Abstract
The structure of pseudorabies virus (PRV) capsids isolated from the nucleus of infected cells and from PRV virions was determined by cryo-electron microscopy (cryo-EM), and compared to herpes simplex virus type 1 (HSV-1) capsids. PRV capsid structures closely resemble those of HSV-1, including distribution of the capsid vertex specific component (CVSC) of HSV-1, which is a heterodimer of the pUL17 and pUL25 proteins. Occupancy of CVSC on all PRV capsids is near 100%, compared to ~50% reported for HSV-1 C-capsids and 25% or less that we measure for HSV-1 A- and B-capsids. A PRV mutant lacking pUL25 does not produce C-capsids and lacks visible CVSC density in the cryo-EM-based reconstruction. A reconstruction of PRV capsids in which green fluorescent protein (GFP) was fused within the N-terminus of pUL25 confirmed previous studies with a similar HSV-1 capsid mutant localizing pUL25 to the CVSC density region that is distal to the penton. However, comparison of the CVSC density in a 9 Ångstrom resolution PRV C-capsid map with the available crystal structure of HSV-1 pUL25 failed to find a satisfactory fit, suggesting either a different fold for PRV pUL25 or a capsid- bound conformation for pUL25 that does not match the X-ray model determined from protein crystallized in solution. The PRV capsid imaged within virions closely resembles C-capsids with the addition of weak but significant density shrouding the pentons that we attribute to tegument proteins. Our results demonstrate significant structure conservation between the PRV and HSV capsids.
Keywords: cryo-electron microscopy, mass spectrometry, CVSC, pUL25, pUL17
Introduction
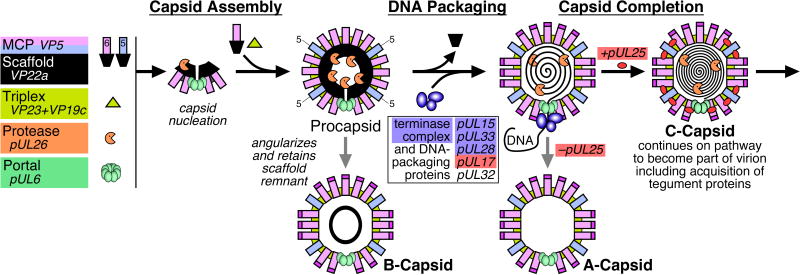
Electron microscopy and biochemical studies have provided important insights into the structure of the herpesvirus capsid, particularly HSV-1, and how it is assembled in the infected cell nucleus (Fig. 1) 1; 2; 3; 4; 5; 6; 7; 8. Three distinct types of capsids – A-, B- and C-capsids – are readily purified from infected cells and a fourth precursor capsid form has also been identified that is analogous to the procapsids of tailed bacteriophages 9. A-, B- and C-capsids share the same shell structure but differ primarily in the material present inside the capsid cavity. C-capsids contain the viral DNA and can mature into infectious virions. They are closely similar, if not identical, in structure and composition to the capsids present in infectious virions 10; 11. In contrast, A- and B-capsids lack DNA and appear to be aberrant dead-end structures although they are occasionally seen inside virion-like particles. The B-capsid cavity retains a cleaved form of the scaffolding protein, VP22a, while the A-capsid cavity lacks significant amounts of either DNA or protein 12. Studies with HSV mutants demonstrate that procapsids are able to package DNA and mature into infectious virions 7; 8; 11; 12; 13; 14; 15.
Figure 1.
Capsid assembly and DNA-packaging pathway for herpesvirus HSV-1.
The clearest views of the HSV-1 capsid structure have come from three-dimensional image reconstructions of cryo-electron microscopy (cryo-EM) data computed to resolutions of 30–8.4 Ångstroms for all four capsid types 16; 17; 18; 19; 20; 21; 22; 23; 24; 25; 26; 27; 28; 29; 30; 31; 32; 33; 34. The icosahedrally symmetric reconstructions show that the capsid is a protein shell approximately 150 Å thick and 1250 Å in diameter. Its major structural features are 162 capsomers arranged on a T=16 icosahedral lattice 35 and composed mainly of the VP5 major capsid protein. 150 hexamers of VP5, called hexons, occupy the capsid edges and faces while each of the 12 vertices are occupied by either a pentamer of VP5 – penton – or in one unique position a dodecamer of the pUL6 portal protein through which the viral genome is packaged and likely also released 27; 36. The capsomers are connected in groups of three by asymmetric molecules called triplexes (320 in all) that lie on the capsid exterior at the base of the capsomer protrusions. Triplexes contain on average one copy of VP19C and two of VP23 33; 37, although triplex composition may prove to vary with position on the capsid shell. The distal tips of the hexons, but not pentons, are capped with the VP26 protein in a 1:1 ratio with the hexon VP5 subunits 25; 26; 30.
A number of essential minor proteins also interact with the procapsid and capsid during assembly and maturation, including the portal complex and the multi-component DNA-packaging machinery. In addition to the portal, only two of these minor capsid proteins remain associated with the capsid – the pUL17 and pUL25 proteins that are organized as a heterodimer to form the capsid vertex specific component (CVSC) 24; 38; 39; 40. The CVSC molecules are found arrayed in five copies around the exterior of each of the capsid vertices, and are essential for maintaining the packaged genome within the capsid, amongst other functions. The DNA is under considerable pressure within the capsid as shown by electron micrographs of HSV-1 C-capsids, for example, where the interduplex spacing is measured at 26 Å, a value comparable to that found in dsDNA bacteriophages and near the theoretical limit for close packing of DNA duplexes 11. The mechanism by which pUL17 and pUL25 act is unknown, although their proximity to the portal vertex is likely to be relevant for retaining DNA while those copies located around penton vertices would presumably carry out other functions, such as signaling readiness for the nuclear egress of mature C-capsids. Our interest is to further characterize the biochemical and structural interactions of pUL17 and pUL25 with each other and the underlying capsid so as to better understand their functions. Further, these proteins present readily accessible targets for interfering with capsid assembly. We have found, however, that purified HSV-1 nucleocapsids include sub-stoichiometric quantities of the CVSC molecules on the capsid surface, hindering our structural studies and we have sought alternative herpesviruses where this problem is avoided.
Pseudorabies virus (PRV) is an α-herpesvirus that infects pigs and many other animals but not primates or humans. Its effects on young pigs and secondary hosts are generally rapid and devastating. PRV is also used to trace neural networks through which it propagates in a retrograde direction compared to synaptic activity. Consequently, much of PRV genetics and biochemistry is known, although no structural work on the capsid has been reported despite the structural proteins having clear parallels with those of HSV-1. The pUL25 protein of HSV-1, for example, can partially complement pUL25-deficient PRV capsids, although the contrary is not true 41. We report here our first results in characterizing PRV capsids, including the capsid composition and structure, and details of CVSC organization. PRV capsids prove to be tractable in our hands for structural studies and are particularly interesting for the comparison with HSV-1 capsids, including differences in retention of CVSC molecules.
Results
Isolation and characterization of PRV capsids
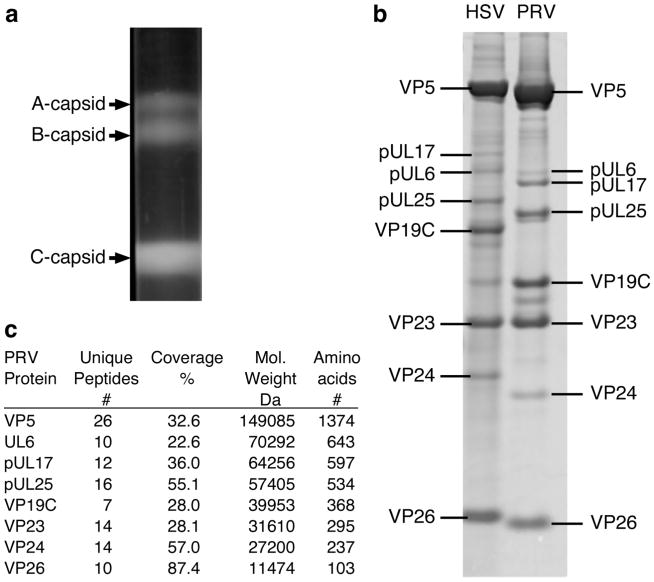
PK15 cells infected with the wild type PRV Becker strain were harvested 24 h postinfection and nuclear lysates were prepared and sedimented through 20 to 50% sucrose gradients (Fig. 2a). Three distinct light-scattering bands were visualized in the gradient positions corresponding to where HSV-1 A, B, and C-capsids typically sediment, with the empty A-capsids being found highest (least dense) on the gradient and DNA-filled C-capsids found lowest (most dense). The gradient was fractionated and the protein composition of the C-capsid fraction was analyzed by SDS-PAGE (Fig. 2b). PRV capsid proteins were identified by mass spectrometry. The major-staining PRV capsid proteins were cut from the gel, trypsin digested and the masses of the resulting peptides determined by MALDI-TOF mass spectrometry for comparison to available databases of viral proteins. As shown in Fig. 2c, MS analysis revealed tryptic peptides from the gel bands that identified the four primary capsid proteins including: the major capsid protein VP5; the two triplex proteins VP19C and VP23; and the hexon associated protein VP26. In addition, four minor capsid-associated proteins were also identified: VP24, which is the protease domain of the pUL26 protein; the pUL6 portal protein; and the pUL17 and pUL25 CVSC proteins.
