Summary
β-barrel membrane proteins are essential for nutrient import, signaling, motility, and survival. In Gram-negative bacteria, the β-barrel assembly machinery (BAM) complex is responsible for the biogenesis of β-barrel membrane proteins, with homologous complexes found in mitochondria and chloroplasts. Here we describe the structure of BamA, the central and essential component of the BAM complex, from two species of bacteria: Neisseria gonorrhoeae and Haemophilus ducreyi. BamA consists of a large periplasmic domain attached to a 16-strand transmembrane β-barrel domain. Three structural features speak to the mechanism by which BamA catalyzes β-barrel assembly. First, the interior cavity is accessible in one BamA structure and conformationally closed in the other. Second, an exterior rim of the β-barrel has a distinctly narrowed hydrophobic surface, locally destabilizing the outer membrane. And third, the β-barrel can undergo lateral opening, evocatively suggesting a route from the interior cavity in BamA into the outer membrane.
Introduction
Membrane proteins serve numerous essential functions and are important therapeutic targets given their surface exposure and critical roles in modulating cellular processes. While the mechanism for membrane integration is well established for α-helical membrane proteins1–4, the mechanism for β-barrel membrane proteins is unknown. β-barrel membrane proteins are only found in the outer membranes of Gram-negative bacteria and mitochondria and chloroplasts, eukaryotic organelles which evolved from bacteria5–10. The machineries catalyzing folding and insertion of β-barrel proteins have been identified and are conserved across species6,9,10.
In Gram-negative bacteria, outer membrane proteins (OMPs) are synthesized in the cytoplasm and transported across the inner membrane into the periplasm by the Sec translocon11. Molecular chaperones then escort nascent OMPs to the inner surface of the outer membrane where they are recognized by the β-barrel assembly machinery (BAM) complex, which consists of the central and essential component called BamA (an OMP itself) and the accessory proteins BamB, BamC, BamD, and/or BamE, all of which are reside in the periplasm and are attached to the membrane via a lipid anchor6,12–16 (Supplementary Figure S1). The periplasmic domain of BamA consists of five polypeptide translocation associated (POTRA) domains that extend from the barrel. Current understanding suggests that the four lipoproteins assemble onto the POTRA scaffold to create a BAM complex consisting of one copy of each protein14,17. Structures have been determined for BamB18–20, BamC20,21, BamD20–23, BamE20,21,24, and the periplasmic (POTRA) domain of BamA25–29. However an understanding of how these proteins coordinate recognition, folding, and membrane insertion of nascent OMPs has been hampered without structural knowledge of the membrane domain of the BAM complex.
To better understand OMP biogenesis, we solved crystal structures of BamA from two bacterial species: Neisseria gonorrhoeae and Haemophilus ducreyi. Two distinct POTRA domain conformations are observed relative to the β-barrel, with the barrel pore either fully accessible from the periplasm or blocked by a POTRA domain. The structures also reveal that BamA has a reduced external hydrophobic surface on one side that could produce local distortions in the outer membrane. Intriguingly, while the H. ducreyi BamA is in a predictable conformation with the first and last β-strands stably zipped with multiple hydrogen bonds, the last β-strand of the N.gonorrhoeae BamA structure is bound to the first by only two hydrogen bonds, with most of the β-strand twisted into the barrel pore. In this conformation, a large access portal provides a direct connection from the periplasm to the lipophilic interior of the outer membrane.
Results
Two structures of BamA
We determined structures for an N-terminally truncated BamA construct from H. ducreyi lacking the first three POTRA domains (HdBamAΔ3) and a full-length BamA construct from N.gonorrhoeae (NgBamA) (Fig. 1a, b, Supplementary Table 1, Supplementary Fig. 2, 3, 4). Both structures include the large C-terminal β-barrel membrane domain (Fig. 1a, b). Despite sequence divergence, each of the POTRA domains retains the conserved β-α-α-β-β fold (Supplementary Fig. 5, 6). In the HdBamAΔ3 structure the POTRA domains extend away from the barrel, allowing full access to the barrel pore from the periplasm. In contrast, the POTRA domains of NgBamA are located in close proximity to the barrel pore, such that POTRA 5 occludes pore access (Fig. 1b, c).
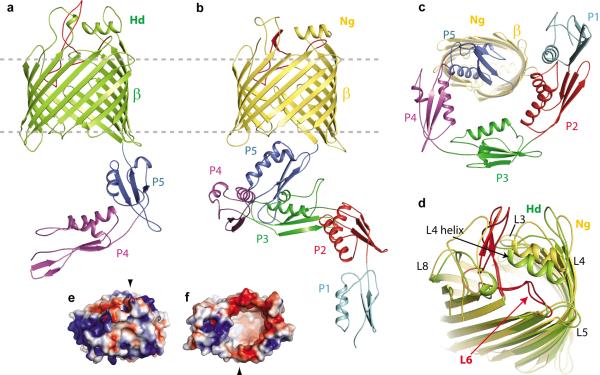
Figure 1. The structure of BamA from the BAM complex.
a. The HdBamAΔ3 crystal structure in cartoon representation showing the β-barrel (green) and POTRA domains 4 and 5 (purple and blue). b. The NgBamA crystal structure showing the β-barrel (gold) and POTRA domains 1–5 (cyan, red, green, purple and blue). c. A periplasmic (bottom) view of the NgBamA crystal structure. d. An alignment of the HdBamAΔ3 (green) and NgBamA (gold) crystal structures highlighting the structural conservation of the extracellular loops and secondary structural elements in loops 4 and 6. e. Electrostatic surface representation of HdBamAΔ3 viewed from the extracellular face and from the inside of the barrel from the periplasmic face (f). Black arrows indicate the locations of strand β-16.
The barrel domains of HdBamAΔ3 and NgBamA each contain 16 antiparallel β-strands, with the first and last strands associating to close the barrel (Fig. 1a, b). Interestingly, the interior of the barrel is almost completely empty, forming a volume of ~13,000 Å3 (Supplementary Fig. 8, 9). The extracellular loops (eL) form a dome over the top of the β-barrel domain, isolating the inside of the barrel from the extracellular space (Fig. 1d). Extracellular loops eL4, eL6 and eL7 contribute significantly to the dome, with minor contributions from eL3 and eL8. Extracellular loop eL4 contains a surface exposed α-helix that sits nearly parallel to the membrane. Although the sequences for this helix vary significantly among BamA homologs, it is structurally conserved (Fig. 1d and Supplementary Fig. 10). The electrostatics of the surface exposed regions in both structures contain a mix of positive and negative patches with a strongly electropositive surface along eL3 and eL6 (Fig. 1e, f) and electropositive charge at the membrane interfaces. The electrostatics of the barrel interior are similar in the two structures, both displaying strongly electronegative surfaces (Fig. 1f and Supplementary Fig. 8, 9).
