Summary
Circadian oscillations in mammalian physiology and behavior are regulated by an endogenous biological clock. Here we show that loss of the PAS protein MOP3 (also known as BMAL1) in mice results in immediate and complete loss of circadian rhythmicity in constant darkness. Additionally, locomotor activity in light–dark (LD) cycles is impaired and activity levels are reduced in Mop3−/− mice. Analysis of Period gene expression in the suprachiasmatic nucleus (SCN) indicates that these behavioral phenotypes arise from loss of circadian function at the molecular level. These results provide genetic evidence that MOP3 is the bona fide heterodimeric partner of mCLOCK. Furthermore, these data demonstrate that MOP3 is a non-redundant and essential component of the circadian pacemaker in mammals.
Introduction
In nearly all organisms, behavioral and physiological processes display 24 hr rhythms that are controlled by circadian pacemakers (Rosbash, 1995; Dunlap, 1999; King and Takahashi, 2000; Wager-Smith and Kay, 2000). Circadian rhythms are regulated by three components: the circadian pacemaker or “clock,” an input mechanism that allows the clock to be reset by environmental stimuli, and an output mechanism that regulates physiological and behavioral processes. In mammals, the master circadian pacemaker is located in the suprachiasmatic nucleus (SCN) of the hypothalamus (Moore, 1997). The SCN controls neural and humoral signals that either drive output rhythms directly or synchronize peripheral oscillators with the day–night cycle (Ralph et al., 1990; Silver et al., 1996; Yamazaki et al., 2000).
Circadian oscillators in Drosophila, Neurospora, and mice appear to have been highly conserved throughout evolution and to involve transcription–translation negative feedback loops (Wilsbacher and Takahashi, 1998; Dunlap, 1999; King and Takahashi, 2000; Wager-Smith and Kay, 2000). The observation that PAS proteins play important roles in maintaining circadian rhythms in most organisms provides support for this idea (Kay, 1997; Reppert, 1998; Gu et al., 2000). Genetic and biochemical evidence demonstrates that the bHLH-PAS protein CLOCK is required for the proper maintenance of circadian rhythms in mammals. In mice, an ENU-induced deletion in the transactivation domain of mCLOCK results in a lengthened free-running period followed by a gradual loss of circadian rhythmicity (Vitaterna et al., 1994; Antoch et al., 1997; King et al., 1997). Much evidence suggests that the heterodimeric partner of CLOCK is another bHLH-PAS protein. Two candidate partners, known as MOP3 and MOP9 (also known as BMAL2), can form heterodimers with CLOCK that can drive transcription from E box elements found in the promoters of circadian-responsive genes and are co-expressed in neurons of the SCN (Gekakis et al., 1998; Hogenesch et al., 1998; Honma et al., 1998; Takahata et al., 1998; Jin et al., 1999; Yu et al., 1999; Hogenesch et al., 2000; Ikeda et al., 2000; Ripperger et al., 2000).
The comparative genetic approach has provided great insight into the workings of the circadian clock and its transcriptional feedback loop. Similar to mice, the circadian oscillator of Drosophila employs a heterodimer of two bHLH-PAS transcription factors known as dCLOCK and dCYCLE (Allada et al., 1998; Darlington et al., 1998; Rutila et al., 1998). The Drosophila CLOCK:CYCLE heterodimer also drives transcription from cognate E box enhancers of circadian-regulated genes such as period (per) and timeless (tim) (Hao et al., 1997; Darlington et al., 1998). The protein products of the per and tim loci act to inhibit the transcriptional activity of the CLOCK:CYCLE complex, thereby closing the feedback loop (Bae et al., 1998; Darlington et al., 1998; Lee et al., 1999). In flies, the product of the cry locus participates in the light-dependent sequestration and degradation of TIM, which allows the resetting of the clock by light (Emery et al., 1998; Stanewsky et al., 1998; Ceriani et al., 1999). Single mutations at each of the dClock, dcycle, tim, per, and doubletime (dbt) loci can cause arrythmicity of locomotor behavior in flies indicating the essential nature of each of these proteins in the function of the clock (Sehgal et al., 1994; Allada et al., 1998; Price et al., 1998; Rutila et al., 1998).
Applying the comparative approach from Drosophila to mammals is informative yet difficult due to the relative complexity of the mammalian genome. Analysis of the mammalian genome indicates that there are often several potential mammalian orthologs for each component of the Drosophila clock. As examples, the mouse and human genomes encode three homologs of per (Per1, Per2, and Per3) and two homologs of cry (Cry1 and Cry2) (reviewed in King and Takahashi, 2000). These and other cases may be an indication of functional redundancy, additional regulatory complexities, or of paralogous genes that have evolved functions outside of biological rhythms in the mammalian system. In particular, the mammalian CRY proteins demonstrate significant differences in the regulation of the Drosophila and mammalian clocks, as mammalian CRY1 and CRY2 appear to possess greater repressor activity of CLOCK than any PER protein. Mice with a double mutation of both mCry1 and mCry2 are arrhythmic upon release into constant darkness and are deficient in entrainment of behavior to light cycles. However, these mice still show that an increase in mPer1 and mPer2 gene expression in response to light indicating that in contrast to Drosophila CRY, mammalian CRY1 and CRY2 are not functioning as photoreceptors (Griffin et al., 1999; Kume et al., 1999; Okamura et al., 1999; van der Horst et al., 1999; Vitaterna et al., 1999). Mice with targeted mutations at the mPer2 and mPer3 loci have also been generated (Zheng et al., 1999; Shearman et al., 2000). The Per2Brdm mice exhibit a shorter rhythm in constant darkness that eventually becomes arrhythmic; whereas, the Per3 null mice maintain a circadian rhythm in constant darkness, but that rhythm is slightly shorter than wild type. The only known mammalian homolog of Drosophila tim appears to be required in mammalian development and may not play a significant role in circadian biology (Gotter et al., 2000). Finally, the tau mutation in hamsters is encoded by casein kinase 1 epsilon, a homolog of the Drosophila circadian gene, dbt (Kloss et al., 1998; Lowrey et al., 2000). Unlike mutants of the core Drosophila clock components, no single-gene mutation in mice results in complete abolition of circadian rhythms.
