Abstract
Triacylglycerol (TAG) analysis and quantification are commonly performed by first obtaining a purified TAG fraction from a total neutral lipid extract using thin-layer chromatography (TLC), and then analyzing the fatty acid composition of the purified TAG fraction by gas chromatography (GC). This process is time-consuming, labor intensive and is not suitable for analysis of small sample sizes or large numbers. A rapid and efficient method for monitoring oil accumulation in algae using high performance liquid chromatography for separation of all lipid classes combined with detection by evaporative light scattering (HPLC–ELSD) was developed and compared to the conventional TLC/GC method. TAG accumulation in two Chlamydomonas reinhardtii (21 gr and CC503) and three Chlorella strains (UTEX 1230, CS01 and UTEX 2229) grown under conditions of nitrogen depletion was measured. The TAG levels were found to be 3–6 % DW (Chlamydomonas strains) and 7–12 % DW (Chlorella strains) respectively by both HPLC–ELSD and TLC/GC methods. HPLC–ELSD resolved the major lipid classes such as carotenoids, TAG, diacylglycerol (DAG), free fatty acids, phospholipids, and galactolipids in a 15-min run. Quantitation of TAG content was based on comparison to calibration curves of trihexadecanoin (16:0 TAG) and trioctadecadienoin (18:2 TAG) and showed linearity from 0.2 to 10 μg. Algal TAG levels >0.5 μg/g DW were detectable by this method. Furthermore TAG content in Chlorella kessleri UTEX 2229 could be detected. TAG as well as DAG and TAG content were estimated at 1.6 % DW by HPLC–ELSD, while it was undetectable by TLC/GC method.
Keywords: TAG quantification, HPLC–ELSD, GC–MS, TLC, Chlamydomonas reinhardtii, Chlorella, Lipid, FAME, Lipid class
Introduction
Microalgae serve as a potential feedstock for biofuel and biodiesel applications due to the ability of some algal strains to accumulate high levels of triacylglycerols (TAG) under various stress conditions [1]. TAG are neutral lipids consisting of a glycerol backbone esterified to three fatty acids. The fatty acid composition of algal TAG is similar to that observed in higher plants, with 16- and 18-carbon fatty acids being predominant [1, 2]. Most conventional fuels including petroleum and diesel contain aliphatic hydrocarbons that are chemically similar to the fatty acid components of TAG [2]. Algal TAG accumulation has been studied under conditions of nutrient limitation and abiotic stress to monitor changes in compound production [1]. These studies include gene expression patterns and their relation to oil accumulation during nitrogen deprivation in C. reinhardtii and Coccomyxa sp. [3], lipid profiling during photoautotrophic and heterotrophic growth in Chlorella zofingiensis [4] and characterization of changes in major lipid droplet proteins in response to nitrogen deprivation in C. reinhardtii [5].
Thin layer chromatography (TLC) and gas chromatography (GC) are the most commonly used analytical methods for lipid characterization and quantification. The combination of methods involves purifying specific lipid classes such as TAG first by TLC, scraping individual bands from the TLC plate, extracting these bands, hydrolyzing the fatty acids to form fatty acid methyl esters (FAME), and finally analyzing the FAME by gas chromatography. The separation of lipid classes by silica TLC [3–5] or silica column [6] requires use of different solvent systems for purification of each lipid class. TAG are well separated from other major lipid classes such as phospholipids and glycolipids in a non-polar solvent such as chloroform, hexane or heptane, however phospholipids and glycolipids cannot be resolved in highly non-polar solvent systems and require more polar solvent systems. After TAG isolation by TLC, intact TAG must be broken down to release the fatty acid components which are converted to FAME by a trans-esterification reaction in preparation for GC analysis. Accurate quantitation by TLC/GC involves a number of purification steps and a trans-esterification reaction which leads to an indirect measurement of the total lipid content. Although the fatty acid compositions by traditional TLC and GC methods are useful for qualitative and quantitative lipid analyses, sample preparation is time-consuming, labor-intensive and not suitable for profiling total lipid compositions in a high-throughput manner.
