Abstract
Like many asymmetrically dividing cells, budding yeast segregates mitotic spindle poles nonrandomly between mother and daughter cells. During metaphase, the spindle positioning protein Kar9 accumulates asymmetrically, localizing specifically to astral microtubules emanating from the old spindle pole body (SPB) and driving its segregation to the bud. Here, we show that the SPB component Nud1/centriolin acts through the mitotic exit network (MEN) to specify asymmetric SPB inheritance. In the absence of MEN signaling, Kar9 asymmetry is unstable and its preference for the old SPB is disrupted. Consistent with this, phosphorylation of Kar9 by the MEN kinases Dbf2 and Dbf20 is not required to break Kar9 symmetry but is instead required to maintain stable association of Kar9 with the old SPB throughout metaphase. We propose that MEN signaling links Kar9 regulation to SPB identity through biasing and stabilizing the age-insensitive, cyclin-B-dependent mechanism of symmetry breaking.
Introduction
Asymmetric cell division is a major mechanism for the generation of cellular diversity in eukaryotes (reviewed in Kaltschmidt and Brand, 2002). During such divisions, the mitotic spindle aligns with the polarity axis of the cell such that polarized factors segregate to only one of the two daughters (reviewed in Siller and Doe, 2009; Barral and Liakopoulos, 2009). The alignment of division and polarity axes with each other requires the two poles of the mitotic spindle to become morphologically and functionally distinct from each other and to respond differently to polarity cues. This differentiation drives their migration to distinct cell ends and, hence, the alignment of the spindle with the cell's polarity axis.
Remarkably, the age of the spindle pole frequently specifies its fate. Indeed, centrioles of higher eukaryotes undergo conservative duplication, leading to formation of an old and a new centriole (reviewed in Azimzadeh and Bornens, 2007; Strnad and Gönczy, 2008). Subsequently, centriole segregation between daughter cells often correlates with their age. For example, stem cells of the Drosophila male germline and the mammalian neocortex inherit the oldest centriole, whereas its daughter centriole segregates to the differentiating cell (Yamashita et al., 2007; Wang et al., 2009). Therefore, centrioles appear to specify pole destiny. How this specification is achieved remains, however, unclear.
Simpler eukaryotes also segregate spindle poles nonrandomly. The budding yeast equivalent of the centrosome, the spindle pole body (SPB), duplicates in a partially conservative manner as well (Adams and Kilmartin, 2000; Yoder et al., 2003), such that one SPB is inherited from the previous mitosis and is composed of mostly old protein, whereas the other SPB is assembled de novo. In unperturbed cells, SPBs segregate nonrandomly, such that the bud inherits the old SPB in virtually all divisions (Pereira et al., 2001).
Much is known about the mechanisms of spindle positioning in yeast, but how and why SPBs become specified remains elusive. In meiosis, the SPB component Nud1, a protein related to mammalian centriolin (Gromley et al., 2003), has been involved in defining SPB identity during the process of spore wall formation (Gordon et al., 2006). How spindle pole identity and inheritance is defined in mitosis is currently unknown. Budding yeast offers a robust framework for the investigation of the mechanism and function of pole inheritance. Prior to spindle assembly, bud emergence determines both the polarity axis and the future cleavage plane of the cell. Therefore, the mitotic spindle must align with the polarity and division axes in order to properly segregate chromosomes between mother and bud. Upon separation of the spindle poles and regardless of its initial position, the old pole migrates toward the bud neck (Pereira et al., 2001). Orientation of the spindle relative to the polarity axis is mediated by the protein Kar9, which interacts indirectly with both actin and microtubules by binding to the microtubule-binding protein Bim1 (Miller et al., 2000; Lee et al., 2000; Korinek et al., 2000), the yeast homolog of EB1, and to type V myosin, Myo2 (Yin et al., 2000). Through these interactions, Kar9 promotes the movement of microtubule plus ends along actin cables toward the bud (Beach et al., 2000; Liakopoulos et al., 2003; Hwang et al., 2003). Kar9's function in the alignment of the spindle with the mother-bud axis relies on the fact that it is recruited to the astral microtubules emanating from only one SPB. Because all actin cables emanate from the bud cortex during metaphase, Kar9 recruitment to only one aster leads to Myo2-dependent pulling and specific orientation of the associated SPB toward the bud (reviewed in Pruyne et al., 2004). Therefore, it is the asymmetry of Kar9 distribution that promotes the alignment of the spindle with the mother-bud axis and orients the old SPB toward the bud (Liakopoulos et al., 2003; Pereira et al., 2001). Although recent studies have shed light on the regulation of Kar9 asymmetry (Liakopoulos et al., 2003; Moore et al., 2006; Moore and Miller, 2007; Leisner et al., 2008; Meednu et al., 2008; Kammerer et al., 2010; Cepeda García et al., 2010), it remains unclear how SPB identity is specified and how Kar9 asymmetry is directed toward the old SPB.
Cells lacking Kar9 align their spindles with the mother-bud axis in early anaphase in a dynein-dependent manner (Adames and Cooper, 2000; Carminati and Stearns, 1997; Grava et al., 2006). Because both the Kar9- and dynein-dependent pathways mediate spindle alignment with the polarity axis, each can compensate for the absence of the other, whereas simultaneous inactivation of both pathways is lethal (Miller and Rose, 1998). However, only the Kar9 pathway specifies SPB inheritance (Pereira et al., 2001; Moore et al., 2006). To better understand this aspect of Kar9 function, we screened for mutations that randomized SPB inheritance similar to disruption of Kar9. Here, we report on one such class of mutants, establishing an unexpected role for the mitotic exit network (MEN) in Kar9-dependent specification of SPBs.
Results
Nud1 and Mitotic Exit Network Mutants Show Defects in SPB Inheritance
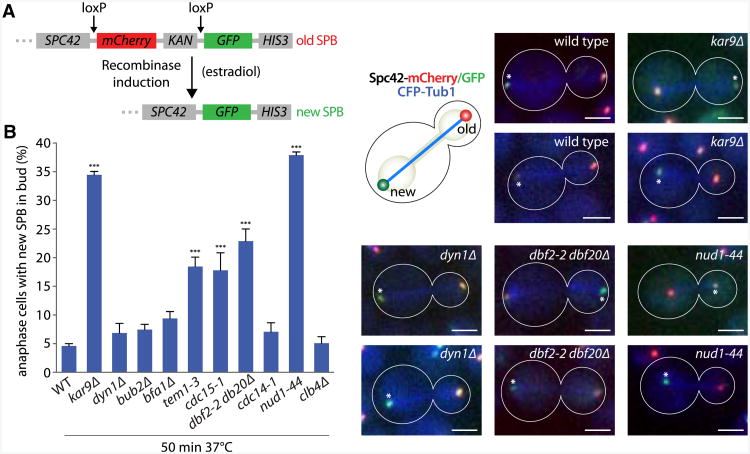
In order to track SPB inheritance, we developed an assay to unambiguously distinguish old and new SPBs (Figure 1A). Similar to Verzijlbergen et al. (2010), this assay uses recombinase-dependent exchange of fluorescent tags, allowing the discrimination of proteins synthesized before and after recombination. A cassette containing the mCherry coding sequence flanked by loxP recombination sites and followed by yeGFP was inserted after the SPC42 locus, a core component of the SPB (Donaldson and Kilmartin, 1996). Upon estradiol treatment, a conditional Cre-recombinase (Lindstrom and Gottschling, 2009) rapidly excises the mCherry sequence and fuses the yeGFP sequence in-frame to SPC42. Therefore, these cells express the Spc42-mCherry fusion protein prior to estradiol treatment and Spc42-yeGFP afterward. Accordingly, two roughly similar mCherry signals reporting the position of both SPBs were detected in untreated metaphase cells (Figure S1 available online). In contrast, the poles of most metaphase spindles were differentially labeled red and green 4 hr after the treatment, with the oldest of the two SPBs showing the highest red and the lowest green intensity.
Figure 1. Nud1 and MEN Mutants Have a SPB Inheritance Defect.
(A) Schematic view of the switchable mCherry/GFP-tag inserted after the SPC42 locus. Cre recombinase is fused to the estradiol-binding domain EBD78, which allows shuttling of Cre to the nucleus upon addition of estradiol. mCherry is flanked by loxP sites to allow excision of mCherry by Cre and switch to GFP.
(B) Percentage of anaphase cells with the new SPB in the bud in cells carrying kar9Δ, dyn1Δ, or temperature-sensitive Nud1 and MEN alleles. Cells were estradiol treated for 4 hr and shifted for 50 min to 37°C before imaging. Stars indicate p values obtained from one-way ANOVA comparing to WT (three clones with total n > 130). Asterisks mark the new SPB. Scale bar, 2 μm. Mean and SD of three clones are shown.
See also Figure S1.