Two conformations of BamA
While the two BamA crystal structures show conserved folds for both the POTRA domains and the β-barrel, there are notable differences between the two species. First is the conformation of the POTRA domains relative to the β-barrel (Fig. 2a). In the NgBamA structure, POTRA 5 sits in proximity to the β-barrel and interacts with periplasmic loops pL3, pL4, pL5 and pL7 to stabilize this closed conformation (Supplementary Fig. 7). Conversely, the POTRA domains of the HdBamAΔ3 structure have undergone a ~70 degree outward swing such that POTRA 5 does not interact with any of the β-barrel periplasmic loops. This could reflect a gating mechanism for regulating access to the interior of the β-barrel.
Figure 2. HdBamA and NgBamA crystal structures reveal conformational changes.
a. Alignment of HdBamAΔ3 (green) and NgBamA (gold) showing open and a closed conformations for the POTRA domains which may serve as a gating mechanism for regulating substrate access to the inside of the β-barrel. b. Compared to HdBamAΔ3 (green), strand β16 is disordered and tucked inside the β-barrel of NgBamA (gold). Arrowheads indicate the location of the C-terminal strand in Hd BamA (black) and NgBamA (red). Membrane view (c) and extracellular view (d) of an alignment of NgBamA and FhaC (gray, PDB code 2QDZ) illustrates conformational differences in the β-barrel and POTRA domains. In FhaC, the N-terminal α-helix (red) and loop 6 occlude the β-barrel preventing free diffusion across the outer membrane, however, in BamA this is accomplished by the extracellular loops that fold over the top of the barrel. Loop 6 (eL6) assumes different conformations between the two structures: e, membrane view; f zoomed view; g, periplasmic view. Unlike FhaC, NgBamA eL6 contains a β-hairpin (dashed circle) and the VRGF/Y motif is located ~18 Å from the periplasmic boundary. Unzipping of this β-hairpin may allow for an extended conformation similar to what is observed for eL6 of FhaC.
A second difference is found at the interface of strands 1 and 16 of the barrel. In the HdBamAΔ3 structure, strands 1 and 16 associate to close the β-barrel with eight hydrogen bonds (Fig. 2b and Supplementary Fig. 7). However in the NgBamA structure, the C-terminal β-strand is twisted and tucked inside the barrel, interacting with strand 1 via only two hydrogen bonds at the extracellular face of the barrel. The NgBamA structure provides the first example of significant destabilization of the C-terminal β-strand, and this conformation would allow access from the interior cavity of BamA to the lipid phase of the outer membrane at the interface of strands 1 and 16.
Comparison of BamA to FhaC
Until now, the only source of structural information for the membrane domain of any member of the Omp85 family of proteins has come from FhaC, which serves as a dedicated toxin translocation pore through the outer membrane of some bacteria30,31. BamA and FhaC share less than 13% sequence identity and function in very different processes, so it is not surprising that the structures differ significantly. First, the RMSDs for the 16-stranded β-barrels are greater than 10 Å due to differing shear numbers (S=20 for FhaC and S=22 for HdBamA and NgBamA) and overall barrel shapes (Fig. 2c). Second, while the extracellular loops of FhaC are in open conformations, as befits a toxin translocation pore, the extracellular loops of HdBamAΔ3 and NgBamA form a closed dome that prevents extracellular access and would limit free diffusion of solutes across the outer membrane (Fig. 2d). Third, the conformation of eL6 differs significantly for FhaC and the two BamA structures. This loop has gained much attention due to its large size and the location of the conserved VRGF/Y motif, with the suggestion that eL6 may extend through the barrel pore to assume at least two conformations, one near the periplasm and another closer to the surface30,32. In both BamA crystal structures, eL6 partially inserts into the barrel pore such that the VRGF/Y motif interacts with β-strands 14–16 about 18 Å away from the periplasmic side of the outer membrane (Fig. 2e and Supplementary Fig. 7). This interaction is mediated by a conserved arginine residue in the barrel pore (R658 in HdBamAΔ3 and R660 in NgBamA). The arginine is stabilized by interactions with conserved barrel residues E696 and D719 in HdBamAΔ3 and E692 and D713 in NgBamA. Extracellular loop eL6 is further stabilized by interactions with a conserved FQF motif located in β strand 16 (Supplementary Fig. 7). In FhaC, eL6 has a similar interaction with the β-barrel, but it extends through the pore, such that the VRGF/Y motif protrudes into the periplasm (Fig. 2f, g).
Modeling and mutagenesis of EcBamA
Since most functional analyses of β-barrel assembly have been undertaken in E. coli, we built a homology model of EcBamA (Supplementary Figs. 10 and 11 and Supplementary Model 1) and mutated residues that are conserved across species or predicted by the model to be functionally relevant (Fig. 3a, Supplementary Table 2). Consistent with previous work highlighting the importance of the VRGF/Y motif33, the R661A mutant exhibited reduced colony growth in LB rich medium, and mutation of the entire VRGF/Y motif was lethal (Fig. 3b, c and Supplementary Table 2). Mutating D740 to arginine was also lethal and the E717A,D740>A double mutant demonstrated minimal growth in shaking culture growth assays in LB media, confirming the proposed electrostatic interaction of the VRGF/Y motif with the inner barrel wall (Fig. 3c). Mutagenesis of potential interactions between the periplasmic loops and POTRA 5 had no effect on cell viability, nor did mutation of the highly conserved FQF motif (Supplementary Table 2, and Supplementary Figs. 10 and 12). Preventing a potential disulfide bond in eL6 had no effect in growth assays, but deletion of the non-conserved E.coli loop insertion (residues 676–700) resulted in reduced colony formation and a slower doubling time compared to wild-type (Fig. 3b, c).
