrality to define two 20-dimensional features, τ1 and τ2, that provide measures of the complexity of the surface ornamentation on each pollen grain. Centrality here refers to the relative importance of the nodes of a network for its local–global connectivity [23].
Figure 3.
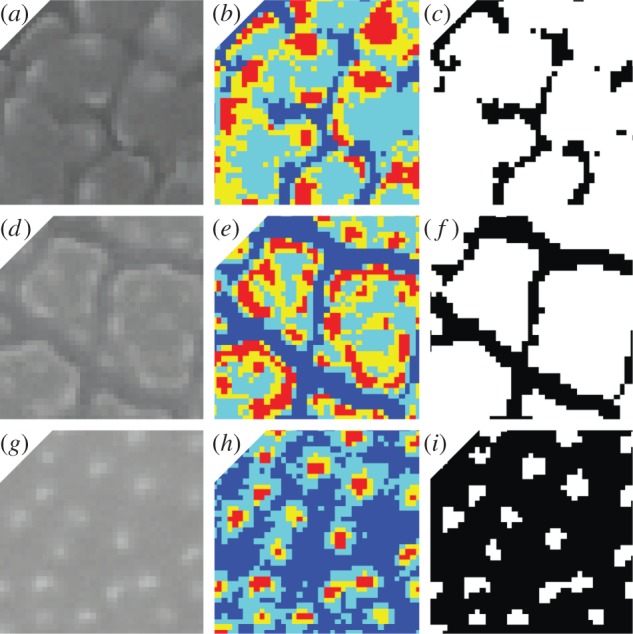
Thumbnails showing the image processing steps that were taken during the construction of features τ1 and τ2. (a–c) show a species with relatively narrow areolae (D. insularis). (d–f) show a species with relatively large areolae (O. hirtellus). (g–i) show a species with scabrate ornamentation consisting of granlua (A. odoratum). (a,d,g) show original ×6000 SEM images of each specimen cropped to 40 × 40 pixels. (b,e,h) show quantized images of each specimen with pixels clustered into four groups from low (dark blue) to high (red) intensity. (c,f) show a binary image with pixels in the cluster of lowest intensity as black pixels. (i) shows a binary image with pixels in the two clusters of lower intensity as black pixels. Binary images in (c,f,i) show black pixels as the background and white pixels as the foreground.
(a). Quantifying the complexity of grass pollen surface ornamentation and construction of features τ1 and τ2
Five windows measuring 40 × 40 pixels were randomly chosen and cropped from each ×6000 SEM image of each specimen [22] within these groups. Minimum variance quantization was used to cluster the pixels in each window into four groups and turn each window into a binary image. For species within group one (figure 2), the pixels in the two clusters of lower intensity were turned into black pixels (background) and those in the two clusters of higher intensity were turned into white pixels (foreground; figure 3i). For species within groups two and three (figure 2), only pixels in the cluster of lowest intensity were turned into black pixels (background) and those in the other three were turned into white pixels (foreground; figure 3c,f). Treating the groups in this way ensured that the primary ornamentation patterns of each species were represented as accurately as possible in the binary images.
We employed the notion of subgraph centrality (SC) in our analysis [23]. For unweighted networks, SC can be defined as follows. For a node v and a non-negative integer ℓ, let μℓ(v) denote the number of closed walks of length ℓ starting at v. Then, the centrality of v is defined as
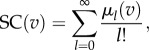 |
4.1 |
a weighted sum that attributes higher importance to shorter walks; that is, to the local connectivity near v. SC(v) can be computed in terms of the eigenvalues and eigenvectors of the adjacency matrix of the network as follows: let v1, … ,vn denote the nodes of the network and let η1, … ,ηn be an orthonormal basis of eigenvectors of the adjacency matrix with associated eigenvalues λ1, … ,λn. Then,
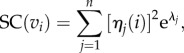 |
4.2 |
where ηj(i) is the ith entry of the vector ηj [23]. This form of SC generalizes to weighted networks, where the adjacency matrix is given by the weights wij. We rank the nodes of the network according to decreasing values of SC.