Here we describe a method using high performance liquid chromatography–evaporative light scattering detector (HPLC–ELSD) that is suitable for comparing major lipid classes in a single injection and can be used to accurately measure TAG content. HPLC–ELSD involves separation of lipids by HPLC, and detection by conversion to small particles in a nitrogen gas stream and collision with light. Separation and quantification of different lipid classes by HPLC–ELSD have been reported in various species. For example, nine TAG from Walnut (Juglans regia L.) were separated and identified by HPLC–ELSD including trilinolein, dilinoleoyl-oleoyl-glycerol and dilinolenoyl-glycerol as the major TAG at 38, 19 and 18 %, respectively [7]. In addition, ten complex lipid classes were separated with a binary gradient system by HPLC–ELSD and glyceroglycolipids including monogalactosyldiacylglycerol (MGDG), digalactosyldiacylglycerol (DGDG) and sulfoquinovosyldiacylglycerol (SQDG) were quantified simultaneously in different plants [8] and 17 neutral and polar lipid classes were separated by HPLC–ELSD using a monolithic silica phase and quantified in four different zooplankton [9]. A 15 min HPLC gradient with hexane based solvent and isopropanol based solvent is used to resolve carotenoids, TAG, DAG and free fatty acids, and the major membrane components: phosphophatidylcholine (PtdCho), MGDG and DGDG. TAG content is estimated based on comparison to calibration curves of trihexadecanoin (16:0 TAG) and trioctadecadienoin (18:2 TAG). C. reinhardtii has been established as the model green microalgae with TAG contents ranging widely from 2 to 20 % of DW in different strains by TLC/GC method [10]. Although HPLC is mostly tandem with UV–VIS or MS, ELSD is more accurate lipid quantification than UV–VIS and more inexpensive and easier than MS. The advantage of using ELSD is to make it possible to quantify not only TAG but other neutral lipids for biofuel or biodiesel at the same run, while it requires several steps and different GC runs by TLC/GC method.
The green microalgae of the genus Chlorella are among the potential candidates of biofuel or biodiesel production due to inherent characteristics of large biomass production and oil accumulation with minimal nutrient requirements [11]. Chlorella cultivation can be readily performed in industrial processes [12, 13]. Reported oil accumulation of Chlorella spans broadly from 3 to 58 % DW, illustrating a wide range of sensitivity by various methods of extraction, analysis and quantification [14–17]. TAG analysis has been reported commonly in Chlorella by gravimetric measurement, or by GC with TLC or column chromatography in C. vulgaris, C. zofingiensis and C. sorokiniana [18–20]. Further means of TAG detection have included Raman spectroscopy in C. sorokiniana [21], thermogravimetric analysis in Chlorella sp. [22] and HPLC–ELSD in C. minutissima [23]. Using HPLC–ELSD, contents of TAG in a variety of algae under different conditions were screened to select a candidate species best suitable for yielding high oil content in optimal conditions.
In this study, we used C. reinhardtii 21 gr (wild type) and CC503 (wall-less) strains as a green alga model system and three Chlorella strains under photoautotrophic and nitrogen deprivation conditions. The total lipid and TAG contents were compared using two different lipid analysis methods, HPLC–ELSD and GC/MS with TLC. C. sorokiniana UTEX 1230, CS-01 and C. kessleri UTEX 2229 were selected according to their phylogenetic relationship with C. variabilis NC64A [24], a model Chlorella that has been extensively characterized. Differences in biomass production among Chlorella species also served as a variable of interest in relationship to lipid production. The HPLC–ELSD method resolves major lipid classes and rivals similar accuracy of quantification of TAG levels in comparison to TLC/GC methods when evaluating the Chlorella strains as well as the Chlamydomonas strains. This method provides a rapid and simple means to detect and quantify algal neutral lipid classes and is an efficient screening tool for metabolic studies of algal lipid accumulation.
