Over the last decade, DNA has been widely employed as a scaffold to form inorganic metallic nanoparticles (MNPs). The unique programmable structure provided by Watson-Crick base pairing and the tunable properties of DNA have been used for growth and positioning of nanoparticle structures. This has enabled new synthetic strategies, such as DNA-metallization,[1-8] DNA-mediated nanoparticle synthesis,[9-15] and DNA-controlled positioning of nanoparticles,[16-17] to create novel, efficient and useful miniaturized optical sensors, electronic devices, circuits and medical theranostic kits. The remarkable molecular recognition properties and self-assembly capabilities of DNA have been intensively utilized for several years. For example, DNA-directed inorganic nanowire particles have been studied for observing conformational changes of double-stranded DNA by addition of metal ions [1]. Moreover, the assembly of nanocrystals of semiconductor materials using DNA as a template has been examined to overcome insulation of DNA in electronic circuits.[18] Thus, the binding affinity of metal ions to oxygen-containing mono/di/tri-phosphate groups and N bonds of bases prevents excessive deposition on DNA with increases in concentration. [19] In addition to its ability to self-assemble, DNA has also been used to produce unique dispersed nanoparticles and to control the positions of these particles on programmable DNA scaffolds. As an example, DNA/RNA sequence, structure and composition have been used to fine tune the size, shape and physicochemical properties of nanoparticles and to generate biocompatible and easily functionalizable nanomaterials. [11-12]
Graphene is an allotrope of carbon composed of one-atom-thick planar sheets of sp2–bonded carbon atoms densely packed in a honeycomb crystal lattice.[20] Since its discovery in 2004, graphene has been used for a broad spectrum of applications based on its unique physical, mechanical and electronic properties. Most recently, graphene oxide (GO), the oxidized form of graphene, has been used in conducting polymers, battery electrodes, heterostructured nanomaterials, electronic and optical sensors, antibacterial materials, and drug and DNA delivery systems.[21] GO has become a unique platform for growth and immobilization of metal nanoparticles to produce water-soluble multicomposite nanostructures. Compared to single MNPs, MNP-GO heterostructures show effective surface enhanced Raman scattering (SERS), catalytic, and antibacterial properties.[22-25]
To date, metal-GO heterostructured nanomaterials have been produced by such techniques as thermal activation,[26-27] teflon line-autoclaving,[28] microwave-assisted synthesis,[24] photochemical synthesis,[29] and electroless deposition,[30] as well as by such chemical reduction methods[31] as in situ reduction of metal precursors[32] and post-deposition of pre-synthesized metal nanoparticles.[33] However, these various synthesis pathways have several major limitations in terms of producing uniformly sized and shaped particles and obtaining enhanced optical, electronic and chemical properties of metal-GO nanocomposites. Moreover, most of the current methods, which use sophisticated experimental procedures and expensive instrumentation, are time-consuming, multistep and labor-intensive processes that require specific conditions for each metal. To address these issues, we report a novel, simple and universal route for the preparation of metal-GO heterostructures with controllable nanoparticle size and shape using double-stranded DNA (dsDNA) as a template. The adsorption of DNA on GO is rationally utilized to create biocompatible and easily functionalizable Ag-GO, Au-GO, Cu-GO, Pt-GO and Au/Cu/Pt-GO heterostructures using the same experimental conditions. The GO used in this work is highly water soluble and the dsDNA adsorbed on surface of GO improved solubility of GO in water. The metal-GO heterostructures stay in water several months without any precipitation.