Feature τ1 is derived from a network having only the background (black) pixels as nodes, and two nodes connected with an edge if they are immediate neighbours to the north, south, east or west. SC was used to rank the pixels of this network. A sequence of 20 expanding subregions of the background was formed, starting with the pixels ranked in the top 5% and adding the next 5% until the entire background of black pixels was covered (figure 4a). For each of the subregions, the number of connected components was calculated, which were recorded in a 20-dimensional feature vector τW. As pixels are added to the shape, the number of connected components may increase or decrease and existing components may coalesce (figure 4a). The last component of the vector τW is the number of connected components of the background. This process was repeated using each of the five randomly cropped 40 × 40 pixel windows from each specimen. The average was taken to obtain a 20-dimensional feature vector τ1.
Figure 4.
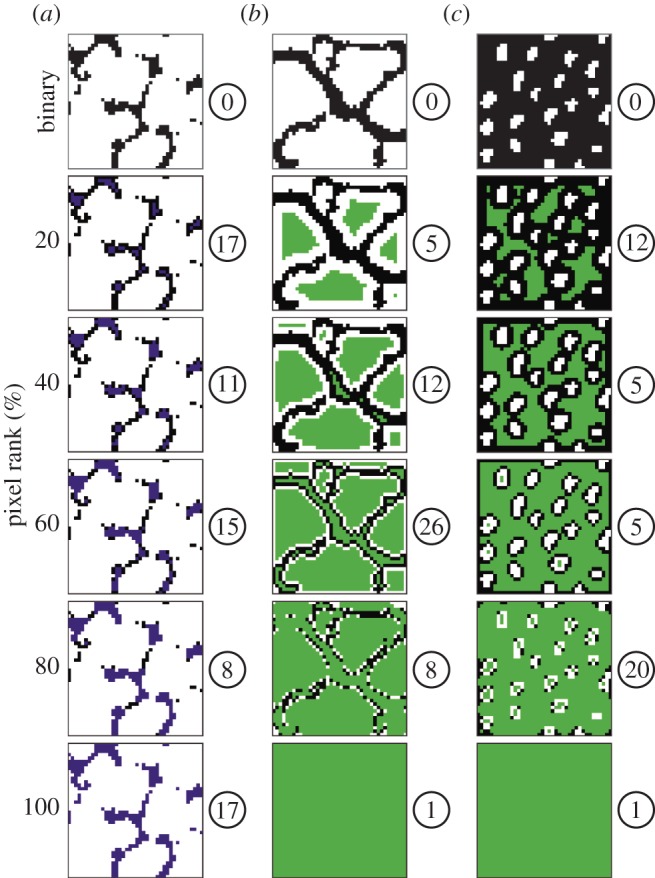
Thumbnails showing examples of expanding subregions and the number of connected components within them. Subregions are displayed as coloured pixels and shown in 20% increments. Examples of three species are shown, each from a different morphological group. (a) D. insularis (Group 3; figure 2 and table 1), (b) O. hirtellus (Group 2; figure 2 and table 1), (c) A. odoratum (Group 1; figure 2, table 1). Binary images at the top of each column correspond to the binary images in figure 3c,f,i, with black pixels as the background and white pixels as the foreground. (a) shows a sequence of expanding subregions within a network having only the background pixels as nodes, which was used to derive feature τ1. Pixels added to this network following SC computations are shown in purple. (b,c) show a sequence of expanding subregions within a weighted network having all pixels as nodes, which was used to derive feature τ2. Pixels added to this network following SC computations are shown in green. The number of connected components present in each subregion is shown in a white circle to the right of each image.