Figure 2.
Characterization of PRV nucleocapsids. (a) Sucrose gradient purification of wild-type PRV-Becker capsids grown on PK-15 pig kidney cells. (b) Comparison of HSV and PRV C- capsid components by SDS-PAGE. (c) PRV capsid proteins from (b) were identified by MALDI- TOF mass spectrometry and are shown with molecular weights and amino acid counts from databases.
PRV pUL25 is required for forming a stable DNA-containing capsid
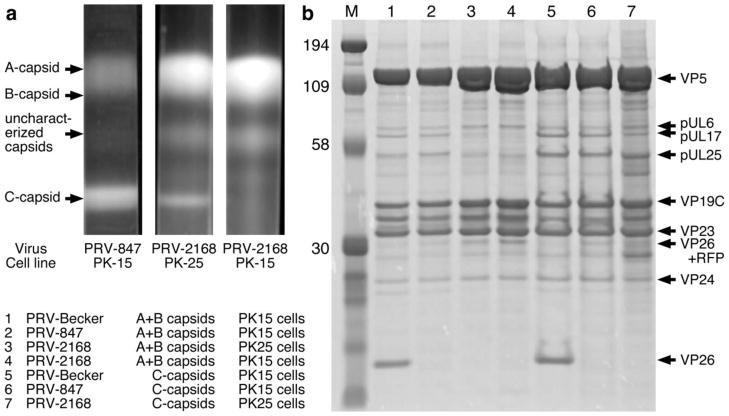
Homologs of the UL25 gene product have been identified in all three subfamilies of Herpesviridae, and we have previously localized pUL25 on the HSV-1 C-capsid to the region of the CVSC protein distal to the adjacent vertex 39. Studies with both HSV and PRV UL25-null mutants have shown that all three capsid forms are present in cells infected with these viruses but the capsids were restricted to the nucleus. Capsid-bound pUL25 is a prerequisite for egress of mature C-capsids from the nucleus with a threshold level required to stabilize the capsid and trigger the nuclear-egress event 40; 42; 43. In order to establish a requirement for PRV pUL25 in the assembly of stable C-capsids, nuclear capsids from wild-type Becker, GS847 (expressing the VP26-RFP fusion protein) and UL25-null mutant GS2168 (parent virus Becker 847) were subjected to sucrose density gradient centrifugation and capsids were detected as visible light-scattering bands. In cells infected with Becker (Fig. 2a) or GS847 (Fig. 3a, left lane), all three capsid forms were detected. In contrast, the gradient for the UL25-null mutant (Fig. 3a, right lane) contained approximately equal numbers of A- and B-capsids that formed one large band on the gradient but no DNA-containing C-capsids were observed. Stable C-capsids were found only when the UL25-null virus was grown on the pUL25-complementing PK25 cells (Fig. 3a, center lane). In addition to the three capsid bands, a fourth band was found with the GS847 and GS2168 samples located in a region between the B- and C-capsids. Although we are not sure of its origin, preliminary cryo-EM and SDS-PAGE studies demonstrated that the band contains intact capsids with a protein composition similar to that of A and B capsids.
Figure 3.
(a) Sucrose gradient purification of PRV capsids. PRV-847 and PRV-2168 have a VP26-RFP fusion, and PRV-2168 additionally lacks the UL25 gene. PK-15 are pig kidney cells, and PK-25 complement for pUL25, allowing the PRV-2168 UL25-null virus to package and retain the dsDNA genome, as indicated by the presence of a C-capsid band. (b) SDS-PAGE analysis of PRV A-, B- and C-capsids isolated from the sucrose gradient shown in (a) with the position of the capsid proteins listed on the right.
The PRV A-, B- and C-capsids isolated from the sucrose gradient were further analyzed by SDS-PAGE (Fig. 3b). The pUL17 and pUL25 proteins appear in a 1:1 ratio for both the combined A+B-capsid and for the C-capsids isolated from PK15 cells infected with Becker or from the GS847 virus. However, more pUL17 and pUL25 are present on the C-capsids compared to the A+B-capsids (Fig. 3b). As expected, the amount of the pUL6 portal protein is constant in all capsid forms. The pUL25 and pUL17 proteins are present on C-capsids of the UL25-null virus (GS2168) when grown on UL25-complementing PK25 cells (low levels) but both are missing when grown on the non-complementing PK15 cells, further supporting the proposal that the two proteins co-assemble on the capsid and consistent with the CVSC molecule being composed of one copy of each of the two proteins, as observed by cryo-EM 38; 39. Interestingly, there was no obvious protein band in the B-capsid samples that could be attributed to the UL26.5 scaffold protein (Fig 3b, lanes 1–4). The protein band located between the two triplex proteins VP19C and VP23 appears to be a likely scaffold protein candidate since it is located in the same region of the gel (between VP19C and VP23) as the HSV-1 scaffold protein.
Copy numbers of pUL25 and pUL17 in PRV capsids
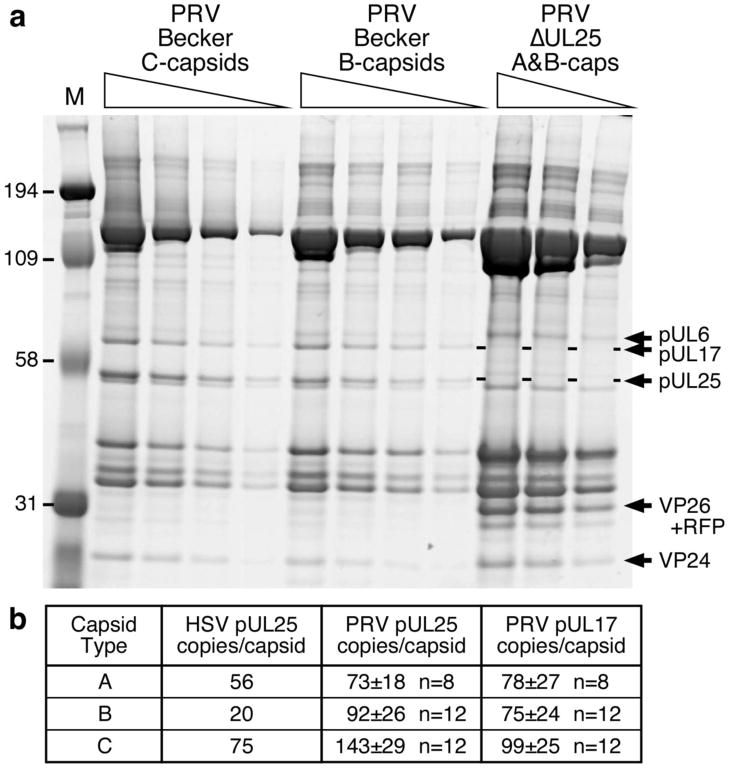
Increasing amounts of the PRV Becker C-capsids and B-capsids, as well as A+B-capsids from the UL25-null virus GS2168, were resolved by SDS-PAGE (Fig. 4a) and the stained gel was then scanned to determine the pUL25 and pUL17 copy numbers per capsid. PRV Becker A-capsids were also analyzed in a similar fashion (data not shown). Quantitation of pUL25 and pUL17 was calibrated against both triplex subunits, VP19C (320 copies per capsid) and VP23 (640 copies per capsid). By the same procedure we obtained a value of 10 copies for pUL6, in reasonable agreement with the expected value of 12. These quantitations yielded pUL25 copy numbers of 143 for C-capsids, 92 for B-capsids, and 73 for A-capsids and for pUL17 the corresponding copy numbers were 99, 75 and 78 (Fig. 4b). The copy number for both pUL25 and pUL17 in C-capsids is more than sufficient to account for the number of CVSC binding sites (60/capsid) observed by cryo-EM for the HSV- 1 C-capsid 24. Furthermore, the relative abundance of pUL25, and hence the CVSC molecule 39, on PRV A- and B-capsids suggests that CVSC is not, in fact, specific to C-capsids as was originally proposed 24.