Materials and Methods
Materials and Growth Conditions
Chlamydomonas. reinhardtii 21 gr (wild type) and CC503 (wall-less) were provided by Dr. K. Horken (University of Nebraska-Lincoln). C. kessleri UTEX 2229 and C. sorokiniana UTEX 1230 were obtained from The Culture Collection of Algae (UTEX) at the University of Texas at Austin. C. sorokiniana CS-01 was provided by Dr. M. Wan (Johns Hopkins University). Chlamydomonas strains were maintained in 100 mL TAP media [25] and Chlorella strains were plated on Bold’s Basal Medium (BBM, [26]) agar plates containing 10 μg/mL tetracycline and 100 μg/mL ampicillin. Liquid cultures of Chlorella were initiated by inoculation of a single isolated colony into 5 mL of BBM. Both species were grown for 14 days at 300 rpm under continuous illumination (160 μmol m−2 s−1) at 25 °C.
Cultures were scaled up to 250-mL flasks and the cells were collected at the stationary phase by centrifugation at 5,000g for 10 min. Each cell pellet was transferred into 3 L of TAP media for Chlamydomonas strains and BBM media for Chlorella strains in a 3-L photo-bioreactor under continuous illumination at 200 μmol m−2 s−1 in a greenhouse environment at 26 ± 2 °C. The 3-L photo-bioreactors were aerated by an air-pump (Whitewater, MTL66) with a flow rate at 1 psi 2.3 cfm in addition to a spinning turbine impeller in the photo-bioreactor set at 180 rpm. After 5 days at the stationary phase of Chlamydomonas strains and 12 days of Chlorella strains, cultures were divided into two 1.5-L aliquots that were centrifuged at 5,000g for 5 min and transferred into either 3 L TAP media or 3 L TAP media without a nitrogen source for Chlamydomonas strains or either 3 L BBM media or 3 L BBM media without a nitrogen source for Chlorella strains. The cultures were maintained for 7 days under the same conditions as above. The algae cells were collected by centrifugation at 5,000g for 5 min. The supernatant was discarded and pellets were stored at −80 °C and lyophilized overnight. The dried algae samples were used for lipid analyses.
Chlorella liquid cultures equal to and >300 mL were grown in photo-bioreactors under continuous illumination at 200 μmol m−2 s−1 in a greenhouse environment at 26 ± 2 °C. After incubation for 14 days, 15 mL cultures were transferred to 300 mL BBM in 500-mL aerated photo-bioreactors. Exponential-phase cells were transferred after 5 days to 3 L BBM in 3-L aerated photo-bioreactors and grown for an additional 7 days. Aeration flow for the 500-mL and 3-L photo-bioreactors were maintained by an air-pump (Whitewater, MTL66) with a flow rate of 1 psi 2.3 cfm in addition to agitation by spinning turbine impellers (180 rpm). The 3 L cultures were again scaled-up and the cells at 4 × 106 cell/mL of each strain were transferred to 80 L BBM into 170-L open aquarium tanks with two Power Head water pumps (1,500 L/h; PETCO King 225) arranged in diagonal corners. Cell growth was measured by monitoring the absorbance at 530 nm and using a hemocytometer. Cells were harvested following a 10-day incubation period during the early stages of stationary phase. 2-L aliquots of cell samples from the aquarium tanks were collected by centrifugation at 5,000g for 5 min. Samples were stored at −20 °C until used for lipid analysis.