As predicted (Pereira et al., 2001; Moore et al., 2006), only very few cells oriented and segregated the new SPB to the bud (4.6% ± 0.7% of anaphase cells; Figure 1B). Cells lacking dynein (dyn1 Δ) segregated the SPBs similar to wild-type (6.9% ± 2.9% of new SPB to the bud), whereas kar9Δ mutant cells missegregated the new SPB to the bud in 34.5% ± 1.0% of the divisions (n > 130). Therefore, our analysis is consistent with a specific role for Kar9 in SPB inheritance (Pereira et al., 2001).
Using this approach, we screened mutations affecting SPB-associated factors for genes involved in SPB inheritance, focusing on components of the SPB's outer plaque. Unexpectedly, this study identified the TEM1 gene (Figure 1B; 18.5% ± 2.8% of the tem1-3 mutant cells segregated the new SPB to the bud at restrictive temperature). Tem1 functions at the top of the MEN, a signaling cascade that controls Cdk1 inactivation and cytokinesis (reviewed in Bardin and Amon, 2001). Tem1 binds and activates the Hippo-related kinase Cdc15, which in turn activates the two LATS-related kinases Dbf2 and Dbf20 (reviewed in Queralt and Uhlmann, 2008). During mitotic exit, Dbf2 and Dbf20 mediate activation of the phosphatase Cdc14 by promoting its release from the nucleolus. Unlike Tem1 inactivation, deletion of the two GTPase-activating proteins (GAPs) Bub2 and Bfa1 did not affect SPB inheritance significantly (7.5% ± 1.6% and 9.4% ± 2.1% of missegregation, respectively). However, the cdc15-1 and dbf2-2 dbf20Δ mutant cells missegregated SPBs in a substantial fraction of anaphases at the restrictive temperature (17.8% ± 5.3% and 22.9% ± 3.7%, respectively, p < 0.001 versus WT), whereas cdc14-1 mutant cells did not (7.1% ± 2.7%). Thus, inactivation of the MEN cascade, but not of Cdc14, caused substantial defects in SPB segregation.
Proper MEN function requires the integrity of the SPB component Nud1/centriolin (Gruneberg et al., 2000). nud1-44 mutant cells showed a strong SPB inheritance defect at the restrictive temperature (37.9% ± 1.0%, p < 0.001 versus WT; Figure 1B), comparable to kar9Δ mutant cells. Thus, our data implicate MEN and the SPB component Nud1 in mitotic SPB inheritance and, hence, in spindle orientation.
Nud1 and MEN Function in the Metaphase Pathway of Spindle Positioning
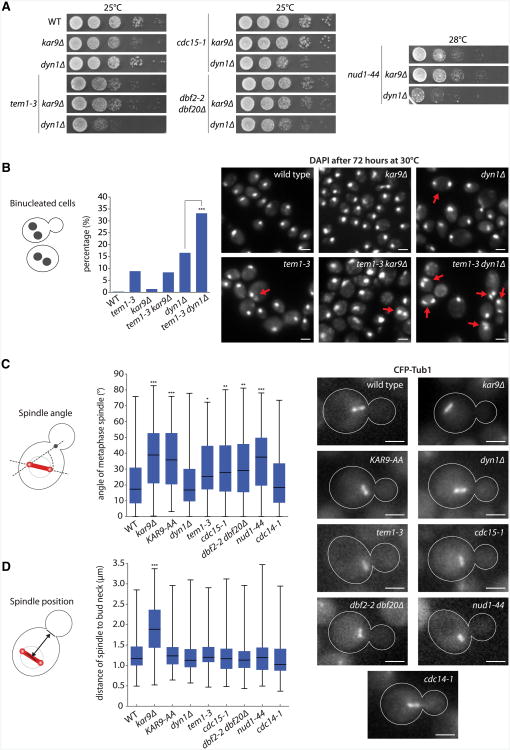
To characterize how Nud1 and MEN contribute to SPB inheritance, we next asked whether MEN influenced either of the Kar9- or dynein-dependent pathways of spindle positioning. Growth assays were performed to test whether the Nud1 and MEN mutations interact genetically with disruptions of either Kar9 or dynein (Figure 2A). Whereas all strains containing Nud1 or MEN mutations were nonviable at 37°C (data not shown), reduced growth at permissive temperature (25°C-28°C) was observed when these mutations were combined with the dyn1 Δ, but not with the kar9Δ mutation. These data suggest that MEN and Nud1 function in the Kar9 pathway of spindle positioning.
Figure 2. Nud1 and MEN Mutants Have a Spindle Alignment Defect.
(A) Genetic interactions between Nud1 and MEN mutations and deletions of the spindle positioning factors KAR9 and DYN1. Strains were grown for 48 hr at 25° C or 28°C.
(B) Percentage of binucleated cells of indicated strains. Cells were grown at semipermissive temperature (30°C) for 72 hr. Stars indicate p values obtained from binomial distribution test on n > 250. Scale bar, 2 µm.
(Cand D) Measurement of angle between the spindle axis and a line connecting distal pole and bud neck (C) and the distance between spindle and bud neck(D) of indicated strains. Cells were shifted to 37°C for 50 min before imaging. Stars indicate p values obtained from one-way ANOVA comparing to WT (three clones with total n > 95). Scale bar, 2 µm.
Consistent with the tem1-3 mutation affecting spindle positioning, a significant fraction of these mutant cells became binucleated (8.9% versus 0.3% in WT, p < 0.001; n > 256) when grown at semipermissive temperature (30°C; Figure 2B). The kar9Δ mutation barely caused binucleation under these conditions (1.4%; n = 422) and did not enhance the effect of the tem1-3 allele (binucleation of tem1-3 kar9Δ cells: 8.4%; n = 287). In contrast, 16.6% of the dyn1 Δ mutant cells were binucleated (n = 259), and this fraction rose to 33.2% (n = 286; p < 0.001) in the tem1-3 dyn1 Δ double mutant. Thus, Tem1 contributed to spindle positioning in a dynein-independent but possibly Kar9-dependent manner.
We therefore asked whether inactivation of Nud1 and MEN perturbed the alignment and migration of metaphase spindles. In wild-type and mutant cells, metaphase spindles were visualized using CFP-tubulin, and their angle with the mother-bud axis was measured (Figure 2C). As expected (Liakopoulos et al., 2003; Kammerer et al., 2010), kar9Δ cells (37.8° ± 2.1°; mean, SEM) and cells expressing a mutant form of Kar9, Kar9-AA, (36.5° ± 2.0°) showed a higher angle than wild-type (21.5° ± 1.7°; p < 0.001). In KAR9-AA, the serines of Kar9's main Cdk1 phosphorylation sites (Ser197 and Ser496) are changed to alanine, causing Kar9 to localize more frequently to both asters and affecting spindle alignment (Liakopoulos et al., 2003; Leisner et al., 2008; Kammerer et al., 2010). Deletion of DYN1 did not affect the alignment of metaphase spindles (21.7° ± 1.6°), whereas inactivation of MEN and Nud1 did (30.6° ± 1.6° in tem1-3 cells, 31.4° ± 1.8° in cdc15-1, 31.6° ± 1.7° in dbf2-2 dbf20Δ, and 36.1° ± 1.8° in nud1-44 cells; compared to WT all p < 0.05). In contrast, the metaphase spindle of the cdc14-1 mutant cells aligned properly (24.5° ± 2.3°). Thus, MEN and Nud1, but not Cdc14, mediate proper alignment of metaphase spindles.
Measurement of the spindle distance to the bud neck (Figure 2D) indicated that only the kar9Δ mutation affected spindle migration to the bud neck. None of the Nud1, MEN, KAR9-AA, or dyn1Δ mutations perturbed this aspect of positioning. Thus, Nud1 and MEN contribute specifically to the alignment of the metaphase spindle with the mother-bud axis. Moreover, the Nud1 and MEN mutants affected spindle positioning very similarly to KAR9-AA.