Figure 4.
Analysis of pUL25 and pUL17 copy number on PRV capsids. (a) The pUL6 protein serves to confirm the quantification made using the triplex proteins, VP19C (320 copies per capsid) and VP23 (640 copies per capsid). (b) Calibrated copy numbers of pUL25 and pUL17 from SDS-PAGE. HSV pUL25 numbers were previously determined 60.
Cryo-EM reconstruction of the PRV C-capsid
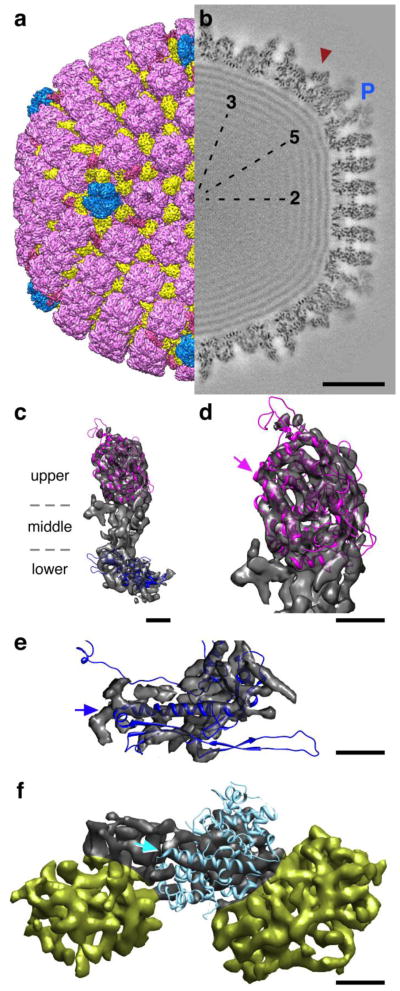
Capsids were isolated from the nuclear fraction of cells infected with wild type PRV Becker at 18 h postinfection. Examination by cryo- EM confirmed that the capsids were highly purified and the resulting micrographs were used to calculate separate reconstructions of A-, B- and C-capsids. In addition, PRV mutant capsids and HSV-1 capsids were also imaged for comparison, as described below. The PRV Becker C-capsid map (Fig. 5) yielded the highest resolution achieved to date for an on-pathway herpesvirus capsid structure of 9.0Å. An HSV-1 B-capsid structure has previously been reported at 8.5 Å resolution 34 although it is an off-pathway form that contains no DNA. The morphology of the PRV Becker C-capsid structure is virtually identical to that of HSV-1, including capsomer positions and shapes, distributions of triplex molecules, capping of hexons but not pentons by VP26, and localization of the capsid vertex-specific component (CVSC) around pentons. Within the capsid, the tightly packed DNA is represented as dense layers spaced 26 Å apart, much as is seen for dsDNA tailed bacteriophages. Perhaps the most interesting departure from other herpesvirus reconstructions to date is the observation of wispy but significant density attached to the pentons (Fig. 5b). We ascribe this density to partial occupancy of tegument proteins, possibly pUL36 (VP1/2), binding to the VP26-free tips of the five major capsid protein subunits that form the penton. This density has not been previously described and while it is too weak for characterizing further, it strongly suggests some capsid tegumentation occurs in the nucleus prior to egress. In support of this is a recent report of a truncated form of pUL36 that binds to capsids in the nucleus and assists in the transfer of DNA-containing capsids to the cytoplasm 44.
Figure 5.
Renditions of the PRV C-capsid reconstruction at 9Å resolution. (a) Surface view with the capsid vertex specific complex (CVSC) density colored red, the pentons blue, hexons purple and the triplex molecules in yellow. (b) A central thin section reveals the CVSC density (red arrowhead) to be as strong as the C-capsid density, indicating full occupancy, as well as additional weaker density shrouding the tips of the penton (blue “P”) that may be tegument proteins. Bar = 200 Å. (c) Attempts to fit atomic models of the HSV-1 VP5 upper domain (purple - PDB Id: 1NO7) and phage HK97 major capsid protein (blue - PDB Id: 1OHG) into the lower domain of a penton subunit (grey transparent surface) are largely similar to the positions determined for the HSV-1 capsid density map 45; 46. No atomic model is available for the middle domain. Bar = 20Å. (d) Close-up view of the VP5 upper domain fit revealing good superposition of helices into tubular density regions, including the long bent helix indicated by the purple arrow. Bar = 20Å. (e) Close up view of the VP5 lower domain fit where the backbone helix of the HK97 model is indicated (blue arrow) and the correspondence is generally consistent in shape although divergent in detail. (f) The HSV-1 pUL25 atomic model (PDB Id: 2F5U), however, does not match the density identified as the CVSC region (grey surface) occupied by pUL25, which has been extracted along with two triplex molecules (yellow surfaces). While one longer helix matches the cryo-EM density, no orientation of the atomic model that accommodates this superposition is feasible, suggesting that the folds may be different between HSV-1 and PRV, or that the fold adopted when crystalizing pUL25 from solution is different from that when pUL25 is bound to capsids. Bar = 20Å.
The paucity of atomic models for the herpesvirus capsid subunits limits attempts to characterize folds and interfaces with a 9 Å-resolution density map. However, several candidate structures from HSV-1 and phage HK97 may be positioned within the VP5 cryo-EM density with some success (Fig 5c). As for the HSV-1 B-capsid density map 45, we find that the crystal structure of the HSV-1 VP5 upper domain convincingly superimposes in the cryo-EM density with good alignment of helices in tubular regions of density (Fig. 5c, purple ribbon representation). Several loops clearly escape the PRV density, as may be expected given the different sources of the structure data. No atomic model exists for the middle or lower domains, but major features of the canonical fold from the dsDNA phage HK97 major capsid protein, gp5, align well with the lower region of the PRV C-capsid density as for HSV-1 46, including the long backbone helix and the triangular axial domain (Fig. 5e, blue ribbon representation). In contrast, the crystal structure of one of the CVSC subunits, pUL25 from HSV-1, does not match features of the density where pUL25 has been previously localized (Fig. 5f). Unfortunately the crystal structure lacks the 133 N-terminal amino acids, and so the position of the GFP insertion between residues 50 and 51 cannot be used as a significant constraint for fitting the model into the density map. It also lacks a number of surface loops that could further define placement within the map. However, a large fraction of the atomic model is α-helical, and should in principle be recognizable in the 9 Å density map, but we failed in our efforts to determine a fit for it. We observe portions of density that could accommodate longer helices than the model contains, suggesting either that the pUL25 protein folds differently when bound to the capsid than when crystalized from solution, or the homologous pUL25 proteins folds differently between HSV-1 and PRV. Given that the PRV pUL25 protein can complement the HSV-1 capsid, and that the two proteins share 47% sequence identity, we favor the second explanation but await further structural work to address this point.
CVSC localization and occupancy
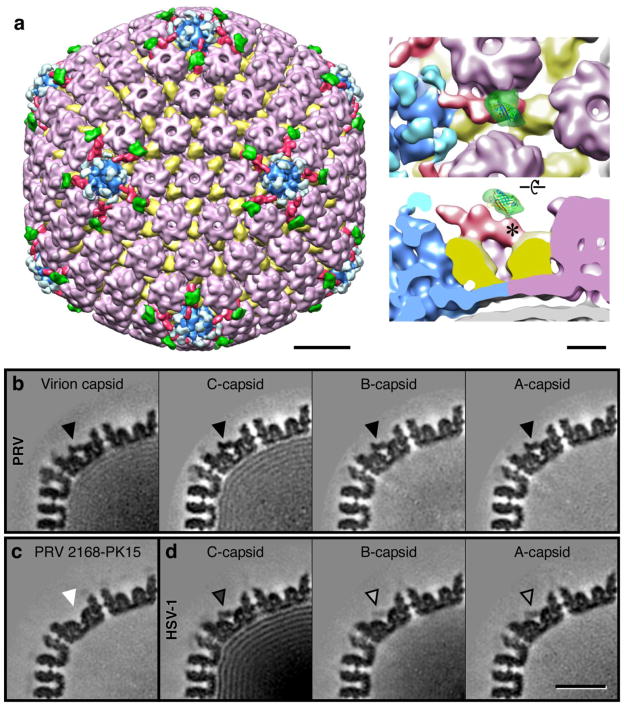
Similar to the HSV-1 C-capsid, the Becker C-capsid map reveals density on the capsid exterior that is adjacent to the vertices and which resembles the CVSC that was first identified (and called C-capsid specific component) by Trus et al 24. Each CVSC density clearly contacts two triplex molecules – that adjacent to the penton, and the next closest – as well as one of the two hexons that it nestles between, much as observed for HSV-1 39. To confirm that PRV pUL25 is similarly located in this CVSC region as observed in HSV-1, we prepared an analogous mutant with pUL25 tagged by insertion of GFP between residues 50 and 51, and visualized the GFP density by cryo-EM (Fig 6a). As was observed with the HSV-1 pUL25-GFP virus 39 and with a PRV pUL25-GFP virus 47, no defect in virus growth was found with our PRV pUL25-GFP expressing virus (data not shown). Reconstruction of the mutant C-capsid reveals extra density above and continuous with the putative CVSC region that is distal to the vertex. The 27-kDa bulk of the GFP tag corresponds to the volume of the extra density and the point of excursion from the surface of CVSC on the wild-type C-capsid map can be pinpointed precisely. This position is identical to the insertion location previously identified for the pUL25-GFP mutant in HSV-1 39, further supporting this localization of the pUL25 protein in the vertex-distal region of the CVSC density and the N-terminal portion of pUL25 being the distal-most part of CVSC. Unlike the HSV-1 density, the GFP-tag in PRV appears to be more constrained and only adopts a single conformation. These observations indicate that pUL25 is a component of the CVSC that anchors pUL17 to the capsid, and is essential for retaining the packaged DNA.