Extraction of Lipids
The dried algae pellets (50, 100, 200 mg) were homogenized using a mortar and pestle with liquid nitrogen and used for the comparison of both TLC/GC and HPLC–ELSD methods. Lipid extraction was performed by a modification of the method described by Bligh and Dyer [27]. Two milliliters of chloroform/methanol (1:2 v/v) containing 0.01 % butyl hydroxyl toluene was added to the ground cells and pooled in a glass tube and repeated three times. Then, 500 μg of tripentadecanoin (15:0 TAG, Nu-Check Prep, MN, USA) was added as an internal standard for quantification of the FAME analysis. The mixture was vortexed at room temperature for 30 min. 2 mL of chloroform and 4 mL of water were added and vortexed. After centrifugation at 1,500g for 5 min, the chloroform phase was collected and the aqueous phase was re-extracted with 5 mL chloroform. This was repeated three times. The pooled chloroform phases were evaporated to dryness under a stream of nitrogen. The total lipid content was determined gravimetrically.
Chromatographic Methods
The separation of lipids by system gold HPLC (Beckman Coulter, CA, USA) with ELSD (Shimadzu, MD, USA) was performed in the gradient mobile phase (Table 1). The Luna 3-μm silica column (100 × 4.60 mm, Phenomenex, CA, USA) was connected with the guard column (silica 4 × 3.0 mm, Phenomenex, CA, USA), which incubated at 50 °C in the column oven. ELSD settings included gain 5, at 30 °C and 350 kPa for nitrogen pressure. The flow rate was 1 mL/min and 20 μL of the samples were injected. The solvent system used hexane based solvent A (hexane/isopropanol/acetic acid, 98.9:1:0.1 v/v/v, all HPLC grade, Fisher Scientific, PA, USA) and isopropanol based solvent B (isopropanol/acetic acid, 99.9:0.1 v/v/v, all HPLC grade, Fisher Scientific, PA, USA). After a 7-min isocratic run with solvent A for elution of neutral lipids, solvent B increased to 95 % for a 1-min wash and then the column was equilibrated by 100 % of solvent A for 6 min (Table 1). Seven TAG including tripentadecanoin, trihexadecanoin (palmitin), triheptadecanoin (margarin), trioctadecanoin (stearin), trioctadecenoin (vaccenin), triocadecadienoin (linolein) and trioctadecatrienoin (alpha-linolenin) (15:0, 16:0, 17:0, 18:0, 18:1, 18:2 and 18:3, Nu-Check Prep MN, USA) in hexane were applied for the reproducibility of peak area and retention times at 1.0 and 5.0 μg (Table 2) and measured from the ranges of 0–15 μg for the calibration curve (Fig. 1a). Then, 1 mL of hexane was added to the dried total lipid samples. SPE silica column (Supelclean LC-Si SPE Tubes 3 mL, SUPELCO Analytical, PA, USA) was equilibrated by hexane and 200 μL of the lipid mixture was added to the equilibrated SPE column. 6 mL of hexane was added and the elution was collected gravitationally after removal of chlorophyll and carotenoid fractions. Samples were stored at −20 °C. 10 mg/mL of TAG, DAG and palmitic acid (TAG, DAG and palmitic acid, Nu-Check Prep MN, USA) were dissolved in hexane. Then, 10 mg/mL of phosphaphatidylcholine (PtdCho) and phosphoethanolamine (PE), 1 mg/mL of MGDG, DGDG and dipalmitoyl glycero trimethyl homoserine (DGTS) (PtdCho, PE, MGDG, DGDG and DGTS, Avanti Polar Lipids, Inc., AL, USA) were dissolved in chloroform/methanol (8:2, v/v) for the authentic standards in Figs. 3a, 5a, 7a. Carotenoids were separated from C. reinhardtii by TLC (silica plate, Sigma-Aldrich) in heptane/diethyl ether/acetic acid (70:30:1, v/v/v, all Fisher Scientific). The yellow carotenoids were scraped and eluted in 3 mL of hexane. The eluted carotenoids were used for the authentic standards in Figs. 3a, 5a, 7a. The percentages of DW were calculated based on the equations from the TAG calibration curves and all the presented data incorporate averages ± standard deviations of three technical replicates.
Table 1.
