Abstract
Tissue-based proteomic approaches (tissue proteomics) are essential for discovering and evaluating biomarkers for personalized medicine. In any proteomics study, the most critical issue is sample extraction and preparation. This problem is especially difficult when recovering proteins from formalin-fixed, paraffin-embedded (FFPE) tissue sections. However, improving and standardizing protein extraction from FFPE tissue is a critical need because of the millions of archival FFPE tissues available in tissue banks worldwide. Recent progress in the application of heat-induced antigen retrieval (AR) principles for protein extraction from FFPE tissue has resulted in a number of published FFPE tissue proteomics studies. However, there is currently no consensus on the optimal protocol for protein extraction from FFPE tissue or accepted standards for quantitative evaluation of the extracts. Standardization is critical to ensure the accurate evaluation of FFPE protein extracts by proteomic methods such as reverse phase protein arrays (RPPA), which is now in clinical use. In our view, complete solubilization of FFPE tissue samples is the best way to achieve the goal of standardizing the recovery of proteins from FFPE tissues. However, further studies are recommended to develop standardized protein extraction methods to ensure quantitative and qualitative reproducibility in the recovery of proteins from FFPE tissues.
Keywords: antigen retrieval, FFPE, Formalin-fixed, paraffin-embedded, proteomics, tissue proteomics, protein extraction, elevated pressure
Introduction
The rapid development of high throughput proteomic techniques has provided a valuable approach for new biomarker discovery in the post-genomic era. It has been suggested that a tissue-based proteomic approach (tissue proteomics) is essential for discovering and evaluating biomarkers for personalized medicine. Several novel terms such as “histoproteomics” [1], “liquid morphology” [2], “proteome maps” [3], and “toponomics” [4] have been proposed to emphasize the increasing use of tissue proteomics in the molecular biomedical research field. In any proteomic study it is accepted that the most critical step is the quantitative recovery of proteins from the sample prior to the downstream analytic method [5]. While protein recovery can be difficult for some fresh or frozen tissues, the problem is especially challenging when the samples are formalin-fixed, paraffin-embedded (FFPE) tissue sections. However, there is a critical need to develop rigorous and reproducible methods to extract proteins from FFPE tissues in view of the large number of archival FFPE tissue banks that have been established worldwide over the last century. These FFPE tissue collections, with their accompanying clinical outcome information, are invaluable resources for translational studies of cancer and other diseases. The availability of modern techniques, such as mass spectrometry (MS), offers the prospect of analyzing literally thousands of proteins in a single assay. An efficient protocol for protein extraction from archival FFPE tissue sections would appear to open the door to a veritable treasure trove of information sequestered in archival tissue banks. In this context, immunohistochemistry (IHC), as applied to the identification of antigens (mainly proteins) in tissue sections, is itself a form of proteomics. The ability to identify proteins in IHC has been greatly enhanced by a simple and effective antigen retrieval (AR) technique. In the AR method, boiling the FFPE tissue sections in water or buffer solutions gives a dramatic enhancement of the IHC signal [6]. This finding has revolutionized the practice of diagnostic IHC to the extent that the relevant published literatures on IHC is divided into the “pre-AR” and “post-AR” eras [7]. With these observations in mind, the merit of evaluating AR methods for extracting proteins from FFPE tissues appears obvious. Indeed, several recent studies have used the principal of heat-induced AR in certain ‘retrieval solutions’ to develop efficient methods to extract proteins from FFPE tissue sections [8–13]. In most of these studies, heat appeared to be the most important factor for achieving a near qualitative and quantitative extraction of proteins from FFPE tissues [12–14]. Currently, most investigators accept that proteins extracted from FFPE tissue using heat-induced AR protocols are suitable for proteomic analysis as evidenced by a reported 40–90% overlap of proteins identified by MS in matched FFPE and fresh tissues obtained from the same specimen[13, 14].
For more than one hundred years, formalin-fixation and paraffin-embedding has served as the standard tissue preparation method in surgical pathology, with routine hematoxylin and eosin stained FFPE tissue sections providing the basis for most pathological diagnosis. These archival tissues, housed in pathology files worldwide, are an invaluable treasure for retrospective research. The recent increase in the use of heat-induced AR protocols for protein extraction from FFPE tissue sections parallels the emergence of the use of AR in IHC two decades ago [13–16]. Further, there is a growing body of literature where AR-based methods are used for the extraction of a variety of molecules (DNA, RNA and proteins) from FFPE tissues. These studies have further extended the utility of archival tissues, and promises to allow the combination of genomics and proteomics with IHC to achieve a true molecular morphology approach to the study of disease [13, 17–22]. So effective have these AR-based approaches become that investigators are now actively exploring the advantages of FFPE tissues for preserving both morphology and molecular diagnostic capability in cell/tissue samples over other methods of sample preparation. [9, 12, 23–32].
Current status of techniques for protein extraction from FFPE tissue
Recently, Tanca, et al [13] published in this journal a comprehensive review on the current status of techniques for extracting proteins from FFPE tissues. In brief, these authors highlighted the following major methodological achievements and remaining hurdles in this area of proteomics. (1) Substantial recovery of full-length proteins from FFPE tissue with reversal of their formaldehyde-induced cross-links has been achieved using a heat-induced AR heating protocol. (2) 2-D polyacrylamide gel electrophoresis (PAGE) analysis using proteins extracted from FFPE tissue sections has been satisfactorily performed by several investigators [13]. (3) Numerous protocols have been published demonstrating a continuing improvement in the efficiency of protein extraction from FFPE tissue. (4) Although it is generally accepted that FFPE tissue proteomic data are dependable enough to be used for biomarker discovery purposes, there remains significant variance between protein profiles from FFPE and matched fresh-frozen tissue sections. In general, FFPE tissue-based proteomics is a work in progress, and several critical issues need refinement and further study. Fowler et al [12] pointed out some challenging issues with respect to proteins extracted from FFPE tissue, such as the potential for selective recovery of proteins from FFPE tissues. Nirmalan et al [33] have emphasized that “the variability observed in extraction efficacies for some membrane proteins emphasizes the need for cautious interpretation of quantitative data from this subset of proteins”. Heaton and Master [34] recently published a comprehensive protocol for extracting peptides from FFPE tissue, describing tissue deparaffinization, cell harvesting by laser-capture microdissection (LCMD), sample extraction, and MS analysis. They highlighted several issues important for standardizing protein extraction and MS analysis, including sample quality. Alkhas et al [35] also reported a standardized procedure for collection-to-injection proteomics of LCMD FFPE uterine tissue. They recovered approximately 100 ng of tryptic peptides from around 600 endometrial tumor cells. Huang et al [36] successfully detected more than 1500 proteins with a confidence level of >70% from LCMD FFPE metastatic and primary melanoma tissue. Though these results show that proteomic analysis of FFPE tissue is possible, many of these studies rely upon LCMD of only a small number of isolated cells. While useful for comparative proteomics and evaluation of known biomarkers, the discovery of new potential biomarkers may require extraction of whole tissue sections.
