Abstract
The human genome contains ~50 genes that were derived from transposable elements or transposons, and many are now integral components of cellular gene expression programs. The human THAP9 gene is related to the Drosophila P-element transposase. Here, we show that human THAP9 can mobilize Drosophila P-elements in both Drosophila and human cells. Chimeric proteins formed between the Drosophila P-element transposase N-terminal THAP DNA binding domain and the C-terminal regions of human THAP9 can also mobilize Drosophila P elements. Our results indicate that human THAP9 is an active DNA transposase that, although “domesticated,” still retains the catalytic activity to mobilize P transposable elements across species.
Transposable elements or transposons are mobile segments of DNA found in both prokaryotic and eukaryotic genomes (1). Large portions of eukaryotic genomes (up to 50% of the human genome) are made up of transposable elements (1–3), and these elements are thought to facilitate genome evolution (1, 4). Mobile elements can undergo “molecular domestication,” where the transposon genes are incorporated into cellular gene expression programs, but are no longer mobile (1, 3, 5–9). They can also evolve cellular DNA recombination functions, such as the V(D)J antigen receptor–recombination system (10, 11).
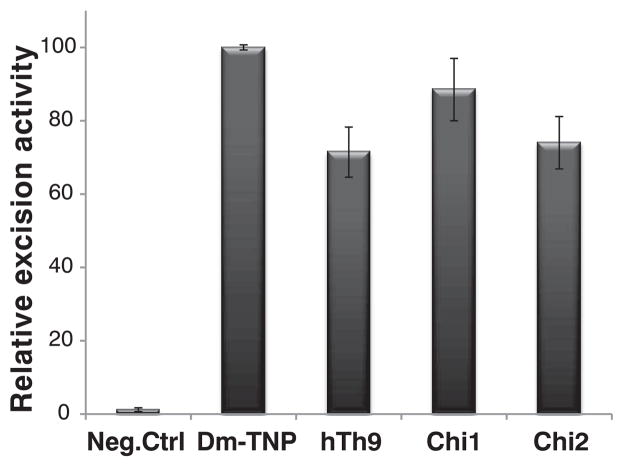
The THAP domain is a C2CH zinc-coordinating DNA binding domain (12). The human genome has 12 THAP domain–containing genes (12). Human THAP9 (hTh9) is homologous with (25% identical with and 40% similar to) (fig. S1) the Drosophila P-element transposase (DmTNP), the founding member of the THAP domain family of DNA binding proteins (fig. S1A) (7, 12, 13). Given the homology and similarities in the DNA binding sites between hTh9 and DmTNP (14), we tested the human THAP9 protein to see if it might be able to mobilize Drosophila P elements in Drosophila cells and human embryonic kidney–293 (HEK293) cells. We used a plasmid-based assay for P-element excision (fig. S3), which relies on scoring P-element–transposase–induced transposon excision events in bacteria (15). When either hTh9 or two different chimeric fusion proteins in which the Drosophila P-element trans-posase N-terminal THAP DNA binding domain was fused to C-terminal portions of human THAP9 (fig. S1B), full-length human THAP9, as well as these chimeras (chimera 1 and chimera 2), gave ~60 to 80% of the P-element excision activity observed with wild-type Drosophila P-element transposase in Drosophila and human cells (Fig. 1 and fig. S4E). Immunoblot analysis indicated that DmTNP, hTh9, and the fusion proteins were expressed at similar levels upon transfection of Drosophila L2 cells with the use of an epitope tag antibody (fig. S2, A and B). No P-element activity was observed in negative-control experiments in which DmTNP or hTh9 expression was lacking (Fig. 1 and fig. S4E). Analysis of plasmid DNA excision products recovered from bacteria indicated that P-element excision had occurred with both human THAP9 and the Drosophila transposase-THAP9 fusion proteins (figs. S4 and S5).
Fig. 1.
Human THAP9 can excise P elements. P-element excision activities generated from a negative-control plasmid (pBluescript empty vector) versus the wild-type P-element transposase, human THAP9, or chimera 1 or chimera 2 expression vectors in Drosophila L2 cells. Values are the average (± SEM) of three independent experiments (n = 3), each done in duplicate, P < 0.05.