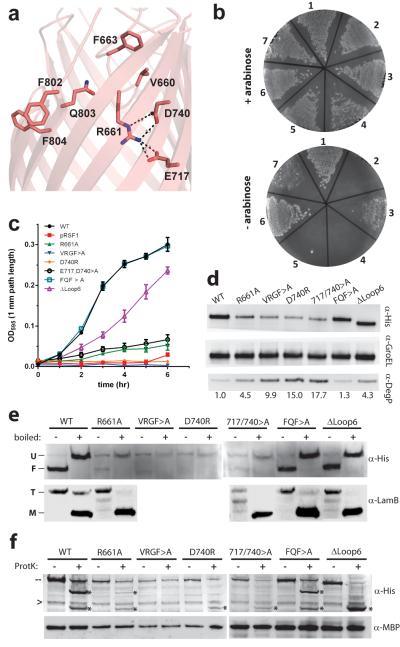
Figure 3. Mutational analysis of E.coli BamA.
a. A homology model of EcBamA showing the conserved VRGF/Y and FQF motifs and putative R661 ionic interactions. b. Colony growth assays of cells reliant upon mutant EcBamA in the absence of arabinose: (1) WT EcBamA, (2) pRSF1 vector control, (3) R661A, (4) VRGF>A, (5) D740R, (6) E717,D740>A, and (7) ΔeL6 (residues 676–700). c. Growth curves of colonies isolated from +arabinose plates and then transferred to −arabinose rich medium. Error bars represent ± standard error of the mean. d. Western blots of EcBamA mutant expression levels (α-His) and DegP up-regulation when grown in −arabinose M63 minimal media. GroEL served as a loading control; numbers represent DegP fold-increases over WT. e. Heat modifiability of EcBamA mutants expressed in LB +arabinose, and of LamB after arabinose wash-out (at the 4 hour time point as in panel c where no growth was observed for the vector control, VRGF>A, or D740R). All His-tagged EcBamA mutants except VRGF>A and D740R showed evidence of folding and LamB trimer formation. f. α-His reactive cleavage products (*) following proteinase K proteolysis of whole cells indicated extracellular accessibility of all EcBamA mutants except VRGF>A. Dashes indicate full-length mutant BamA; arrowheads indicate a nonspecific α-His-HRP reactive protein. Maltose binding protein (MBP) was used to control for outer membrane integrity. All data shown is representative of at least three independent experiments.
Investigating these growth phenotypes, we found that R661A, VRGF>A, D740R, and E717,D740>A BamA mutants exhibited low expression levels compared to WT (Fig. 3d). Additionally, expression of these mutants and the ΔLoop6 deletion mutant resulted in significant up-regulation of the quality control protein DegP. Functional analysis of BamA mutants showed that those which supported reduced growth in LB shaking culture still supported LamB expression and trimer formation. E717,D740>A mediated LamB folding, however, appeared reduced (Fig. 3e).
Finally, we studied the biogenesis of these BamA mutants (Fig. 3e, 3f). When expressed in the presence of endogenous BamA, all mutants except VRGF>A and D740R showed an indication of folding with heat modifiable gel-shifts. Proteinase K cleavage products were observed for all mutants except VRGF>A, demonstrating exposure to the extracellular space and thus OM insertion for all other constructs. Taken together, these results suggest that the interaction of R661 with the interior barrel wall is important for proper folding of BamA, while the unique eL6 insertion may play a role in the efficiency of E. coli BamA.
BamA distorts the outer membrane
Compared to other OMPs, the hydrophobic belt of the BamA β-barrel is significantly reduced in width along the C-terminal strand (~9Å) compared to the opposite side of the barrel (~20 Å) (Supplementary Fig. 13). Hypothesizing this could destabilize the local membrane environment, we used molecular dynamics (MD) simulations to investigate the effects of BamA on membrane stability. Lipid order within the membrane was assessed by looking at the order parameter (SCD) for the lipid tails in simulations of NgBamA as well as FhaC and BtuB34 controls. For NgBamA, lipids close to the C-terminal strand (approximately 15–25 lipids within 12 Å of residue 788) had ~3-fold decrease in order compared to those along the opposite side of the β-barrel (within 12 Å of residue 531) while only a marginal difference was observed for analogous positions on FhaC and BtuB (Fig. 4a and Supplementary Fig. 14). Furthermore, by looking at the mass density of lipid glycerol groups as a measure for membrane thickness, we found that for NgBamA the membrane thickness near the C-terminal strand (centered at residue 788) was 16 Å less than along the opposite side of the barrel, while for the reference FhaC, no decrease was seen (Fig. 4b and Supplementary Fig 15). This dramatic decrease in lipid order and membrane thickness near the C-terminal strand of NgBamA leads us to suggest that one function of the β-barrel is to prime the membrane for OMP insertion.
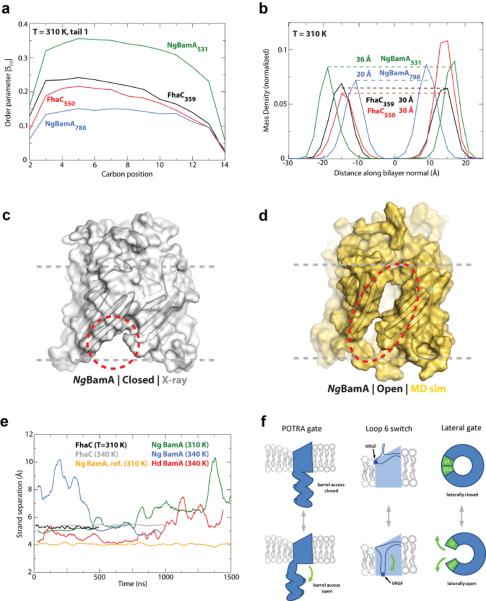
Figure 4. BamA primes the membrane for OMP insertion.
Molecular dynamics (MD) simulations investigated the stability of the BamA β-barrel. a. SCD values, a measure of lipid order, were decreased near NgBamA strand β16 (centered at residue 788) compared to the opposite side of the β-barrel (centered at residue 531). Minimal differences were observed for FhaC comparing analogous sites. b. MD analysis revealed that the β-barrel of NgBamA imparts a thinning of the membrane by 16 Å near strand β16 (centered at residue 788) when compared to the opposite side of the barrel (centered at residue 531), whereas, no difference was observed for FhaC. The MD simulations also indicated that the interaction between strands β1 and β16 is significantly destabilized, allowing the β-barrel to undergo a lateral opening (c and d, closed and open states, respectively). e. Quantification of the separation between strands β1 and β16 shows that both NgBamA (green) and HdBamA (red) structures have the propensity to open, unlike FhaC (black and grey). As reference, no change was observed between strands β13 and β14 for NgBamA (orange). f. Summary of the putative conformational switches of BamA based on structural and computational analysis. The first is the conformational gate of the POTRA domains (membrane view), the second is the conformational switch of loop 6 from a resting state (observed in our crystal structures) to a putative activated state as observed in FhaC, potentially representing the protease-sensitive state observed by Rigel et al.32, and the third is the lateral opening event (extracellular surface view).