Multiple mammalian homologs of CLOCK and CYCLE also exist. The mammalian genome encodes the originally identified CLOCK protein and a close homolog known as NPAS2 (also known as MOP4), as well as two close homologs of dCYCLE, MOP3, and MOP9 (Hogenesch et al., 1997, 2000; Ikeda and Nomura, 1997; Zhou et al., 1997; Ikeda et al., 2000). The functional and orthologous nature of the murine and Drosophila Clock genes are clearly supported by the genetic data showing their respective roles in circadian rhythms (Vitaterna et al., 1994; Antoch et al., 1997; King et al., 1997; Allada et al., 1998). The recent observation that an Npas2 null allele does not influence this biology is an indication mClock and mNpas2 are not functionally redundant (Garcia et al., 2000). The idea that hMOP3 is the functional ortholog of dCYCLE is primarily circumstantial and has yet to be demonstrated convincingly by genetic means. This question has taken on increased significance since the recent identification of MOP9 (Hogenesch et al., 2000; Ikeda et al., 2000). The low-level expression of mMOP9 in the SCN leads to the possibility that MOP3 and MOP9 are functionally redundant. Alternatively, it is possible that these two proteins exert differential regulation on the circadian feedback mechanism, or that either one or both of these two proteins plays no role in mammalian circadian biology.
Results
Loss of MOP3 Results in Arrhythmic Behavior under Free-Running Conditions
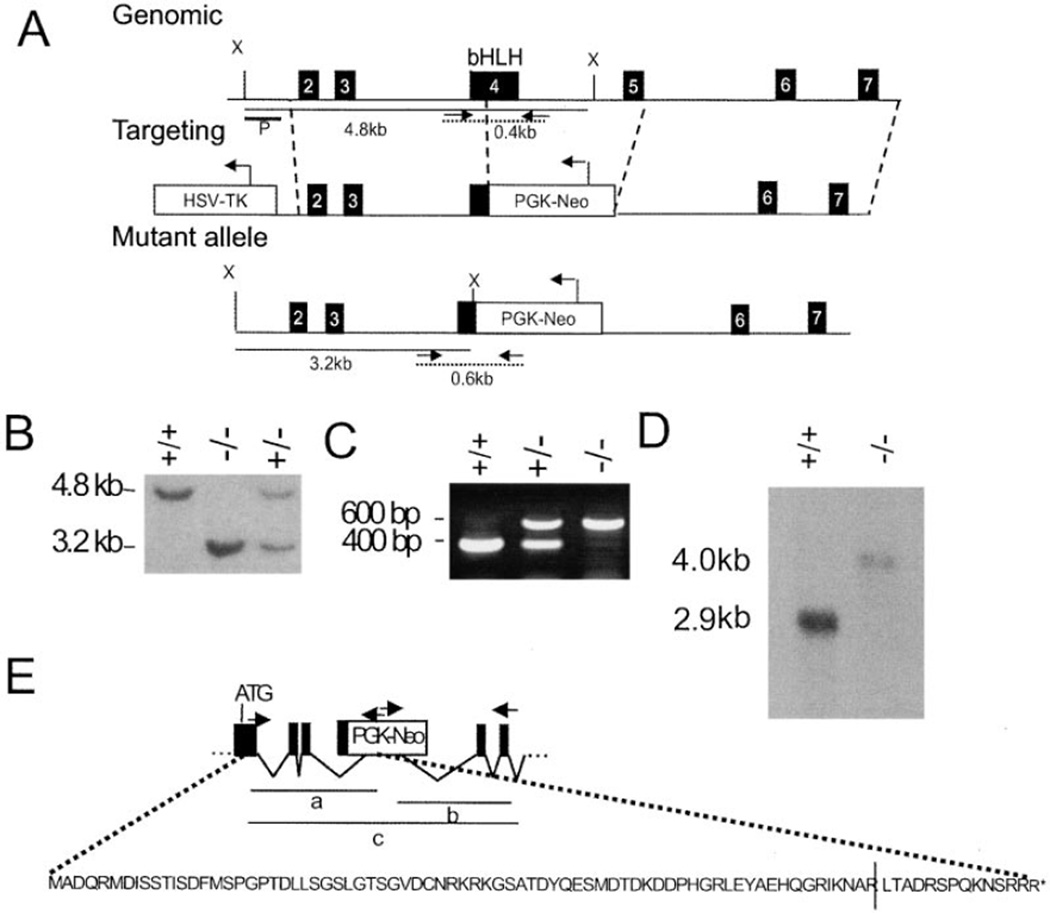
In an effort to understand the role of these two putative CYCLE homologs in murine circadian rhythmicity, we generated and characterized a null allele at the mMop3 locus. A targeted disruption of mMop3 was generated in GS-1 embryonic stem cells by replacing the helix-loop-helix domain within exon 4 and all of exon 5 with a neomycin-resistance (Neo) gene cassette (Figure 1A). No lethality was associated with the targeted mMop3 allele, as null mice were born with the expected wild-mice do not express circadian rhythms of locomotor type:heterozygote:null ratio (43:83:45). A larger mRNA transcript from the mutant allele is present and is expressed at low levels compared to the wild-type mMop3 mRNA (Figure 1D). To ensure no significant protein could be expressed from the targeted Mop3 allele, we used three sets of RT-PCR primers to amplify the predominant transcript from total brain RNA. We observed that the major transcript from the targeted allele uses cryptic splice acceptor and donor sites within the Neo cassette.
Figure 1. Generation of mMop3−/− Mice.
(A) Schematic diagram of the region surrounding the bHLH domain of mMop3, the targeting construct, and the resulting mutant allele. Exon numbers reflect known coding exons with exon one being the furthest 5' known start site for MOP3 protein. Dashed lines indicate the regions of homology used for homologous recombination. Solid lines indicate fragment sizes detected by probe (P) following XbaI (X) digest of genomic DNA. Dotted lines represent the fragment sizes generated by PCR genotyping of wild-type and mutant alleles. (B) Southern blot of mouse tail biopsies showing bands of 4.8 kb and 3.2 kb indicating the presence of the wild-type and mutant alleles, respectively. (C) PCR genotyping of tail biopsies showing bands of 400 bp and 600 bp indicating the presence of the wild-type and mutant alleles. (D) Northern blot of mRNA extracted from whole brain tissue. (E) Schematic of the predicted amino acid sequence of isolated transcript from the Mop3 mutant allele. Black arrows show position of RT-PCR primers and a, b, and c correpond to the three isolated fragments. All three fragments corresponded to a single splice variant that would result in the truncation of the protein 15 amino acids after the splice junction between exon-3 and NeoR sequence (vertical bar).
The predicted protein sequence shown in Figure 1E truncates 15 amino acids following the splice site between exon 3 and the NeoR cassette. This mutant protein product contains no known functional domains as it truncates prior to the inclusion of the basic region in exon 4, and thus is unlikely to form a functional protein. This analysis and the lack of any phenotype in the Mop3+/− mice (see below) argue strongly that our targeting strategy generated a null allele at mMop3.