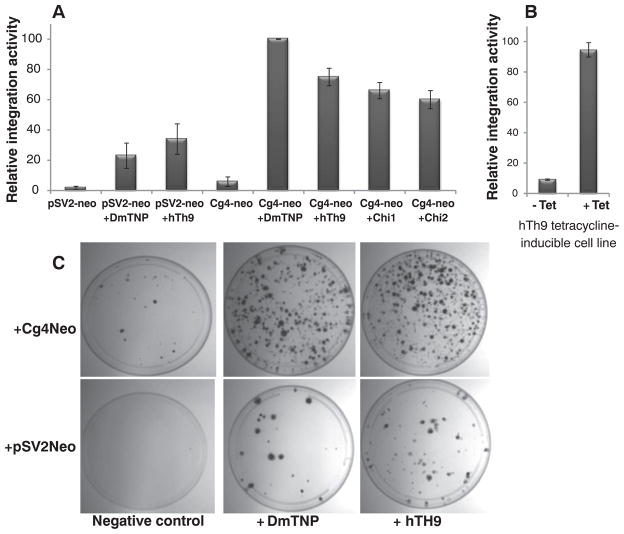
Next, we tested whether human THAP9 could carry out transposition of a genetically marked Drosophila P element from a plasmid into the human genome in HEK293 cells. We used an assay for integration in which the Cg4 P-element vector carried an SV40 promoter–neomycin phosphotransferase fusion (Cg4-neo) that can confer G418 resistance in mammalian cells upon transposition into human chromosomes (fig. S6) (16). Upon transfection of the DmTNP or hTh9 expression vectors into human HEK293 cells along with Cg4-neo and subsequent G418 selection, many colonies were obtained (Fig. 2, A and C). [On average, ~50 to 75 colonies were obtained per 10-cm plate with hTh9 or DmTNP) (details in fig. S7).] To examine the levels of random integration, independent of P element–mediated transposition, the integration assay was also carried out with the pSV2-neo reporter plasmid, which contains an SV40 promoter–neomycin phosphotransferase fusion gene but lacks the P-element transposon termini. Transfection of pSV2-neo along with the DmTNP or hTh9 expression vectors gave rise to fewer G418-resistant colonies, by a factor of 3 to 5, than those obtained in the experiments performed with P-element–transposon–containing Cg4-neo and DmTNP or hTh9 (Fig. 2, B and C, and fig. S7). These observations imply that the DmTNP and hTh9 proteins can nick DNA, independent of having P-element termini, which would lead to elevated gene transfer via DNA linearization. A similar observation was made for the SET domain and mariner transposase fusion gene–containing protein (SETMAR or Metnase protein), but this protein is inactive for transposition of HsMAR transposons (8, 9). However, most important, the presence of P-element termini on Cg4-neo enhanced the DNA integration activity of both DmTNP and hTh9 3 to 5 times above background, which suggested transpositional DNA integration. Many G418-resistant colonies were also obtained (fig. S7) when Cg4-neo was transfected into a stable HEK293 cell line induced to express a tetracycline-inducible human THAP9 gene (Fig. 2B).
Fig. 2.
Human THAP9 can transpose P elements. (A) A comparison of the P-element transposition activities of a negative control plasmid (pBluescript empty vector) versus the P-element transposase, human THAP9, chimera 1 or chimera 2 expression vectors cotransfected with Cg4-neo or pSV2-neo in HEK293 cells. (B) A comparison of P-element transposition activity after Cg4-neo was transfected into a tetracycline-inducible, stable HEK293 cell line expressing human THAP9. Values are the average (±SEM) of five independent experiments (n = 5), each done in duplicate, P < 0.05. (C) Crystal-violet staining of colonies obtained after G418 selection of HEK293 cells cotransfected with the Cg4-neo reporter plasmid or pSV2-neo along with a negative-control plasmid (pBluescript empty vector), P-element transposase, or human THAP9 expression vectors.
To analyze the nature of the DNA integration events in the G418-resistant colonies, genomic DNA was isolated from individual colonies obtained from DmTNP or hTh9 cotransfections with Cg4-neo, and the sites of P-element insertion were characterized by splinkerette polymerase chain reaction (PCR) (17, 18) followed by DNA sequencing. DNA sequence analysis of PCR integration sites identified distinct integration sites with novel 8–base pair (bp) target-site duplications (TSDs) for individual integration events into the human genome that had occurred with both DmTNP and hTh9 (Table 1 and tables S1 and S2). Taken together, these data indicate that human THAP9 actively integrates genetically marked Drosophila P-element vectors into human cells by transposition.
Table 1.
DNA sequence analysis of P-element integration sites and identification of unique TSDs. Sequencing was performed from both the 5′ and 3′ ends of a P-element integration site. The exact locations of the integrations into a specific human chromosome, as well as nearby genes, are indicated.