Comparison of the HdBamAΔ3 and NgBamA structures revealed a possible gating mechanism involving POTRA 5 that could regulate substrate access to the inside of the β-barrel. To investigate pore access and barrel stability, we monitored the effect of the C-terminal strand on the stability of the β-barrel (Fig. 4c, d and Supplementary Fig. 16). Simulations unexpectedly demonstrated a lateral opening event in the β-barrel of both structures via separation of the first and last β-strands. For HdBamAΔ3, whose structure contained an ordered C-terminal strand and POTRA 5 oriented away from the β-barrel, the separation between strands β1 and β16 ranged from 4 Å (X-ray structure) to 7.5 Å (MD simulation) and became more destabilized as the simulation progressed, with the largest openings occurring toward the end of the experiment (Fig. 4e and Supplementary Fig. 16). In contrast, strand separation in NgBamA, which had a disordered C-terminal strand and POTRA 5 interacting with the periplasmic loops of the β-barrel, was almost immediate and ranged from 5 Å (X-ray structure) to 10 Å (MD simulation), with a larger separation than observed for HdBamAΔ3 at the same temperature (Fig. 4c, d, e, Supplementary Fig. 16, and Supplementary Video 1). NgBamA simulation performed at a lower temperature (310 K) exhibited the same lateral opening delayed by approximately 1 microsecond. Control simulations of FhaC and BtuB revealed no such lateral opening at any temperature over the same time scales. Similarly, the average distance between strands 13 and 14 in NgBamA was stable during the simulation (Fig. 4e). Lateral openings in a β-barrel have only been observed in three other structures: FadL35, PagP36, and OmpW37, all of which transport hydrophobic molecules. Surprisingly, not only did we observe an opening between strands β1 and β16 in the simulations for both HdBamAΔ3 and NgBamA, but also a closing event such that once opened, the β-barrel did not fully unfold as might be expected. Stabilization of the β-barrel may be attributed to both the intimate interactions between the extracellular loops and specific interactions between eL6 with the opposite side of the barrel via the conserved VRGF/Y motif.
Concluding remarks
In summary, we find that BamA can perturb the OM in at least two ways: by a reduced hydrophobic surface near β-strand 16 that results in decreased lipid order and membrane thickness, and by transient separation of β-strands 1 and 16 that produces a lateral opening in the barrel. Taken together with movements of the POTRA domains, a highly dynamic membrane environment is created by BamA in the immediate vicinity of the BAM complex. Elegant biophysical analysis of OMP biogenesis in vitro has suggested that some β-barrels can be folded in the periplasm prior to insertion into the OM38,39, but it was unclear how these barrels could insert into the lipid bilayer and whether this is the only folding mechanism utilized by OMPs. Our studies reveal structural features of BamA that would catalyze the entry of β-barrels into the outer membrane, and we envision two possible mechanisms (Supplementary Video 2). The first mechanism utilizes the hypothetical conformational switch of loop 6, the POTRA gating motion, and the lateral opening event working in concert to thread nascent OMPs through the β-barrel of BamA directly into the outer membrane (Fig. 4f). This mechanism would use the exposed strands of BamA (from the lateral opening event) as a template to initiate barrel formation by β-augmentation, forming a transient BamA-OMP complex. The OMP would continue to fold until it eventually buds off from BamA and is released into the outer membrane. More complex OMPs would favor this BamA-assisted approach. However, simpler OMPs that can readily fold into membranes might use a second mechanism independent of the β-barrel domain of BamA. Here, nascent OMPs may be trafficked into close proximity of the outer membrane by interactions with the POTRA domains of BamA for direct insertion into the locally destabilized membrane. Whether similar mechanisms are observed in eukaryotic systems remains to be determined but our improved model for the mitochondrial homolog Sam50/Tob55 will assist future studies (Supplementary Fig. 17, 18 and Supplementary Model 2). Our work represents a significant step forward in understanding bacterial OMP membrane insertion, and future investigations will determine whether insight gained from BamA represents a universal mechanism for the biogenesis of all β-barrel membrane proteins.
Methods
Cloning, expression, and purification of HdBamAΔ3
HdBamAΔ3 from Haemophilus ducreyi (strain 35000HP / ATCC 700724) was subcloned into a modified pET20b vector (EMD Millipore) containing an N-terminal pelB signal sequence, a 10X-His tag and a TEV site starting with residue Y262. Expression was performed in BL21(DE3) cells at 20°C without induction in TB-media supplemented with 100 μg/mL carbenicillin. For purification, cells were resuspended in lysis buffer (50 mM Tris-HCl, pH 7.5, 200 mM NaCl, 1 mM MgCl2, 10 μg /ml DNaseI, 100 μg/ml 4-(2-Aminoethyl) benzenesulfonyl fluoride (AEBSF)) and lysed by two passages through an Emulsiflex C3 (Avestin) homogenizer at 4°C. The lysate was centrifuged at 12,000×g for 10 minutes to remove unlysed cells and the supernatant was incubated with 2% Triton X-100 for 30 minutes at room temperature. The mixture was centrifuged at 160,000×g for 90 minutes at 4°C. The membrane pellets were resuspended in 50 mM Tris-HCl, pH 7.5, 200 mM NaCl, 20 mM imidazole and solubilized by constant stirring in 5% Elugent for 16 hours at 4°C. Solubilized membranes were centrifuged at 265,000×g for 60 minutes at 4°C and the supernatant filtered and applied to a 15-ml Ni-NTA column (Qiagen). HdBamAΔ3 was eluted using 250 mM imidazole. Peak fractions were pooled, concentrated, dialyzed and treated with TEV-His protease overnight at 4° C. To remove uncleaved protein and the TEV-His protease, the protein was re-applied to a 2nd Ni-NTA column, concentrated and then applied to an S-300HR sephacryl size exclusion column (GE Healthcare) using 20 mM Tris-HCl, pH 7.5, 200 mM NaCl, 10 mM sodium citrate, 10 mM lithium sulfate, 0.8% C8E4 and 0.02% NaN3. Peak fractions were verified using SDS-PAGE. Selenomethionine substituted protein was prepared using the same protocol as previously described48.
Cloning, expression, and purification of NgBamA
NgBamA from Neisseria gonorrhoeae (strain ATCC 700825 / FA 1090) was subcloned into the pET20b vector (EMD Millipore) containing an N-terminal pelB signal sequence, a 10X-His tag and a TEV site starting with residue F23. Expression was performed in BL21(DE3) cells at 20°C without induction in TB-media supplemented with 100 μg/mL carbenicillin. Purification was performed as described for HdBamAΔ3.