Based upon the above observations, it is clear that there are several major topics that must be addressed to assure the scientific accuracy of FFPE tissue proteomics. (1)What factors affect efficient and reproducible extraction of proteins from FFPE tissue? (2) What is the optimal protocol for extracting proteins from FFPE tissues that is both reproducible and effective? (3) Is it necessary to establish a universal standard for the evaluation of the quality and quantity of proteins extracted from FFPE tissues? (4) To avoid potential bias due to preferential extraction of proteins by a single protocol, are multiple extraction steps using a range of pH values, buffers or heating conditions necessary to achieve complete protein coverage? Therefore, it is clear that further study along the lines of standardization of protein extraction is required at this time.
Factors affecting extraction of proteins from FFPE tissue
While the results of recent proteomics studies are encouraging, there are a number of challenges that must be addressed to develop better and more reproducible techniques for extracting and identifying proteins useful for proteomic analysis from archival FFPE tissue. These issues include selective recovery of protein, incomplete recovery of protein, incomplete reversal of formaldehyde modifications, and protein degradation. The extraction and identification by MS of proteins from FFPE tissues has been hampered by the deleterious effects of formaldehyde-induced protein modifications and cross-links that are formed during tissue fixation and subsequent histological processing. Three types of formaldehyde-induced chemical modifications have been identified in proteins and model peptides: (a) methylol (hydroxymethyl) adducts, (b) Schiff’s bases, and (c) stable methylene bridges [37, 38]. Formaldehyde can react with lysine, cysteine, arginine, tryptophan, histidine, and the N-terminal amine to form methylol adducts. The methylol adduct can subsequently undergo a dehydration reaction to form a Schiff’s base, which is seen most frequently in lysine and tryptophan residues. Additionally, the protein N-terminal amine can be converted to a stable 4-imidazolidione adduct [37] and a Mannich reaction can occur between adducted tyrosine and arginine residues in close spatial proximity [39]. Intramolecular protein cross-links (methylene bridges) have been reported in both model peptides [38] and whole proteins, such as insulin [37].
In addition to the primary covalent modifications caused by formalin fixation, other groups have studied the effects of tissue processing and long term storage on the recovery of proteins from FFPE tissue. Xie et al showed that when tissues were inadequately dehydrated during tissue processing, the retained water resulted in reduced antigenicity and that exposure to high humidity resulted in protein degradation during storage [40]. They also found that fixation time had little effect on protein antigenicity, but that protein integrity was improved by storing FFPE tissues at low temperatures (4°C). In contrast, Wolff et al found that total protein recoverable from FFPE tissues decreased with fixation time, with yields dropping by ~20% for every 24 hours in formalin [41]. Xie et al [40] concluded that oxidation did not appear to contribute to protein degradation in FFPE tissues; however, Sprung et al [42] found that methionine oxidation in archival FFPE tissue increased from 17% after 1 year to 25% after 10 years of storage. They also observed a decreased representation of proteins with C-terminal lysine residues with increased storage time. Scicchitano et al [43] compared protein recovery from matched liver tissue prepared as OCT-frozen specimens versus FFPE tissue blocks and FFPE slides. Recovery of proteins from LCMD liver samples isolated from FFPE slides was close to 90% compared to OCT-frozen tissue. For whole liver tissue sections, protein recovery dropped to ~42% with greater variability between specimens. Scicchitano et al also observed that no intact proteins >120 kDa were detected by SDS-PAGE and Western blot suggesting that hydrolysis of the peptide backbone may occur in FFPE tissue [43]. Similar findings were reported by Fowler et al [44].
Balgley et al [45] investigated the effect of archival storage time on the proteome analysis of nine uterine leiomyomas dating from 1990 to 2002 using transient capillary isotachophoresis/capillary zone electrophoresis-coupled linear ion-trap mass spectrometry. Evaluation of the results by the Pearson coefficient indicated that the leiomyoma specimens correlated strongly with each other, but not by archival age. Analysis of the mass spectrometry data by k-means clustering also failed to reveal a strong effect of archival age on the proteome. However, when individual proteins were analyzed, k-means clustering suggested that low abundance proteins are more difficult to retrieve as the tissue blocks aged beyond 10 years. A shotgun proteomic analysis of colon adenoma FFPE tissues carried out by Sprung et al [42] also concluded that the proteome did not change significantly for archival storage times of up to 10 years. In contrast to these findings, Wolff et al found that protein recovery from FFPE colon tissue decreased with age, with 42% less protein recoverable from specimens dating from 1990 relative to specimens from 2010 [41].
In summary, there is no clear consensus at this time on the effect of fixation time, tissue processing, and archival storage time on the proteome recovered from FFPE tissues. Caution must be used when comparing studies that estimate the success of protein extraction from FFPE tissues by comparing the overlap of proteins identified in FFPE tissue extracts with those from matched fresh or frozen tissue. Even for matched fresh tissue specimens, protein overlap can vary significantly due to variations in sample preparation, instrument performance, and the type of mass spectrometer used in the analysis [45]. An additional complication is that some studies report results that include proteins identified with a single constituent peptide while other studies will only include proteins identified by ≥2 peptides. Typically, the overlap between proteins recovered from FFPE tissues and their matching fresh or frozen counterparts drops by 30–50% when single peptide identifications are excluded [42].