Nud1 and men affect kar9 distribution on metaphase spindles
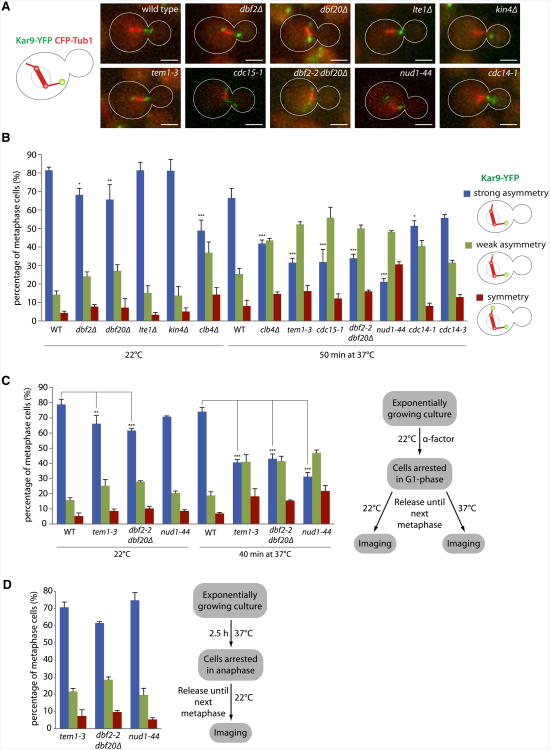
The KAR9-AA mutation impairs Cdk1-dependent phosphorylation and affects Kar9 distribution (Liakopoulos et al., 2003; Moore and Miller, 2007). Thus, we investigated whether Nud1 or MEN inactivation also affected the localization of Kar9. Time-lapse movies of 100 s (10 frames) were recorded, and the distribution of Kar9-YFP was scored in wild-type and mutant cells coexpressing CFP-Tub1 (Figure 3A). All metaphase cells were sorted in either one of the three following classes. “Strongly asymmetric” cells carried Kar9 exclusively on one side of the spindle throughout the movie. In “symmetric” cells, roughly equal amounts of Kar9 appeared on both sides of the spindle over the course of the movie, and “weakly asymmetric” cells showed intermediate Kar9 localization. At 22°C, 81.4% ± 1.7% of wild-type metaphase cells fell in the strongly asymmetric class (Figure 3B). This value was reduced in dbf2Δ and dbf20Δ mutant cells (68.2% ± 3.5% and 65.6% ± 8.0%, respectively, p < 0.05 and p < 0.01 versus WT) and even more in the tem1-3, cdc15-1 single-mutant and dbf2-2 dbf20Δ double-mutant cells shifted for 50 min to 37°C (31.6% ± 2.3%, 31.9% ± 6.8%, and 33.9% ± 2.2% of strong asymmetry, respectively, p < 0.001 versus 66.5% ± 5.3% for WT at the same temperature). The effect was strongest in the nud1-44 mutant cells (21.2% ± 1.7% of cells with strong Kar9 asymmetry, p < 0.001). By comparison, inactivation of the B-type cyclin Clb4, already known to control Kar9 asymmetry, caused a decrease of the strongly asymmetric category to 41.9% ± 1.9% of the meta-phase population at 37°C. The phenotype of the cdc14-1 and cdc14-3 mutant cells was much less pronounced (51.4% ± 2.8% and 55.6% ± 1.9% of strong asymmetry, respectively), consistent with Cdc14 playing a rather marginal role in Kar9 regulation, spindle positioning, and SPB inheritance. Deletion of LTE1 and KIN4, which both act upstream of MEN, had no effect on Kar9 localization. Taken together, these data indicate that the SPB component Nud1 and the MEN proteins Tem1, Cdc15, and Dbf2/20 contribute to the control of Kar9 distribution on metaphase spindles. Measurement of Kar9 fluorescence on still images confirmed these conclusions (Figure S2A).
Figure 3. Kar9 Asymmetry Is under Control of Nud1 and MEN in Metaphase.
(A) Cells expressing temperature-sensitive alleles of Nud1 and MEN proteins while carrying Kar9-YFP and CFP-Tub1. Scale bar, 2 µm.
(B) Quantification of (A) as described in the text. Cells were shifted to37°Cfor50min before imaging if indicated. Stars indicate p values obtained from one-way ANOVA comparing “strong asymmetry” to WT (three clones with total n > 130). Mean and SD of three clones are shown.
(c) G1 arrest of Nud1 and MEN mutant cells and release into metaphase at either 22°C or 37°C for 40 min. Quantification as in (B). Mean and SD of three clones are shown.
(D) Anaphase arrest of Nud1 and MEN mutant cells by shifting to 37°C for 2.5 hr and release into metaphase at 22°C. Quantification as in (B). Mean and SD of three clones are shown.
See also Figure S2.
In order to test whether Nud1 and MEN controlled Kar9 distribution indirectly through their action in the previous mitosis or directly during metaphase, we next arrested cells in the G1 phase with α factor and released them at the restrictive temperature (Figure 3C). After 40 min at 37°C, i.e., when most of the cells had entered their first metaphase without passing through anaphase, 74.2% ± 2.9% of wild-type cells showed strongly asymmetric Kar9, whereas this value dropped in the tem1-3 and nud1-44 single-mutant and dbf2-2 dbf20Δ double-mutant cells (40.7% ± 1.9%, 31.2% ± 2.8%, and 43.1% ± 3.1%, respectively, p < 0.001 versus WT). When the same cells were released at permissive temperature (22°C), only mild effects were observed (66.2% ± 5.4%, 70.8% ± 0.6%, and 61.7% ± 1.3% strong asymmetry, respectively, versus 78.9% ± 3.5% for WT). Thus, MEN inactivation at the onset of mitosis was sufficient to affect the distribution of Kar9 in metaphase.
Conversely, when the same mutant cells were arrested in anaphase by shifting them to 37°C for 2.5 hr and released until the next metaphase at 22°C (Figure 3D), no Kar9 defects were observed compared to cells kept throughout at 22°C (Figure 3C), indicating that defects of Nud1 and MEN in the previous cycle do not affect Kar9 localization once activity is restored. Thus, Nud1 and MEN promote Kar9 asymmetry directly in metaphase.
Beyond its role in MEN, Nud1 also mediates microtubule attachment to the SPB (Gruneberg et al., 2000), and therefore, microtubule organization defects could explain the phenotypes of the nud1-44 cells. To determine whether this was the case, we analyzed microtubule organization in cells carrying nud1-44 and GFP-Tub1. Although some anaphase cells showed detached cytoplasmic microtubules (16.5% versus 2.9% in WT, n > 68; Figure S2B), microtubule attachment remained unaffected in the metaphase of nud1-44 cells (1.0% of cells with detached aMTs versus 1.7% in WT, n > 98). Thus, Nud1 functions in SPB inheritance independently of its role in microtubule attachment.
MEN Does Not Act on Kar9 Asymmetry through Cdk1/Clb4, SUMO, or the SAC
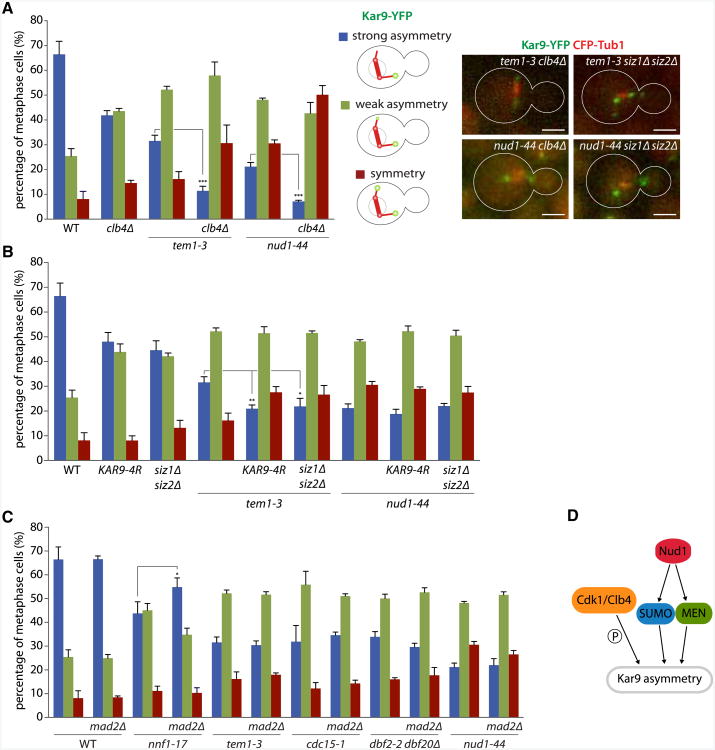
Both sumoylation and Cdk1/Clb4-dependent phosphorylation of Kar9 control Kar9 distribution independently of each other (Leisner et al., 2008). Thus, we asked whether MEN functioned in one of these processes. First, characterization of Kar9-YFP distribution showed enhanced phenotypes in the tem1-3 clb4Δ and nud1-44 clb4Δ double mutant (11.4% ± 1.8% and 7.2% ± 0.5% strong asymmetry, respectively), compared to tem1-3, nud1-44, and clb4Δ single-mutant cells (Figure 4A; p < 0.001). Thus, both Tem1 and Nud1 regulate Kar9 asymmetry in parallel to Cdk1/Clb4.
Figure 4. MEN Acts on Kar9 Asymmetry Independently of Cdk1, SUMO, and SAC Activation.