We hypothesized that if Mop3 and Mop9 were not functionally redundant in the circadian system, then a null mutation at the Mop3 locus would significantly affect the circadian system in mice. Therefore, we examined the wheel-running activity of wild-type, Mop3+/−, and Mop3−/− mice by entraining the animals to a schedule of 12 hr light–12 hr dark (LD 12:12) for 14 days and then transferring them to constant darkness (DD). In constant darkness, wild-type and Mop3+/− mice expressed robust circadian rhythms of activity (Figures 2A and 2B, and data not shown; Table 1). Average free running periods in the wild-type and Mop3+/− mice were 23.63 hr and 23.68 hr, respectively. Spectral analysis to using Fast Fourier transform (FFT) detected high-amplitude periodicity in the circadian range (18–30 hr) for both genotypes (Table 1).A 6hr light pulse administered at CT16 elicited an average phase delay of 3.47 : 1.17 hr in wild-type mice (Figures 2A and 2B). In contrast, Mop3−/− activity in constant darkness (Figures 2C and 2D). Spectral analysis did not detect periodicity in the circadian range for the Mop3−/− mice (Table 1). Moreover, a 6 hr light pulse administered on the 22nd day in constant darkness had no significant effect on locomotor behavior in Mop3−/− mice and did not lead to phase shifts or obvious suppression of activity, as was observed in wild-type mice (Figures 2C and 2D).
Figure 2.
Wheel-Running Activity in Wild-Type and Mop3−/− Mice (A–D) Representative activity records of individual wild-type (A and B) and Mop3−/− (C and D) littermates are presented in double-plotted format. The bar above the activity record shows the light–dark cycle. As indicated to the right of each record, animals were individually housed in a light–dark (LD) cycle for 14 days and then transferred constant darkness (DD). On day 22 in DD, animals received a 6 hr light pulse (300 lux) given at CT16 (indicated by the arrow). 10 days after the light pulse, animals were returned to LD for 18 days and then released into constant darkness for 3 days.
Table 1.
Phenotypic Characteristics of Mop3−/− Mice
| Characteristic§ | Wild-Type | Mop3+/− | Mop3−/− | ANOVA p Value |
|---|---|---|---|---|
| Free running period (hr) | 23.64 ± 0.10 | 23.64 ± 0.12 | No rhythm | 0.81 |
| Phase shift (hr) | −3.72 ± 0.39 | ND | NA | NA |
| FFT circadian amplitude (% total variance) | 16.50 ± 1.83 | 14.56 ± 1.94 | 0.26 ± 0.04* | 1.00 × 10−6 |
| FFT ultradian amplitude (% total variance) | 1.93 ± 0.20 | 2.32 ± 0.40 | 3.29 ± 0.60 | 0.097 |
| Activity level (wheel counts) | ||||
| LD: Total activity OPer 24 hr, days 4–13 | 37,767 ± 2,980 | 30,077 ± 3,860 | 10,079 ± 2,072* | 1.00 × 10−6 |
| % Activity in light phase, days 4–13 | 4.17 ± 1.00 | 4.67 ± 2.48 | 28.62 ± 5.52* | 0.00018 |
| DD: Total activity per 24 hr, days 1–20 | 35,773 ± 5,236 | 28,425 ± 4,640 | 18,424 ± 1,678* | 0.021 |
| Total activity per 24 hr, days 22–31 (post-light pulse) | 35,070 ± 4,611 | ND | 10,260 ± 1,872* | 0.00032 |
| Phase of activity onset in LD, days 4–13 (hr) | ||||
| Phase angle relative to light-off (hr) | 0.21 ± 0.14 | −0.12 ± 0.06 | −2.35 ± 0.72* | 0.0029 |
| Variance of phase angle relative to light-off (hr) | 0.34 | 0.20 | 7.68* | 5.29 × 10−8¶ |
Values are presented as mean ± SEM except variance. Effects of genotype were analyzed by a Generalized Model (GLM) ANOVA by using NCSS (Kayesville, UT) in all cases except variance, in which an F ratio statistical analysis was used. Free running period was determined using χ2 periodogram for days 1–20 in constant darkness. Phase shift is in response to a 6 hr (~300 lux) phase shift on day 22 in constant darkness. FFT, Fast Fourier transform. FFT circadian amplitude values represent the peak relative amplitude in the circadian range (18–30 hr) normalized a total variance of 100%. LD; 12 hr light–12 hours dark cycle. DD; constant darkness. Phase angle relative to lights-off represents the average time difference between activity onset and lights-off as determined by 24 hr average activity profile. Total activity was measured as total wheel counts per 24 hr period; light phase refers to the light-on portion of the LD cycle.
Wild-type, n = 7; Mop3+/− n = 5; and Mop3−/−, n = 7 for free running period, FFT peak period, and total activity in DD. For total activity in LD: Wild-type, n = 18; Mop3+/− n = 5; and Mop3−/−, n = 17. For phase of activity onset in LD: Wild-type, n = 18; Mop3+/− n = 5; and Mop3−/−, n = 15 (due to two animals that failed to entrain). ND: not determined; NA: not applicable.
Significantly different values as indicated by GLM ANOVA or
F test.
The FFT analyses revealed residual, low-amplitude periodicities in the 5–12 hr ultradian range in Mop3−/− mice (Figures 2C and 2D; Table 1) under DD conditions. Ultradian rhythms have been documented in animals with disrupted circadian rhythms, such as Clock/Clock mice, SCN-lesioned mice, and SCN-lesioned voles (Gerkema et al., 1990; Schwartz and Zimmerman, 1991; Vitaterna et al., 1994), but such rhythms appear not to be driven by the circadian oscillator (Lehmann, 1977; Gerkema et al., 1993). An oscillatory mechanism will produce a negative value using serial correlation coefficient analyses of activity bout length (Pittendrigh and Daan, 1976). The average serial correlation coefficient values for activity bouts were 0.00223 ± 0.096 in wild-type mice, −0.186 ± 0.152 in Mop3+/− mice, and 0.185 ±0.072 in Mop3−/− mice. These values indicate that the ultradian bouts are not generated by an oscillatory mechanism. Furthermore, FFT analysis detected low-amplitude ultradian periodocities in the wild-type and Mop3+/− animals that were not significantly different from those in Mop3−/− mice (Table 1). The Mop3 mutant allele is recessive, as the phenotype of Mop3+/− mice is not different from that of wild-type mice (Table 1).