Crystallization and data collection
For crystallization, native HdBamAΔ3 and NgBamA were concentrated to 10 mg/ml and sparse matrix screening was performed using a TTP Labtech Mosquito crystallization robot using hanging drop vapor diffusion and plates incubated at 21°C. While detergent screening, LCP screening, and bicelle screening were all performed, only bicelle crystallization40 produced well diffracting crystals leading to structure determination. Here, 40 uL of protein solution was mixed with 10 uL of 35% bicelle mixture (DMPC:CHAPSO at 2.8:1 ratio)41, mixed and incubated on ice for at least 30 min prior to setting trays. The best native crystals for HdBamAΔ3 were grown from 100 mM Na-citrate, 100 mM HEPES 7.5, 12% MPD. Selenomethionine substituted crystals of HdBamAΔ3 were crystallized using the same conditions as for native. The best crystals for NgBamA were grown from 0.1 M NaCl, 0.1 M K-phosphate 7.0, 32% PEG 300, and 200 mM Na-malonate. Crystals were harvested directly from the crystallization drops and native data were collected at the SER-CAT (ID) and the GM/CA-CAT (ID-D) beamlines of the Advanced Photon Source of Argonne National Laboratory. Data collection for Se-SAD phasing of the HdBamAΔ3 structure was performed at the I02 beamline of the Diamond Light Source. All data were processed using either HKL2000 or XDS. A summary of the data collection statistics can be found in Supplementary Table 1.
Structure determination
For HdBamAΔ3 and NgBamA native datasets, molecular replacement attempts using the known structures of the POTRA domains (PDB codes 2QCZ, 3EFC, 2QDF, and 3QB6) and a model for the β-barrel domain using FhaC (PDB code 2QDZ) were unsuccessful. While we were unable to grow selenomethionine substituted NgBamA in large enough quantities for crystallization, we were able to express selenomethionine substituted HdBamAΔ3 and grew crystals using the native conditions. We were able to use these crystals to collect a 2.91 angstrom SAD dataset at the selenium peak wavelength. AutoSol (PHENIX)49 was then used to locate the selenium sites, finding 7 out of 8 possible sites and producing a density modified map which we could build an initial model into. Density for POTRA 4 was largely disordered, explaining why the methionine in this domain was not useful for phasing. However, since POTRA 4 mediates crystal packing, rigid body refinement was used to optimally place this domain, which was built without side chains. We then used this initial structure to solve the native HdBamAΔ3 structure to 2.9 angstroms resolution with R/Rfree values of 0.22/0.27. The NgBamA crystal structure was then solved by molecular replacement in Phaser-MR (PHENIX)49 using search models based on the β-barrel domain of HdBamAΔ3 and the reported POTRA domains of EcBamA (PDB codes 2QCZ, 3EFC, 2QDF, and 3QB6). Here, only the β-barrel domain and POTRAs 1 and 2 could be placed by molecular replacement (no solutions for POTRAs 3, 4, or 5). However, POTRAs 3, 4, and 5 were then manually placed based on weak difference density and subsequent rigid body refinement to optimize the domain positions. NgBamA was solved to a final resolution of 3.2 Å with R/Rfree values of 0.23/0.28. All model building was performed using COOT44,45 and subsequent refinement done in PHENIX49. RMSD analysis was performed within PyMOL (Schrödinger). We would like to note that the conformational switch between the POTRA domains of HdBamAΔ3 (open) and NgBamA (closed) is supported by previous studies, however, we cannot exclude this observation being due to a crystallization artifact. Therefore, further studies are needed to validate this hypothesis. For all structures, figures were made with PyMOL (Schrödinger) or Chimera50 and annotated and finalized with Adobe Illustrator.
Homology modeling
For the BamA homology model from E. coli (EcBamA), a pairwise sequence alignment of BamA from E. coli and H. ducreyi was performed using ClustalW51. The alignment was then input into CHAINSAW (CCP4)42,43 along with the structure of HdBamAΔ3 to produce an initial model of EcBamA. Insertions were then added manually within COOT44,45 and a disulfide bond restraint added within loop 6 between residues Cys690 and Cys700 (not present in either HdBamA or NgBamA). The POTRA domains (from CHAINSAW) were then replaced with the known structures (PDB codes 2QDF and 3QB6) and each POTRA domain aligned according to the NgBamA full length structure. Missing side chains were added using Deepview/Swiss-PdbViewer52 and final model minimization performed using Chiron46. For the Sam50 homology model from S. cerevisiae (ScSam50), a pairwise sequence alignment of the β-barrel of HdBamA was performed for all Sam50 homologs shown in Supplementary Figure 18 using ClustalW51 by restricting the alignment to the last ~430 residues, with the most convincing alignment being for S. cerevisiae based on an even spread of identities and similarities. The alignment was then fed into CHAINSAW (CCP4)42,43 along with the structure of HdBamAΔ3 to produce an initial model of the β-barrel of ScSam50. Insertions were then added manually within COOT44,45. The N-terminal domain of ScSam50 was then modeled as a single POTRA domain based on secondary structure predictions (Supplementary Figures 18 and 19) which indicated only a single POTRA domain containing the conserved β-α-α-β-β fold. Here, the model for the ScSam50 POTRA domain was built manually within COOT44,45 and secondary structure elements aligned to POTRA 1 of EcBamA to retain the conserved overall fold. The models for the β-barrel and the POTRA domain of ScSam50 were then aligned to the β-barrel and POTRA 5 of the NgBamA crystal structure, merged into a single model, missing side chains added using Deepview/Swiss-PdbViewer52 and final model minimization performed using Chiron46.
Molecular dynamics simulations
Systems containing the full-length NgBamA and HdBamA, i.e., with 5 and 2 POTRA domains, respectively, were first constructed by placing the protein in a DMPE lipid bilayer, used as an outer membrane mimic as done previously34,53. The resulting protein-membrane complex was then solvated and K+ and Cl- ions added to a concentration of 150 mM; the NgBamA system contained 240,000 atoms and HdBamA 206,000 atoms. Equilibration of the systems was carried out in stages for 30 ns using the simulation program NAMD54. From these equilibrated systems, new ones were constructed in which the POTRA domains were truncated at residue 417 in NgBamA and 419 in HdBamA. Such truncation was necessary in order to fit the limitations of the Anton supercomputer47. Anton makes simulations on the time scale of μs routinely accessible, and was used for all production simulations reported here. The CHARMM27/CMAP force field for proteins47,55, TIP3P for water56, and CHARMM36 for lipids57 were used. The simulations were run in the NPT ensemble, thereby allowing the membrane to expand in response to BamA opening.