(A and B) Quantification of Kar9 asymmetry on cells carrying tem1-3 ornud1-44 in combination with mutations affecting either Cdk1/Clb4 (A) or SUMO (B). All cells were shifted to 37°C for 50 min before imaging. Quantification as in Figure 3B. Stars indicate p values obtained from one-way ANOVA comparing “strong asymmetry” (three clones with total n > 110). Scale bar, 2 μm. Mean and SD of three clones are shown.
(C) Quantification of Kar9 asymmetry on cells carrying MEN mutations combined with MAD2 or mad2Δ as in Figure 3B. Mean and SD of three clones are shown.
(D) Schematic drawing of the three genetic pathways controlling Kar9 asymmetry.
Similarly, the tem1-3 KAR9-4R double mutant and tem1-3 siz1 Δ siz2Δ triple mutants showed an enhanced phenotype (20.9% ± 1.5% and 21.9% ± 3.3% strong asymmetry, respectively; Figure 4B) compared to tem1-3 and KAR9-4R single and siz1Δ siz2Δ double mutants (Leisner et al., 2008; p < 0.05) and were quantitatively similar to that of the nud1-44 single-mutant cells. KAR9-4R encodes a nonsumoylatable form of Kar9, whereas Siz1 and Siz2 are the SUMO ligases involved in Kar9 sumoylation. In contrast, neither of the nud1-44 KAR9-4R double and nud1-44 siz1Δ siz2Δ triple mutants showed a stronger phenotype than the nud1-44 single-mutant cells. Together, these data indicate that MEN acts independently of SUMO on Kar9, whereas the SPB component Nud1 acts upstream of both MEN and SUMO for the control of Kar9 distribution. These data also explain why Kar9 localizes more symmetrically in nud1-44 than in MEN mutant cells.
Activation of the spindle assembly checkpoint (SAC) also makes the metaphase distribution of Kar9 more symmetric (Leisner et al., 2008). Inactivation of the essential kinetochore component Nnf1 activates SAC and decreases Kar9 asymmetry (43.8% ± 4.9% strong asymmetry in nnf1-17 mutant cells compared to 66.5% ± 5.3% for WT grown at 37°C for 50 min, p < 0.001; Figure 4C). This effect is partially rescued by removing the checkpoint protein Mad2 (54.8% ± 3.9%, p < 0.05). However, MAD2 inactivation failed to restore Kar9 distribution in the tem1-3, cdc15-1, dbf2-2 dbf20Δ, and nud1-44 mutant cells. Thus, in Nud1 or MEN mutant cells, SAC activation is not the cause of Kar9 symmetry. Together, these data establish that Nud1 and MEN act independently of Cdk1/Clb4, SUMO, and SAC in the control of Kar9 distribution.
Dbf2 and Dbf20 Act Directly on Kar9
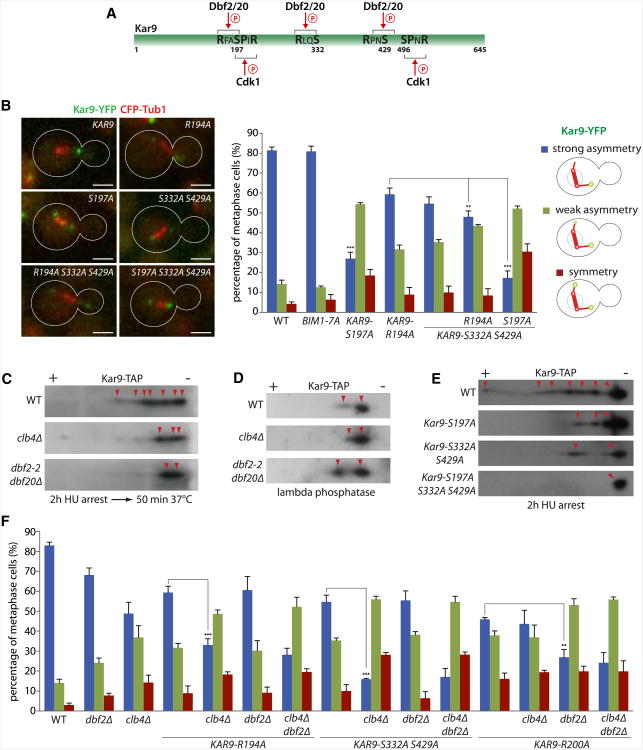
Based on these observations, we rationalized that MEN may directly target Kar9 pathway components. Analysis of Kar9 primary sequence indicates the existence of three Dbf2/Dbf20 consensus sites (RXXS, Mah et al., 2005) at Ser197, Ser332, and Ser429 (Figure 5A). Thus, both Cdk1/Clb4 and Dbf2/Dbf20 may phosphorylate Kar9, with potentially one target residue in common (Ser197). Likewise, Kar9's partner Bim1 contains four Dbf2/20 consensus sites, which are phosphorylated in vivo (Zimniak et al., 2009).
Figure 5. Dbf2/20-Dependent Phosphorylation of Kar9 Promotes Its Asymmetry in Metaphase.
(A) Schematic drawing of Kar9 and its consensus sites for Dbf2/20 and Cdk1.
(B) Kar9 localization in cells expressing Kar9 or Bim1 with Dbf2/20 consensus sites mutated. Quantification as in Figure 3B. Stars indicate p values obtained from one-way ANOVA comparing “strong asymmetry” to WT (three clones with total n > 130). Scale bar, 2 μm. Mean and SD of three clones are shown.
(C-E) Two-dimensional gel electrophoresis on lysates of cells carrying Kar9-TAP at their endogenous locus in wild-type, dbf2-2 dbf20Δ, or clb4Δ mutant background without (C) or with (D) addition of λ-phosphatase. (E) Same analysis for cells carrying consensus site mutations of Kar9. All strains were arrested with hydroxyurea for 2 hr and for (C) shifted to 37°C for 50 min before harvesting. Different isoforms are indicated with red arrows.
(F) Quantification of Kar9 asymmetry on cells expressing Kar9 consensus site mutations combined with dbf2Δ and clb4Δ as in Figure 3B. Stars indicate p values obtained from one-way ANOVA comparing “strong asymmetry” (three clones with total n > 135). Mean and SD of three clones are shown.
To test whether Dbf2/20 targeted Kar9 and Bim1, we mutated their target motifs and asked whether this affected Kar9 distribution. Mutating all Dbf2/Dbf20 consensus sites of Bim1 (BIM1-7A) did not affect Kar9 distribution. Thus, Bim1 is not a relevant target (Figure 5B). However, the S197A mutation on Kar9 (KAR9-S197A) strongly affected Kar9 distribution (strong asymmetry: 27.0% ± 3.3% versus 81.4% ± 1.7% for WT, p < 0.001). To test whether Dbf2/20 phosphorylated this site, we mutated the arginine critical for Dbf2/20 targeting in –3 position (KAR9-R194A). This mutation partially affected Kar9 asymmetry (59.4% ± 3.2% strong asymmetry). Simultaneously mutating the two other putative Dbf2/20 target sites (S332A and S429A) also decreased Kar9 asymmetry (54.7% ± 3.4%), and this was additive with R194A(KAR9-R194A S332A S429A: 48.0% ± 3.0% of strong asymmetry, p < 0.01 compared to KAR9-R194A). Ablating both Cdk1 and Dbf2/20 sites (KAR9-S197A S332A S429A) led to the strongest phenotype (17.4% ± 3.5% of strong asymmetry). Together, these data suggest that Kar9 asymmetry requires its phosphorylation by both Cdk1 and Dbf2/20.
To directly test whether Dbf2 and Dbf20 are required for Kar9 phosphorylation in vivo, extracts of cells expressing Kar9-TAP from the endogenous locus and arrested in metaphase with hydroxyurea (S phase and metaphase overlap in budding yeast) were assayed by two-dimensional gel electrophoresis and western blotting. In wild-type cells, seven or more Kar9 isoforms were detected (Figure 5C), the most acidic ones being sensitive to phosphatase treatment (Figure 5D). Kar9 from extracts of the clb4Δ single-mutant and dbf2-2 dbf20Δ double-mutant cells migrated as a hypophosphorylated protein, with dbf2-2 dbf20Δ cells producing the stronger effect. The Kar9 proteins lacking the Cdk1-Dbf2/20 site (Kar9-S197A), the Dbf2/20 consensus sites alone (Kar9-S332A S429A), or all of them (Kar9-S197A S332A S429A) were also hypophosphorylated (Figure 5E), with the triple mutant showing the strongest effect. Thus, full phosphorylation of Kar9 during metaphase requires Cdk1/Clb4 and Dbf2/20 functions, as well as Ser197, 332, and 429 of Kar9.