Figure 6.
Localization of the PRV pUL25 protein and occupancy of the CVSC molecule on different capsid types of PRV and HSV-1, estimated from the portions shown of central thin sections through cryo-EM density maps. (a) Surface view of the PRV C-capsid that has the GFP sequence inserted between residues 50 and 51 in pUL25. The CVSC density is colored red, and the extra density in green is ascribed to the GFP tag. Bar = 200Å. Close up views at right, showing the site (*) where the GFP density contacts the CVSC molecule, much as observed in HSV-1 39. Bar = 50Å. CVSC occupancy as revealed by central sections through density maps of (b) the PRV virion capsid and C-, B- and A-capsids as marked, with the CVSC density indicated by an arrowhead; (c) PRV 2168 grown on PK15 cells; and (d) HSV-1 C-, B- and A-capsids as marked. Bar = 250Å. For PRV A-, B-, and C- capsids, and virion capsids, the strength of the CVSC density is comparable to that of the capsid, whereas for HSV-1, the CVSC density is only about 55% of the C-capsid density, and below 25% for the A- and B-capsids. The PRV 2168 UL25-null mutant completely lacks CVSC density when grown on PK15 cells, but the CVSC is present when pUL25 is supplied by PK25 cells (data not shown).
Occupancy of the CVSC region appears to be considerably higher for PRV capsids than for HSV-1 capsids, and we made a systematic comparison between them and with a pUL25-null mutant of PRV (Fig 6b–d). The PRV Becker C-capsid reconstruction shows the CVSC density to have the same strength as that of the capsid, indicating that occupancy of the CVSC binding sites around the vertices is 100%. This is considerably higher than the ~55% estimated for HSV-1 C-capsids by comparing density levels of the CVSC region to the background (0%) and to the strongest densities in the capsid (100%) 24. Presumably the PRV capsids retain a higher proportion of CVSC during the preparation procedures for cryo-EM, or the CVSC molecules bind more strongly to PRV capsids than to HSV-1 capsids. The reconstruction calculated from B-capsids found in the PRV Becker C-capsid sample reveals recognizable but weaker CVSC density, and we estimate occupancy at ~55%, indicating that on average only 30 of the 60 peri-pentonal sites are occupied by CVSC. The significant presence of the CVSC density in all capsid types prompted us to modify the name to CVSC 38, indicating specificity to vertices rather than to just C-capsids as was previously proposed 24. In comparison, the GS2168 B-capsid that lacks pUL25 has a complete absence of density in this location (Fig. 6c), but the CVSC is restored when grown on the complimenting PK25 cells (data not shown). Further, in reexamining CVSC occupancy on HSV-1 capsids from samples we reconstructed, we observe higher levels (Fig. 6d) than reported previously 24, further suggesting that CVSC may be dislodged relatively easily during purification of nucleocapsids.
Structure of capsids in PRV virions
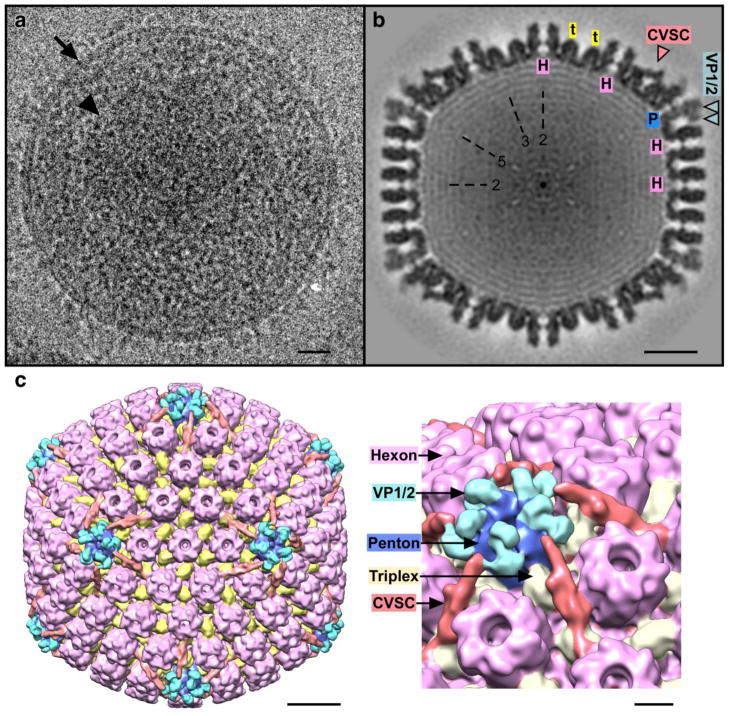
We also imaged virions of PRV by cryo-EM and calculated an icosahedral reconstruction of the capsid to compare with the nucleocapsid structures, particularly the C-capsid. Although this kind of analysis is compelling for studying the fully mature capsid, virions are less tractable than purified capsids for several reasons. For one, the larger area occupied by virions effectively reduces the yield of capsids per electron micrograph resulting in a smaller dataset of particles. Further signal is lost to reduction of capsid density contrast due to shrouding by the deep tegument protein layer, membrane and glycoproteins, as well as by the greater ice thickness required to embed the larger virions (Fig 7a). Nonetheless, capsids could be readily identified in the micrographs and the resulting icosahedrally-averaged density map, although at a modest resolution of 22.6 Å, is remarkable for the detail in the capsid density and for the low background noise surrounding the capsid despite the dense environment (Fig. 7b–c). The main capsid elements are strongly represented, appearing similar to the PRV and HSV-1 C-capsids, although the additional density observed above pentons on the PRV C-capsid pentons is considerably more striking in the virion capsid. We expect to further exploit fully enveloped capsids such as these in studying mutants of capsid surface proteins, such as pUL25 and pUL17, for which we observe a tendency to fall off nucleocapsids during purification for structural studies.
Figure 7.
(a) Cryo-EM image of a PRV virion, with the membrane well-resolved (arrow) and the capsid (arrowhead) visible within the surrounding tegument layers. Bar = 200Å. (b) Central thin section through the virion capsid reconstruction revealing both the CVSC density and heavy shrouding of penton tips by what may be VP1/2 tegument proteins (blue arrowheads). “H” indicates hexons; “P” pentons; “t” triplexes; and “2”, “3”, and “5” indicate symmetry axes. Bar = 200Å. (c) Surface view of the virion capsid reconstruction with the CVSC density in red and the putative tegument density in light blue and located on top of dark blue pentons. Hexons including the VP26 tip density are colored purple and the triplexes are yellow. Bar = 200Å. Close up view at right - bar = 50Å.
Discussion
Using a combination of molecular genetics, biochemical and structural techniques, we have shown that the capsids of α-herpesviruses PRV and HSV-1 are highly similar in the subunit compositions and in their morphologies at the molecular level. We have focused on the CVSC molecule, finding that it is arranged identically on the PRV capsid as for HSV-1, and that the PRV pUL25 protein is a constitutive member that functions to retain packaged DNA within the capsid. The similarities between the PRV and HSV-1 capsids are striking, and our observation of greater stability for the PRV C-capsid compared to HSV-1 suggests that it may be a more useful target for studying α-herpesvirus structure. The report that HSV-1 pUL25 partially complements PRV capsids highlights a significant conservation of structure and function for this protein and its interaction with the herpesvirus capsid 41. In characterizing PRV capsid structures further, we expected to generate a ‘baseline’ capsid structure of at least 10Å resolution or better for assessing differences with other capsid forms and with HSV-1 capsids that will ultimately inform on the details of capsid assembly, maturation and function.