HPLC gradient condition of mobile phase
| Time (min) | Percentage of solvent | Flow-rate (mL/min) | |
|---|---|---|---|
| A | B | ||
| 0 | 100 | 0 | 1 |
| 7 | 100 | 0 | 1 |
| 8 | 5 | 95 | 1 |
| 9 | 100 | 0 | 1 |
| 15 | 100 | 0 | 1 |
A 98.9 % hexane, 1.0 % isopropanol, 0.1 % acetic acid, B 99.9 % isopropanol, 0.1 % acetic acid
Table 2.
Reproducibility of peak areas and retention times of TAG standards injected 12 times
| TAG | Conc. (μg) | Retention time (min) | Peak area | ||||
|---|---|---|---|---|---|---|---|
| Ave. | SD | % RSD | Ave. | SD | % RSD | ||
| 15:0 | 1.0 | 1.92 | 0.03 | 0.9 | 875,942 | 46,138 | 5.3 |
| 5.0 | 1.95 | 0.01 | 0.4 | 9,278,987 | 281,635 | 3.0 | |
| 16:0 | 1.0 | 1.94 | 0.02 | 0.8 | 805,907 | 57,103 | 7.1 |
| 5.0 | 1.97 | 0.02 | 0.8 | 8,580,230 | 277,905 | 3.2 | |
| 17:0 | 1.0 | 1.96 | 0.02 | 0.6 | 706,495 | 53,524 | 7.0 |
| 5.0 | 1.95 | 0.03 | 1.0 | 7,517,341 | 277,102 | 3.7 | |
| 18:0 | 1.0 | 1.91 | 0.01 | 0.4 | 772,662 | 54,400 | 7.0 |
| 5.0 | 1.91 | 0.02 | 0.7 | 4,764,890 | 275,968 | 5.8 | |
| 18:1 | 1.0 | 1.92 | 0.03 | 1.1 | 814,658 | 40,017 | 4.9 |
| 5.0 | 1.92 | 0.01 | 0.5 | 9,152,411 | 424,981 | 4.6 | |
| 18:2 | 1.0 | 1.93 | 0.03 | 0.7 | 918,516 | 44,708 | 4.9 |
| 5.0 | 1.94 | 0.02 | 0.6 | 8,625,403 | 356,563 | 4.1 | |
| 18:3 | 1.0 | 1.92 | 0.02 | 0.7 | 928,705 | 59,874 | 6.4 |
| 5.0 | 1.92 | 0.02 | 0.9 | 9,576,351 | 527,472 | 5.5 | |
Conc. concentration, Ave. average, SD standard deviation, RSD ratio of SD and Ave
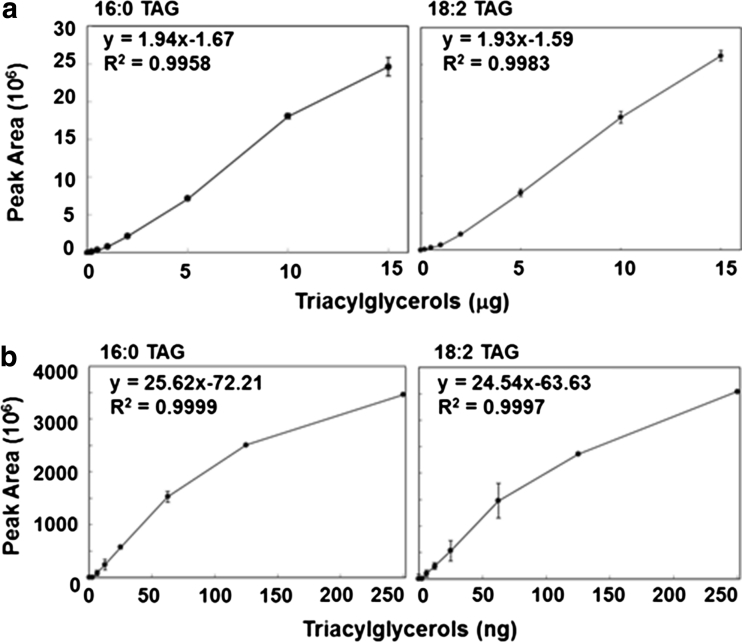
Fig. 1.