Studies with tissue surrogates, a model system in which one or more proteins form a solid protein plug when treated with formaldehyde, highlighted several of these issues. Recovery of proteins from these model tissue surrogates was generally modest when the tissue surrogates were extracted using a method based upon heat-induce AR originally reported by Shi et al [9]. The protein extraction efficiency was approximately 26% for a multi-protein tissue surrogate heated in a water bath at 80 to 100 °C using a Tris buffer with 2% (w/v) sodium dodecyl sulfate [46]. Additionally, studies with multi-protein tissue surrogates revealed extraction bias, meaning that the composition of the solubilized proteins did not match that of the corresponding tissue surrogate [10, 46]. Protein extracts from surrogates containing carbonic anhydrase:lysozyme (2:1 mole/mole) appeared to contain disproportionate percentages of lysozyme. Approximately 72% of total protein by mass corresponded to monomeric lysozyme, while monomeric carbonic anhydrase and a band of the correct size for a lysozyme:carbonic anhydrase hetero-dimer accounted for 19% and 3.5%, respectively [10]. Further studies with a complex multi-protein FFPE tissue surrogate comprised of hen egg-white lysozyme, bovine carbonic anhydrase, bovine ribonuclease A, bovine serum albumin, and equine myoglobin (55:15:15:10:5 wt%) showed similar bias. As summarized in Table 1, when the multi-protein tissue surrogate was subjected to a commonly used extraction protocol (heating in a water bath in Tris buffer) [9, 10], only two of the five proteins were identified by gel electrophoresis or MS. The percentages of false protein identification by MS for this extraction condition was also 42%, compared to only 3.3% for the native, unfixed protein mixture [46].
Table 1.
LC/MS analysis for a 5-protein FFPE tissue surrogate
| Condition | % protein recovery | Protein Identification by LC/ MS
|
|||||
|---|---|---|---|---|---|---|---|
| % sequence Coverage*
| |||||||
| % False protein ID’s ** | Lysozyme | Carbonic Anhydrase | RNAse A | BSA | Myoglobin | ||
|
| |||||||
| Native Protein mixture | 100% | 3.3 ± 0.6 | 66% | 56% | 63% | 54% | 38% |
| FFPE; extracted in a water bath | 26% | 42 ± 4.0 | 15% | n.d. | 7% | n.d. | n.d. |
| FFPE, extracted with high pressure | 96% | 7.8 ± 1.5 | 69% | 36% | 59% | 26% | 28% |
Multi-protein FFPE tissue surrogates were extracted at 100°C for 30 min followed by 80°C for 2 h in a water bath at atmospheric pressure or with high pressure (40,000 psi) in 50 mM Tris-HCl, 2% (w/v) SDS buffer, pH 4. The extracts were washed extensively, and digested overnight with trypsin at 37°C in 50 mM NH4HCO3, pH 7.9 with 20% acetonitrile (v/v) before MS.
%Sequence coverage: percent of theoretical tryptic peptides identified by LC/MS/MS.
The false identification rate was determined as percentage of proteins incorrectly identified for spectra with scores ≥10.5, for 2 technical replicates. n.d. – none detected.
Finally, it has been established that elevated temperature is required to provide the energy necessary to reverse formaldehyde-induced protein adducts and cross-links. Such heat treatment can produces pH -dependent protein modifications. At acidic pH, these potential modifications are deamination of glutamine and asparagine residues, and peptide bond hydrolysis on the carboxylate side of aspartic acid residues [47]. The MS analysis of a lysozyme tissue surrogate recovered following treatment at 80°C and a pressure of 43,500 psi suggests that deamination is not a significant problem under these conditions. However, lower molecular weight hydrolysis products were identified as peptide fragments produced by aspartic acid hydrolysis [11]. At neutral pH, deamination of glutamine and asparagine residues can occur along with thiol-catalyzed disulfide exchange. At alkaline pH, β-elimination of cystine residues can occur, which involves a heterolytic cleavage of the disulfide bond to form dehydroalanine and thiocysteine residues. Dehydoalanine can then react with the ε-amino groups of lysine residues to form a lysinoalanine cross-link [48]. Many of these themally-induced modifications can be accounted for by including them in the peptide database search, as was done for the lysozyme tissue surrogate discussed above [11]. However, deamination of glutamine and asparagine yields glutamate and aspartate, respectively; changes that would not be detected by a typical peptide database searched.
Is complete protein solubilization from FFPE tissues required for standardization?
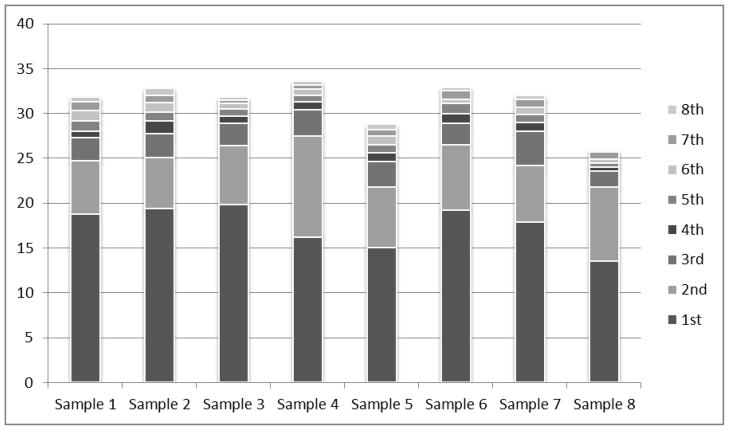
Our previous experience with frozen cell or tissue samples revealed that such samples were completely solubilized following the protein extraction step. However, when extracting proteins from FFPE tissue sections a residual tissue pellet invariably remained following the extraction process. We recognized that some proteins may remain in the residual tissue pellet, and inferred that this may be the major reason why the yields of protein extracted from FFPE tissue are always lower than comparable extracts obtained from fresh-frozen tissue. In reviewing the literature, few people appear to have noted this critical issue. Recently, our research group attempted to standardize protein extraction from FFPE tissue sections by using 8 sequential extraction steps to achieve complete solubilization of whole FFPE tissue sections. This was repeated for 8 serial tissue sections (replicates) taken from a single FFPE swine liver tissue block. For all replicates, tissue sections were extracted in 20 mM Tris-HCl, pH 7, 2% (w/v) SDS, using our previously reported heat-based AR protocol [9]. After each protein extraction step, the remaining pellet was resuspended in the extraction buffer and re-heated. All 8 protein extracts were quantified separately and then pooled. Figure 1 shows the results of the sequential protein extractions for the 8 tissue replicates [49]. Between replicates, we found that the total pooled protein yields were similar, although variable protein yields could be seen after each of the 8 individual extraction steps. In addition, we investigated to see if there were any qualitative differences in the proteins recovered by the 8 sequential extraction steps. 1-D PAGE showed that a similar pattern of protein bands were present in the first two serial extracts (not shown), although the weaker intensity of the bands in the second extracts were due to lower amount of proteins extracted as shown in Figure 1. However, further detailed studies will be required to determine if there are any qualitative differences in the proteins recovered in each of the 8 sequential extractions relative to the final pooled extract.