Occupancy of the CVSC density region varies considerably between PRV capsid types, and between PRV and HSV capsids of the same type. In addition, the CVSC is also only found surrounding pentons on capsids and not elsewhere on the capsid surface in icosahedrally averaged reconstructions. This variation suggests that the CVSC molecule has constraints on recognizing potential binding sites, and little flexibility to adjust to non-peripentonal locations where (for example) the surface curvature may be different, or to capsids where the absence of tightly packaged DNA may also affect capsid shape. In our hands, the HSV-1 C-capsid includes a higher proportion of the CVSC molecule than was reported previously 24, suggesting that it may be easily lost during specimen purification. Indeed few herpesvirus capsid reconstructions prior to 2007 revealed any CVSC density, and in several cases it was assigned as tegument rather than capsid structural proteins 45; 48; 49.
The CVSC density is generally higher for PRV capsids than HSV-1 and this is particularly significant for the on-pathway C-capsids with nearly 100% occupancy for PRV (as for capsids in virions) compared to 55% for HSV-1. The higher occupancy in PRV may reflect tighter binding of the CVSC to the capsid than for HSV-1, which may be an indication that pUL25 in PRV plays a more significant role in stabilizing the capsid in addition to its demonstrated function in retaining packaged DNA. We also find more intact C-capsids in our PRV preparations while many HSV-1 C-capsids appear to have lost their cargo of DNA, also suggesting that the PRV C-capsid is more stable, which is an advantage for structural studies. The function of stabilizing the capsid is a global mechanism, and pentons appear to be particularly in need of reinforcing as they are susceptible to dislodging, presumably due to the higher local curvature of the capsid surface at the vertices as well as the larger angle between penton subunits (72°) compared to hexon subunits (60°). The other function of retaining packaged DNA is local to the portal vertex, where the viral DNA enters and exits the capsid, and occupancy of the CVSC around the portal vertex may be critical to this function. Stabilization of pentamers, therefore, may be a function of CVSC that varies in importance amongst herpesviruses, while its role in retaining the genome at the portal vertex is crucial for all. Unfortunately the imposition of icosahedral symmetry does not allow the portal vertex to be visualized in our reconstructions where it is outweighed by the signal of the other 11 penton-bearing vertices. In future, the portal vertex may be identified by labeling the portal protein with a small tag (gold particle) that distinguishes it in cryo-electron micrographs from the similarly-sized pentons, thus allowing alignment of portal vertices for generating non-symmetric structures where any corresponding conformational variation of the CVSC molecule may be visualized at that vertex.
We note that a discrepancy exists between the total complement of CVSC subunits per capsid observed in the cryo-EM reconstructions and that estimated by SDS-PAGE. In the reconstructions, 5 copies around each of 12 vertices accounts for 60 copies each of pUL25 and pUL17 on the PRV C-capsids, while calibrated SDS-PAGE readings show 143 copies of pUL25 and 99 of pUL17. The pUL17 protein is found on HSV-1 UL25-null capsids but they can only be detected by Western blot analysis and are not visible on stained gels.40 We expect that there would be a low amount of UL17 on the PRV UL25-null capsids but we do not have a PRV pUL17 antibody to do the Western blot analysis. The whereabouts of the excess of pUL25 and pUL17 beyond those copies visualized by cryo-EM is unclear, but considering that the capsid includes many triplexes and hexons in spatial arrangements similar to those around the vertices that bind CVSC, we suppose that some CVSC or pUL25 monomers may bind these alternate locations weakly and randomly such that their signal does not survive the extensive averaging with unoccupied sites that is a consequence of the icosahedral reconstruction procedure. Alternatively, or in addition, copies of pUL25 and pUL17 may reside in the tegument layer, as has been previously suggested for pUL17 50.
An unexpected outcome of this work was our failure to fit the HSV-1 pUL25 subunit atomic model into the 9 Å-resolution PRV C-capsid density map, despite full occupancy of the CVSC density on the capsid. Although the pUL25 crystal structure is compact, it contains significant α-helix content that we would expect to see well enough resolved in the cryo-EM density map for reasonable attempts at fitting, despite absence of the N-terminal 132 residues, ~25% of the total pUL25 protein, as well as 5 surface loops in the atomic model. Our conclusion is that there is likely an inconsistency between the HSV pUL25 crystal structure and the pUL25 density revealed on the PRV C-capsid, either due to differences between the HSV-1 and PRV pUL25 protein folds, or due to an alteration in the structure of pUL25 on binding the capsid compared to the form into which it crystallizes from solution. The density map is insufficient to trace the α-carbon backbone of pUL25, or to distinguish its limits with the adjacent pUL17 subunit, and further progress on this aspect of the capsid structure will require higher resolution as well as localization of tags inserted into the regions of the protein included in the crystal structure. The missing surface loops suggest themselves as likely places to attempt insertions due to their obvious accessibility and flexibility, and the gaps are short enough to still provide powerful constrains on the locations of insertion sites.
This study demonstrates the utility of PRV as a complement to HSV-1 for studying the structure and function of capsids during assembly and maturation, particularly for its higher retention of packaged DNA and the CVSC surface molecule. High occupancy of capsid components is essential for improving the resolution of these features and detailing subunit folds and interfaces that govern their functions. The successful structural analysis of the PRV capsid within virions extends visualization of assembly intermediates to the final mature capsid and has already revealed that some specific interactions are made between the capsid and tegument that are not well visualized in nucleocapsids. Icosahedrally symmetric reconstructions of these virion capsids may be expected to provide a useful higher-resolution complement to cryo-tomographic maps of the entire asymmetric virion, and may be improved in future by collecting larger datasets. Future work will also examine the conformation of the CVSC at the portal vertex where its interaction with the portal is currently unknown due to the imposition of icosahedral symmetry in the models. Efforts towards reducing symmetry and ultimately dispensing with it altogether will be crucially important for understanding these aspects of the function of pUL25 and pUL17.
Materials and Methods
Viruses and Cells
The wild type PRV-Becker virus was obtained from Greg Smith (Northwestern University) and propagated as previously described 51. In addition Dr. Smith kindly provided us with a PRV-Becker recombinant encoding the VP26 capsid protein fused to the monomeric red fluorescent protein (mRFP1; PRV-GS847) and a virus carrying both mRFP1-VP26 and a deletion in the UL25 gene (PRV-GS2168) 51; 52. African green monkey kidney cells (Vero) and Pig kidney epithelial cells (PK15) were used for propagation of PRV stocks that did not require complementation. PK15-UL25 complementing cell line was required to propagate PRV-GS2168 virus 51. The pUL25-GFP (vFH455) PRV virus was generated by recombination of a PRV genome maintained within a bacterial artificial chromosome (BAC). The PRV BAC clone in GS1783 bacteria (gifted by G. Smith) and mutagenesis was performed using the two-step bacteriophage Red-mediated homologous recombination system 53, as previously described 42. The following primers were used to amplify the green fluorescent protein (GFP) gene from peGFP-in with GFP inserted in-frame between amino acids 50 and 51 of the UL25 open reading frame (5-GGC TTC AGC GAG GGC CTC GAC GCG CGC CTC GCG CTG GCG CAC GCG AAC GCG ATG GTG AGC AAG GGC GAG GA-3′; and 5′-CGC GCC CGC GGC CAT GGC GTT GTC CAG CGC CGC GGC GGC CGC GCG GCG GCG CTT GTA CAG CTC GTC CAT GCC G-3′). To recover the recombinant virus, FH455 BAC DNA was transfected into Vero cells, and recombinant viruses harvested from the transfected cell lysates were plaque purified on Vero cells.