Mop3−/− Mice Have Altered Activity on Light–Dark Cycles and Reduced Total Activity Levels
In addition to its extreme effect on circadian activity in constant darkness, the Mop3 mutation also has an unexpected effect on activity in the presence of a light cycle (Figure 2C). Upon examination of wheel running activity in light–dark cycles, all wild-type and Mop3+/− mice began wheel-running within 0.5 hr of lights-off. In contrast, in Mop3−/− mice, the phase of activity onset was highly variable with respect to lights-off (Figures 2C and 2D; Table 1). A 24 hr average activity profile confirmed that of 17 Mop3−/− mice, only 5 began activity within 0.5 hr of lights-off and 2 failed to entrain altogether. The anticipatory phase of the activity rhythm of MOP3−/− mice strongly suggests that “masking” alone cannot account for the rhythmic behavior in light–dark cycles. Furthermore, distribution of activity during the light–dark cycle is significantly altered in Mop3−/− mice: 28% of wheel-running activity in Mop3−/− mice occurs during the light phase, in contrast to 4.2% and 4.1% of activity in the light phase of wild-type and Mop3+/− mice, respectively (Table 1). The Mop3−/− behavior in LD 12:12 cycles could be attributed to poor entrainment of the circadian pacemaker, because the null mice have rhythmic activity that anticipates lights-out. However, we cannot rule out the possibility that “masking” contributes to the activity pattern in response to light. Such acute effects by light and other environmental cues on behavior have been documented in SCN lesioned Syrian hamsters and rats (Wachulec et al., 1997; Redin and Mrosovsky, 1999a, 1999b).
In addition to the effects of the Mop3−/− mutation on circadian rhythmicity, the Mop3 mutant mice also displayed lower average levels of wheel-running activity as compared to wild-type mice both in a light cycle and in constant darkness (Table 1). Interestingly, such decreases in total activity are not found in Clock/Clock, mPer2Brdm1, or Cry1−/−, Cry2−/− mutant mice (M. H. Vitaterna, personal communication; Vitaterna et al., 1994; Zheng et al., 1999). Taken together, these results indicate that mMop3 is indeed a core clock component that is essential for the generation and maintenance of circadian rhythms in constant conditions, normal entrainment of behavior to LD cycles, as well as normal activity levels in both LD and DD conditions. Moreover, the Mop3−/− mutation represents an example of a single gene mutation in mammals that causes immediate loss of circadian rhythmicity in constant environmental conditions.
Molecular Analysis of Circadian Regulated Genes in the SCN and Liver of Mop3−/− Mice
The effects of the Mop3−/− mutation on circadian behavior could be the result of disruptions in either the central pacemaker or peripheral output pathways. Therefore, we examined the expression of two known CLOCK-regulated genes in the SCN, mPer1 and mPer2 using in situ hybridization. In Clock/Clock mutant mice, mPer1 and mPer2 expression in the SCN is blunted and nonrhythmic (Jin et al., 1999); thus, we hypothesized that if MOP3 is the true transcriptional partner of CLOCK, expression of these genes would be altered in Mop3−/− mice as well. Wild type and Mop3−/− mice were sacrificed every 4 hr from 58 hr to 82 hr in constant darkness (3rd and 4th cycles in constant darkness) (Jin et al., 1999). Wild-type animals expressed a robust rhythm of mPer1 and mPer2 expression (Figures 3E and 3J). Peak expression of mPer1 occurred at 66 hr in constant darkness, which corresponds to CT6 (Figures 3A and 3B). Peak expression of mPer2 in wild-type animals occurred at 70 hr in constant darkness, or CT10 (Figures 3F and 3G). In contrast, expression levels of both mPer1 and mPer2 were not rhythmic and were near baseline levels in Mop3−/− mice (Figures 3C–3E and 3H–3J). The extremely low expression of mPer1 and mPer2 in the SCN of Mop3−/− mice provides direct genetic evidence that MOP3 is required for the positive regulation of gene expression in the mammalian circadian pacemaker.
Figure 3. mPer1 and mPer2 Expression in the SCN of Wild-Type and Mop3−/− Mice.
Coronal brain sections from mice sacrificed every 4 hr from 54 hr to 82 hr in constant darkness were hybridized with mPer1 and mPer2 riboprobes. (A–E) mPer1 in situ hybridization. (A) Wild-type mPer1 hybridization at 66 hr, which corresponds to CT6; (B) wild-type mPer1 hybridization at 78 hr, or CT18; (C) Mop3−/− mPer1 hybridization at 66 hr; (D) Mop3−/− mPer1 hybridization at 78 hr; (E) time course of mPer1 expression in the SCN of wild-type (filled circles) and Mop3−/− (open circles) mice. (F–J) mPer2 in situ hybridization. (F) Wild-type mPer2 hybridization at 70 hr, which corresponds to CT10; (G) wild-type mPer2 hybridization at 82 hr, or CT22; (H) Mop3−/− mPer2 hybridization at 70 hr; (I) Mop3−/− mPer2 hybridization at 82 hr; (J) time course of mPer2 expression in the SCN of wild-type (filled circles) and Mop3−/− (open circles) mice. The bar at the top indicates subjective night in black and subjective day in gray. Asterisks indicate significant differences between wild-type and Mop3−/− mice at the times shown (mPer1 GLM ANOVA, F(7,35) 16.41, p < 1.0 × 10−6; mPer2 GLM ANOVA, F(7,35) 18.61, p < 1.0 × 10−6, Tukey-Kramer posthoc comparison p < 0.05).