To determine the functional relationships between the kinases and the target sites, we combined Kar9 alleles with kinase mutations (Figure 5F). Combining KAR9-R194A with the clb4Δ disruption led to increased asymmetry phenotypes compared to both single-mutant strains (33.1% ± 3.2% strong asymmetry versus 59.4% ± 3.2% and 48.8% ± 5.7%, respectively, p < 0.001), whereas combining it with dbf2Δ showed no additive effect. Thus, the R194A mutation prevented the action of Dbf2/20, but not of Clb4. Similarly, combining KAR9-S332A S429A caused additive effects with clb4Δ, but not with dbf2Δ. We conclude that Ser197, 332, and 429 are targets of Dbf2/Dbf20-dependent phosphorylation.
The mutation R200A is predicted to affect phosphorylation of Ser197 by Cdk1 while retaining the consensus motif for Dbf2/20. Accordingly, cells carrying this mutation showed a similar defect in Kar9 asymmetry as clb4Δ mutant cells (46.0% ± 0.9% and 48.8% ± 5.7%, respectively; Figure 5F). This effect was enhanced upon deletion of DBF2, but not in combination with the clb4Δ mutation (27.0% ± 3.9% and 43.6% ± 3.8%, respectively). Thus, both Cdk1 and Dbf2/20 target Ser197.
Nud1 and MEN Control SPB Specification
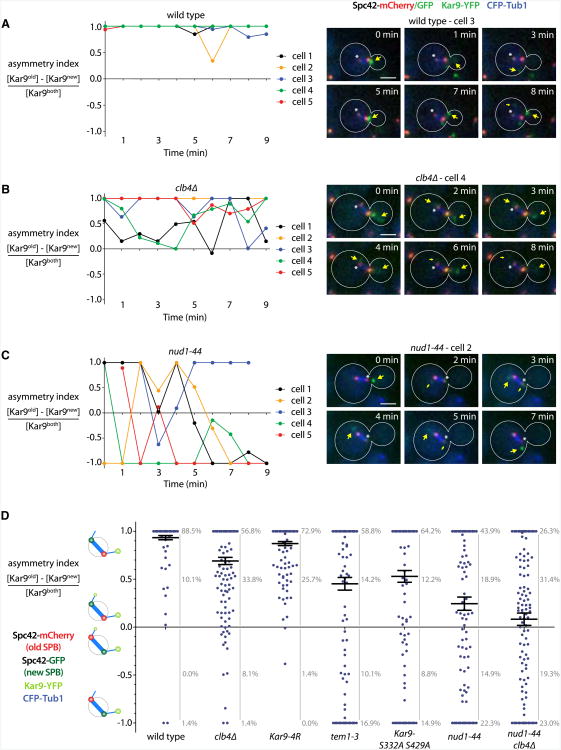
Together, our data indicate that Cdk1, the SPB protein Nud1, and the MEN cascade control Kar9 distribution on metaphase spindles through modification of Kar9. However, these data failed to explain why Nud1 and MEN specifically affected SPB inheritance, whereas Cdk1 did not (Figure 1B). To address this question, 10 min movies (1 frame/min) of cells expressing Spc42 fused to the switchable mCherry/GFP tag (see Figure 1 A) along with Kar9-YFP and CFP-Tub1 were recorded, and Kar9 fluorescence was measured on each side of the spindle over time. Because these cells coexpressed CFP-tubulin, cytoplasmic Kar9-YFP and the GFP-labeled new SPB were easily distinguished (see also Figure S1). Using these measurements, we derived an asymmetry index, dividing the difference between Kar9 fluorescence associated with new and old SPBs by the total Kar9 fluorescence (Figure 6A). The value of this index can adopt values from +1 when Kar9 is found solely on the old SPB to 0 when Kar9 localizes equally to both poles and to −1 when Kar9 is solely on the new SPB. Thus, +1 and −1 express that Kar9 is fully asymmetric, and intermediary values indicate that Kar9 is fully or partially symmetric, and their sign indicates the orientation of the bias relative to old and new SPB. Following the value of this index over time in cells of different genotypes leads to the following observations.
Figure 6. Nud1 and MEN Activity Specifies SPB Identity and Stabilizes Kar9 on the Old Pole.
(A–C) Quantification of Kar9 asymmetry orientation relative to old and new SPB performed on cells expressing Spc42 fused to a switchable mCherry/GFP-tag as well as Kar9-YFP and CFP-Tub1. Wild-type (A), clb4Δ (B), and nud1-44 (C) mutant cells were imaged for 10 min, and Kar9 fluorescence was measured for each of the ten time points. An asymmetry index was defined as described in the text. For each strain, five representative cells are shown in the graph. Asterisks mark the new SPB, and yellow arrows indicate Kar9 fluorescence. Scale bar, 2 μm.
(D) Asymmetry indices obtained in (A)–(C) for wild-type or mutant strains were pooled (>17 cells, 148 total time points each). Mean and SE are shown.
See also Figure S1.
First, the asymmetry index varied little over time in the wild-type cells, remaining at +1 in 88.5% of the frames (Figures 6A and 6D). Loss of asymmetry was a rare event (frequency = 0.08/min), and asymmetry was very rarely oriented toward the new SPB (negative index values in only 1.4% of the frames). Accordingly, Kar9 asymmetry almost never switched from one pole to the other in a given cell (switch from +1 to −1 or vice versa at 0.014/min).
Second, in the clb4Δ mutant cells (Figures 6B and 6D), Kar9 was fully asymmetric in only 58.2% of the frames. Incursions in partial or full symmetry were long and lasted up to 9 min (2.8 ± 2.5 min in average, n = 22, compared to 1.2 ± 0.4 min in wild-type, n = 14). However, once asymmetry was established, it was lost only slightly more frequently than in wild-type cells (frequency = 0.13/min). Negative asymmetry values occurred slightly more frequently than in wild-type but remained a minority (9.5% of the frames), and asymmetry switched as rarely between poles as in wild-type (0.014/min). The mutation KAR9-4R, preventing sumoylation, led to similar although less pronounced effects (Figure 6D). Thus, sumoylation and Cdk1-dependent phosphorylation contribute primarily to the breakage of Kar9 symmetry and are largely dispensable for its orientation toward the old SPB.
Third, Kar9 was fully asymmetric 66.2% of the time in the nud1-44 mutant cells (Figures 6C and 6D), and the intermediary periods lasted 1.8 ± 1.8 min on average (n = 28). Hence, compared to the clb4Δ mutant, the nud1-44 mutant cells are only mildly defective in symmetry breaking. In contrast, the nud1-44 mutant cells showed a two to three times higher frequency of asymmetry loss (0.26/min) thanwild-type and clb4Δ mutant cells, indicating that it was defective in asymmetry maintenance. Most remarkably, the index value was frequently negative (37.2% of the frames), and asymmetry switched between poles eight times more frequently than in wild-type and clb4Δ mutant cells (0.108/min). Accordingly, among the fully asymmetric time points, asymmetry was oriented toward the new SPB in 33.0% of the cases (1.7% in wild-type and 2.4% in clb4Δ mutant cells). Therefore, a pattern frequently observed in these cells corresponds to asymmetry being lost and soon reestablished—often, however, toward the opposite pole (Figure 6C). Thus, the main phenotype of nud1-44 mutant cells is the inability to choose the old pole and stabilize asymmetry toward it. As expected, the KAR9-S332A S429A and tem1-3 mutations lead to milder but, in essence, similar phenotypes as the nud1-44mutation (Figure6D).
Fourth, the phenotype of the nud1-44 clb4Δ double mutant (Figure 6D) combined and slightly enhanced those of the single mutants: the cells lingered in a symmetric state about as long as clb4Δ cells (2.6 ± 2.5 min, 50.7% of frames with intermediary index), whereas negative indices were similarly frequent as in the nud1-44 cells (42.3% of frames versus 37.2% in nud1-44 and 9.5% in clb4Δ). Thus, Clb4 promotes Kar9 asymmetry in the nud1-44 mutant, whereas Nud1 orients and stabilizes the Kar9 bias toward the old SPB in clb4Δ mutant cells. Together, ourdata indicate that Nud1 specifies the identity of SPBs, leading to the stabilization of Kar9 asymmetry toward the old one. MEN-dependent phosphorylation of Kar9 mediates this stabilization.