Capsid purification
Capsids were purified as described previously 42. Briefly, Vero cells (1.5 × 108) were infected overnight (18 h at 37°C) at an MOI of 5 PFU/cell. Infected cells were harvested, rinsed with PBS and resuspended in 20mM Tris pH 7.5 plus protease inhibitors (Roche), adjusted to 1% Triton X-100 and incubated for 30 minutes on ice. The resulting nuclei were harvested by low-speed centrifugation, resuspended in 10 ml TNE (500mM NaCl, 10mM Tris, 1mM EDTA, pH 7.5), and then sonicated to lyse the nuclei. The nuclear lysate was then cleared by low speed centrifugation and the resulting supernatant was layered on top of 20–50% sucrose (in TNE) gradients (SW41 rotor at 24000 rpm for 1hr). The position of A-, B- and C-capsids were observed as light scattering bands with A capsids being found highest (least dense) on the gradient and C-capsids found lowest (dense fractions). The fractions were collected using a Beckman Fraction Recovery system (Beckman catalog # 34890). Fractions containing A-, B- and C-capsids were separately combined and then precipitated by adding an equal volume of 16% trichloroacetic acid (TCA). Because the A and B capsid bands from the PRV-GS847 and PRV-GS2168 viruses ran close together, the A and B capsid fractions were combined prior to TCA precipitation. Pellets were resuspended in 100 μL of 2x PAGE loading buffer (Invitrogen) supplemented with 0.4M Tris-base. The capsids proteins were visualized on 4–12% SDS-PAGE gels that were stained with Imperial Blue (Pierce).
Virion Purification
Vero cells (1.5 × 108) were infected overnight (18 h at 37°C) with PRV or HSV-1 at an MOI of 5 PFU/cell. Infected cells were scraped into the cell medium and 5M NaCl was added to a final concentration of 0.5 M NaCl. Cells were pelleted and the media was transferred to SW-28 rotor tubes and virions were pelleted out of the medium by centrifugation at 20K RPM for 35 minutes. The resulting pellet was resuspended in TNE buffer + protease inhibitors and the sample was layered on top of a 20–50% sucrose (in TNE) gradient (SW41 rotor at 24000 rpm for 1hr). The gradient was fractionated using a Beckman Fraction Recovery system and the virion fractions were pooled, resuspended in TNE buffer and the virions were pelleted (SW41 rotor at 24000 rpm for 1hr). The virions were resuspended in low salt TNE buffer (150 mM NaCl, 10mM Tris, 1mM EDTA, pH 7.5) for cryo-EM studies.
Capsid protein copy numbers
Per-capsid copy numbers for the pUL17 and pUL25 proteins were determined by quantitative measurement of stained bands in SDS-polyacrylamide gels of purified wild type PRV Becker A-, B- and C-capsids. Gels were loaded with 40, 20, 10 and 5 μL of the combined capsid fractions and after electrophoresis the gels were stained with Imperial Blue. After de-staining, the gels were scanned on a Typhoon 9400 scanner [GE Healthcare] and the intensity of the individual protein bands was used to calculate the pUL25 and pUL17 capsid copy numbers based on the staining of the two triplex proteins, VP19C and VP23, assuming 640 copies of VP19C and 320 copies of VP23 per capsid. The pUL25 and pUL17 capsid copy numbers along with the standard deviation were based on the average from multiple capsid preps; n = 8 for the B and C capsids and n = 6 for the A capsids.
MALDI-TOF MS analysis
Analyses were carried out beginning with Coomassie-stained protein bands excised from an SDS-polyacrylamide gel. PRV-Becker and HSV-1 KOS C capsid proteins were separated on 4–12% SDS-PAGE gels and visualized as above by staining with Imperial Blue. In-gel digestion of polyacrylamide plugs obtained from the PRV Becker samples was carried out. Gel plugs were placed in 1.5mL microcentrifuge tubes that had been rinsed with methanol and air dried. The gel slices were de-stained by rinsing each in 75mM Ammonium Bicarbonate in 40% ethanol, and then rinsed three times in 400 μL of 50% acetonitrile in 25mM ammonium bicarbonate at room temperature with shaking for 15 minutes each rinse before dehydrating by rinsing in 100% acetonitrile at room temperature with shaking for 10 minutes and subsequent drying by speed vacuum. The samples were then resuspended in 50 μL of 25mM Ammonium Bicarbonate and 10 μL of trypsin (20 μg/ml) and incubated at 37°C overnight. The next morning the trypsin solution from each gel slice was removed and transferred to new clean 1.5mL microcentrifuge tubes. The gel slice was treated twice with 25 μL 50% Acetonitrile, 2.5% TFA (trifluoroacetic acid) at 37°C with vigorous shaking for 15 minutes. The two extractions were combined with the original trypsin solution and dried in spend vac. The dried peptides were mixed with 5 μL of α-cyano-4-hydroxycinnamic acid (CHCA), and 0.5 μL was spotted onto the target for MALDI-TOF analysis. MALDI-TOF was performed using the 4700 Proteomics Analyzer (ABI, Foster City, CA). Mass spectra were individually calibrated by using internal trypsin peaks with Data Explorer software available from ABI. Proteins were identified using Protein Prospector (University of California, San Francisco; http://prospector.ucsf.edu/) set to a mass accuracy of +/−50 ppm to compare unknown mass fingerprints to those of known proteins in the NCBI database.
Cryo-electron microscopy
3ul of highly purified capsid samples were placed on Quantifoil (Quantifoil Micro Tools GmbH, Jena, Germany) or regular lacey carbon-coated copper grids, mounted on an FEI Vitrobot (FEI Co., Hillsboro OR) where the grid was blotted to remove excess sample and then plunge-frozen into a 2:1 mixture of liquid ethane:propane 54. Frozen grids were maintained at liquid nitrogen temperature during transfer onto a Gatan 626 cryo-holder (Gatan, Pleasanton CA) and imaging in an FEI T20 FEG cryo-electron microscope. The microscope was operated at 200kV with nominal magnification of 29,000x (calibrated at 30,700x) and low dose methods were used to minimize electron beam damage to the sample. Defocus was varied between 1.0 to 3.0 μm, and exposures of 0.5 to 1.0 second were made on Kodak SO163 film that was subsequently developed for 12 minutes in full strength D19.
For the PRV A-, B-, and C-capsid samples, Quantifoil grids were similarly prepared but imaged on film in an FEI Polara microscope operating at 300 kV and at a magnification of 39,000x. For imaging the PRV-C capsid sample, an early version of a semi-automated imaging procedure developed by Felix de Haas (FEI Europe Apps Lab, Eindhoven) was used to move the microscope stage between the regularly positioned holes in the carbon support film after defining the spacing and directions of the hole array. Movement of the stage by dead-reckoning was sufficiently accurate over 7×7 areas of holes for each exposure to include at least 2/3 of the area of targeted hole, and the stage movement was followed by a 2 minute delay for mechanical stabilization. This approach has the benefit of a simple and quick set-up followed by automated collection of the image data.
Micrographs were digitized on a Nikon Super Coolscan 9000 (Nikon USA, Melville, NY) or on an inverted Nikon microscope controlled by the Nikon Elements software and with an automated stage feeding system developed in-house. The Nikon 9000 has a fixed scan rate of 6.35μm, corresponding to 2.1 Å/pixel at the sample for the TF20 images, and 1.6 Å/pixel for the Polara images, while the inverted microscope effective pixel size was 6.60μm, corresponding to 2.2 Å/pixel at the sample for the TF20 images. Particle images were boxed out of the digitized images using the “x3dpreprocess” program 55 and estimates of defocus were made manually with the BSOFT suite 56. Initial models were calculated for each sample using the RMC (Random Model Calculations) procedure 57 and refinements and reconstructions were calculated with the AUTO3DEM package 58. Reconstruction details and EMDB ID’s are shown in Table 1. Surface views of density maps were prepared in USCF Chimera 59 using thresholds estimated to enclose ~100% of the expected protein complement. CVSC and GFP densities were colored according to boundaries estimated by comparison with density maps lacking the molecules.
Table 1.
Details of PRV and HSV-1 capsid reconstructions. Resolution estimates were made within radial shells including all capsid density but excluding background and internal DNA density, and identifying the point where Fourier Shell Correlation (FSC) curves dropped below the 0.5 correlation coefficient level. The PRV 2168 virus does not make C-capsids on PK15 cells. Density maps were deposited with the EMDataBank (http://www.emdatabank.org/) and assigned the accession ID numbers as shown.
| Sample | Micrographs # | Particle Counts | Resolution (Å) | EMDB ID | |
|---|---|---|---|---|---|
| Total | Selected | ||||
| PRV C-capsids | 535 | 15016 | 11908 | 9.0 | EMD-5650 |
| PRV B-capsids | 47 | 1234 | 1111 | 15.9 | EMD-5652 |
| PRV A-capsids | 48 | 1915 | 1816 | 14.1 | EMD-5654 |
| PRV virion capsids | 150 | 3091 | 2647 | 22.6 | EMD-5655 |
| PRV 2168 PK15 B-capsids | 41 | 7861 | 5870 | 14.7 | EMD-5656 |
| PRV pUL25-gfp C-capsids | 97 | 3407 | 2872 | 23.2 | EMD-5657 |
| HSV-1 C-capsids | 48 | 4107 | 3687 | 16.7 | EMD-5659 |
| HSV-1 B-capsids | 48 | 1935 | 1846 | 16.9 | EMD-5660 |
| HSV-1 A-capsids | 48 | 1163 | 1093 | 16.9 | EMD-5661 |
Molecular modeling
Atomic models of the HSV-1 VP5 major capsid protein’s upper domain (PDB ID: 1NO7), the HSV-1 pUL25 protein (PDB: 2F5U) lacking 133 N-terminal residues, and the phage HK97 gp5 mature major capsid protein from the complete capsid (PDB: 1OHG) were placed into the cryo-EM density map of the PRV C-capsid using USCF Chimera 59. Initial placements were manual, following previous fits into a HSV-1 B-capsid map for the VP5 upper domain 45 and the HK97 gp5 protein 46, and these were refined as solid bodies within Chimera. The pUL25 model simply could not be fit manually or computationally as a solid body, and the resolution appears insufficient for the large-scale refolding that would appear necessary to accommodate the cryo-EM density.