| Unique 8-bp TSD | Human chromosome integrated into | Coordinates of integration | Nearby features |
|---|---|---|---|
| Cg4 TSD (20) | |||
| GTCTGGCC | |||
| Drosophila P-element transposase | |||
| GTGGCCAT | 17 | 4627461 | Zinc finger protein 232 and ubiquitin carboxyl-terminal hydrolase 6 |
| GTCTGCCA | 20 | 16559372 | Kinesin-like protein KIF16B isoform 1 and U2 small nuclear ribonucleoprotein B |
| GTGTTCGA | 21 | 3044319 | Ubiquitin carboxyl-terminal hydrolase 25 and coxsackievirus and adenovirus receptor isoform 5 precursor |
| TCTGCCTT | 11 | 25665157 | BH3-like motif-containing cell death inducer and ubiquitin-associated and Src homology 3 (SH3) domain–containing protein B |
| GTCGGCCT | 3 | 61490435 | Receptor-type tyrosine-protein phosphatase gamma |
| GGATCTCG | 9 | 25168146 | Izumo sperm-egg fusion protein 3 and tumor suppressor candidate gene 1 protein |
| Human THAP9 | |||
| TCGGCCTG | 14 | 68818627 | Leucine-rich repeat transmembrane protein FLRT2 precursor and galactocerebrosidase isoform d |
| GTCTCTCT | 11 | 18181696 | FCH and double SH3 domains protein 2 and P2Y purinoceptor 2 |
| GTCTGCCT | X | 6771218 | Glutamate receptor 3 isoform 1 precursor, glutamate receptor 3 isoform 2 precursor |
| GTGTCTGC | 11 | 24531338 | Teneurin-4 and protein FAM181B |
The studies reported here indicate that the human THAP9 gene encodes an active DNA transposase that can mobilize Drosophila P-element transposons in Drosophila and human cells. It will be interesting to investigate the physiological relevance of THAP9’s transposition function and to find out if any THAP9 recombination signal DNA elements can be found in the human genome. This is the first report, beyond the V(D)J recombination system, of an active DNA transposase in the human genome. P element–like transposons and THAP9-related genes are not restricted to Drosophila or related insect species but are widely distributed in eukaryotic genomes like Ciona (sea squirt), zebrafish, chicken, and Trichomonas vaginalis (a parasitic protozoan) (7, 19). The THAP9 gene is absent and has apparently been lost from sequenced rodent genomes (6). Although many of the human transposase–related genes are derived from DNA transposons (43 of 47) (2), most have not been characterized, with the exception of the V(D)J recombinase RAG1 and RAG2 (10, 11) and the SETMAR (Metnase) protein (8). It is possible that other human genes of this class, besides THAP9, may also encode active DNA transposases.
Supplementary Material
Acknowledgments
We thank J. M. Taliaferro, J. Aspden, and M. Francis for comments and discussion; R. Rawat for technical assistance; and C. J. Potter ( Johns Hopkins Medical School) for helpful suggestions about the splinkerette PCR experiments. This work was supported by NIH grants R01GM48862, R01GM61987, R01 GM097352, R01GM104385, and R01GM094890. S.M. and D.C.R. conceived the experiments. S.M., A.S., and D.C.R. performed cell culture and molecular biological experiments and analyzed data. S.M. and D.C.R. wrote the paper.
Footnotes
The authors declare no competing financial interests.
Requests for materials should be addressed to the corresponding author.
References and Notes
- 1.Feschotte C, Pritham EJ. Annu Rev Genet. 2007;41:331. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lander ES, et al. Nature. 2001;409:860. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Schaack S, Gilbert C, Feschotte C. Trends Ecol Evol. 2010;25:537. doi: 10.1016/j.tree.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kazazian HH., Jr Science. 2004;303:1626. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 5.Feschotte C. Nat Rev Genet. 2008;9:397. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hammer SE, Strehl S, Hagemann S. Mol Biol Evol. 2005;22:833. doi: 10.1093/molbev/msi068. [DOI] [PubMed] [Google Scholar]
- 7.Quesneville H, Nouaud D, Anxolabehere D. Mol Biol Evol. 2005;22:741. doi: 10.1093/molbev/msi064. [DOI] [PubMed] [Google Scholar]
- 8.Shaheen M, Williamson E, Nickoloff J, Lee SH, Hromas R. Genetica. 2010;138:559. doi: 10.1007/s10709-010-9452-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee SH, et al. Proc Natl Acad Sci USA. 2005;102:18075. doi: 10.1073/pnas.0503676102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. Annu Rev Immunol. 2000;18:495. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 11.Gellert M. Annu Rev Biochem. 2002;71:101. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 12.Roussigne M, et al. Trends Biochem Sci. 2003;28:66. doi: 10.1016/S0968-0004(02)00013-0. [DOI] [PubMed] [Google Scholar]
- 13.Hagemann S, Pinsker W. Mol Biol Evol. 2001;18:1979. doi: 10.1093/oxfordjournals.molbev.a003739. [DOI] [PubMed] [Google Scholar]
- 14.Sabogal A, Lyubimov AY, Corn JE, Berger JM, Rio DC. Nat Struct Mol Biol. 2010;17:117. doi: 10.1038/nsmb.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mul YM, Rio DC. EMBO J. 1997;16:4441. doi: 10.1093/emboj/16.14.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Southern PJ, Berg P. J Mol Appl Genet. 1982;1:327. [PubMed] [Google Scholar]
- 17.Horn C, et al. Nat Genet. 2007;39:933. doi: 10.1038/ng0807-933. [DOI] [PubMed] [Google Scholar]
- 18.Potter CJ, Luo L. PLoS ONE. 2010;5:e10168. doi: 10.1371/journal.pone.0010168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carlton JM, et al. Science. 2007;315:207. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beall EL, Rio DC. Genes Dev. 1996;10:921. doi: 10.1101/gad.10.8.921. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.