Feature τ2 is derived from a weighted network having all 1600 pixels of each 40 × 40 window as nodes, and two nodes connected with an edge using the same neighbouring relation as for feature τ1. Edges connecting two foreground (white) pixels or two background (black) pixels received weight 1, whereas background–foreground transition edges received weight α, 0 < α < 1. Experiments indicate that τ2 remains essentially unchanged for α ≤ 0.01, so we used α = 0.01 in all calculations. As for feature τ1, SC was used to rank the pixels of this network and a sequence of 20 expanding subregions of the 40 × 40 window was formed (figure 4b,c). The number of connected components was calculated at each stage of the sequence (figure 4b,c), which was recorded in a 20-dimensional feature vector. This process was repeated using each of the five randomly cropped 40 × 40 pixel windows from each specimen. The average was taken to obtain a 20-dimensional feature vector τ2.
(b). Species classification of grass pollen
To optimize the combination of ι (sculptural element size) and ν (sculptural element density) with the feature τ derived from centrality, we allow a scaling factor a > 0, and use the 22-dimensional feature vector ϒ = (aι, aν, τ) to classify species. For each group, we experimented with a range of values of a, and selected the value that yielded the highest classification accuracy. We employed either feature τ1 or feature τ2 depending on which feature yielded the highest classification accuracy. Feature vector ϒ was reduced to three dimensions using principal component analysis.
As S. tenuifolia clusters alone using features ι and ν, we only further subdivide species in groups one, two and three (figure 2). Classification of species within these groups was validated with a leave-one-out experiment using a k-nearest-neighbour classifier. The choice of k was based on classification performance. Within group one, 78 out of 100 specimens were classified correctly at the species level (78%) using feature τ2, a = 15.8 and k = 5. For this group, feature τ2 quantifies the complexity of the patterning formed by the granula on the pollen surface (figure 4c). (Typically, for complex patterning, the vector τ will have several larger values that reflect the presence of numerous connected components. In many cases, the values of τ also show an oscillatory behaviour indicating that many new connected components are created and existing ones get merged as more pixels are added to the count.) Within group two, 50 out of 60 specimens were classified correctly (83%) using feature τ2, a = 8 and k = 6. For this group, feature τ2 quantifies the complexity of the patterning formed by the negative reticulum and the areolae (figure 4b). Within group three, 50 out of 60 specimens were classified correctly (83%) using feature τ1, a = 5 and k = 3. For this group, feature τ1 quantifies the complexity of the patterning formed by the negative reticulum (figure 4a).
Our classification of the 12 species investigated is shown schematically as a tree in figure 5. At the first level, we use features ι and ν to cluster the 12 species into groups that have similar patterns of surface ornamentation (figure 5). At the second level, we use the 22-dimensional feature vector ϒ to classify species within groups one, two and three (figure 5). Classification errors are introduced at the first and second levels of the classification. A leave-one-out experiment with all 240 specimens yielded a classification accuracy of 77.5% (see the electronic supplementary material, figure S1). Given that leave-one-out cross-validation was used to estimate model parameters and the predictive performance of k-nearest-neighbour classifiers, the performance rates reported in the paper are subject to the general limitations of the cross-validation procedure, as estimates so obtained may exhibit considerable variation (cf. [24]).
Figure 5.
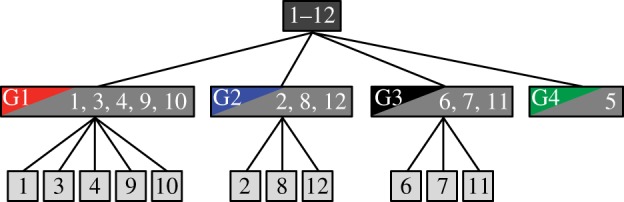
Tree showing the classification of the 12 species of grass investigated in this study. The uppermost dark grey box represents all 12 species. The medium grey boxes at the first level of the tree show the 12 species clustered into four groups, labelled G1–4, and colour coded as for figure 2. The light grey boxes at the second level of the tree show the classification of species within each cluster. See table 1 for species codes.