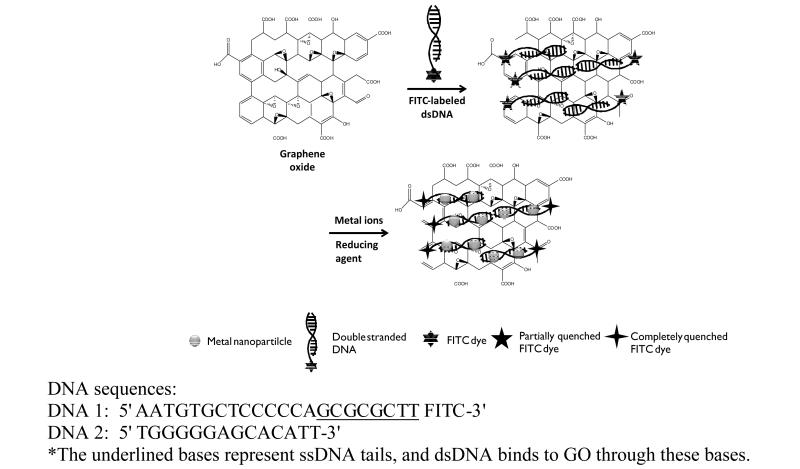
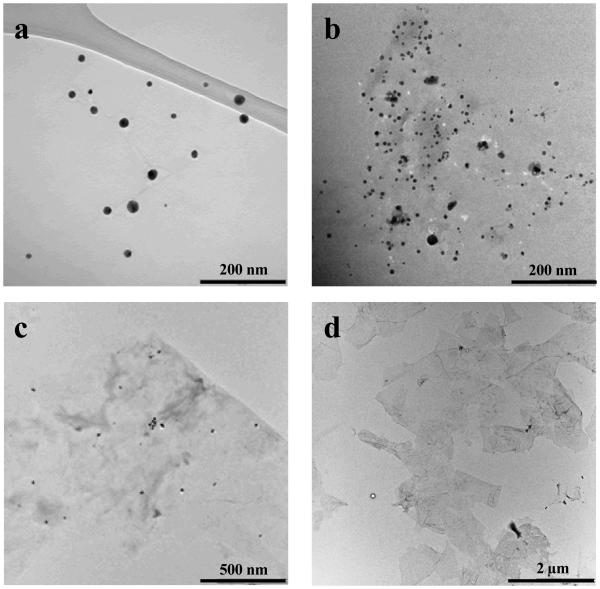
The large surface of GO containing several functional groups and domains (e.g., uncharged polar hydroxyl and epoxide groups, charged hydrophilic carboxylate groups located at edges, and π bonds, including sp2 electrons and hydrophobic graphenic domains) makes GO an ideal platform for GO-biofunctional group interactions. In particular, utilization of DNA adsorption on GO has enabled new applications and synthesis opportunities. Single-stranded DNA (ssDNA) shows preferential binding over dsDNA onto the GO surface via the exposed aromatic rings holding sp2 electrons, leading to π-π stacking between ssDNA and GO.[34] However, when ssDNA is hybridized with its complementary DNA to form dsDNA, insufficient sp2 electrons are available on the dsDNA aromatic groups to enable π-π stacking between the aromatic groups of dsDNA and aromatic groups of GO. In our approach, the fluorophore–terminated ssDNA tail was used to track binding of our designed dsDNA on GO surfaces, as shown above (scheme 1). The quenching of the fluorophore (FITC) located on the 3' prime end of DNA-1 verified the adsorption on the GO surface via the unhybridized extension bases of DNA-1. Uniformly sized Au, Ag, Cu and Pt nanoparticles and Au/Cu/Pt alloy structures were produced by chemical reduction of cationic metal ions (Au3+, Ag1+, Cu2+, Pt4+) bound to and accumulated on the main groove of dsDNA adsorped on GO surface (scheme 1). The reaction temperature, reaction time, stirring rate, concentration of each metal ion (100 μM) and reductant (500 μM) were kept constant to synthesize each MNP-GO hybrid structure. The TEM images of Au-GO and Ag-GO samples (Figures 1a and 1b) show well-ordered nanoparticle distributions of 16 nm and 12 nm, respectively. A small number of larger nanoparticles can also be seen in the TEM images. These can be attributed to extended reaction time causing aggregative growth in the metallic nanoparticle formation experiment. Extending the reaction time limits the otherwise free space between metal ions, then the strong van der Waals interactions between metal nanoparticles causes them to agglomerate. In that case, we are lack of generating single dispersed nanoparticles. No formation of MNPs occurs during the 30 min reaction time at room temperature in the absence of dsDNA (Figure 1c). It is proven that dsDNA is required for metal nanoparticle nucleation and growth (Figure S1). The TEM image of pristine GO is included for comparison (Figure 1d). In our approach, the dsDNA provides a homogeneous distribution of Au and Ag nanoparticles on the GO surface, whereas previous studies without dsDNA using different techniques and experimental conditions resulted in nanoparticles on the randomly distributed pre-existing functional groups on the GO surface.[23, 27]
Scheme 1.
Schematic illustration of the synthesis of MNP@dsDNA-GO composites.
Figure 1.
TEM images of (a) ~16 nm Au NPs, (b) ~12 nm Ag NPs deposited onto dsDNA- GO, (c) control experiment Ag-GO without DNA template and (d) pristine GO without DNA.