Final system sizes were 110,000 (dimensions of 116×113×84 Å3) and 85,000 atoms (102×106×79 Å3) for NgBamA and HdBamA, respectively. The number of lipids for each system was 406 (NgBamA) and 304 (HdBamA), while the number of water molecules was 20,000 (NgBamA) and 15,000 (HdBamA). Proteins were at least 50–60 Å from their periodic images in the membrane plane and 15 Å along the normal axis. As controls, FhaC and the vitamin B12 transporter BtuB were also simulated using the same parameters as the BamA systems. The POTRA domains of FhaC were removed as done for BamA, such that the N-terminal helix (residues 1–30) and the barrel (residues 207 to 554) were retained. The missing extracellular loop (residues 384 to 397) was modeled and inserted into the structure. The size of the final FhaC system was 83,000 atoms with dimensions 92×94×96 Å3; for BtuB the size was 71,000 atoms (83×81×103 Å3). The FhaC and BtuB systems contained 251 and 173 lipids and 16,000 and 14,000 water molecules, respectively.
Two temperatures were used for NgBamA (310 K and 340 K), FhaC (310 K and 340 K), and BtuB (325 K and 353 K) for two reasons: first, each temperature permits a different phase of the membrane (gel-like at 310 K and fluid at 340 K) to be explored, and second, the probability of fluctuations in the barrel opening are expected to be increased at a higher temperature. Temperatures of 353–490 K have previously been validated for peptide-membrane partitioning studies and were found to not significantly affect the systems' thermodynamic properties58. Regardless of the temperature, the FhaC and BtuB barrels were found to be stable, whereas the NgBamA was not. HdBamA was apparently more stable than NgBamA at 340 K, although barrel opening was nonetheless observed after ~1 μs. Two temperatures were used for NgBamA (310 K and 340 K), FhaC (310 K and 340 K), and BtuB (325 K and 353 K) for two reasons. First, each temperature permits a different phase of the membrane (gel-like at 310 K and fluid at 340 K) to be explored, and second, the probability of fluctuations in the putative lateral opening of the β-barrel are expected to be increased at a higher temperature. We note that the gel-like membrane at 310 K is likely more representative of the expected low fluidity for the outer membrane. The total time for all Anton simulations is 7.5 μs.
Functional assessment of E.coli BamA mutants
For functional analysis of mutations of E. coli BamA (EcBamA), the coding sequence for EcBamA containing an N-terminal pelB signal sequence and 10X-His tag was placed into the pRSF-1b vector (EMD Millipore). Mutants were prepared using standard site-directed mutagenesis protocols (primer sequences available upon request). JCM-166 cells, whose endogenous BamA is under the control of an arabinose promoter13, were transformed via electroporation with pRSF1 vectors coding for a mutant BamA under kanamycin selection, and were then plated on LB+agar plates with 50 μg/mL kanamycin (kan) in the presence or absence of 0.1% L-(+)-arabinose and grown for 12–15 hrs at 37°C.
BamA mutants which exhibited decreased colony formation compared to WT were further analyzed in culture growth assays. Colonies on a 0.1% arabinose plate were isolated and grown in LB media plus 50 μg/mL kanamycin at 37°C. The expression of endogenous BamA on the plus-arabinose plates allowed at least some initial growth of all mutant transformants, and after 3–4 hours all cultures were washed in PBS and diluted in fresh LB+kan to an OD600 of ~0.04. Cultures were incubated shaking at 37°C, with small aliquots taken for triplicate OD595 measurements on a BioRad iMark Microplate Reader every hour. Preliminary experiments demonstrated that those mutants that grew similar to wild type on plates also grew similar to wild type in growth curves. Additional images from our colony growth assays and preliminary growth curves are available upon request.
For Western Blot analysis of mutant BamA and DegP expression levels, colonies grown on LB agar + kan + 0.1% arabinose were isolated and grown in the absence of arabinose in M63 minimal media59 with 0.4% glycerol, 0.1% casamino acids, and 1 mM MgSO4 supplements plus 50 μg/mL kan at 30°C. Cells were harvested at an OD600 of 0.5–1.0, boiled at 99°C for 10 minutes and lysates were resolved with NuPAGE (Invitrogen). Antibodies used were: α-polyHistine Horseradish Peroxidase (anti-His-HRP) Conjugate, 1:6000 (Sigma); α-LamB, 1:2000 (kindly provided by R. Misra); α-DegP, 1:5000 (kindly provided by J. Beckwith); α-GroEL, 1:20000 (Sigma); α-MBP, 1:5000 (NEB); α-mouse HRP, 1:15000 (Sigma); α-rabbit HRP, 1:15000 (Sigma). Antibody staining was visualized with the ECL Prime kit (GE Healthcare).
Heat modifiability assays
To determine whether the BamA mutants were folded properly, heat modifiability assays were performed using whole cells or whole cell lysates. Transformed cells were cultured overnight at 30°C in LB + 0.1% arabinose, permitting folding and OM incorporation by the endogenous BamA. To prepare whole cell lysates, cells were centrifuged, resuspended in 1X PBS, 1% β-dodecylmaltoside (DDM), 10 mM EDTA, 10 ug/mL lysozyme, supplemented with AEBSF and DNaseI, and rocked at room temperature for 15 mins before being centrifuged again for 10 mins at 15,000 rpm using a microcentrifuge. SDS-loading buffer was then added to either whole cells (an estimated 2 × 107 cells) or lysates and either boiled (0.5% or 1% SDS in loading buffer) or left at room temperature (0.1% SDS in loading buffer) for 5–10 mins. The samples were then separated using NativePage 4–12% gels (Invitrogen) with 1X MES SDS-PAGE running buffer by running the gels for 60 mins at 150 volts (constant) at 4°C. Transfer to PVDF membrane was performed using the iBlot system (Invitrogen) and anti-HIS-HRP (Sigma) antibodies were used for western blot analysis and imaged using an ImageQuant LAS 4000 imaging system (GE Healthcare).
Proteinase K digestion assays
To access the surface exposure of BamA and mutants, proteinase K digestion assays were performed on whole cells as has been reported previously32. Briefly, colonies on an LB + 0.1% arabinose agar plate were grown in LB + 0.1% arabinose liquid culture overnight at 30°C. 0.5–1.0 mL of cells at OD600 of 1.0 were centrifuged and resuspended in 20 mM Tris-HCl 7.4, 100 mM NaCl. Then, 0.5 mg/mL proteinase K was added and incubated at 37°C for 30 minutes.