Figure 1.
Sequential Protein Extraction. Protein concentrations (μg/μl) were measured after each sequential extraction step and after the extracts were pooled. Eight serial tissue sections (identified as samples 1–8) from a single 40 micron thick FFPE swine liver tissue block were each extracted in 8 sequential steps by heating in 20 mM Tris-HCL with 2% (w/v) SDS. Each colored bar shows how much protein in μg/μl was extracted during each separate extraction step. Note that no single extraction step resulted in complete protein solubilization resulting in significant variability among the 8 sequential extractions. However, pooling all 8 extracts resulted in a comparable total protein concentration between replicates (P=0.9527). Use of animal tissue has been approved by USC Animal Care Unit.
Studies of the type described above are critically important to ensure the development of protein extraction methods for FFPE tissue sections that are devoid of qualitative protein extraction bias [10, 12] and that yield accurate single protein and total protein quantitation. Such qualitative and quantitative accuracy is essential for the analysis of FFPE protein extracts by proteomic methods such as RPPA [28, 50] and quantitative IHC [14]. Accordingly, improved methods for standardizing protein extraction from FFPE tissue sections are critical if we wish to exploit FFPE tissue archives as a source for retrospective proteomic studies. In the preliminary study described above, we found that a sequential extraction procedure resulted in complete solubilization of FFPE tissue sections, although it is a time consuming procedure [49].
Recent studies regarding improving efficiency to reach the goal of complete solubiliztion
Nirmalan et al [33] developed an effective protocol using heat-induced AR in combination with Laemmli buffer to achieve an average protein extraction efficiency of 63% from FFPE tissue sections. However, it was pointed out that the temperature extraction regime necessary for formaldehyde-induced cross-link reversal also causes membrane protein aggregation. They emphasized the need for protocol refinement and cautious interpretation of quantitative data for this subset of proteins. Several modified protocols, based on a combination of chemical adjuvants and physical treatment, have recently been developed that improve the efficiency of protein extraction from FFPE tissue sections [1, 12, 30, 33]. We tested several buffer additives, including glycine, Tween-20, Triton X-100, dish soap, DTT, or guanidine-HCl in various concentrations, either singly or in various combinations. The protein extraction efficiency under each protocol was determined [49], with the goal of establishing a more efficient protocol for protein extraction from FFPE tissue. In addition, we also tested several mechanical homogenization procedures including mortar and pestle and/or ultrasound to break up the cells or tissue prior to protein extraction. Although these procedures did improve extraction efficiency, no one protocol or buffer completely solubilized the FFPE tissue sections [49].
Jiang et al [30] reported that a buffer containing 40 mM Tris, 6 M guanidine-HCl, and 65 mM DTT, pH 8.2 combined with boiling yielded complete protein solubilization of FFPE tissue sections. However, further study involving analysis of the extracted proteins is needed to validate this protocol. Although this study demonstrated that higher yields of proteins could be achieved [30], extensive post-extraction cleanup before performing downstream analysis, such as MS, is required. In our hands, protein analysis was complicated by a significant loss of protein after the ethanol precipitation step of the Jiang protocol (from 42.1μg/μl to 5.2 μg/μl) [49]. Finally, Murphy et al [51], reported that the incubation of FFPE tissue sections at 37°C in a Tris-HCl buffer, pH 8, containing EDTA and guanidine-HCl achieved complete solubilization of the tissue.
Elevated pressure improves the extraction of proteins recovered from FFPE tissue
Using proteins in aqueous solution, Rait et al demonstrated that the majority of protein formaldehyde adducts and cross-links were consistently reversed with mild heating following the removal of excess formaldehyde by dialysis [17, 18]. Subsequent studies by Fowler et al showed that the ethanol dehydration step of histology caused most formaldehyde-treated proteins to adopt conformations enriched in β-sheets, leading to the formation of a dense network of intermolecular formaldehyde cross-links [52]. These results suggested that the difficulty of re-hydrating the dense network of formaldehyde cross-linked proteins is likely the primary impediment to recovering proteins from FFPE tissue.
The addition of elevated pressure (45,000 psi) to augment heat treatment dramatically improved both the protein extraction efficiency (from 60 to 100%) and the reversal of formaldehyde-induced protein cross-links [11]. A similar result was observed with a 5-protein tissue surrogate [46]. As discussed above, when the tissue surrogate was heated in a water bath at atmospheric pressure, protein recovery was modest and only 2 of the 5 proteins in the tissue surrogate were correctly identified by MS. However, when the 5-protein tissue surrogate was heated under elevated pressure (40,000 psi) no extraction bias was observed as the proteomic analysis of the extracted proteins by LC/MS was virtually identical to that of the native protein solution (Table 1) [46]. Subsequent experiments with FFPE mouse liver were equally encouraging. When FFPE liver sections were extracted using a combination of heat and elevated pressure (40,000 psi), there was a 4-fold increase in protein extraction efficiency, a 3-fold increase in the extraction of intact proteins, and up to a 30-fold increase in the number of non-redundant proteins identified by MS, compared to FFPE tissue extracted with heat alone. More importantly, the number of non-redundant proteins identified in the FFPE tissue was nearly identical to that of matched fresh-frozen sample [44]. 1 y-old FFPE liver was then extracted in Tris buffer, pH 8, containing SDS and DTT, using conditions previously reported by Ostasiewicz et al [53]. The tissue sections were heated with or without elevated pressure, and the extraction efficiency and MS results for the 1 year-old samples are summarized in Table 2. These results, combined with mechanistic studies, indicated that elevated hydrostatic pressure hydrates the inner core of the proteins, induces protein unfolding, and reduces the size of the protein aggregates, thus allowing full access of the formaldehyde adducts and cross-links to the reversal buffer [46]. Elevated hydrostatic pressure has potential as an adjuvant method that can be applied to existing or future FFPE protein extraction protocols. While it remains to be shown that this method is useful with every protein extraction protocol, elevated pressure acts through purely physical means and should be compatible with FFPE protein extraction buffers of any pH and containing any detergent, protein denaturant, or other additive [11]. This should allow its integration into a wide range of protein extraction protocols for MS-based proteomics with little or no alteration to downstream sample preparation and analysis.