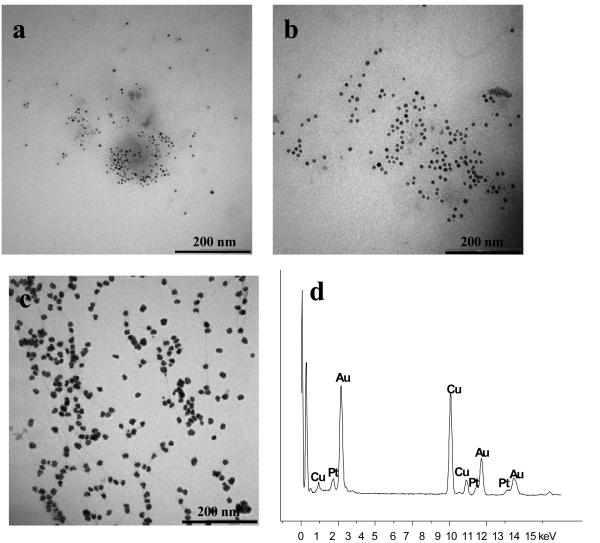
TEM images of Cu and Pt NPs (Figures 2a and 2b) on the dsDNA-GO surface demonstrate highly uniform and controlled size: 13 nm for Cu and ~10 nm for Pt. Use of optimum concentration ratio between Cu2+, Pt4+ and DNA (100 to 1, Cu2+ or Pt4+/DNA) gave rise to nearly mono-distributed, spherically shaped nanoparticles and led to perfect alignment of nanoparticles on the GO surface. The high binding affinity of cationic metal ions to dsDNA causes metallic nanoparticles to form on dsDNA, instead of on random locations on the GO surface. Thus, the use of DNA as the template plays an essential role for nucleation, stabilization and growth of MNPs on the GO surface. Moreover, no study has reported alloy-GO hybrid nanostructure formation. We used the same concentration (100 μM) of three different metal ions, Au3+, Cu2+ and Pt4+, to prepare an alloy structure on dsDNA-GO. No individual Au, Cu and Pt nanoparticles can be observed in the TEM images (Figure 2c) because all metal ions preferentially reacted with dsDNA, rather the functional groups on GO surface. Au, Cu and Pt elemental composition of Au/Cu/Pt alloy-GO can be identified on the EDX spectrum via line scan technique (Figure 2d).
Figure 2.
TEM images of (a) ~13 nm Cu NPs, (b) ~10 nm Pt NPs grown on dsDNA-GO, (c) ~20 nm Au/Cu/Pt alloy NPs grown on dsDNA-GO with sodium ascorbate and (d) Au, Cu and Pt elemental analysis of Au/Cu/Pt alloy-GO composite.
Previous studies have shown that nucleic acids bind metal ions via different types of interactions [9]. The phosphate and base regions of DNA are equally responsible for binding Cu2+ ions, while the nucleobase moiety of DNA preferentially binds Au3+ and Pt4+ noble metal ions. [10] This fact allowed us to accumulate metal ions into the major groove of DNA containing phosphate and base moieties. Reduction and formation order of metal ions among Au3+, Cu2+ and Pt4+ ions was determined based on their Fermi levels and their binding affinities to DNA.[35] Thus, Au3+ was first reduced and then bound to DNA, while Cu2+ and Pt4+ showed second and third reduction and binding affinities.
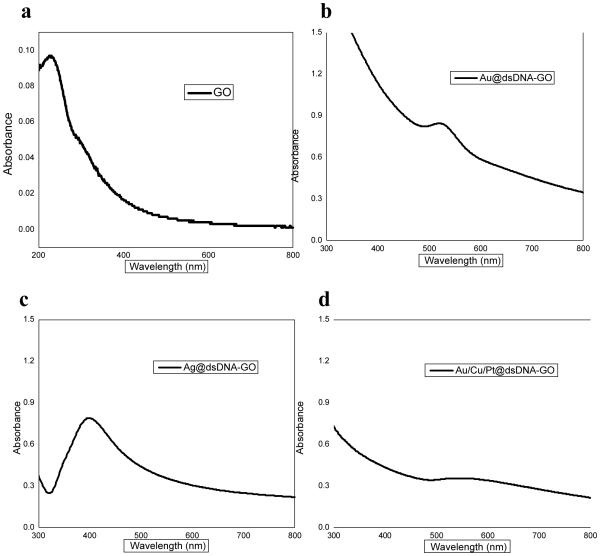
DNA-mediated growth of different types of MNPs on the GO surface was studied by UV/Vis absorption and fluorescence intensity measurements. The UV-Vis spectra of the DNA-mediated MNP-GO nanocomposite are shown (Figure 3), as well as pristine GO. The spectrum of pristine GO contains a peak at around 230 nm (Figure 3a). The UV-Vis absorption spectra of aqueous solutions of Ag@dsDNA-GO and Au@dsDNA-GO complexes indicate the formation of Ag and Au NPs on GO with corresponding peaks at 400 nm and 530 nm, respectively (Figures 3b and 3c). The alloy-GO nanostructure (Au/Cu/Pt@ dsDNA-GO) also exhibits a broad peak around 580 nm which may be an indication of the formation of alloy nanoparticles (combination of Au3+, Cu2+ and Pt4+ metal ions) (Figure 3d).
Figure 3.
UV-Vis spectra of aqueous solutions of (a) GO only , (b) Ag@dsDNA-GO, (c) Au@dsDNA-GO and (d) Au/Cu/Pt@ dsDNA-GO nanostructures. Note the different absorbance full scales.