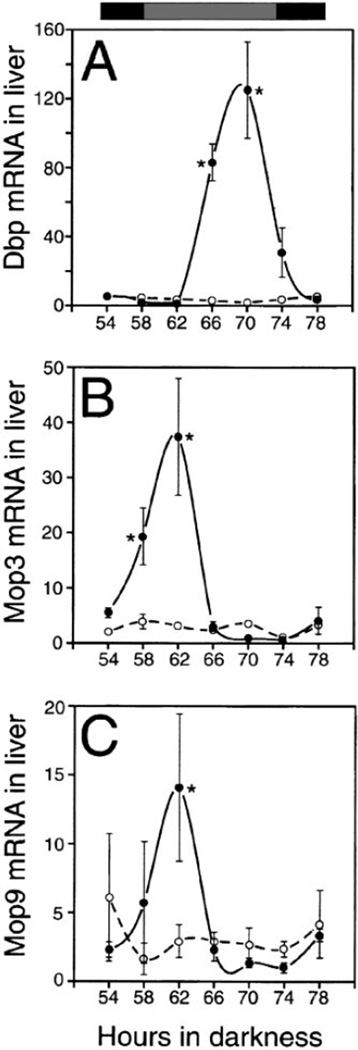
To determine if the Mop3-dependent regulation extends to peripheral oscillators, we examined the expression of albumin D-element binding protein (mDbp) in liver. We chose mDbp because it is under circadian regulation but is not a part of the central feedback mechanism (Lopez-Molina et al., 1997). The regulation of mDbp in peripheral tissues appears to be due to direct transcriptional control by CLOCK through E box enhancer elements (Ripperger et al., 2000). Furthermore, mDbp null mutant mice show abnormalities in circadian behavior without disruption of the core oscillator in the SCN (Lopez-Molina et al., 1997). In the liver of Mop3 wild-type mice, mDbp mRNA displays a robust rhythm that peaks at 70 hr in DD, or CT10, whereas in Mop3−/− mice mDbp levels remain low and non-cycling at all times (Figure 4A). The disruption of circadian-regulated gene expression in both the SCN and the periphery of Mop3−/− mice demonstrates that MOP3 is essential for the generation of circadian rhythms at the molecular level in the central pacemaker as well as in peripheral oscillators.
Figure 4. Analysis of mDbp, mMop3, and mMop9 mRNA in Peripheral Tissues of Mop3−/− Mice.
Analysis of mRNA levels analyzed using TaqMan technology™ from livers taken over a time course of 54–82 hr in constant darkness from wild-type animals (filled circles) and null animals (open circles). The bar at the top indicates subjective night in black and subjective day in gray. (A) Expression of mDbp. Asterisks indicate significant differences between wild-type and Mop3−/− mice at the times shown (GLM ANOVA, F(7,34) = 9.36, p = 5.0 × 10−5, Tukey-Kramer posthoc comparison p ≤0.05). (B) Expression of mMop3. Asterisks indicate significant differences between wild-type and Mop3−/− mice at the times shown (GLM ANOVA, F(7,34) 3.52, p 0.012, Tukey-Kramer posthoc comparison p < 0.05). (C) Expression of mMop9. Asterisks indicate significant differences between wild-type and Mop3−/− mice at the times shown (GLM ANOVA, F(7,34) 2.75, p 0.036, Tukey-Kramer posthoc comparison p < 0.05).
The combined analysis of behavior and gene expression in Mop3−/− mice shows that the role of Mop3 in circadian rhythm generation is not redundant; indeed, the close homolog MOP9 is insufficient to rescue this function of MOP3. To better understand Mop9 and Mop3 circadian regulation, we examined their expression levels in the liver from wild-type and Mop3−/− animals. In wild-type animals, both the Mop9 transcript and the Mop3 transcript cycle in the liver with peak expression at CT2. In the Mop3−/− mutants, however, the message levels of Mop9 and the mutant Mop3 were low and not cyclic (Figures 4B and 4C). In the simplest model, these data suggest that the MOP3-CLOCK protein complex positively activates both genes. Whether or not MOP9 plays a role in the circadian system remains unclear.
Discussion
The results presented here suggest that the Mop3 locus is of critical importance at all levels of the mammalian circadian system. The results presented here also indicate that the MOP9 homolog is not functionally redundant with MOP3 but is dependent on MOP3 for circadian regulation. At the level of the circadian pacemaker, we provide genetic evidence that the MOP3 protein exerts its effects on the molecular feedback loop by acting as a positive regulator of gene expression. Given the effects of the dominant-negative Clock mutation, the effects of the Mop3 null allele on Per gene expression are consistent with an activator role of the CLOCK-MOP3 transcription factor complex (King et al., 1997; Gekakis et al., 1998). The observation that Mop3−/− mice do not entrain to a light cycle provides genetic evidence that MOP3 plays a functional role in the input pathway. In support of this idea, a recent study shows that MOP3 (BMAL1) protein levels display a circadian rhythm in the SCN that peaks at night and are reduced upon light stimulus (Tamaru et al., 2000). Finally, the critical role of MOP3 in the circadian output pathway is clearly supported by the arrhythmicity of transcriptional output gene mDBP in the liver of Mop3 mutant mice. Furthermore, Mop3−/− mice show decreased activity levels both in a light cycle and in constant darkness indicating that MOP3 may play a role in behavioral outputs beyond its role in generating behavioral rhythms.
The effect of a null mutation of mMop3 on locomotor activity in constant dark conditions is similar to the phenotype seen in Cryptochrome 1 and 2 mutant mice. However, on light–dark cycles, Cry1−/− Cry2−/− double mutant mice do not show anticipatory activity as was seen in MOP3−/− mice (Vitaterna et al., 1999; van der Horst et al., 1999). Furthermore, the Cry1−/− Cry2−/− double mutant mice also respond behaviorally to light pulses by immediately stopping activity at light onset and immediately starting activity at light offset. Such strong“masking” effects of light were not observed in MOP3−/− mice.
The effect of a Mop3 mutation on the level of activity is a novel phenotype that has not been reported in Clock, mPer2Brdm, and Cry1−/− Cry2−/− double mutant mice (Vitaterna et al., 1994; Zheng et al., 1999; M. H. Vitaterna, personal communication). Reduced activity has also been reported in mice with a targeted mutation at the dbp locus, a circadian regulated transcription factor that we show here is controlled by MOP3 (Lopez-Molina et al., 1997). It is not clear from our results, however, whether the reduced activity phenotype in the Mop3−/− mice is caused by the loss of circadian rhythmicity or whether it may represent a novel role of MOP3 in regulation of metabolic or behavioral outputs. In conclusion, these data represent an example of a single-gene mutation that causes complete abolition of both behavioral and molecular circadian rhythms as well as disruptions in both input (entrainment) and output (activity level) pathways in mammals. Such widespread phenotypic consequences on the circadian system of mice argue that MOP3 rests near the top of the circadian gene hierarchy in mammals.