Kar9 Asymmetry May Reinforce SPB Specification
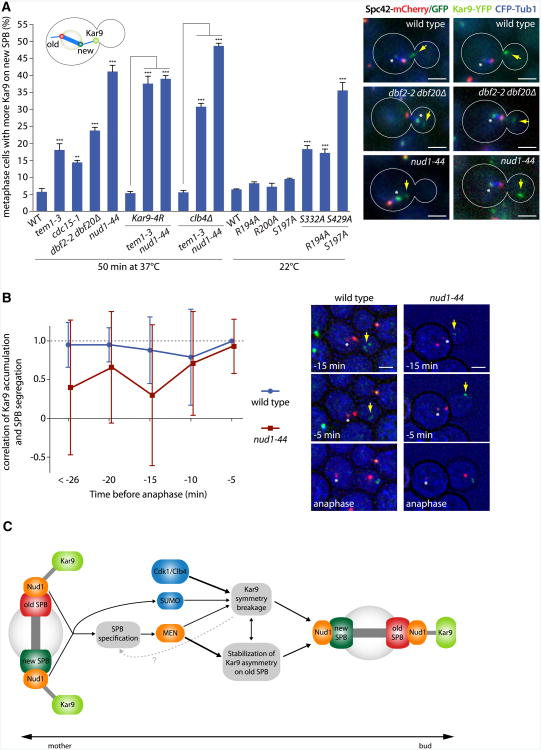
The effect of combining mutations further clarified the relationships between Kar9 asymmetry and SPB specification. Using 100 s long movies (10 frames) of cells expressing the switchable Spc42-mCherry/GFP tag, we characterized the frequency at which Kar9 asymmetry was misoriented in single and double mutants (Figure 7A). Analysis of the single mutants confirmed that Nud1 and MEN play a prominent role in SPB specification. However, combination of nud1-44 or tem1-3 with clb4Δ randomized Kar9 orientation further. The same was observed when combining KAR9-4R with the tem1-3 mutation, but not with the nud1-44 mutation, consistent with sumoylation acting in parallel to MEN and downstream of Nud1. Similarly, KAR9-S332A S429A mutant cells directed Kar9 asymmetry toward the new SPB as frequently as MEN mutants. Ablation of Ser197, which had no effect on its own, strongly potentiated this effect (KAR9-S197A S332A S429A). These observations suggest that, although SPB specification is largely separable from Clb4- and SUMO-dependent establishment of Kar9 asymmetry, this last process feeds back into and reinforces the specification of SPB identity.
Figure 7. Specification of SPB Identity by Nud1 and MEN Directs the Old Pole to the Bud.
(A) Cells expressing Spc42 fused to a switchable mCherry/GFP-tag as well as Kar9-YFP and CFP-Tub1. Graph shows percentage of metaphase cells carrying more Kar9 on aMTs emanating from the new SPB. All cells were treated for 4 hr with estradiol, and strains were shifted for 50 min to 37°C if indicated. Stars indicate p values obtained from one-way ANOVA comparing to WT or as indicated (three clones with total n > 125). Asterisks mark the new SPB, and yellow arrows indicate Kar9 fluorescence. Scale bar, 2 μm. Mean and SD of three clones are shown.
(B) Correlation between the SPB with the highest Kar9 level in metaphase and the SPB migrating into the bud in anaphase. Correlation was determined for each time point prior to anaphase onset (>17 cells, > 169 total time points). Asterisks mark the new SPB, and yellow arrows indicate Kar9 fluorescence. Scale bar, 2 μm.
(C) The genetic pathways controlling Kar9 asymmetry, spindle alignment, and SPB inheritance in metaphase.
See also Figure S1.
Kar9 Asymmetry Drives SPB Inheritance
Throughout our data, the orientation of Kar9 bias toward old and new SPBs correlated well with SPB inheritance. The clb4Δ mutant cells oriented Kar9 bias toward the old SPB (Figure 6D and 7A) and segregated it efficiently to the bud (Figure 1B), whereas the nud1-44 and MEN mutations randomized Kar9 asymmetry between SPBs and affected their inheritance. Thus, Kar9 distribution seemed to govern SPB inheritance. To test this idea more directly, we imaged nud1-44 mutant and wild-type cells labeled for Kar9 and Spc42 at 5 min intervals as they progressed from metaphase into anaphase (Figure 7B). For each time point, we correlated the orientation of Kar9 asymmetry toward the old and new SPB in metaphase with SPB segregation to the bud in anaphase. For wild-type, correlation was high throughout metaphase but maximal and with the lowest standard deviation (correlation = 1.0 ± 0.0) at the last time point prior to anaphase onset. In nud1-44 mutant cells, in which Kar9 asymmetry switched between poles, Kar9 asymmetry and SPB inheritance poorly correlated through most of metaphase. Yet, right prior to anaphase onset, the correlation raised to 0.93 ± 0.35. Thus, Kar9 distribution at anaphase onset defined which SPB enters the bud, even in the nud1-44 mutant cells. We conclude that Nud1 and MEN control SPB inheritance through the regulation of Kar9 distribution.
Discussion
By demonstrating that Nud1 and MEN control Kar9 recruitment to the aster of the old SPB and segregation of that SPB to the bud, our data establish three points. First, they implicate the conserved SPB/centriole component Nud1/centriolin as a major determinant of pole specification in mitotic cells and therefore establish that the specification of SPB identity during spindle orientation is driven by the SPBs themselves, reminiscent of SPB specification during spore wall formation (Gordon et al., 2006). Second, Kar9 asymmetry is the result of converging pathways, each acting at least in part through modification of Kar9 itself. Third, MEN functions already in metaphase, much earlier than previously assumed.
An Unexpected Function For The Mitotic Exit Network In Metaphase Spindle Positioning
The current models of MEN function suggest that the Tem1 GTPase, the most upstream component of the cascade, is activated in late mitosis to promote mitotic exit in cells with a correctly positioned spindle (Caydasi et al., 2010). However, the exact mechanisms of Tem1 activation are not yet fully understood. Based on the following observations, our study indicates that MEN is actually active already in metaphase.
First, MEN inactivation perturbs Kar9 distribution between the asters of the metaphase spindle, causing defects in spindle alignment and orientation with the division axis of the cell. Strikingly, these phenotypes were observed in the first metaphase after MEN inactivation, whereas no defect in Kar9 asymmetry was observed when MEN was inactivated in the previous cycle and subsequently reactivated. Thus, MEN activity is required in early mitosis for spindle orientation.
Second, MEN activity was required for phosphorylation of Kar9 in metaphase cells. These phosphorylation events required the integrity of the three Dbf2/20 consensus sites in Kar9 (Ser197, Ser332, and Ser429). Mutating these serines to alanine phenocopied MEN inactivation in metaphase cells and showed no additive effect when combined with MEN mutations. Our data therefore indicate that the MEN kinases Dbf2 and Dbf20 phosphorylate Kar9 on Ser197, Ser332, and Ser429 during metaphase to promote proper orientation of the spindle (Figure 7C). The interpretation that MEN is already active in early mitosis is consistent with preanaphase activity reported for the SIN pathway, the fission yeast equivalent of the MEN (Hachet and Simanis, 2008). Therefore, it will be interesting to understand how MEN combines functions as distinct as the specification of SPB identity in metaphase and mitotic exit in late anaphase.
Balancing the RolesofMEN before andduring Anaphase
An important question raised by our findings is how mitotic exit is restrained when MEN activity arises in metaphase? Two non-reciprocally exclusive models could explain this apparent conundrum.
On the one hand, MEN activation might be less of a trigger for mitotic exit than a permissive condition. Mitotic exit itself could be caused by another signal that allows MEN activity to be translated into Cdc14 activation. For instance, MEN activity might be unable to promote Cdc14 activation and mitotic exit as long as Cdk1 activity is high and the anaphase-promoting complex (APC/C) remains inactive, for example, by preventing MEN from reaching its nuclear targets Net1 and Cdc14. In support of this model, Cdc14 had little or no relevance to MEN function during metaphase. Moreover, recent results have indicated that Cdk1 indeed prevents Dbf2 from promoting mitotic exit (Koönig et al., 2010). The observation that Cdc14 early anaphase release (FEAR) and the activity of the anaphase-promoting complex are both required for progression toward mitotic exit (Stegmeier et al., 2002; Sullivan and Uhlmann, 2003; Wäsch and Cross, 2002) are fully consistent with this model.
On the other hand, MEN activity might be thresholded such that its activity in metaphase is sufficient for spindle orientation, but not to promote mitotic exit. This possibility is supported by the observation that Dbf2 kinase activity is low in metaphase and highest in late anaphase (Visintin and Amon, 2001). Furthermore, regulators of mitotic exit, such as Lte1 and Kin4, had no effect on spindle positioning in metaphase. Thus, these proteins might specifically function in mitotic exit by controlling the increase of MEN activity during anaphase.
The role of MEN in Kar9 asymmetry appears to be largely independent of the phosphatase Cdc14. Indeed, the MEN kinases Dbf2/20 phosphorylate Kar9 directly, and the cdc14-alleles show only mild phenotypes. However, Cdc14 can dephosphorylate Kar9 in vivo and in vitro (Bloom et al., 2011). Thus, some contribution of Cdc14 to Kar9 asymmetry, perhaps redundantly with another phosphatase, cannot be excluded.
How Do Nud1 and MEN Function in Spindle Asymmetry?