If dye modified-ssDNA is used, it is well known that GO significantly quenches fluorescence of the dye. In contrast to ssDNA, dsDNA does not lie flat on the GO surface through π-π stacking, because aromatic rings (bases) are blocked, as noted in the discussion above, and the outer charged phosphate groups have considerably lower affinity for adsorption onto the GO surface. In dsDNA, the spaces (minor and major grooves) are found between strands. When the backbones of strands are far away from each other, wider spaces called major grooves occur. Proteins and metal ions bind at the major grooves, because the fringes of the nucleobases are more accessible there. In our approach, we used dsDNA with an ssDNA tail to grow MNPs on the major grooves and to show partial quenching of FITC dye on the 3'-end of the ssDNA tail.
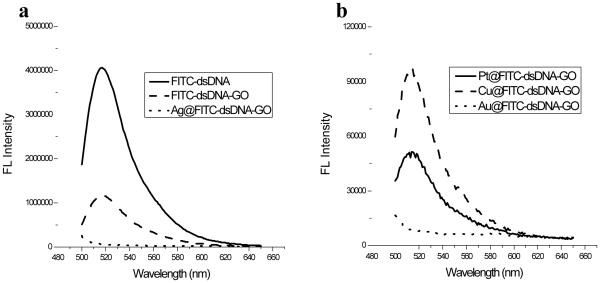
Fluorescence measurements were studied with scan range from 480 nm to 650 nm for all fluorescent spectra and the excitation point was fixed to 480 nm and emission was set 520 nm through 650 nm presented in (Figure 4a). We calculated quenching efficiency of GO and Ag NP for the first quenching and the second quenching respectively by using the formula of QE1 = (1-β1) × 100% and QE2 = (1-β2) × 100%, where β1 is the ratio of ?uorescence quenched by GO and β2 is the ratio of fluorescence quenched by GO and Ag NPs to the original ?uorescence of FITC labeled DNA in solution.[36] The FITC-labeled dsDNA shows intense fluorescence emission in solution, so it was measured at the specific point (at 521 nm) of emission wavelength for the first fluorescence data but after binding to GO, fluorescence intensity of FITC in mixture was measured to generate second fluorescence data at this specific wavelength. The quenching efficiency (QE1) of GO, around 65-70%, was calculated by subtracting second fluorescence data from first fluorescence data. The incomplete quenching may be attributed to adsorption of dsDNA in a tilted position and lack of direct contact between FITC-dsDNA and the surface of the GO sheets. In the presence of Ag NPs formed on dsDNA-tempated GO surfaces, the fluorescence intensity of FITC in final mixture was measured to generate third fluorescence data, The quenching efficiency (QE2) of Ag@GO composite, around ~95-98%, was also calculated by subtracting third fluorescence data from first fluorescence data. It is demonstrated on fluorescence spectra that the second quenching effect caused by Ag NP grown on dsDNA. It is proven that the fluorescence quenching spectra of FITC-labeled dsDNA were generated based on binding position of it on surface of GO and distatance of it to Ag NPs grown on DNA. The ssDNA tail containing 8 bases and FITC dye corresponds to approximately ~ 3.06 nm and dsDNA in which 12 nm sized Ag NP grown containing 14 bases is length of ~ 4.76 nm. The 12 nm size of Ag NP is already close enough to quench FITC dye in whole DNA system. When the two molecules are in close proximity (donor and acceptor molecules), energy transfer occurs between these molecules by electron transfer. In our system, Ag NPs formed on dsDNA serve as photon acceptors to quench the fluorescence of FITC dye located on the 3'-end. The quenching efficiency is associated with type, size, and shape of the nanoparticles, and is also strongly dependent upon the distance between the nanoparticles and fluorophores. Au NPs were more effective quenchers than Pt and Cu MNPs (Figure 4b). This may result from the structural nature of gold NP surfaces and strong localized surface plasmon of Au NP. The emission wavelength of FITC dye overlaps with surface plasmon band of Au NPs, and then more effective quenching may take place with Au NP compared to Cu and Pt NPs. Hence, the high molar extinction coefficients and broad energy bandwidth make Au NP highly effective quencher.
Figure 4.
Fluorescence emission spectra of (a) FITC-modified dsDNA probe, FITC-dsDNA-GO, and Ag@dsDNA-GO; (b) Au@dsDNA-GO, Cu@dsDNA-GO and Pt@ dsDNA-GO nanostructures Note the different emission full scales.