Experimental Procedures
Generation of Mop3−/− Mice
A 7.5 kb region of homology to MOP3 genomic DNA was isolated from BAC DNA (Genome Systems) by long-range PCR. A Neomycin resistance cassette (Neo) was inserted in reverse orientation to the Mop3 allele into the exon containing the bHLH region deleting DNA from the middle of that exon until just 3' of the next coding exon. Homologous recombinants were isolated following electroporation of 10 µg targeting construct into GS1 ES cells (Genome Systems). 200 clones surviving G418/Gancycolivir double selection were screened by Southern blot for homologous recombination using a 300 bp probe (PL1225) to a region just outside the 5' end of the targeting construct on XbaI digest. Heterozygous mice were obtained from these cells by standard methods. Animal care and use procedures were approved by University of Wisconsin, Madison institutional guidelines. Southern blot analysis was performed on genomic DNA from tail biopsies to detect transmission of the targeted allele. Subsequent to the first generation, genotyping was performed using multiplex PCR with OL2646, CCACCAAGCCCAG CAACTCA; OL2647, ATTCGGCCCCCTATCTTCTGC; and OL278, TCGCCTTCTATCGCCTTCTTGACG. OL2626 and OL2647 amplify a 400 bp band corresponding to the wild-type allele. PCR was per-formed on genomic DNA from tail biopsies for 40 cycles of 95°C, 15 s: 60°C, 15s: 72°C, 1 min in 1× PCR buffer (Promega) containing 3.5 mM MgCl2. The region in which OL2647 was designed is deleted in the targeted allele therefore in the presence of the targeted allele OL2646 amplifies a 600 bp band with OL278 (Neomycin specific) as a reverse primer. For expression analysis, polyA mRNA was isolated from brains of wild-type, Mop3+/−, and Mop3−/− mice killed at ZT-23 (ZT: Zeitgeber time) using Trizol (Gibco). Northern blot hybridization on 5 µg mRNA was performed using a 210 bp DNA probe specific for a region in the 3' end of MOP3 coding sequence. Primers used for RT-PCR were: Fragment a OL949, 5'-TGGCAGACCAGAGAATCG and OL278, 5'-TCGCCTTCTATCGCCTTCTTGACG; fragment b, OL662, 5'-GCGCGAGCCCCTGATGCTC and OL489 5'-TTA GGATGCAGGTAGTCAAACA; fragment c, OL949 and OL489.
Behavioral Analysis
Mice were singly housed in cages equipped with running wheels on an LD 12:12 cycle for 2 weeks before being released into constant darkness. Animal care and use procedures were approved by Northwestern University institutional guidelines. A subset of wild-type and Mop3−/− mice (n 7 each genotype) received a 6 hr light pulse (300 lux) on day 22 in DD. Activity data were collected as previously described (Vitaterna et al., 1994; Antoch et al., 1997; King et al., 1997). X2 periodogram, Fast Fourier transform (FFT), least squares fit of activity onset, and total activity analyses were performed using ClockLab (Actimetrics, Evanston, IL). X2 periodogram was performed for days 1–20 in constant darkness at 1 min resolution over the 5–36 hr period range. FFT was performed for days 1–20 in constant darkness as an amplitude spectrum normalized over the frequency range of 0 to 1 cycles/hour. Circadian time or CT indicates the phase of the animal’s endogenous circadian rhythm while in free-running conditions, where CT0 marks the beginning of the subjective day and CT12 marks the beginning of the subjective night. Serial correlation coefficients of activity bout length were performed within ClockLab (Actimetrics, Evanston, IL) for the first 20 days in DD with maximum gap length 90 min and threshold of 20 counts/minute. As a control, we also calculated the average serial correlation coefficient for circadian activity onsets in the wild type and Mop3+ /− mice for the first 20 days in constant darkness. The average value of −0.519 ±0.067 in wild type (Student’s t test; p = 0.00025) indicates that the daily onset of activity in constant conditions is generated by a (circadian) oscillatory mechanism. Because the Mop3−/− mice do not have a discernable circadian onset of activity, such an analysis cannot be performed on mutant mice. For analysis of entrainment to Light:Dark cycling of days 4–13 in LD, the 9 day average activities (wheel revolutions) were plotted against the 24 hr period using ClockLab (Actimetrics, Evanston, IL). Onset of activity was designated as the phase at which 20% of the peak activity was reached.
Gene Expression Analysis
Animals were sacrificed every 4 hr under infrared light via cervical dislocation beginning at 54 hr in constant darkness (third cycle in constant darkness). Brains were dissected under dim red light, frozen on dry ice, and stored at −80° C before sectioning. Peripheral tissues were then collected under white light, frozen on dry ice, and stored at −80° C before RNA extraction. In situ hybridization procedures were performed as described (Vitaterna et al., 1999). Riboprobes for mPer1 and mPer2 were generated from nucleotides 474–856 (GenBank AF099229) and nucleotides 243–701 (GenBank AF035830), respectively. Exposure time was 14 days, and hybridization signal was quantified as described (Vitaterna et al., 1999). For liver expression analysis, total liver RNA was isolated using Trizol; followed with DNase treatment using Amp-grade DNase (Gibco). Three micrograms of total RNA was reverse transcribed using Superscript II (Gibco) and 25 ng total RNA equivalent was analyzed by TaqMan technology (PE-Applied Biosystems) (Heid et al., 1996) using an ABI7700 (Perkin Elmer). Primers and probes used for mMop3, mDbp, and mMop9 were as follows:
mMop3 forward, 5'-CCAAGAAAGTATGGACACAGACAAA-3'
mMop3 reverse, 5'-GCATTCTTGATCCTTCCTTGGT-3'
mMop3 probe, 5'-TGACCCTCATGGAAGGTTAGAATATGCA GAAC-3'
mDbp forward, 5'-CGTGGAGGTGCTTAATGACCTTT-3'
mDbp reverse, 5'-CATGGCCTGGAATGCTTGA-3'
mDbp probe, 5'-AACCTGATCCCGCTGATCTCGCC-3'
mMop9 forward, 5'-TGGTGCCTTCGTGACTCTGA-3'
mMop9 reverse, 5'-GTTGACAGACACAATGTACTCCAGC-3'
mMop9 probe, 5'-TTCAGCTTCACAAACCCTTGGACCAAAG-3'
The obtained CT (cycle number at which amplification threshold of detection is reached) values were normalized to Rodent GADPH (PE Applied Biosystems) expression by the Δ Δ Ct method using trough wild-type levels as the calibrator value. The mean Δ ΔCT was converted to relative expression values by the equation 2−Δ Δ CT and a range was calculated by 2−(Δ Δ CT +StdevΔ Δ CT). Values shown are 2−Δ Ct (± range of 2−Δ Δ CT).
Acknowledgments
We thank Martha H. Vitaterna for assistance with the ANOVA analyses and Ravi Allada for helpful discussions. This work was supported by The National Institutes of Health Grants P30-CA07175 and ES05703. C. A. Bradfield was supported through a Burroughs Well-come Scholarship in Toxicology. M. Bunger was supported by a Mary Engsberg Fellowship in Cancer Research. L. D. Wilsbacher is a Fellow in the Medical Scientist Training Program. M. C. Simon and J. S. Takahashi are Investigators in the Howard Hughes Medical Institute.