Phenylmethanesulfonylfluoride (PMSF) was then added to a final concentration of 10 mM and the cells were centrifuged again and supernatant removed. PMSF was then added again and then cells resuspended in 1X LDS loading buffer, boiled and separated using NuPAGE analysis. Transfer to PVDF membrane was performed using the iBlot system (Invitrogen) and anti-HIS-HRP (Sigma) and anti-MBP (NEB) antibodies were used for Western blot analysis and imaged using an ImageQuant LAS 4000 imaging system (GE Healthcare).
Supplementary Material
Acknowledgments
We thank Harris Bernstein and Raffaele Ieva for providing JCM-166 cells, John Beckwith and Rajeev Misra for providing antibodies, and Ann Marie Stanley and Travis Barnard for discussions and comments on the manuscript. NN, AK, NCE, HC, and SKB are supported by the Intramural Research Program of the NIH, National Institute of Diabetes and Digestive and Kidney Diseases. We thank the respective staffs at the Southeast Regional Collaborative Access Team (SER-CAT) and General Medicine and Cancer Institute's Collaborative Access Team (GM/CA-CAT) beamlines at the Advanced Photon Source, Argonne National Laboratory and the Diamond Light Source for their assistance during data collection. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38 (SER-CAT), and by the US Department of Energy, Basic Energy Sciences, Office of Science, under contract No. DE-AC02-06CH11357 (GM/CA-CAT). Anton computer time was provided by the National Resource for Biomedical Supercomputing and the Pittsburgh Supercomputing Center through Grant RC2GM093307 from the NIH, using a machine donated by DE Shaw Research. TL is an Australian Research Council (ARC) Federation Fellow and acknowledges support from ARC Discovery Project (DP120101878) and ARC Linkage International Grant (LX0776170).
Footnotes
Supplementary Information is linked to the online version of the paper at www.Nature.com/nature.
Author Contributions
NN, HC, NCE, and SKB cloned, expressed, and purified HdBamAΔ3 and NgBamA. PL performed data collection for experimental phasing. NN crystallized and solved the HdBamAΔ3 and NgBamA crystal structures. NN and AK performed the homology modeling and functional assays. JG designed, conducted and analyzed the MD simulations. NN, AK, and SKB analyzed and discussed all data. TL and SKB conceived and designed the original project. NN, AK, TL and SKB wrote the manuscript.
Author Information
Coordinates and structure factors for HdBamAΔ3 and NgBamA are deposited in the Protein Data Bank (accession codes 4K3C, 4K3B). The authors declare no competing financial interests.
References
- 1.Dalbey RE, Wang P, Kuhn A. Assembly of bacterial inner membrane proteins. Annu Rev Biochem. 2011;80:161–187. doi: 10.1146/annurev-biochem-060409-092524. [DOI] [PubMed] [Google Scholar]
- 2.White SH, von Heijne G. How translocons select transmembrane helices. Annu Rev Biophys. 2008;37:23–42. doi: 10.1146/annurev.biophys.37.032807.125904. [DOI] [PubMed] [Google Scholar]
- 3.du Plessis DJ, Nouwen N, Driessen AJ. The Sec translocase. Biochim Biophys Acta. 2011;1808:851–865. doi: 10.1016/j.bbamem.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 4.Osborne AR, Rapoport TA, van den Berg B. Protein translocation by the Sec61/SecY channel. Annu Rev Cell Dev Biol. 2005;21:529–550. doi: 10.1146/annurev.cellbio.21.012704.133214. [DOI] [PubMed] [Google Scholar]
- 5.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walther DM, Rapaport D, Tommassen J. Biogenesis of beta-barrel membrane proteins in bacteria and eukaryotes: evolutionary conservation and divergence. Cell Mol Life Sci. 2009;66:2789–2804. doi: 10.1007/s00018-009-0029-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Webb CT, Heinz E, Lithgow T. Evolution of the beta-barrel assembly machinery. Trends Microbiol. 2012;20:612–620. doi: 10.1016/j.tim.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 8.Paschen SA, Neupert W, Rapaport D. Biogenesis of beta-barrel membrane proteins of mitochondria. Trends Biochem Sci. 2005;30:575–582. doi: 10.1016/j.tibs.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 9.Jiang JH, Tong J, Tan KS, Gabriel K. From Evolution to Pathogenesis: The Link Between beta-Barrel Assembly Machineries in the Outer Membrane of Mitochondria and Gram-Negative Bacteria. Int J Mol Sci. 2012;13:8038–8050. doi: 10.3390/ijms13078038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tommassen J. Assembly of outer-membrane proteins in bacteria and mitochondria. Microbiology. 2010;156:2587–2596. doi: 10.1099/mic.0.042689-0. [DOI] [PubMed] [Google Scholar]
- 11.Pugsley AP. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knowles TJ, Scott-Tucker A, Overduin M, Henderson IR. Membrane protein architects: the role of the BAM complex in outer membrane protein assembly. Nat Rev Microbiol. 2009;7:206–214. doi: 10.1038/nrmicro2069. [DOI] [PubMed] [Google Scholar]
- 13.Wu T, et al. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 14.Hagan CL, Silhavy TJ, Kahne D. beta-Barrel membrane protein assembly by the Bam complex. Annu Rev Biochem. 2011;80:189–210. doi: 10.1146/annurev-biochem-061408-144611. [DOI] [PubMed] [Google Scholar]
- 15.Ricci DP, Silhavy TJ. The Bam machine: A molecular cooper. Biochim Biophys Acta. 2012;1818:1067–1084. doi: 10.1016/j.bbamem.2011.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rigel NW, Silhavy TJ. Making a beta-barrel: assembly of outer membrane proteins in Gram-negative bacteria. Curr Opin Microbiol. 2012;15:189–193. doi: 10.1016/j.mib.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagan CL, Kim S, Kahne D. Reconstitution of outer membrane protein assembly from purified components. Science. 2010;328:890–892. doi: 10.1126/science.1188919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noinaj N, Fairman JW, Buchanan SK. The Crystal Structure of BamB Suggests Interactions with BamA and Its Role within the BAM Complex. J Mol Biol. 2011;407:248–260. doi: 10.1016/j.jmb.2011.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heuck A, Schleiffer A, Clausen T. Augmenting beta-Augmentation: Structural Basis of How BamB Binds BamA and May Support Folding of Outer Membrane Proteins. J Mol Biol. 2011;406:659–666. doi: 10.1016/j.jmb.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 20.Kim KH, Aulakh S, Paetzel M. Crystal structure of beta-barrel assembly machinery BamCD protein complex. J Biol Chem. 2011;286:39116–39121. doi: 10.1074/jbc.M111.298166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albrecht R, Zeth K. Structural basis of outer membrane protein biogenesis in bacteria. J Biol Chem. 2011;286:27792–27803. doi: 10.1074/jbc.M111.238931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sandoval CM, Baker SL, Jansen K, Metzner SI, Sousa MC. Crystal structure of BamD: an essential component of the beta-Barrel assembly machinery of gram-negative bacteria. J Mol Biol. 2011;409:348–357. doi: 10.1016/j.jmb.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong C, Hou HF, Yang X, Shen YQ, Dong YH. Structure of Escherichia coli BamD and its functional implications in outer membrane protein assembly. Acta Crystallogr D Biol Crystallogr. 2012;68:95–101. doi: 10.1107/S0907444911051031. [DOI] [PubMed] [Google Scholar]