We used DNA-directed Ag-GO hybrid nanostructures to develop a sensitive, simple and reliable strategy for detection of dopamine. The large surface area of dsDNA-GO decorated with Ag NPs is a highly effective platform to capture nanomolar levels of dopamine from solution and to observe the amount of adsorbed dopamine using zeta potential measurements (shown in Figure S2 of Supporting Information). The zeta-potential is an electrostatic potential caused by charges around a particle’s surface, and the zeta-potential value gives a highly sensitive way to measure the charge density of colloidal particles and to detect any modification resulting from a change of charge density on the surfaces of particles. GO (20 μg/mL) is negatively charged with a −26.41 mV zeta potential due to the presence of hydrophilic carboxyl groups (data not shown). The adsorption of dsDNA, composed of a negatively charged phosphate backbone, on the GO surface results in a more negative zeta-potential, −34.05 mV. The zeta potential becomes even more negative with Ag NPs, −49.32 mV. Addition of dopamine, a positively charged small biomolecule, at different concentrations ranging from 1 nM to 60 μM results in the formation of Ag-catechol bonds and less negative zeta potentials. The color of Ag@dsDNA-GO nanostructures in aqueous solution becomes darker with addition of dopamine (photos shown in Figure S3 of Supporting Information). The addition of diffrent concentration of dopamine (1 nM, 60 nM, 600 nM and 60 μM) into Ag@dsDNA-GO solution made it highly viscous and resulted in the decrease on mobility of Ag@dsDNA-GO colloids (−3.33 MU, −3.01 MU, −2.93 MU, −2,29 MU). Consequently, the zeta potential cannot be measured accurately above 300 μM DA due to quite low mobility, −0.16 MU. Mobility, i.e., the ratio of velocity to applied electric field, is a major parameter in the equation to accurately measure zeta potential values.[37] We think that excess amounts of dopamine bound to the surface of Ag NPs by a catechol reaction and also adsorbed on the back surface of GO via π-π stacking lead to the decreased mobility of Ag@dsDNA-GO composites. In addition to that, the addtion of DA to an aqueous solution of Ag@dsDNA-GO increases the absorbance of the Ag NP becuase more DA molecules bind to surface of Ag NP and stiblize it (UV-Vis graph shown in Figure S4 of Supporting Information). The characteristic absorption peak of Ag NP on GO exhibits a single and very broad absorption peak in around 410 nm wavelength, which is the frequency of the surface plasmon oscillations for Ag NP. As the concentration of dopamine is raised from 1 nM to 60 nM, absorption peak of Ag NPs becomes narrower and the absorbance increases and displays slightly blue shift. Surprisingly, after addition of 600 nM of dopamine, the absorbance values start decrease, and with addition of 60 μM of dopamine, the absorbance is low and broad.[38] The increase in DA concentration causes that DA saturates surface of Ag NPs and excess DA nonspeficicly adsorbs on surfaces of GO, then caused decrease of mobility of Ag@dsDNA-GO complex and some aggregation in solution. The highly vicous Ag@dsDNA-GO complex does not produce proper optical absorbance value. The generated absrobance values are consistent with Figures S2 and S3.
To examine the specificity of our system to dopamine, we tested several other analytes including positively charged metal ions and peptides such as Cu2+, Zn2+, glutamine, lysine, glycine, cystamine dihydrochloride, arginine, nicotine, uric acid and ascorbic acid. The dramatic charge change was only observed in the presence of dopamine, whereas there was no significant charge change in the presence of other substances (shown in Figure S5 of Supporting Information). This selectivity is strongly related to highly specific catechol interaction occurring between Ag NP and diols groups of dopamine. The negligible charge changes in presence of some other analytes may be caused by nonspecific binding between some analytes and Ag NP or GO.
Ag NPs have been well practically used as effective and universal germicidal against to virus and bacteria. The common proposed mechanism of toxicity of Ag NPs against bacteria is that Ag NPs can react with bacteria surface and cause direct and severe damage to bacterial membrane. The combined toxic effect of Ag NP itself and silver ions (Ag+) released from Ag NP deactivate enzyme for cell respiratory and replication and induce the cell death. [39-42] Although Ag NPs display effective antibacterial properties towards variety types of bacteria, the aggregation of Ag NPs when in reacting with bacteria lowers antibacterial effect. To overcome this major drawback, the combination of GO and Ag heterostructure was developed to generate highly enhanced antibacterial effect compared to bare Ag NPs.
Ag@dsDNA-GO heterostructures display excellent antibacterial properties, which are attributed to the synergistic effect between graphene oxide and Ag NPs.[43-45] Use of Ag@dsDNA-GO composites introduces three promising advantages, compared to Ag NPs free in solution: (1) GO acts as a stabilizer to prevent, for the most part, agglomeration of Ag NPs and to maintain the high surface reactivity of Ag NPs; (2) the antibacterial activity of Ag@dsDNA-GO composites is enhanced by the synergistic effect between Ag NPs and GO; (3) the Ag@dsDNA-GO composites may have stronger adsorption properties due to the negatively charged DNA, which may increase adhesion between Ag-GO composites and the bacterial cell membrane.[46] The X.perforans bacteria cells before and after treatment with 20 ppm of Ag@dsDNA@GO composites were observed with TEM images (Figure S6). The bacterial cell is adsorbed on surface Ag@dsDNA@GO and Ag NPs react with bacterial membrane to cause irreversible changes on morphology membrane and finally induced cell death. Experimental results proved that Ag@dsDNA-GO nanostructures possessed very effective antibacterial properties towards Xanthomonas perforans (X.perforans), as shown by the bacterial cell viability assay (Figure S7). Ag NPs kill bacteria by reacting with the bacterial cell wall.[47] However, single-component Ag NPs grown on dsDNA showed a much lower antibacterial effect than Ag@dsDNA-GO nanostructures. When they are in contact with bacterial cells, Ag NPs may aggregate, thereby decreasing their active surface areas and activity. Although plane-GO itself shows antibacterial effect against to some type of bacteria [48], GO does not have any antibacterial effects on X.perforans.