References
- Allada R, White NE, So WV, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- Antoch MP, Song EJ, Chang AM, Vitaterna MH, Zhao Y, Wilsbacher LD, Sangoram AM, King DP, Pinto LH, Takahashi JS. Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell. 1997;89:655–667. doi: 10.1016/s0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae K, Lee C, Sidote D, Chuang KY, Edery I. Circadian regulation of a Drosophila homolog of the mammalian Clock gene: PER and TIM function as positive regulators. Mol. Cell. Biol. 1998;18:6142–6151. doi: 10.1128/mcb.18.10.6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceriani MF, Darlington TK, Staknis D, Mas P, Petti AA, Weitz CJ, Kay SA. Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science. 1999;285:553–556. doi: 10.1126/science.285.5427.553. [DOI] [PubMed] [Google Scholar]
- Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TDL, Weitz CJ, Takahashi JS, Kay SA. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Emery P, So WV, Kaneko M, Hall JC, Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95:669–679. doi: 10.1016/s0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- Garcia JA, Zhang D, Estill SJ, Michnoff C, Rutter J, Reick M, Scott K, Diaz-Arrastia R, McKnight SL. Impaired cued and contextual memory in NPAS2-deficient mice. Science. 2000;288:2226–2230. doi: 10.1126/science.288.5474.2226. [DOI] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- Gerkema MP, Daan S, Wilbrink M, Hop MW, van der Leest F. Phase control of ultradian feeding rhythms in the common vole (Microtus arvalis): the roles of light and the circadian system. J. Biol. Rhythms. 1993;8:151–171. doi: 10.1177/074873049300800205. [DOI] [PubMed] [Google Scholar]
- Gerkema MP, Groos GA, Daan S. Differential elimination of circadian and ultradian rhythmicity by hypothalamic lesions in the common vole, Microtus arvalis. J. Biol. Rhythms. 1990;5:81–95. doi: 10.1177/074873049000500201. [DOI] [PubMed] [Google Scholar]
- Gotter AL, Manganaro T, Weaver DR, Kolakowski LF, Jr, Possidente B, Sriram S, MacLaughlin DT, Reppert SM. A time-less function for mouse timeless. Nat. Neurosci. 2000;3:755–756. doi: 10.1038/77653. [DOI] [PubMed] [Google Scholar]
- Griffin EA, Jr, Staknis D, Weitz CJ. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science. 1999;286:768–771. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- Gu Y-Z, Hogenesch J, Bradfield C. In Annu. Rev. Pharmacol. Toxicol. San Diego, CA: Academic Press; 2000. The PAS superfamily: sensors of environmental and developmental signals; pp. 519–561. [DOI] [PubMed] [Google Scholar]
- Hao H, Allen DL, Hardin PE. A circadian enhancer mediates PER-dependent mRNA cycling in Drosophila melanogaster. Mol. Cell. Biol. 1997;17:3687–3693. doi: 10.1128/mcb.17.7.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- Hogenesch J, Gu Y-Z, Moran S, Shimomura K, Radcliffe L, Takahashi J, Bradfield C. The basic-helix-loop-helix-PAS protein MOP9 is a brain specific heterodimeric partner of circadian and hypoxia factors. J. Neurosci. 2000;20:1–5. doi: 10.1523/JNEUROSCI.20-13-j0002.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenesch JB, Chan WK, Jackiw VH, Brown RC, Gu YZ, Pray-Grant M, Perdew GH, Bradfield CA. Characterization of a subset of the basic-helix-loop-helix-PAS superfamily that interacts with components of the dioxin signaling pathway. J. Biol. Chem. 1997;272:8581–8593. doi: 10.1074/jbc.272.13.8581. [DOI] [PubMed] [Google Scholar]
- Hogenesch JB, Gu Y-Z, Jain S, Bradfield CA. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc. Natl. Acad. Sci. USA. 1998;95:5474–5479. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma S, Ikeda M, Abe H, Tanahashi Y, Namihira M, Honma K, Nomura M. Circadian oscillation of BMAL1, a partner of a mammalian clock gene Clock, in rat suprachiasmatic nucleus. Biochem. Biophys. Res. Comm. 1998;250:83–87. doi: 10.1006/bbrc.1998.9275. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Nomura M. cDNA cloning and tissue-specific expression of a novel basic helix-loop-helix/PAS protein (BMAL1) and identification of alternatively spliced variants with alternative translation initiation site usage. Biochem. Biophys. Res. Comm. 1997;233:258–264. doi: 10.1006/bbrc.1997.6371. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Yu W, Hirai M, Ebisawa T, Honma S, Yoshimura K, Honma K, Nomura M. cDNA cloning of a novel bHLH-PAS trascription factor superfamily gene, BMAL2: its mRNA expression, subcellular distribution, and chromosomal localization. Biochem. Biophys. Res. Comm. 2000;275:493–502. doi: 10.1006/bbrc.2000.3248. [DOI] [PubMed] [Google Scholar]
- Jin X, Shearman LP, Weaver DR, Zylka MJ, de Vries GJ, Reppert SM. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell. 1999;96:57–68. doi: 10.1016/s0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- Kay SA. PAS, present, and future: clues to the origins of circadian clocks. Science. 1997;276:753–754. doi: 10.1126/science.276.5313.753. [DOI] [PubMed] [Google Scholar]
- King DP, Takahashi JS. Molecular genetics of circadian rhythms in mammals. Annu. Rev. Neurosci. 2000;23:713–742. doi: 10.1146/annurev.neuro.23.1.713. [DOI] [PubMed] [Google Scholar]
- King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TD, Vitaterna MH, Kornhauser JM, Lowrey PL, Turek FW, Takahashi JS. Positional cloning of the mouse circadian Clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS, Young MW. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Ie. Cell. 1998;94:97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- Lee C, Bae K, Edery I. PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK-CYC/dBMAL1 heterodimer without disrupting formation of the heterodimer: a basis for circadian transcription. Mol. Cell. Biol. 1999;19:5316–5325. doi: 10.1128/mcb.19.8.5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann U. Stochastic principles in the temporal control of activity behaviour. Interntl. J. Chronobiol. 1977;4:223–266. [PubMed] [Google Scholar]