Our data indicate that Nud1 and MEN function in SPB specification by orienting Kar9 asymmetry toward the old SPB. They identify two distinct mechanisms for the stable accumulation of Kar9 on the aster of the old SPB (Figure 7C). Cdk1/Clb4 and sumoylation promote the breakage of Kar9 symmetry, mostly irrespective of the age of the SPB. In parallel, Nud1 and MEN functions add directionality to the system by biasing and stabilizing Kar9 asymmetry toward the old SPB. As a consequence, cells lacking Nud1 or MEN function are able to establish Kar9 asymmetry but fail to reliably and stably choose one of the two SPBs. Together, the interplay of these two processes ensures both the robustness of Kar9 asymmetry and the nonrandom inheritance of SPBs. Because Kar9 itself is a major target of Nud1 and MEN in the process of spindle positioning, these data suggest that asymmetry in MEN activity must be an early event in SPB specification. Whether and how MEN activity is biased toward one of the two SPBs during metaphase will be an important question to address in the future. Likewise, it will be interesting to determine how Nud1 regulates the MEN. Our data suggest that it might directly activate Tem1, possibly even by functioning as nucleotide exchange factor (GEF) for this GTPase.
SPBs/Centrosomes Control Their Own Specification
Similar to many stem cells, budding yeast does not segregate its spindle poles randomly. Previous studies concluded that historical cues directed the old SPB to the bud. Carrying more microtubules than the new one, the old SPB is more likely to interact with Myo2 and to amplify Kar9 asymmetry through interactions with the cell cortex (Cepeda-García et al., 2010). Indeed, bud cortical proteins contribute to Kar9 asymmetry. However, the same proteins function in MEN regulation to control the asymmetric distribution of Bfa1 and Bub2 between SPBs (Monje-Casas and Amon, 2009). We show that the nud1-44 mutation perturbs SPB specification and inheritance without affecting microtubule anchorage to the SPB. Thus, our data establish that signals emanating from the SPB itself specify the fate of the SPBs, the cortex contributing probably rather secondary signals to the amplification and stabilization of SPB specification. Accordingly, cortex mutants have a weaker phenotype than nud1-44 mutant cells (Cepeda-García et al., 2010).
The idea that centrosomes control their own specification is supported by recent observations (Conduit and Raff, 2010; Januschke et al., 2011). In Drosophila neuroblasts, the centro-some containing the grandmother centriole is first decorated with numerous microtubules. However, it sheds its pericentriolar material shortly after formation of the new centrosome around the mother centriole. As a consequence, in these stem cells, the old centrosome segregates to the differentiating cells, and the renewing stem cell inherits the new centrosome. Thus, as in yeast, the specification of the fate of the spindle poles and centriole inheritance in Drosophila neuroblasts is not simply determined by microtubule coverage but, rather, by centro-some-associated signals. The fact that Nud1 is related to the centriolar protein centriolin suggests that this last protein might play a central and conserved role in pole specification. Finally, though our observations do not clarify the biological relevance of segregating spindle poles in such a tightly controlled manner, they open avenues for addressing this fascinating question in the future.
Experimental Procedures
Strains and Growth Conditions
Media and genetic methods as described Guthrie and Fink (1991). Yeast strains are listed in Table S1; all strains were derived from S288C or were backcrossed 3×. Fluorescent or TAP-tagged proteins were tagged at endogenous loci (Knop et al., 1999). CFP-Tub1 was inserted at the URA3 and gdp:Cre-EBD78 at the LEU2 loci. The switchable mCherry/GFP-cassette is described in Extended Experimental Procedures. Kar9 alleles were constructed by site-directed mutagenesis (pfu-Turbo, Stratagene) and verified by PCR and sequencing after integration. Deletions were performed as described (Knop et al., 1999). For cell-cycle arrests, cells (OD = 0.6) were incubated in medium containing 0.1 M hydroxyurea for 2 hr or 10 μg/ml α factor for 2.5 hr. Fluorophore switch was induced with estradiol 4 hr before imaging.
Fluorescence Microscopy
One-hundred second and 10 min time-lapse microscopy were performed using an Olympus BX51 microscope, TILLVision software (TILLPhotonics). For the localization of GFP-, YFP-, and CFP-labeled proteins and DAPI, Z stacks of five layers (step size 0.3 μm) and maximum projections were used. Fluorescence microscopy was performed with a monochromator Poly-chromIV aslight source and a CCD camera(Imago, TillPhotonics). Sixty minute time-lapse microscopy was performed on a Personal Deltavision microscope, with Z stacks of 10 layers (step size 0.5 μm). Images were analyzed with ImageJ. DAPI staining was done by ethanol fixation for 20 min and incubation in DAPI medium (1 μg/ml) for 5 min.
Two-Dimensional SDS-PAGE
Two-dimensional gel electrophoresis and western blotting were performed on cells expressing Kar9-TAP from its endogenous locus as described (Vogel et al., 2001). For a detailed protocol, see Extended Experimental Procedures.
Supplementary Material
Acknowledgments
We thank S. Baldi, M. Bayer, A. Bolognesi, S. Buvelot-Frei, F. Caudron, L. Clay, A. Denoth, A. Farcas, M. Faty, P. Meraldi, M. Peter, J. Saarikangas, and C. Weirich for discussion and critical reading of the manuscript and N. Paget and R. Visintin for technical help. We also thank M. Charbonneau, S. Piatti, and F. Uhlmann for strains and reagents and the Light Microscopy Centre of the ETH Zurich for technical support. M.H. and Y.B. were supported by ETHZ and grants of ERC and SNF to Y.B. D.C. and J.V. were supported by CIHR grant MOP-64404 to J.V. D.L. and D.E.G. were supported by NIH grant AG023779 to D.E.G.
Footnotes
Supplemental Information includes Extended Experimental Procedures, two figures, and one table and can be found with this article online at doi:10.1016/j.cell.2012.01.041.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adames NR, Cooper JA. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J Cell Biol. 2000;149:863–874. doi: 10.1083/jcb.149.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams IR, Kilmartin JV. Spindle pole body duplication: a model for centrosome duplication? Trends Cell Biol. 2000;10:329–335. doi: 10.1016/s0962-8924(00)01798-0. [DOI] [PubMed] [Google Scholar]
- Azimzadeh J, Bornens M. Structure and duplication of the centrosome. J Cell Sci. 2007;120:2139–2142. doi: 10.1242/jcs.005231. [DOI] [PubMed] [Google Scholar]
- Bardin AJ, Amon A. Men and sin: what's the difference? Nat Rev Mol Cell Biol. 2001;2:815–826. doi: 10.1038/35099020. [DOI] [PubMed] [Google Scholar]
- Barral Y, Liakopoulos D. Role of spindle asymmetry in cellular dynamics. Int Rev Cell Mol Biol. 2009;278:149–213. doi: 10.1016/S1937-6448(09)78004-9. [DOI] [PubMed] [Google Scholar]
- Beach DL, Thibodeaux J, Maddox P, Yeh E, Bloom K. The role of the proteins Kar9 and Myo2 in orienting the mitotic spindle of budding yeast. Curr Biol. 2000;10:1497–1506. doi: 10.1016/s0960-9822(00)00837-x. [DOI] [PubMed] [Google Scholar]
- Bloom J, Cristea IM, Procko AL, Lubkov V, Chait BT, Snyder M, Cross FR. Global analysisof Cdc14 phosphatase reveals diverse roles in mitotic processes. J Biol Chem. 2011;286:5434–5445. doi: 10.1074/jbc.M110.205054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carminati JL, Stearns T. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J Cell Biol. 1997;138:629–641. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caydasi AK, Ibrahim B, Pereira G. Monitoring spindle orienta tion: Spindle position checkpoint in charge. Cell Div. 2010;5:28. doi: 10.1186/1747-1028-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepeda-García C, Delgehyr N, Ortiz MA, ten Hoopen R, Zhiteneva A, Segal M. Actin-mediated delivery of astral microtubules instructs Kar9p asymmetric loading to the bud-ward spindle pole. Mol Biol Cell. 2010;21:2685–2695. doi: 10.1091/mbc.E10-03-0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit PT, Raff JW. Cnn dynamics drive centrosome size asymmetry to ensure daughter centriole retention in Drosophila neuroblasts. Curr Biol. 2010;20:2187–2192. doi: 10.1016/j.cub.2010.11.055. [DOI] [PubMed] [Google Scholar]