In conclusion, we have developed a novel technique to synthesize DNA-directed metal nanoparticles on GO. The simultaneous synthesis and functionalization achieved by our approach can be easily applied to produce different types of metal-based GO hybrid nanostructures, including Au-GO, Ag-GO, Cu-GO, Pt-GO and Au/Cu/Pt-GO composites. The experimental conditions and parameters are not dependent upon the types of metal ion precursors used to form nanoparticles. The synthesis of nanoparticles takes less than 2 hours and can be successfully and simply carried out under ambient conditions. This approach represents a time- and cost-saving route for the preparation of DNA-mediated MNPs on GO.
The DNA-guided Ag-GO nanostructures were used to design a dopamine quantitation method based on zeta potential measurements. Initial investigation of antibacterial activity indicated that Ag@dsDNA-GO nanostructures significantly decreased the viability of X.perforans. Further antibacterial studies are currently underway.
Experimental Section
Chemical and materials, the detailed synthesis procedure of DNA and instrumentation and characterization are given in the supporting information.
Synthesis of DNA-guided Ag NPs on GO
The synthesis of DNA-guided Ag NPs was accomplished through reduction of AgNO3 with NaBH4. DNA-1: 5' AAT GTG CTC CCC CA GCGCGCTT FITC-3' and DNA-2: 5' TGG GGG AGC ACA TT-3' were mixed together at the same concentration (1μM) to obtain double-stranded DNA (dsDNA) via hybridization in 10mM HEPES buffer (50mM NaNO3, pH 7.5) for 30 min. Then, graphene oxide (GO) at a final concentration of 20 μg/mL was added to the DNA solution for adsorption of dsDNA on the GO. After 30 min incubation, the mixture was centrifuged, and excess DNA in the supernatant was decanted. The precipitated DNA-GO was redispersed in 1 mL of 10 mM HEPES buffer, followed by addition of AgNO3 to the DNA-GO mixture under vigorous stirring (final concentration of AgNO3 is 100 μM in mixture of DNA-GO and AgNO3). After 5 minutes of incubation, freshly prepared reducing agent solution of 500 μM NaBH4 was injected dropwise into the mixture containing DNA, GO and AgNO3 for 30 min under vigorously stirring. The resultant mixture was washed twice with HEPES buffer and centrifuged at 12000 rpm for 15 min to remove free Ag NPs. The final products were redispersed in HEPES buffer for further characterization.
Au@dsDNA-templated GO
Gold (III) chloride trihydrate was used with sodium citrate to form Au NPs on dsDNA-GO. HAuCl4.3H2O solution was added to the dsDNA-GO mixture with final concentration of 100 μM Au3+, and the resulting mixture was stirred for 5 minutes. The freshly prepared reducing agent solution of 500 μM sodium citrate was added in a dropwise manner for 30 min with vigorous stirring. Purification was achieved in the manner described above.
Cu@dsDNA-templated GO and Pt@dsDNA-templated GO
The protocol given above for the formation of Ag and Au NPs was followed, except CuSO4 and PtCl4 were separately reduced on dsDNA-GO using sodium L-ascorbate as the reductant. The final concentrations of metal ions (Cu2+ or Pt4+) and reducing agent in mixtures were adjusted to 100 μM and 500 μM, respectively.
Au/Cu/Pt@dsDNA-templated GO
Au3+, Cu2+ and Pt4+ solutions, at 100 μM each metal ion, were simultaneously added into the dsDNA-GO mixture stated above with vigorous stirring. After 10 minutes incubation, reducing agent of 500 μM sodium L-ascorbate was added dropwise to the mixture for 30 minutes of stirring.
Supplementary Material
Acknowledgements
We are grateful to the Interdisciplinary Center for Biotechnology Research (ICBR) at the University of Florida. We also sincerely appreciate Dr. Kathryn Williams for her help with manuscript preparation. We acknowledge the Ministry of National Education, Republic of Turkey, for financial support to Ismail Ocsoy with full fellowship during his PhD studies at the University of Florida. This work is supported by grants awarded by the National Key Scientific Program of China (2011CB911000), the Foundation for Innovative Research Groups of NSFC (Grant 21221003), China National Instrumentation Program 2011YQ03012412 and by the National Institutes of Health (GM066137, GM079359 and CA133086).
Footnotes
Supporting Information
Supporting information is available from Wiley Online Library or from the author.