Table 2.
MS analysis of FFPE and matched fresh-frozen mouse liver tissue. 1-year old FFPE mouse liver was homogenized in extraction buffer and heated with or without elevated pressure. Fresh-frozen tissue was extracted either at atmospheric pressure using the indicated extraction conditions, or on ice for 2.5 h
| Tissue | Pressure (psi) | Buffer a | Extraction conditions | %Protein Extraction b | Unique Peptide IDs | Unique Protein IDs |
|---|---|---|---|---|---|---|
| Frozen, 1 y | 14.7 | Tris/DTT | 95 °C, 3 min | 100% | 5872 | 3415 |
| FFPE, 1 y | 14.7 | Tris/DTT | 95 °C, 1h | 18% | 107 | 107 |
| FFPE, 1 y | 40,000 | Tris/DTT | 95 °C, 1h | 79% | 5180 | 3492 |
Extraction buffer: 100 mM Tris-HCl, pH 8, 100 mM DTT, 2% (w/v) SDS [40].
The amount of protein extracted from fresh frozen tissue was set to 100%.
Comments and Future Directions
Protein extraction from FFPE tissue sections has been widely studied in recent years [13] prompted by the fact that protein extracts from FFPE tissue are an invaluable resource for the retrospective study of disease progression and response to therapy, which broadens the scope of proteomic analysis in both basic and clinical research [9, 12, 27–31, 42]. However, to date there is neither a universal protocol for protein extraction from FFPE tissues, nor any basic principle of standardization that is acceptable for practice. Standardization in this context is an important issue, particularly with respect to the measurement of single protein concentrations within total FFPE protein extracts or the relative quantitation of proteins from multiple FFPE tissues for biomarker discovery [50]. Since there is currently no method that can measure the amount of protein in FFPE tissues prior to sample extraction, it is critical that tissue samples be completely solubilized for accurate measurement of either total protein or a single protein within the extract. Consequently, complete solubilization of FFPE tissue samples, as part of any protein extraction procedure, is reasonably recommended as the best way to achieve the goal of standardization for tissue proteomics. Giavalisco et al [54] reported a three step sequential procedure for the extraction of total protein from plant tissue using different chemicals or chaotropes. This method achieved a more complete protein profile than a single extraction procedure. Most recently, Ericsson and Nister [55] coined the concept of “maximal extraction and solubilization of proteins from tissue” as an approach for the standardization of sample preparation for MS analysis. Because there is no way to quantitate the initial amount of protein in a tissue sample, it was recommended to use the percentage of tissue solubilization as a measure of protein extraction efficiency. Unfortunately, most published protocols fail to include a measure of the percentage of tissue solubilized by their protein extraction procedure. A further complication pointed out by Nirmalan et al [33] is that the estimation of total protein concentration in FFPE tissue extracts can be inaccurate, although some studies have developed approaches to improve the accuracy of this measurement [56]. Also, from mechanistic studies with tissue surrogates, it appears that protein solubilization and the reversal of formaldehyde adducts are uncoupled events; specifically, methods that maximize protein solubilization should not be assumed to also maximize reversal of formaldehyde-induced protein cross-links [46]. For maximum protein retrieval, methodologies that address both of these issues, such as the use of elevated pressure and heat, should improve the proteomic analysis of FFPE tissue. These approaches, together with rigorous biostatistics and methodology to control the overall coefficient of variance in a study [45] will help to standardize protein extraction from FFPE tissue.
Further study is clearly needed to develop more efficient protocols that achieve the goal of complete solubilization of FFPE tissue during the protein extraction step. Studies that explore the use of different chemical adjuvants, tissue homogenization methods, heating protocols, and the use of homogeneous or inhomogeneous sequential extraction methods are likely to accomplish this goal. The recently developed elevated pressure-assisted method is a powerful approach for improving tissue solubilization using existing protein extraction protocols. The only drawback may be the requirement for instrumentation that may limit its wide application though some commercial instrumentation for high-pressure extraction is available.
In conclusion, FFPE tissue proteomics is a timely topic in this era of personalized medicine and the search for disease biomarkers. Therefore, standardization of FFPE tissue extraction is a critical issue that needs further study to clarify the following issues:
How does sample treatment before and after formalin-fixation and paraffin-embedding affect MS results?
Does partial solubilization of FFPE tissue sections lead to extraction bias resulting in a misrepresentation of the tissue proteome?
Is complete solubilization of FFPE tissue sections required to standardize both the accurate quantification of individual proteins in a single tissue section or to accurately compare protein content between different FFPE tissue sections?
What are the optimal components and properties of extraction buffers used for FFPE tissues? These elements can include ionic strength, pH, detergents, chaotropic agents, protein denaturants, protein stabilizers, and protein reducing agents.
What are the optimal extraction conditions for FFPE tissues? These conditions can include tissue grinding, freeze/thaw, sonication, pressure, and heat.
Is there a “universal” extraction method that works for all FFPE tissues or must the extraction method be optimized for a particular FFPE tissue type?
Can complete solubilization of FFPE tissue sections be achieved using a single extraction step or are multiple extraction steps required?
If multiple extraction steps are required, is a single extraction buffer sufficient (homogeneous extraction) or are a series of extraction buffers of different composition and properties required (heterogeneous extraction)?