5. Comparison with an alternative texture descriptor and human subjects
As a baseline for the species-level classification accuracy of our approach, we evaluated a texture descriptor based on the modelled statistical distribution of brightness values in local image patches. This approach is common in computational image analysis and has been used previously for classifying materials [25] and detection of boundaries between textured regions [26].
Each cropped ×6000 SEM image of grass pollen grain was down-sampled to a resolution of 120 × 120. Each 5 × 5 pixel window centred at every pixel location within the image (excluding boundaries) was quantized by subtracting the mean brightness of the patch and assigning it to the closest element in a pre-computed dictionary of canonical appearances. We used a generic dictionary containing 75 elements learned from a set of consumer photographs by minimizing sparse reconstruction error (graciously provided by Ren & Bo [27]). The relative frequencies of these appearances were stored in a 75-bin histogram and left-out samples were classified using k-nearest-neighbour with an L1 distance (cityblock). k = 3 was selected by cross-validation. A histogram recording the frequencies of the different patch appearances in a sample image was used as the texture descriptor for the sample. k-nearest-neighbour classification using this histogram descriptor with k = 3 and leave-one-out cross-validation yielded a final species-level classification accuracy of 85.8% (see the electronic supplementary material, figure S2).
We next measured the ability of seven human subjects to classify the same SEM images of grass pollen that were used for our computational image analyses (see the electronic supplementary material, dataset S2). Each subject was provided with a reference library containing six images of each species of grass pollen, grouped and labelled by species. Each subject was then provided with a set of 120 unlabelled images of grass pollen containing 10 images of each of the 12 species. Two of these 10 images were duplicates, and one of these 10 images also appeared in the reference library. Images were classified by assigning each image to one of the 12 species. The images and classification key are provided in the electronic supplementary material, dataset S3.
The classification accuracy of the seven human subjects ranged from 68.33 to 81.67% (average 75.48%; figure 6). However, classification consistency between subjects was low; only 28.33% of the specimens were classified correctly by all seven subjects. This consistency falls to 21.82% when the visually distinctive S. tenuifolia is excluded from the analysis.
Figure 6.
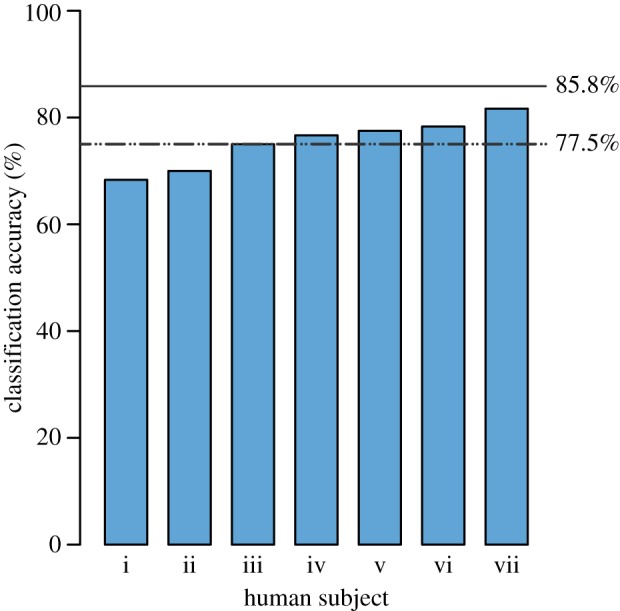
Bar chart showing the accuracy of seven human subjects asked to classify SEM images of grass pollen. Lower dashed line shows the accuracy of the algorithmic approach that we have developed in this paper (77.5%). Upper solid line shows the accuracy of a texture descriptor based on histograms of local quantized image patches (85.8%).