References
- 1.Dittmer WU, Simmel FC. Applied Physics Letters. 2004;85:4. [Google Scholar]
- 2.Schreiber R, Kempter S, Holler S, Schüller V, Schiffels D, Simmel SS, Nickels PC, Liedl T. Small. 2011;7:1795–1799. doi: 10.1002/smll.201100465. [DOI] [PubMed] [Google Scholar]
- 3.Tan SJ, Campolongo MJ, Luo D, Cheng W. Nat Nano. 2011;6:268–276. doi: 10.1038/nnano.2011.49. [DOI] [PubMed] [Google Scholar]
- 4.Pal S, Varghese R, Deng Z, Zhao Z, Kumar A, Yan H, Liu Y. Angewandte Chemie International Edition. 2011;50:4176–4179. doi: 10.1002/anie.201007529. [DOI] [PubMed] [Google Scholar]
- 5.Geng Y, Liu J, Pound E, Gyawali S, Harb JN, Woolley AT. J. Mater. Chem. 2011;21:12126–12131. [Google Scholar]
- 6.Guo W, Yuan J, Dong Q, Wang E. J. Am. Chem. Soc. 2010;132:932–934. doi: 10.1021/ja907075s. [DOI] [PubMed] [Google Scholar]
- 7.Sharma J, Yeh H, Yoo H, Werner JH, Martinez JS. Chem. Commun. 2011;47:2294–2296. doi: 10.1039/c0cc03711g. [DOI] [PubMed] [Google Scholar]
- 8.Gourine AV, Llaudet E, Dale N, Spyer KM. Nature. 2005;436:108–111. doi: 10.1038/nature03690. [DOI] [PubMed] [Google Scholar]
- 9.Berti L, Burley GA. Nat Nano. 2008;3:81–87. doi: 10.1038/nnano.2007.460. [DOI] [PubMed] [Google Scholar]
- 10.Izatt RM, Christensen JJ, Rytting JH. Chem. Rev. 1971;71:439–481. doi: 10.1021/cr60273a002. [DOI] [PubMed] [Google Scholar]
- 11.Gugliotti LA, Feldheim DL, Eaton BE. Science. 2004;304:850–852. doi: 10.1126/science.1095678. [DOI] [PubMed] [Google Scholar]
- 12.Ma N, Dooley CJ, Kelley SO. J. Am. Chem. Soc. 2006;128:12598–12599. doi: 10.1021/ja0638962. [DOI] [PubMed] [Google Scholar]
- 13.Hinds S, Taft BJ, Levina L, Sukhovatkin V, Dooley CJ, Roy MD, MacNeil DD, Sargent EH, Kelley SOJ. Am. Chem. Soc. 2006;128:64–65. doi: 10.1021/ja057002+. [DOI] [PubMed] [Google Scholar]
- 14.Coffer JL, Bigham SR, Pinizzotto RF, Yang H. Nanotechnology. 1992;3:69. [Google Scholar]
- 15.Bigham SR, Coffer JL. Colloids Surf. Physicochem. Eng. Aspects. 1995;95:211–219. [Google Scholar]
- 16.Rothemund PWK. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 17.Pal S, Deng Z, Wang H, Zou S, Liu Y, Yan H. J. Am. Chem. Soc. 2011;133:17606–17609. doi: 10.1021/ja207898r. [DOI] [PubMed] [Google Scholar]
- 18.Pu S, Zinchenko A, Murata S. Nanotechnology. 2011;22:375604. doi: 10.1088/0957-4484/22/37/375604. [DOI] [PubMed] [Google Scholar]
- 19.Liu J, Geng Y, Pound E, Gyawali S, Ashton JR, Hickey J, Woolley AT, Harb JN. ACS Nano. 2011;5:2240–2247. doi: 10.1021/nn1035075. [DOI] [PubMed] [Google Scholar]
- 20.Geim AK, Novoselov KS. Nature Materials. 2007;6:183–191. doi: 10.1038/nmat1849. [DOI] [PubMed] [Google Scholar]
- 21.Sanchez VC, Jachak A, Hurt RH, Kane AB. Chem. Res. Toxicol. 2011;25:15–34. doi: 10.1021/tx200339h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu C, Wang X. Small. 2009;5:2212–2217. doi: 10.1002/smll.200900548. [DOI] [PubMed] [Google Scholar]
- 23.Tang X, Cao Z, Zhang H, Liu J, Yu Z. Chem. Commun. 2011;47:3084–3086. doi: 10.1039/c0cc05613h. [DOI] [PubMed] [Google Scholar]
- 24.Liu X, Pan L, Lv T, Zhu G, Sun Z, Sun C. J. Mater. Chem. 2007;17:2679–2694. [Google Scholar]
- 25.Williams G, Seger B, Kamat PV. ACS Nano. 2008;2:1487–1491. doi: 10.1021/nn800251f. [DOI] [PubMed] [Google Scholar]
- 26.Xu W, Zhang L, Li J, Lu Y, Li H, Ma Y, Wang W, Yu S. J. Mater. Chem. 2011;21:4593–4597. [Google Scholar]