- 24.Knowles TJ, et al. Structure and function of BamE within the outer membrane and the beta-barrel assembly machine. EMBO Rep. 2011;12:123–128. doi: 10.1038/embor.2010.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gatzeva-Topalova PZ, Walton TA, Sousa MC. Crystal structure of YaeT: conformational flexibility and substrate recognition. Structure. 2008;16:1873–1881. doi: 10.1016/j.str.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim S, et al. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317:961–964. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H, et al. High-resolution structure of a new crystal form of BamA POTRA4-5 from Escherichia coli. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67:734–738. doi: 10.1107/S1744309111014254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gatzeva-Topalova PZ, Warner LR, Pardi A, Sousa MC. Structure and flexibility of the complete periplasmic domain of BamA: the protein insertion machine of the outer membrane. Structure. 2010;18:1492–1501. doi: 10.1016/j.str.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knowles TJ, et al. Fold and function of polypeptide transport-associated domains responsible for delivering unfolded proteins to membranes. Mol Microbiol. 2008;68:1216–1227. doi: 10.1111/j.1365-2958.2008.06225.x. [DOI] [PubMed] [Google Scholar]
- 30.Clantin B, et al. Structure of the membrane protein FhaC: a member of the Omp85-TpsB transporter superfamily. Science. 2007;317:957–961. doi: 10.1126/science.1143860. [DOI] [PubMed] [Google Scholar]
- 31.Fan E, Fiedler S, Jacob-Dubuisson F, Muller M. Two-partner secretion of gram-negative bacteria: a single beta-barrel protein enables transport across the outer membrane. J Biol Chem. 2012;287:2591–2599. doi: 10.1074/jbc.M111.293068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rigel NW, Ricci DP, Silhavy TJ. Conformation-specific labeling of BamA and suppressor analysis suggest a cyclic mechanism for beta-barrel assembly in Escherichia coli. Proc Natl Acad Sci U S A. 2013;110:5151–5156. doi: 10.1073/pnas.1302662110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leonard-Rivera M, Misra R. Conserved residues of the putative L6 loop of Escherichia coli BamA play a critical role in the assembly of beta-barrel outer membrane proteins, including that of BamA itself. J Bacteriol. 2012;194:4662–4668. doi: 10.1128/JB.00825-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gumbart J, Wiener MC, Tajkhorshid E. Coupling of calcium and substrate binding through loop alignment in the outer-membrane transporter BtuB. J Mol Biol. 2009;393:1129–1142. doi: 10.1016/j.jmb.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hearn EM, Patel DR, Lepore BW, Indic M, van den Berg B. Transmembrane passage of hydrophobic compounds through a protein channel wall. Nature. 2009;458:367–370. doi: 10.1038/nature07678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cuesta-Seijo JA, et al. PagP crystallized from SDS/cosolvent reveals the route for phospholipid access to the hydrocarbon ruler. Structure. 2010;18:1210–1219. doi: 10.1016/j.str.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hong H, Patel DR, Tamm LK, van den Berg B. The outer membrane protein OmpW forms an eight-stranded beta-barrel with a hydrophobic channel. J Biol Chem. 2006;281:7568–7577. doi: 10.1074/jbc.M512365200. [DOI] [PubMed] [Google Scholar]
- 38.Tamm LK, Hong H, Liang B. Folding and assembly of beta-barrel membrane proteins. Biochim Biophys Acta. 2004;1666:250–263. doi: 10.1016/j.bbamem.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 39.Tamm LK, Arora A, Kleinschmidt JH. Structure and assembly of beta-barrel membrane proteins. J Biol Chem. 2001;276:32399–32402. doi: 10.1074/jbc.R100021200. [DOI] [PubMed] [Google Scholar]
- 40.Agah S, Faham S. Crystallization of membrane proteins in bicelles. Methods Mol Biol. 2012;914:3–16. doi: 10.1007/978-1-62703-023-6_1. [DOI] [PubMed] [Google Scholar]
- 41.Ujwal R, Bowie JU. Crystallizing membrane proteins using lipidic bicelles. Methods. 2011;55:337–341. doi: 10.1016/j.ymeth.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 43.Stein N. CHAINSAW: a program for mutating pdb files used as templates in molecular replacement. J. Appl. Cryst. 2008;41:641–643. [Google Scholar]
- 44.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramachandran S, Kota P, Ding F, Dokholyan NV. Automated minimization of steric clashes in protein structures. Proteins. 2011;79:261–270. doi: 10.1002/prot.22879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shaw DE, et al. Anton, a special-purpose machine for molecular dynamics simulation. Commun. ACM. 2008;51:91–97. [Google Scholar]
- 48.Fairman JW, et al. Crystal structures of the outer membrane domain of intimin and invasin from enterohemorrhagic E. coli and enteropathogenic Y. pseudotuberculosis. Structure. 2012;20:1233–1243. doi: 10.1016/j.str.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 50.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 51.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 52.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 53.Barnard TJ, et al. Molecular basis for the activation of a catalytic asparagine residue in a self-cleaving bacterial autotransporter. J Mol Biol. 2012;415:128–142. doi: 10.1016/j.jmb.2011.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mackerell AD, Jr., Feig M, Brooks CL., 3rd. Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 56.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics. 1983;79:926–935. [Google Scholar]
- 57.Klauda JB, et al. Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. The Journal of Physical Chemistry B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ulmschneider JP, Smith JC, White SH, Ulmschneider MB. In silico partitioning and transmembrane insertion of hydrophobic peptides under equilibrium conditions. J Am Chem Soc. 2011;133:15487–15495. doi: 10.1021/ja204042f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Elbing K, Brent R. Media preparation and bacteriological tools. Curr Protoc Mol Biol. 2002;Chapter 1(Unit 1 1) doi: 10.1002/0471142727.mb0101s59. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.