6. Discussion
Quantitative image-based analyses can enumerate the small, subtle and diverse morphological differences among taxa that may be observed by the expert, but cannot easily be conveyed by the terminology used to describe pollen and spores [18]. This has been a significant barrier to the classification of grass pollen [5,13,14]. For example, the patterns of grass pollen surface ornamentation revealed by SEM have been used to define morphotypes that contain many species of grass [13,14]. However, these morphotypes form a continuum without clearly defined boundaries between them [13], and this is partly responsible for their limited use in palynological studies of grass [5,13,14]. They include the Hordeum-type, Triticum-type, Avena-type and Setaria-type [14]. The Setaria-type, for example, ‘is characterized by extensive field-like [areolae] of irregularly polygonal outlines. Their bulging surface is studded with very small pointed spinules, in most cases, (3–)5–8(–10) [sic]’ ([14], p.139). Our analyses demonstrate how the morphological characters that are described by such terminology can be quantified and used to classify grass pollen.
We have attempted to develop and use features that can be directly related to biologically significant characteristics of grass pollen (figures 1, 3 and 4). For example, the clustering step in our classification uses features related to sculptural element size and density (figures 1 and 2). The four morphological groups that are produced are consistent with visual perception of surface ornamentation (figure 2) and this step is analogous to the description of grass pollen morphotypes [5,13,14], but using measurements of morphology rather than qualitative descriptions.
Our approach is rooted in the accurate description of morphology, and achieves classification results that are comparable to the results of manual human classification (figure 6) and a more conventional computational image analysis based on the distribution of brightness values in local image patches ([25,26]; see the electronic supplementary material, figures S1 and S2). These comparisons highlight that there are a variety of analytical techniques that could be used to classify grass pollen once sufficient morphological information has been recovered from individual specimens. However, in our experiments with human subjects, classification accuracy comes at the expense of consistency, highlighting that the taxonomic resolution of the pollen and spore record can be reduced by disagreement among multiple analysts [18,28]. The patch appearance histogram approach resulted in higher identification accuracies for 10 of the 12 species, but this increase in accuracy (approx. 8% on average) comes at the expense of interpretability. The histogram counts encode the image appearance in a distributed way and, in contrast to the features we developed in this paper (figures 1 and 4), are not easily understood in terms of basic morphological features of pollen grains. We anticipate that the use of features that can be directly related to biologically significant morphological characters will be critical in the interpretation of fossil samples, where extant reference specimens are unavailable, and all possible morphologies are not known. Interpretability is important in this context, as character-based features will produce unsupervised clusters that are more intuitively understood by palynological experts. This is not always true of the supervised learning approaches popular in automated pollen classification, which generally provide low interpretability of biological features [18,29].
Additionally, the development of quantitative measures of shape and ornamentation has wide applications in other branches of the biological sciences that rely on visual inspection for classification of phenotypic differences. The methods to quantify morphology that we have developed in this paper have immediate utility for the classification of other groups of organisms for which the patterns of surface ornamentation are an important taxonomic character, such as diatoms and ostracods, as well as the potential to quantify the complexity of other biological structures, such as venation in leaves and insect wings.
7. Conclusion
In this paper, we have used a combination of high-resolution microscopy and computational image analyses to classify 12 species of modern grass pollen. We have classified these species by developing features that quantify the size and density of sculptural elements (figure 1) and measure the complexity of the surface ornamentation that they form (figures 3 and 4). These features can be understood in terms of the basic morphological features of the grass pollen exine. In our experiments using this algorithmic method, 186 out of 240 specimens were classified correctly, yielding a classification accuracy of 77.5%. We also compared a baseline texture classification approach using histograms of local quantized image patches [25,26], which yielded an accuracy of 85.8% but provides low interpretability. Seven human subjects achieved classification accuracies between 68.33 and 81.67% (figure 6) on a subset of these images. However, classification consistency between subjects was low, and just 28.33% of the specimens were correctly classified by all subjects.
Our results support the view that a combination of high-resolution microscopy and computational image analyses can generate classifications at fine taxonomic levels that are beyond the capability of human experts [18,28]. This approach has the potential to dramatically increase the taxonomic resolution of pollen and spore records of ancient vegetation, which will in turn expand the range and depth of hypotheses that can be tested using the fossil record [18,19].