- Donaldson AD, Kilmartin JV. Spc42p: a phosphorylated component of the S. cerevisiae spindle pole body (SPD) withan essential func tion during SPB duplication. J Cell Biol. 1996;132:887–901. doi: 10.1083/jcb.132.5.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon O, Taxis C, Keller PJ, Benjak A, Stelzer EH, Simchen G, Knop M. Nud1p, the yeast homolog of Centriolin, regulates spindle pole body inheritance in meiosis. EMBO J. 2006;25:3856–3868. doi: 10.1038/sj.emboj.7601254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grava S, Schaerer F, Faty M, Philippsen P, Barral Y. Asymmetric recruitment of dyneinto spindle poles and microtubules promotes proper spindle orientation in yeast. Dev Cell. 2006;10:425–439. doi: 10.1016/j.devcel.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Gromley A, Jurczyk A, Sillibourne J, Halilovic E, Mogensen M, Grois-man I, Blomberg M, Doxsey S. A novel human protein of the maternal centriole is required for the final stages of cytokinesis and entry into S phase. J Cell Biol. 2003;161:535–545. doi: 10.1083/jcb.200301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruneberg U, Campbell K, Simpson C, Grindlay J, Schiebel E. Nud1p links astral microtubule organization and the control of exit from mitosis. EMBO J. 2000;19:6475–6488. doi: 10.1093/emboj/19.23.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology. New York: Academic Press; 1991. [Google Scholar]
- Hachet O, Simanis V. Mid1p/anillin and the septation initiation network orchestrate contractile ring assembly for cytokinesis. Genes Dev. 2008;22:3205–3216. doi: 10.1101/gad.1697208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang E, Kusch J, Barral Y, Huffaker TC. Spindle orientation in Saccharomyces cerevisiae depends on the transport of microtubule ends along polarized actin cables. J Cell Biol. 2003;161:483–488. doi: 10.1083/jcb.200302030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke J, Llamazares S, Reina J, Gonzalez C. Drosophila neuroblasts retain the daughter centrosome. Nat Commun. 2011;2:243. doi: 10.1038/ncomms1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltschmidt JA, Brand AH. Asymmetric cell division: microtu-bule dynamics and spindle asymmetry. J Cell Sci. 2002;115:2257–2264. doi: 10.1242/jcs.115.11.2257. [DOI] [PubMed] [Google Scholar]
- Kammerer D, Stevermann L, Liakopoulos D. Ubiquitylation regulates interactions of astral microtubules with the cleavage apparatus. Curr Biol. 2010;20:1233–1243. doi: 10.1016/j.cub.2010.05.064. [DOI] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15(10B):963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Koönig C, Maekawa H, Schiebel E. Mutual regulation of cyclin-dependent kinase and the mitotic exit network. J Cell Biol. 2010;188:351–368. doi: 10.1083/jcb.200911128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek WS, Copeland MJ, Chaudhuri A, Chant J. Molecular linkage underlying microtubule orientation toward cortical sites in yeast. Science. 2000;287:2257–2259. doi: 10.1126/science.287.5461.2257. [DOI] [PubMed] [Google Scholar]
- Lee L, Tirnauer JS, Li J, Schuyler SC, Liu JY, Pellman D. Positioning of the mitotic spindle by a cortical-microtubule capture mechanism. Science. 2000;287:2260–2262. doi: 10.1126/science.287.5461.2260. [DOI] [PubMed] [Google Scholar]
- Leisner C, Kammerer D, Denoth A, Britschi M, Barral Y, Liakopou-los D. Regulation of mitotic spindle asymmetry by SUMO and the spindle-assembly checkpoint in yeast. Curr Biol. 2008;18:1249–1255. doi: 10.1016/j.cub.2008.07.091. [DOI] [PubMed] [Google Scholar]
- Liakopoulos D, Kusch J, Grava S, Vogel J, Barral Y. Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell. 2003;112:561–574. doi: 10.1016/s0092-8674(03)00119-3. [DOI] [PubMed] [Google Scholar]
- Lindstrom DL, Gottschling DE. The mother enrichment program: a genetic system for facile replicative life span analysis in Saccharo-myces cerevisiae. Genetics. 2009;183:413–422. doi: 10.1534/genetics.109.106229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah AS, Elia AE, Devgan G, Ptacek J, Schutkowski M, Snyder M, Yaffe MB, Deshaies RJ. Substrate specificity analysis of protein kinase complex Dbf2-Mob1 by peptide library and proteome array screening. BMC Biochem. 2005;6:22. doi: 10.1186/1471-2091-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meednu N, Hoops H, D'silva S, Pogorzala L, Wood S, Farkas D, Sorrentino M, Sia E, Meluh P, Miller RK. The spindle positioning protein Kar9p interacts with the sumoylation machinery in Saccharomyces cerevisiae. Genetics. 2008;180:2033–2055. doi: 10.1534/genetics.108.095042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RK, Rose MD. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J Cell Biol. 1998;140:377–390. doi: 10.1083/jcb.140.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RK, Cheng SC, Rose MD. Bim1p/Yeb1p mediates the Kar9p-dependent cortical attachment of cytoplasmic microtubules. Mol Biol Cell. 2000;11:2949–2959. doi: 10.1091/mbc.11.9.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monje-Casas F, Amon A. Cell polarity determinants establish asymmetry in MEN signaling. Dev Cell. 2009;16:132–145. doi: 10.1016/j.devcel.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JK, Miller RK. The cyclin-dependent kinase Cdc28p regulates multiple aspects of Kar9p function in yeast. Mol Biol Cell. 2007;18:1187–1202. doi: 10.1091/mbc.E06-04-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JK, D'silva S, Miller RK. The CLIP-170 homologue Bik1p promotes the phosphorylation and asymmetric localization of Kar9p. Mol Biol Cell. 2006;17:178–191. doi: 10.1091/mbc.E05-06-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G, Tanaka TU, Nasmyth K, Schiebel E. Modes of spindle pole body inheritance and segregation of the Bfa1p-Bub2p checkpoint protein complex. EMBO J. 2001;20:6359–6370. doi: 10.1093/emboj/20.22.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D, Legesse-Miller A, Gao L, Dong Y, Bretscher A. Mechanisms of polarized growth and organelle segregation in yeast. Annu Rev Cell Dev Biol. 2004;20:559–591. doi: 10.1146/annurev.cellbio.20.010403.103108. [DOI] [PubMed] [Google Scholar]
- Queralt E, Uhlmann F. Cdk-counteracting phosphatases unlock mitotic exit. Curr Opin Cell Biol. 2008;20:661–668. doi: 10.1016/j.ceb.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller KH, Doe CQ. Spindle orientation during asymmetric cell division. Nat Cell Biol. 2009;11:365–374. doi: 10.1038/ncb0409-365. [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Visintin R, Amon A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 2002;108:207–220. doi: 10.1016/s0092-8674(02)00618-9. [DOI] [PubMed] [Google Scholar]
- Strnad P, Goönczy P. Mechanisms of procentriole formation. Trends Cell Biol. 2008;18:389–396. doi: 10.1016/j.tcb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Sullivan M, Uhlmann F. A non-proteolytic function of separase links the onset of anaphase to mitotic exit. Nat Cell Biol. 2003;5:249–254. doi: 10.1038/ncb940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzijlbergen KF, Menendez-Benito V, van Welsem T, van Deventer SJ, Lindstrom DL, Ovaa H, Neefjes J, Gottschling DE, van Leeuwen F. Recombination-induced tag exchange to track old and new proteins. Proc Natl Acad Sci USA. 2010;107:64–68. doi: 10.1073/pnas.0911164107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R, Amon A. Regulation of the mitotic exit protein kinases Cdc15 and Dbf2. Mol Biol Cell. 2001;12:2961–2974. doi: 10.1091/mbc.12.10.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Drapkin B, Oomen J, Beach D, Bloom K, Snyder M. Phosphorylation of gamma-tubulin regulates microtubule organization in budding yeast. Dev Cell. 2001;1:621–631. doi: 10.1016/s1534-5807(01)00073-9. [DOI] [PubMed] [Google Scholar]
- Wang X, Tsai JW, Imai JH, Lian WN, Vallee RB, Shi SH. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature. 2009;461:947–955. doi: 10.1038/nature08435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wäsch R, Cross FR. APC-dependent proteolysis of the mitotic cyclin Clb2 is essential for mitotic exit. Nature. 2002;418:556–562. doi: 10.1038/nature00856. [DOI] [PubMed] [Google Scholar]
- Yamashita YM, Mahowald AP, Perlin JR, Fuller MT. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science. 2007;315:518–521. doi: 10.1126/science.1134910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Pruyne D, Huffaker TC, Bretscher A. Myosin V orien tates the mitotic spindle in yeast. Nature. 2000;406:1013–1015. doi: 10.1038/35023024. [DOI] [PubMed] [Google Scholar]
- Yoder TJ, Pearson CG, Bloom K, Davis TN. The Saccharo myces cerevisiae spindle pole body is a dynamic structure. Mol Biol Cell. 2003;14:3494–3505. doi: 10.1091/mbc.E02-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimniak T, Stengl K, Mechtler K, Westermann S. Phosphoregulation of the budding yeast EB1 homologue Bim1p by Aurora/Ipl1p. J Cell Biol. 2009;186:379–391. doi: 10.1083/jcb.200901036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.