Point Summary.
We characterize the structure of pseudorabies virus (PRV) capsids by cryoEM
PRV and herpes simplex virus type 1 (HSV-1) capsids appear to be very similar
The CVSC molecule and pUL25 protein have higher occupancy on PRV capsids than HSV-1
The HSV-1 pUL25 crystal structure is inconsistent with the PRV cryoEM density
PRV capsids offer better stability than HSV-1 for structural studies
Acknowledgments
We would like to thank Dr. Greg Smith for his kind gift of PRV viruses and complementing cell line PK25, and Dr. Josh Yoder for helping to fit models into the PRV C-capsid density map. We thank Jacob Jantzi and Krystal LaBarge for assisting with aspects of cryo-EM analyses, and Dr. Jay Carroll for his help with the mass spectrometry studies. Felix de Haas of FEI kindly allowed us to use an early version of his “Xilefon” package for automatic data acquisition. This work was supported by NIH grant 1R01A1089803-01A1 to J.F.C and F.L.H and R56AI060836 to F.L.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Steven A, Spear PG. Herpesvirus capsid assembly and envelopment. In: Burnett R, WC, Garcea R, editors. Structural Biology of Viruses. Oxford University Press; New York: 1996. pp. 312–351. [Google Scholar]
- 2.Rixon F. Structure and assembly of herpesviruses. Seminars in Virology. 1993;4:135– 144. [Google Scholar]
- 3.Harper D. Herpsvirus assembly proteins. Rev Med Virol. 1994;4:119–128. [Google Scholar]
- 4.Roizman B. Herpesviridae. In: Fields BN, DMK, Howley PM, Chanock RM, Melnick JL, Monath TP, Roizman B, Straus SE, editors. Fields Virology. Lippincott Raven; Philadelphia: 1996. pp. 2221–2230. [Google Scholar]
- 5.Roizman B, Sears AE. Herpes simplex viruses and their replication. In: Fields BN, DMK, Howley PM, Chanock RM, Melnick JL, Monath TP, Roizman B, Straus SE, editors. Fields Virology L. Lippincott Raven; Philadelphia: 1996. pp. 2231–2295. [Google Scholar]
- 6.Dargan D. The structure and assembly of herpesviruses. In: Horne JHaR., editor. Electron Microscopy of Proteins. Vol. 5. Academic Press; London: 1986. pp. 359–437. [Google Scholar]
- 7.Homa FL, Brown JC. Capsid assembly and DNA packaging in herpes simplex virus. Rev Med Virol. 1997;7:107–122. doi: 10.1002/(sici)1099-1654(199707)7:2<107::aid-rmv191>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 8.Brown JC, McVoy MA, Homa FL. Packaging DNA into herpesvirus capsids. In: Bogner E, AH, editors. Structure-function relationships of human pathogenic viruses. Kluwer Acacdemic/Plenum Publishers; New York: 2002. pp. 111–155. [Google Scholar]
- 9.Trus BL, Booy FP, Newcomb WW, Brown JC, Homa FL, Thomsen DR, Steven AC. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J Mol Biol. 1996;263:447–62. doi: 10.1016/s0022-2836(96)80018-0. [DOI] [PubMed] [Google Scholar]
- 10.Perdue ML, Cohen JC, Randall CC, O’Callaghan DJ. Biochemical studies of the maturation of herpesvirus nucleocapsid species. Virology. 1976;74:194–208. [PubMed] [Google Scholar]
- 11.Booy FP, Newcomb WW, Trus BL, Brown JC, Baker TS, Steven AC. Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell. 1991;64:1007–15. doi: 10.1016/0092-8674(91)90324-r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newcomb WW, Brown JC. Structure of the herpes simplex virus capsid: effects of extraction with guanidine hydrochloride and partial reconstitution of extracted capsids. J Virol. 1991;65:613–20. doi: 10.1128/jvi.65.2.613-620.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibson W, Roizman B. Proteins specified by herpes simplex virus. 8. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J Virol. 1972;10:1044–52. doi: 10.1128/jvi.10.5.1044-1052.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McNab AR, Desai P, Person S, Roof LL, Thomsen DR, Newcomb WW, Brown JC, Homa FL. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J Virol. 1998;72:1060–70. doi: 10.1128/jvi.72.2.1060-1070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sherman G, Bachenheimer SL. Characterization of intranuclear capsids made by ts morphogenic mutants of HSV-1. Virology. 1988;163:471–80. doi: 10.1016/0042-6822(88)90288-7. [DOI] [PubMed] [Google Scholar]
- 16.Conway JF, Trus BL, Booy FP, Newcomb WW, Brown JC, Steven AC. Visualization of three-dimensional density maps reconstructed from cryoelectron micrographs of viral capsids. J Struct Biol. 1996;116:200–8. doi: 10.1006/jsbi.1996.0031. [DOI] [PubMed] [Google Scholar]
- 17.Conway JF, Trus BL, Booy FP, Newcomb WW, Brown JC, Steven AC. The effects of radiation damage on the structure of frozen hydrated HSV-1 capsids. J Struct Biol. 1993;111:222–33. doi: 10.1006/jsbi.1993.1052. [DOI] [PubMed] [Google Scholar]
- 18.Spencer JV, Trus BL, Booy FP, Steven AC, Newcomb WW, Brown JC. Structure of the herpes simplex virus capsid: peptide A862-H880 of the major capsid protein is displayed on the rim of the capsomer protrusions. Virology. 1997;228:229–35. doi: 10.1006/viro.1996.8392. [DOI] [PubMed] [Google Scholar]
- 19.Trus BL, Gibson W, Cheng N, Steven AC. Capsid structure of simian cytomegalovirus from cryoelectron microscopy: evidence for tegument attachment sites. J Virol. 1999;73:2181–92. doi: 10.1128/jvi.73.3.2181-2192.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trus BL, Heymann JB, Nealon K, Cheng N, Newcomb WW, Brown JC, Kedes DH, Steven AC. Capsid structure of Kaposi’s sarcoma-associated herpesvirus, a gammaherpesvirus, compared to those of an alphaherpesvirus, herpes simplex virus type 1, and a betaherpesvirus, cytomegalovirus. J Virol. 2001;75:2879–90. doi: 10.1128/JVI.75.6.2879-2890.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng N, Trus BL, Belnap DM, Newcomb WW, Brown JC, Steven AC. Handedness of the herpes simplex virus capsid and procapsid. J Virol. 2002;76:7855–9. doi: 10.1128/JVI.76.15.7855-7859.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grunewald K, Desai P, Winkler DC, Heymann JB, Belnap DM, Baumeister W, Steven AC. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science. 2003;302:1396–8. doi: 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- 23.Cardone G, Winkler DC, Trus BL, Cheng N, Heuser JE, Newcomb WW, Brown JC, Steven AC. Visualization of the herpes simplex virus portal in situ by cryo-electron tomography. Virology. 2007;361:426–34. doi: 10.1016/j.virol.2006.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trus BL, Newcomb WW, Cheng N, Cardone G, Marekov L, Homa FL, Brown JC, Steven AC. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-Filled HSV-1 capsids. Mol Cell. 2007;26:479–89. doi: 10.1016/j.molcel.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Booy FP, Trus BL, Newcomb WW, Brown JC, Conway JF, Steven AC. Finding a needle in a haystack: detection of a small protein (the 12-kDa VP26) in a large complex (the 200-MDa capsid of herpes simplex virus) Proc Natl Acad Sci U S A. 1994;91:5652–6. doi: 10.1073/pnas.91.12.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trus BL, Homa FL, Booy FP, Newcomb WW, Thomsen DR, Cheng N, Brown JC, Steven AC. Herpes simplex virus capsids assembled in insect cells infected with recombinant baculoviruses: structural authenticity and localization of VP26. J Virol. 1995;69:7362–6. doi: 10.1128/jvi.69.11.7362-7366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trus BL, Cheng N, Newcomb WW, Homa FL, Brown JC, Steven AC. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J Virol. 2004;78:12668–71. doi: 10.1128/JVI.78.22.12668-12671.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schrag JD, Prasad BV, Rixon FJ, Chiu W. Three-dimensional structure of the HSV1 nucleocapsid. Cell. 1989;56:651–60. doi: 10.1016/0092-8674(89)90587-4. [DOI] [PubMed] [Google Scholar]