If multiple extraction steps are used, is optimal proteomic analysis achieved by analyzing the individual extracts or by analyzing the pooled extracts?
Is quantitative recover of integral membrane proteins from FFPE tissues possible?
How effective is enzyme digestion prior to MS analysis when using an FFPE tissue extract with a visible residual pellet and could enzymatic digestion abrogate the need for complete solubilization of FFPE tissue sections?
How do formalin-induce chemical modifications and thermally-induced protein degradation affect MS database searches and protein identification, and can they be compensated for in search software?
Would standardizing methods for quantifying proteins solubilized from FFPE tissues and methods used for mass spectral analysis and informatics identification of FFPE constituent proteins improve quantitative reproducibility?
Would improved methods for the extraction and analysis of proteins from FFPE tissues result in definitive conclusions about the effect of tissue processing and archival storage time on the proteomic analysis?
Acknowledgments
This project has been funded, in part, with federal funds under grant NIH grant 1 R42 CA122715 and 1R21 CA134359 from the NCI Innovative Molecular Analysis Technologies (IMAT) Program, and the Veterans Health Administration under a Merit Review award. We greatly appreciated Ms. Leslie K. Garcia, HT, for her excellent assistance with protein extraction. Mass spectrometry was conducted at Calibrant Biosystems, Inc. by Drs. X. Fang, C. Lee and Ms. F. He. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, or the Veterans Health Administration nor does the mention of trade names, commercial products, or organization(s) imply endorsement by the United States Government.
Abbreviations
- AR
antigen retrieval
- FFPE
formalin-fixed, paraffin-embedded
- DTT
dithiothreitol
- IHC
immunohistochemistry
- MS
Mass Spectrometry
- PAGE
polyacrylamide gel electrophoresis
- RPPA
reverse phase protein arrays,
Footnotes
Conflict of interest
The authors report no conflicts of interest
References
- 1.Matsuda KM, Chung JY, Hewitt SM. Histo-proteomic profiling of formalin-fixed, paraffin-embedded tissue. Expert Rev Proteomics. 2010;7:227–237. doi: 10.1586/epr.09.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker KF, Taylor CR. “Liquid morphology”: Immunochemical analysis of proteins extracted from formalin-fixed paraffin-embedded tissues: Combining proteomics with immunohistochemistry. Appl Immunohistochem Mol Morphol. 2011;19:1–9. doi: 10.1097/PAI.0b013e3181f50883. [DOI] [PubMed] [Google Scholar]
- 3.Ahrens CH, Brunner E, Qeli E, Basler K, Aebersold R. Generating and navigating proteome maps using mass spectrometry. Nat Rev Mol Cell Biol. 2010;11:789–801. doi: 10.1038/nrm2973. [DOI] [PubMed] [Google Scholar]
- 4.Pierre S, Scholich K. Toponomics: studying protein-protein interactions and protein networks in intact tissue. Mol Biosyst. 2010;6:641–647. doi: 10.1039/b910653g. [DOI] [PubMed] [Google Scholar]
- 5.Saravanan RS, Rose JK. A critical evaluation of sample extraction techniques for enhanced proteomic analysis of recalcitrant plant tissues. Proteomics. 2004;4:2522–2532. doi: 10.1002/pmic.200300789. [DOI] [PubMed] [Google Scholar]
- 6.Shi SR, Key ME, Kalra KL. Antigen retrieval in formalin-fixed, paraffin-embedded tissues: an enhancement method for immunohistochemical staining based on microwave oven heating of tissue sections. J Histochem Cytochem. 1991;39:741–748. doi: 10.1177/39.6.1709656. [DOI] [PubMed] [Google Scholar]
- 7.Gown AM. Unmasking the mysteries of antigen or epitope retrieval and formalin fixation. Am J Clin Pathol. 2004;121:172–174. doi: 10.1309/9G5F-Y3U3-QB4R-15DR. [DOI] [PubMed] [Google Scholar]
- 8.Ikeda K, Monden T, Kanoh T, Tsujie M, et al. Extraction and analysis of diagnostically useful proteins from formalin-fixed, paraffin-embedded tissue sections. J Histochem Cytochem. 1998;46:397–403. doi: 10.1177/002215549804600314. [DOI] [PubMed] [Google Scholar]
- 9.Shi SR, Liu C, Balgley BM, Lee C, Taylor CR. Protein extraction from formalin-fixed, paraffin-embedded tissue sections: quality evaluation by mass spectrometry. J Histochem Cytochem. 2006;54:739–743. doi: 10.1369/jhc.5B6851.2006. [DOI] [PubMed] [Google Scholar]
- 10.Fowler CB, Cunningham RE, O’Leary TJ, Mason JT. “Tissue surrogates” as a model for archival formalin-fixed paraffin-embedded tissues. Lab Invest. 2007;87:836–846. doi: 10.1038/labinvest.3700596. [DOI] [PubMed] [Google Scholar]
- 11.Fowler CB, Cunningham RE, Waybright TJ, Blonder J, et al. Elevated hydrostatic pressure promotes protein recovery from formalin-fixed, paraffin-embedded tissue surrogates. Lab Invest. 2008;88:185–195. doi: 10.1038/labinvest.3700708. [DOI] [PubMed] [Google Scholar]
- 12.Fowler CB, O’Leary TJ, Mason JT, Shi SR, Taylor CR. Techniques of Protein Extraction from FFPE Tissue/Cells for Mass Spectrometry. In: Shi SR, Taylor CR, editors. Antigen Retrieval Immunohistochemistry Based Research and Diagnostics. John Wiley & Sons; Hoboken, NJ: 2010. pp. 335–346. [Google Scholar]
- 13.Tanca A, Pagnozzi D, Addis MF. Setting proteins free: progresses and achievements in proteomics of formalin-fixed, paraffin-embedded tissues. Proteomics Clin Appl. 2011;6:7–21. doi: 10.1002/prca.201100044. [DOI] [PubMed] [Google Scholar]