- 27.Pasricha R, Gupta S, Srivastava AK. Small. 2009;5:2253–2259. doi: 10.1002/smll.200900726. [DOI] [PubMed] [Google Scholar]
- 28.Cao A, Liu Z, Chu S, Wu M, Ye Z, Cai Z, Chang Y, Wang S, Gong Q, Liu Y. Adv Mater. 2010;22:103–106. doi: 10.1002/adma.200901920. [DOI] [PubMed] [Google Scholar]
- 29.Monn G, Park Y, Kim W, Choi W. Carbon. 2011;49:3454–3462. [Google Scholar]
- 30.Liu X, Mao J, Liu P, Wei X. Carbon. 2011;49:477–483. [Google Scholar]
- 31.Liu J, Fu S, Yuan B, Li Y, Deng Z. J. Am. Chem. Soc. 2010;132:7279–7281. doi: 10.1021/ja100938r. [DOI] [PubMed] [Google Scholar]
- 32.Das MR, Sarma RK, Saikia R, Kale VS, Shelke MV, Sengupta P. Colloids and Surfaces B: Biointerfaces. 2011;83:16–22. doi: 10.1016/j.colsurfb.2010.10.033. [DOI] [PubMed] [Google Scholar]
- 33.Huang J, Zhang L, Chen B, Ji N, Chen F, Zhang Y, Zhang Z. Nanoscale. 2010;2:2733–2738. doi: 10.1039/c0nr00473a. [DOI] [PubMed] [Google Scholar]
- 34.Lu C, Yang H, Zhu C, Chen X, Chen G. Angewandte Chemie International Edition. 2009;48:4785–4787. doi: 10.1002/anie.200901479. [DOI] [PubMed] [Google Scholar]
- 35.Wang C, Tian W, Ding Y, Ma Y, Wang ZL, Markovic NM, Stamenkovic VR, Daimon H, Sun S. J. Am. Chem. Soc. 2010;132:6524–6529. doi: 10.1021/ja101305x. [DOI] [PubMed] [Google Scholar]
- 36.Wang Z, Huang P, Bhirde A, Jin A, Ma Y, Niu G, Neamati N, Chen X. Chem. Commun. 2012;48:9768–9770. doi: 10.1039/c2cc31974h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sze A, Erickson D, Ren L, Li D. Journal of Colloid and Interface Science. 2003;261:402–410. doi: 10.1016/S0021-9797(03)00142-5. [DOI] [PubMed] [Google Scholar]
- 38.Baron R, Zayats M, Willner I. Anal. Chem. 2005;77:1566–1571. doi: 10.1021/ac048691v. [DOI] [PubMed] [Google Scholar]
- 39.Elechiguerra J, Burt JL, Morones JR, Camacho-Bragado C, Gao X, Lara HH, Yacaman M. Journal of Nanobiotechnology. 2005;3:1–6. doi: 10.1186/1477-3155-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morones JR, Elechiguerra JL, Camacho A, Holt K, Kouri JB, Ramírez JT, Yacaman MJ. Nanotechnology. 2005;16:2346–2353. doi: 10.1088/0957-4484/16/10/059. [DOI] [PubMed] [Google Scholar]
- 41.Panáček A, Kvítek L, Prucek R, Kolář M, Večeřová R, Pizúrová N, Sharma VK, Nevěčná TJ, Zbořil R. J. Phys. Chem. B. 2006;110:16248–16253. doi: 10.1021/jp063826h. [DOI] [PubMed] [Google Scholar]
- 42.Sotiriou GA, Pratsinis SE. Environmental Science & Technology. 2010;44(14):5649–5654. doi: 10.1021/es101072s. [DOI] [PubMed] [Google Scholar]
- 43.Liu L, Liu J, Wang Y, Yan X, Sun DD. New J. Chem. 2011;35:1418–1423. [Google Scholar]
- 44.Ma J, Zhang J, Xiong Z, Yong Y, Zhao XS. Journal of Materials Chemistry. 2011;21:3350–3352. [Google Scholar]
- 45.Shen J, Shi M, Li N, Yan B, Ma H, Hu Y, Ye M. Nano Res. 2010;3:339–349. [Google Scholar]
- 46.Xu W, Zhang L, Li J, Lu Y, Li H, Ma Y, Wang W, Yu S. J. Mater. Chem. 2011;21:4593–4597. [Google Scholar]
- 47.Lago VD, Oliveira LF, Goncalves KA, Kobarg J, Cardoso MB. J. Mater. Chem. 2011;21:12267–12273. [Google Scholar]
- 48.Kholmanov IN, Stoller MD, Edgeworth J, Lee WH, Li H, Lee J, Barnhart C, Potts JR, Piner R, Akinwande D, Barrick JE, Ruoff RS. ACS Nano. 2012;6:5157–5163. doi: 10.1021/nn300852f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.