Acknowledgements
Michael Urban provided the specimens of grass pollen used in this paper. Margaret Collinson provided advice on the preparation of pollen grains for SEM. Claire Belcher, Sarah Baker, Derek Haselhorst, Jacqueline Rodriguez, Shivangi Tiwari and Cassandra Wesseln produced the human baseline data. We thank our two anonymous reviewers for their comments and Carlos Jaramillo for encouraging us to pursue the problem of grass pollen classification. This research was carried out in part in the Frederick Seitz Materials Laboratory Central Facilities, University of Illinois.
Data accessibility
SEM images of grass pollen and files used to test human classification performance: University of Illinois IDEALS digital archive (https://www.ideals.illinois.edu/handle/2142/43358).
Funding statement
We acknowledge funding from the National Science Foundation (DBI-1052997 to S.W.P., DBI-102942 to W.M. and DBI-1053036 to C.C.F.). L.M. was partly supported by a Marie Curie International Incoming Fellowship within the 7th European Community Framework Programme (PIIF-GA-2012-328245).
References
- 1.Jackson ST, Booth RT. 2007. Validation of pollen studies. In Encyclopaedia of quaternary sciences (ed. Elias SA.), pp. 2413–2422 Amsterdam, The Netherlands: Elsevier [Google Scholar]
- 2.Grass Phylogeny Working Group II 2012. New grass phylogeny resolves deep evolutionary relationships and discovers C4 origins. New Phytol. 193, 304–312 (doi:10.1111/j.1469-8137.2011.03972.x) [DOI] [PubMed] [Google Scholar]
- 3.Freeling M. 2001. Grasses as a single genetic system. Reassessment 2001. Plant Physiol. 125, 1191–1197 (doi:10.1104/pp.125.3.1191) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wodehouse RP. 1935. Pollen grains. New York, NY: McGraw-Hill [Google Scholar]
- 5.Fægri K, Kaland PE, Krzywinski K. 1992. Textbook of pollen analysis by Knut Fægri & Johs Iversen. Chicester, UK: Wiley [Google Scholar]
- 6.Strömberg CAE. 2011. Evolution of grasses and grassland ecosystems. Annu. Rev. Earth Planet. Sci. 39, 517–544 (doi:10.1146/annurev-earth-040809-152402) [Google Scholar]
- 7.Piperno DR. 2006. Phytoliths: a comprehensive guide for archaeologists and paleoecologists. New York, NY: AltaMira [Google Scholar]
- 8.Joly C, Barille L, Barreau M, Mancheron A, Visset L. 2007. Grain and annulus diameter as criteria for distinguishing pollen grains of cereals from wild grasses. Rev. Palaeobot. Palynol. 146, 221–233 (doi:10.1016/j.revpalbo.2007.04.003) [Google Scholar]
- 9.Holst I, Moreno JE, Piperno DR. 2007. Identification of teosinte, maize, and Tripsacum in Mesoamerica by using pollen, starch grains, and phytoliths. Proc. Natl Acad. Sci. USA 104, 17 608–17 613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schüler L, Behling H. 2011. Poaceae pollen grain size as a tool to distinguish past grasslands in South America: a new methodological approach. Veg. Hist. Archaeobot. 20, 83–96 (doi:10.1007/s00334-010-0265-z) [Google Scholar]
- 11.Flenley JR, King SM. 1984. Late Quaternary pollen records from Easter Island. Nature 307, 47–50 (doi:10.1038/307047a0) [Google Scholar]
- 12.Andersen ST, Bertelsen F. 1972. Scanning electron microscope studies of pollen of cereals and other grasses. Grana 12, 79–86 (doi:10.1080/00173137209428830) [Google Scholar]
- 13.Page JS. 1978. A scanning electron microscope survey of grass pollen. Kew Bull. 32, 313–319 (doi:10.2307/4117102) [Google Scholar]