- 14.Shi SR, Shi Y, Taylor CR. Antigen retrieval immunohistochemistry: review and future prospects in research and diagnosis over two decades. J Histochem Cytochem. 2011;59:13–32. doi: 10.1369/jhc.2010.957191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi SR, Balgley BM, Taylor CR. Symbiosis of Immunohistochemistry and Proteomics: Marching to A New Era. In: Shi SR, Taylor CR, editors. Antigen Retrieval Immunohistochemistry Based Research and Diagnostics. John Wiley & Sons; Hoboken, NJ: 2010. pp. 391–397. [Google Scholar]
- 16.Shi SR, Taylor CR. Extended Application of Antigen Retrieval Technique in Immunohistochemistry and In Situ Hybridization. In: Shi SR, Taylor CR, editors. Antigen Retrieval Immunohistochemistry Based Research and Diagnostics. John Wiley & Sons; Hoboken, NJ: 2010. pp. 25–45. [Google Scholar]
- 17.Rait VK, O’Leary TJ, Mason JT. Modeling formalin fixation and antigen retrieval with bovine pancreatic ribonuclease A: I-structural and functional alterations. Lab Invest. 2004;84:292–299. doi: 10.1038/labinvest.3700045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rait VK, Xu L, O’Leary TJ, Mason JT. Modeling formalin fixation and antigen retrieval with bovine pancreatic RNase A II. Interrelationship of cross-linking, immunoreactivity, and heat treatment. Lab Invest. 2004;84:300–306. doi: 10.1038/labinvest.3700041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi SR, Taylor CR. Antigen Retrieval Immunohistochemistry Based Research and Diagnostics. John Wiley & Sons; Hoboken, NJ: 2010. [Google Scholar]
- 20.D’Amico F, Skarmoutsou E, Stivala F. State of the art in antigen retrieval for immunohistochemistry. J Immunol Methods. 2009;341:1–18. doi: 10.1016/j.jim.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 21.O’Leary TJ, Fowler CB, Evers DL, Mason JT. Protein fixation and antigen retrieval: chemical studies. Biotech Histochem. 2009;84:217–221. doi: 10.3109/10520290903039086. [DOI] [PubMed] [Google Scholar]
- 22.Yamashita S. Heat-induces antigen retrieval: mechanisms and application to histochemistry. Prog Histochem Cytochem. 2007;41:141–200. doi: 10.1016/j.proghi.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 23.Frank TS, Svoboda-Newman SM, Hsi ED. Comparison of methods for extracting DNA from formalin-fixed paraffin sections for nonisotopic PCR. Diagn Mol Pathol. 1996;5:220–224. doi: 10.1097/00019606-199609000-00012. [DOI] [PubMed] [Google Scholar]
- 24.Masuda N, Ohnishi T, Kawamoto S, Monden M, Okubo K. Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res. 1999;27:4436–4443. doi: 10.1093/nar/27.22.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi SR, Cote RJ, Wu L, Liu C, et al. DNA extraction from archival formalin-fixed, paraffin-embedded tissue sections based on the antigen retrieval principle: heating under the influence of pH. J Histochem Cytochem. 2002;50:1005–1011. doi: 10.1177/002215540205000802. [DOI] [PubMed] [Google Scholar]
- 26.Shi SR, Taylor CR. Extraction of DNA/RNA from Formalin-Fixed, Paraffin-embedded Tissue Based on the Antigen Retrieval Principle. In: Shi SR, Taylor CR, editors. Antigen Retrieval Immunohistochemistry Based Research and Diagnostics. John Wiley & Sons; Hoboken, NJ: 2010. pp. 47–71. [Google Scholar]
- 27.Addis MF, Tanca A, Pagnozzi D, Crobu S, et al. Generation of high-quality protein extracts from formalin-fixed, paraffin-embedded tissues. Proteomics. 2009;9:3815–3823. doi: 10.1002/pmic.200800971. [DOI] [PubMed] [Google Scholar]
- 28.Becker KF, Schott C, Hipp S, Metzger V, et al. Quantitative protein analysis from formalin-fixed tissues: implications for translational clinical research and nanoscale molecular diagnosis. J Pathol. 2007;211:370–378. doi: 10.1002/path.2107. [DOI] [PubMed] [Google Scholar]
- 29.Guo T, Wang W, Rudnick PA, Song T, et al. Proteome analysis of microdissected formalin-fixed and paraffin-embedded tissue specimens. J Histochem Cytochem. 2007;55:763–772. doi: 10.1369/jhc.7A7177.2007. [DOI] [PubMed] [Google Scholar]
- 30.Jiang X, Feng S, Tian R, Ye M, Zou H. Development of efficient protein extraction methods for shotgun proteome analysis of formalin-fixed tissues. J Proteome Res. 2007;6:1038–1047. doi: 10.1021/pr0605318. [DOI] [PubMed] [Google Scholar]
- 31.Palmer-Toy DE, Krastins B, Sarracino DA, Nadol JB, Jr, Merchant SN. Efficient method for the proteomic analysis of fixed and embedded tissues. J Proteome Res. 2005;4:2404–2411. doi: 10.1021/pr050208p. [DOI] [PubMed] [Google Scholar]
- 32.Sato Y, Sugie R, Tsuchiya B, Kameya T, et al. Comparison of the DNA extraction methods for polymerase chain reaction amplification from formalin-fixed and paraffin-embedded tissues. Diagn Mol Pathol. 2001;10:265–271. doi: 10.1097/00019606-200112000-00009. [DOI] [PubMed] [Google Scholar]
- 33.Nirmalan NJ, Harnden P, Selby PJ, Banks RE. Development and validation of a novel protein extraction methodology for quantitation of protein expression in formalin-fixed paraffin-embedded tissues using western blotting. J Pathol. 2009;217:497–506. doi: 10.1002/path.2504. [DOI] [PubMed] [Google Scholar]
- 34.Heaton KJ, Master SR. Peptide extraction from formalin-fixed paraffin-embedded tissue. Curr Protoc Protein Sci. 2011;65:23.5.1–23.5.19. doi: 10.1002/0471140864.ps2305s65. [DOI] [PubMed] [Google Scholar]
- 35.Alkhas A, Hood BL, Oliver K, Teng PN, et al. Standardization of a sample preparation and analytical workflow for proteomics of archival endometrial cancer tissue. J Proteome Res. 2011;10:5264–5271. doi: 10.1021/pr2007736. [DOI] [PubMed] [Google Scholar]