- Lopez-Molina L, Conquet F, Dubois-Dauphin M, Schibler U. The DBP gene is expressed according to a circadian rhythm in the suprachiasmatic nucleus and influences circadian behavior. EMBO J. 1997;16:6762–6771. doi: 10.1093/emboj/16.22.6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey PL, Shimomura K, Antoch MP, Yamazaki S, Zemenides PD, Ralph MR, Menaker M, Takahashi JS. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science. 2000;288:483–491. doi: 10.1126/science.288.5465.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RY. Circadian rhythms: basic neurobiology and clinical applications. Annu. Rev. of Med. 1997;48:253–266. doi: 10.1146/annurev.med.48.1.253. [DOI] [PubMed] [Google Scholar]
- Okamura H, Miyake S, Sumi Y, Yamaguchi S, Yasui A, Muijtjens M, Hoeijmakers JH, van der Horst GT. Photic induction of mPer1 and mPer2 in cry-deficient mice lacking a biological clock. Science. 1999;286:2531–2534. doi: 10.1126/science.286.5449.2531. [DOI] [PubMed] [Google Scholar]
- Pittendrigh CS, Daan S. A functional analysis of circadian pacemakers in nocturnal rodents. J. Comp. Physiol. A. 1976;106:223–355. [Google Scholar]
- Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- Ralph MR, Foster RG, Davis FC, Menaker M. Transplanted suprachiasmatic nucleus determines circadian period. Science. 1990;247:975–978. doi: 10.1126/science.2305266. [DOI] [PubMed] [Google Scholar]
- Redin U, Morosovsky N. Masking of locomotor activity in hamsters. J. Comp. Physiol. A. 1999a;184:429–437. doi: 10.1007/s003590050342. [DOI] [PubMed] [Google Scholar]
- Redin U, Morosovsky N. Masking by light in hamsters with SCN lesions. J. Comp. Physiol. A. 1999b;184:439–448. doi: 10.1007/s003590050343. [DOI] [PubMed] [Google Scholar]
- Reppert SM. A clockwork explosion. Neuron. 1998;21:1–4. doi: 10.1016/s0896-6273(00)80234-2. [DOI] [PubMed] [Google Scholar]
- Ripperger JA, Shearman LP, Reppert SM, Schibler U. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev. 2000;14:679–689. [PMC free article] [PubMed] [Google Scholar]
- Rosbash M. Molecular control of circadian rhythms. Curr. Opin. Genet. Dev. 1995;5:662–668. doi: 10.1016/0959-437x(95)80037-9. [DOI] [PubMed] [Google Scholar]
- Rutila JE, Suri V, Le M, So WV, Rosbash M, Hall J. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell. 1998;93:805–814. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- Schwartz WJ, Zimmerman P. Lesions of the suprachiasmatic nucleus disrupt circadian locomotor rhythms in the mouse. Physiol. Behav. 1991;49:1283–1287. doi: 10.1016/0031-9384(91)90364-t. [DOI] [PubMed] [Google Scholar]
- Sehgal A, Price JL, Man B, Young MW. Loss of circadian behavioral rhythms of per RNA oscillations in the Drosophila mutant timeless. Science. 1994;263:1603–1606. doi: 10.1126/science.8128246. [DOI] [PubMed] [Google Scholar]
- Shearman LP, Jin X, Lee C, Reppert SM, Weaver DR. Targeted disruption of the mPer3 gene: subtle effects on the circadian clock function. Mol. Cell. Biol. 2000;20:6269–6275. doi: 10.1128/mcb.20.17.6269-6275.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver R, LeSauter J, Tresco PA, Lehman MN. A diffusible coupling signal from the transplanted suprachiasmatic nucleus controlling circadian locomotor rhythms. Nature. 1996;382:810–813. doi: 10.1038/382810a0. [DOI] [PubMed] [Google Scholar]
- Stanewsky R, Kaneko M, Emery P, Beretta B, Wager-Smith K, Kay SA, Rosbash M, Hall JC. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell. 1998;95:681–692. doi: 10.1016/s0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- Takahata S, Sogawa K, Kobayashi A, Ema M, Mimura J, Ozaki N, Fujii-Kuriyama Y. Transcriptionally active heterodimer formation of an Arnt-like PAS protein, Arnt3, with HIF-1a, HLF, and clock. Biochem. Biophys. Res. Comm. 1998;248:789–794. doi: 10.1006/bbrc.1998.9012. [DOI] [PubMed] [Google Scholar]
- Tamaru T, Isojima Y, Yamada T, Okada M, Nagai K, Takamatsu K. Light and glutamate-induced degradation of the circadian oscillating protein BMAL1 during the mammalian clock resetting. J. Neurosci. 2000;20:7525–7530. doi: 10.1523/JNEUROSCI.20-20-07525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D, Buijs R, Bootsma D, Hoeijmakers JH, Yasui A. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD, Dove WF, Pinto LH, Turek FW, Takahashi JS. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, Hitomi K, Thresher RJ, Ishikawa T, Miyazaki J, Takahashi JS, Sancar A. Differential regulation of mammalian period genes and circadian rhythmicity by Cryptochromes 1 and 2. Proc. Natl. Acad. Sci. USA. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachulec M, Hua L, Tanaka H, Paleso E, Satinoff E. Suprachiasmatic nuclei lesions do not eliminate homoestatic thermoregulatory responses in rats. J. Biol. Rhythms. 1997;12:226–234. doi: 10.1177/074873049701200304. [DOI] [PubMed] [Google Scholar]
- Wager-Smith K, Kay SA. Circadian rhythm genetics: from flies to mice to humans. Nature. 2000;26:23–27. doi: 10.1038/79134. [DOI] [PubMed] [Google Scholar]
- Wilsbacher LD, Takahashi JS. Circadian rhythms: molecular basis of the clock. Curr. Opin. Genet. Dev. 1998;8:595–602. doi: 10.1016/s0959-437x(98)80017-8. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- Yu W, Ikeda M, Abe H, Honma S, Ebisawa T, Yamauchi T, Honma K, Nomura M. Characterization of three splice variants and genomic organization of the mouse BMAL1 gene. Biochem. Biophys. Res. Comm. 1999;260:760–767. doi: 10.1006/bbrc.1999.0970. [DOI] [PubMed] [Google Scholar]
- Zheng B, Larkin DW, Albrecht U, Sun ZS, Sage M, Eichele G, Lee CC, Bradley A. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature. 1999;400:169–173. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]
- Zhou YD, Barnard M, Tian H, Li X, Ring HZ, Francke U, Shelton J, Richardson J, Russell DW, McKnight SL. Molecular characterization of two mammalian bHLH-PAS domain proteins selectively expressed in the central nervous system. Proc. Natl. Acad. Sci. USA. 1997;94:713–718. doi: 10.1073/pnas.94.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]