- 14.Köhler E, Lange E. 1979. A contribution to distinguishing cereal from wild grass pollen grains by LM and SEM. Grana 18, 133–140 (doi:10.1080/00173137909424973) [Google Scholar]
- 15.Peltre G, Cerceau-Larrival MT, Hideux M, Abadie M, David B. 1987. Scanning and transmission electron microscopy related to immunochemical analysis of grass pollen. Grana 26, 158–170 (doi:10.1080/00173138709429945) [Google Scholar]
- 16.Chaturvedi M, Datta K, Nair PKK. 1998. Pollen morphology of Oryza (Poaceae). Grana 37, 79–86 (doi:10.1080/00173139809362647) [Google Scholar]
- 17.Jacobs SWL, Everett J. (eds) 2000. Grasses: systematics and evolution. Melbourne, Australia: CSIRO [Google Scholar]
- 18.Punyasena SW, Tcheng DK, Wesseln C, Mueller PG. 2012. Classifying black and white spruce using layered machine learning. New Phytol. 196, 937–944 (doi:10.1111/j.1469-8137.2012.04291.x) [DOI] [PubMed] [Google Scholar]
- 19.Sivaguru M, Mander L, Fried G, Punyasena SW. 2012. Capturing the surface texture and shape of pollen: a comparison of microscopy techniques. PLoS ONE 7, e39129 (doi:10.1371/journal.pone.0039129) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Punt W, Hoen PP, Blackmore S, Nilsson Le Thomas A. 2007. Glossary of pollen and spore terminology. Rev. Palaeobot. Palynol. 143, 1–81 (doi:10.1016/j.revpalbo.2006.06.008) [Google Scholar]
- 21.Pothen A, Fan C-J. 1990. Computing the block triangular form of a sparse matrix. ACM Trans. Math. Softw. 16, 303–324 (doi:10.1145/98267.98287) [Google Scholar]
- 22.Heckbert PS. 1982. Color image quantization for frame buffer display. Comput. Graph. 16, 297–307 (doi:10.1145/965145.801294) [Google Scholar]
- 23.Estrada E, Rodríguez-Velázquez JA. 2005. Subgraph centrality in complex networks. Phys. Rev. E71, 056103. [DOI] [PubMed] [Google Scholar]
- 24.Efron B, Tibshirani R. 1997. Improvements on cross-validation: the 632+ bootstrap method. J. Am. Statist. Assoc. 92, 548–560 [Google Scholar]
- 25.Leung T, Malik J. 2001. Representing and recognizing the visual appearance of materials using three-dimensional textons. Int. J. Comput. Vision 43, 29–44 (doi:10.1023/A:1011126920638) [Google Scholar]
- 26.Martin DR, Fowlkes CC, Malik J. 2004. Learning to detect natural image boundaries using local brightness, color, and texture cues. IEEE Trans. Pattern Anal. Mach. Intell. 26, 530–549 (doi:10.1109/TPAMI.2004.1273918) [DOI] [PubMed] [Google Scholar]
- 27.Ren X, Bo L. 2012. Discriminatively trained sparse code gradients for contour detection. In Advances in neural information processing systems 25 (eds Bartlett P, Pereira FCN, Burges CJC, Bottou L, Weinberger KQ.), pp. 1–9 La Jolla, CA: Salk Institute, Neural Information Processing Systems Foundation [Google Scholar]
- 28.MacLeod N, Benfield MC, Culverhouse PF. 2010. Time to automate identification. Nature 467, 154–155 (doi:10.1038/467154a) [DOI] [PubMed] [Google Scholar]
- 29.Holt K, Allen G, Hodgson R, Marsland S, Flenley J. 2011. Progress towards an automated trainable pollen location and classifier system for use in the palynology laboratory. Rev. Palaeobot. Palynol. 167, 175–183 (doi:10.1016/j.revpalbo.2011.08.006) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
SEM images of grass pollen and files used to test human classification performance: University of Illinois IDEALS digital archive (https://www.ideals.illinois.edu/handle/2142/43358).