- 29.Zhou ZH, Prasad BV, Jakana J, Rixon FJ, Chiu W. Protein subunit structures in the herpes simplex virus A-capsid determined from 400 kV spot-scan electron cryomicroscopy. J Mol Biol. 1994;242:456–69. doi: 10.1006/jmbi.1994.1594. [DOI] [PubMed] [Google Scholar]
- 30.Zhou ZH, He J, Jakana J, Tatman JD, Rixon FJ, Chiu W. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat Struct Biol. 1995;2:1026–30. doi: 10.1038/nsb1195-1026. [DOI] [PubMed] [Google Scholar]
- 31.Heymann JB, Cheng N, Newcomb WW, Trus BL, Brown JC, Steven AC. Dynamics of herpes simplex virus capsid maturation visualized by time-lapse cryo-electron microscopy. Nat Struct Biol. 2003;10:334–41. doi: 10.1038/nsb922. [DOI] [PubMed] [Google Scholar]
- 32.Baker ML, Jiang W, Bowman BR, Zhou ZH, Quiocho FA, Rixon FJ, Chiu W. Architecture of the herpes simplex virus major capsid protein derived from structural bioinformatics. J Mol Biol. 2003;331:447–56. doi: 10.1016/s0022-2836(03)00696-x. [DOI] [PubMed] [Google Scholar]
- 33.Newcomb WW, Trus BL, Booy FP, Steven AC, Wall JS, Brown JC. Structure of the herpes simplex virus capsid. Molecular composition of the pentons and the triplexes. J Mol Biol. 1993;232:499–511. doi: 10.1006/jmbi.1993.1406. [DOI] [PubMed] [Google Scholar]
- 34.Zhou ZH, Dougherty M, Jakana J, He J, Rixon FJ, Chiu W. Seeing the herpesvirus capsid at 8.5 A. Science. 2000;288:877–80. doi: 10.1126/science.288.5467.877. [DOI] [PubMed] [Google Scholar]
- 35.Caspar DL, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 36.Newcomb WW, Juhas RM, Thomsen DR, Homa FL, Burch AD, Weller SK, Brown JC. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J Virol. 2001;75:10923–32. doi: 10.1128/JVI.75.22.10923-10932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spencer JV, Newcomb WW, Thomsen DR, Homa FL, Brown JC. Assembly of the herpes simplex virus capsid: preformed triplexes bind to the nascent capsid. J Virol. 1998;72:3944–51. doi: 10.1128/jvi.72.5.3944-3951.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toropova K, Huffman JB, Homa FL, Conway JF. The Herpes Simplex Virus 1 UL17 Protein Is the Second Constituent of the Capsid Vertex-Specific Component Required for DNA Packaging and Retention. Journal of Virology. 2011;85:7513–22. doi: 10.1128/JVI.00837-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conway JF, Cockrell SK, Copeland AM, Newcomb WW, Brown JC, Homa FL. Labeling and localization of the herpes simplex virus capsid protein UL25 and its interaction with the two triplexes closest to the penton. J Mol Biol. 2010;397:575–86. doi: 10.1016/j.jmb.2010.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cockrell SK, Huffman JB, Toropova K, Conway JF, Homa FL. Residues of the UL25 Protein of Herpes Simplex Virus That Are Required for Its Stable Interaction with Capsids. J Virol. 2011;85:4875–4887. doi: 10.1128/JVI.00242-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuhn J, Leege T, Klupp BG, Granzow H, Fuchs W, Mettenleiter TC. Partial functional complementation of a pseudorabies virus UL25 deletion mutant by herpes simplex virus type 1 pUL25 indicates overlapping functions of alphaherpesvirus pUL25 proteins. J Virol. 2008;82:5725–34. doi: 10.1128/JVI.02441-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cockrell SK, Sanchez ME, Erazo A, Homa FL. Role of the UL25 protein in herpes simplex virus DNA encapsidation. J Virol. 2009;83:47–57. doi: 10.1128/JVI.01889-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klupp BG, Granzow H, Keil GM, Mettenleiter TC. The capsid-associated UL25 protein of the alphaherpesvirus pseudorabies virus is nonessential for cleavage and encapsidation of genomic DNA but is required for nuclear egress of capsids. J Virol. 2006;80:6235–46. doi: 10.1128/JVI.02662-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leelawong M, Lee JI, Smith GA. Nuclear egress of pseudorabies virus capsids is enhanced by a subspecies of the large tegument protein that is lost upon cytoplasmic maturation. J Virol. 2012;86:6303–14. doi: 10.1128/JVI.07051-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bowman BR, Baker ML, Rixon FJ, Chiu W, Quiocho FA. Structure of the herpesvirus major capsid protein. EMBO J. 2003;22:757–65. doi: 10.1093/emboj/cdg086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baker ML, Jiang W, Rixon FJ, Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J Virol. 2005;79:14967–14970. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bohannon KP, Sollars PJ, Pickard GE, Smith GA. Fusion of a fluorescent protein to the pUL25 minor capsid protein of pseudorabies virus allows live-cell capsid imaging with negligible impact on infection. J Gen Virol. 2012;93:124–9. doi: 10.1099/vir.0.036145-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen DH, Jakana J, McNab D, Mitchell J, Zhou ZH, Dougherty M, Chiu W, Rixon FJ. The pattern of tegument-capsid interaction in the herpes simplex virus type 1 virion is not influenced by the small hexon-associated protein VP26. J Virol. 2001;75:11863–7. doi: 10.1128/JVI.75.23.11863-11867.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou ZH, Chen DH, Jakana J, Rixon FJ, Chiu W. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J Virol. 1999;73:3210–8. doi: 10.1128/jvi.73.4.3210-3218.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thurlow JK, Rixon FJ, Murphy M, Targett-Adams P, Hughes M, Preston VG. The herpes simplex virus type 1 DNA packaging protein UL17 is a virion protein that is present in both the capsid and the tegument compartments. J Virol. 2005;79:150–8. doi: 10.1128/JVI.79.1.150-158.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coller KE, Lee JI, Ueda A, Smith GA. The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25 and VP1/2 proteins. J Virol. 2007;81:11790–7. doi: 10.1128/JVI.01113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith GA, Pomeranz L, Gross SP, Enquist LW. Local modulation of plus-end transport targets herpesvirus entry and egress in sensory axons. Proc Natl Acad Sci U S A. 2004;101:16034–9. doi: 10.1073/pnas.0404686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tischer BK, von Einem J, Kaufer B, Osterrieder N. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques. 2006;40:191–7. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- 54.Tivol WF, Briegel A, Jensen GJ. An improved cryogen for plunge freezing. Microsc Microanal. 2008;14:375–9. doi: 10.1017/S1431927608080781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Conway JF, Steven AC. Methods for reconstructing density maps of “single” particles from cryoelectron micrographs to subnanometer resolution. J Struct Biol. 1999;128:106–18. doi: 10.1006/jsbi.1999.4168. [DOI] [PubMed] [Google Scholar]
- 56.Heymann JB, Belnap DM. Bsoft: Image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 57.Yan X, Dryden KA, Tang J, Baker TS. Ab initio random model method facilitates 3D reconstruction of icosahedral particles. J Struct Biol. 2007;157:211–25. doi: 10.1016/j.jsb.2006.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yan X, Sinkovits RS, Baker TS. AUTO3DEM-an automated and high throughput program for image reconstruction of icosahedral particles. J Struct Biol. 2007;157:73–82. doi: 10.1016/j.jsb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 60.Newcomb WW, Homa FL, Brown JC. Herpes simplex virus capsid structure: DNA packaging protein UL25 is located on the external surface of the capsid near the vertices. J Virol. 2006;80:6286–94. doi: 10.1128/JVI.02648-05. [DOI] [PMC free article] [PubMed] [Google Scholar]