- 36.Huang SK, Darfler MM, Nicholl MB, You J, et al. LC/MS-based quantitative proteomic analysis of paraffin-embedded archival melanomas reveals potential proteomic biomarkers associated with metastasis. PLoS One. 2009;4:e4430. doi: 10.1371/journal.pone.0004430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Metz B, Kersten GFA, Baart GJ, de Jong A, et al. Identification of formaldehyde-induced modifications in proteins: reactions with insulin. Bioconjug Chem. 2006;17:815–822. doi: 10.1021/bc050340f. [DOI] [PubMed] [Google Scholar]
- 38.Metz B, Kersten GFA, Hoogerhout P, Brugghe HF, et al. Identification of formaldehyde-induced modifications in proteins: reactions with model peptides. J Biol Chem. 2004;279:6235–6243. doi: 10.1074/jbc.M310752200. [DOI] [PubMed] [Google Scholar]
- 39.Sompuram SRVK, Messana E, Bogen SA. A Molecular Mechanism of Formalin Fixation and Antigen Retrieval. Am J Clin Pathol. 2004;121:190–199. doi: 10.1309/BRN7-CTX1-E84N-WWPL. [DOI] [PubMed] [Google Scholar]
- 40.Xie R, Chung JY, Ylaya K, Williams RL, et al. Factors influencing the degradation of archival formalin-fixed paraffin-embedded tissue sections. J Histochem Cytochem. 2011;59:356–365. doi: 10.1369/0022155411398488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolff C, Schott C, Porschewski P, Reischauer B, Becker KF. Successful protein extraction from over-fixed and long-term stored formalin-fixed tissues. PLoS One. 2011;6:e16353. doi: 10.1371/journal.pone.0016353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sprung RW, Jr, Brock JW, Tanksley JP, Li M, et al. Equivalence of protein inventories obtained from formalin-fixed paraffin-embedded and frozen tissue in multidimensional liquid chromatography-tandem mass spectrometry shotgun proteomic analysis. Mol Cell Proteomics. 2009;8:1988–1998. doi: 10.1074/mcp.M800518-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scicchitano MS, Dalmas DA, Boyce RW, Thomas HC, Frazier KS. Protein extraction from formalin-fixed, paraffin-embedded tissue enables robust proteomic profiles by mass spectrometry. J Histochem Cytochem. 2009;57:849–860. doi: 10.1369/jhc.2009.953497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fowler CB, Waybright TJ, Veenstra TD, O’Leary TJ, Mason JT. Pressure-assisted protein extraction: a novel method for recovering proteins from archival tissue for proteomic analysis. J Proteome Res. 2012;11:2602–2608. doi: 10.1021/pr201005t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balgley BM, Guo T, Zhao K, Fang X, et al. Evaluation of archival time on shotgun proteomics of formalin-fixed and paraffin-embedded tissues. J Proteome Res. 2009;8:917–925. doi: 10.1021/pr800503u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fowler CB, Chesnick IE, Moore CD, O’Leary TJ, Mason JT. Elevated pressure improves the extraction and identification of proteins recovered from formalin-fixed, paraffin-embedded tissue surrogates. PLoS One. 2010;5:e14253. doi: 10.1371/journal.pone.0014253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zale SE, Klibanov AM. Why does ribonuclease irreversibly inactivate at high temperatures? Biochemistry. 1986;25:5432–5444. doi: 10.1021/bi00367a014. [DOI] [PubMed] [Google Scholar]
- 48.Volkin DB, Klibanov AM. Thermal destruction processes in proteins involving cysteine residues. J Biol Chem. 1987;262:2945–2950. [PubMed] [Google Scholar]
- 49.Shi SR, Fang X, Garcia L, He F, Taylor CR. Standardization of protein extraction from formalin-fixed, paraffin-embedded tissue sections: An approach of complete solubilization. Poster presentation at The 62nd Annual Meeting of the Histochemical Society; Woods Hole, MA, USA. March 27–31: 2011. [Google Scholar]
- 50.Berg D, Hipp S, Malinowsky K, Bollner C, Becker KF. Molecular profiling of signalling pathways in formalin-fixed and paraffin-embedded cancer tissues. Eur J Cancer. 2010;46:47–55. doi: 10.1016/j.ejca.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 51.Murphy CL, Eulitz M, Hrncic R, Sletten K, et al. Chemical typing of amyloid protein contained in formalin-fixed paraffin-embedded biopsy specimens. Am J Clin Pathol. 2001;116:135–142. doi: 10.1309/TWBM-8L4E-VK22-FRH5. [DOI] [PubMed] [Google Scholar]
- 52.Fowler CB, O’Leary TJ, Mason JT. Modeling formalin fixation and histological processing with bovine ribonuclease A: Effects of ethanol dehydration on reversal of formaldehyde-induced cross-links. Lab Invest. 2008;88:785–791. doi: 10.1038/labinvest.2008.43. [DOI] [PubMed] [Google Scholar]
- 53.Ostasiewicz P, Zielinska DF, Mann M, Wisniewski JR. Proteome, phosphoproteome, and N-glycoproteome are quantitatively preserved in formalin-fixed paraffin-embedded tissue and analyzable by high-resolution mass spectrometry. J Proteome Res. 2010;9:3688–3700. doi: 10.1021/pr100234w. [DOI] [PubMed] [Google Scholar]
- 54.Giavalisco P, Nordhoff E, Lehrach H, Gobom J, Klose J. Extraction of proteins from plant tissues for two-dimensional electrophoresis analysis. Electrophoresis. 2003;24:207–216. doi: 10.1002/elps.200390016. [DOI] [PubMed] [Google Scholar]
- 55.Ericsson C, Nister M. Protein extraction from solid tissue. Methods Mol Biol. 2011;675:307–312. doi: 10.1007/978-1-59745-423-0_17. [DOI] [PubMed] [Google Scholar]
- 56.Etherton TD, Thompson EH, Allen CE. Improved techniques for studies of adipocyte cellularity and metabolism. J Lipid Res. 1977;18:552–557. [PubMed] [Google Scholar]