Comparison of calibration curves and their equations of trihexadecanoin (16:0 TAG) and trioctadecadienoin (18:2 TAG) between HPLC–ELSD (a) and GC–MS (b)
Fig. 3.
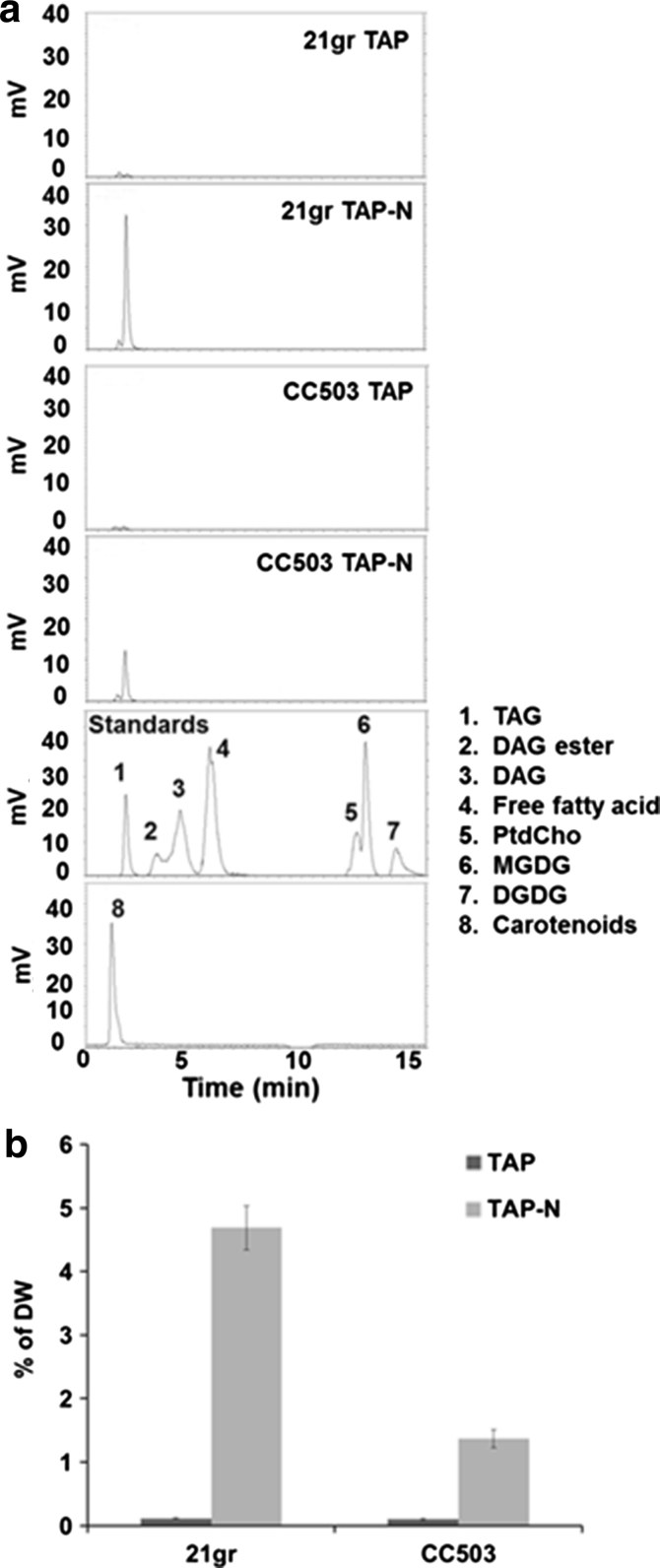
a Separation of TAG from other neutral-, polar-lipids and glycolipids in C. reinhardtii 21 gr and CC503 under TAP and TAP-N conditions and authentic standards by HPLC–ELSD. 1 triacylglycerols (TAG, 1 μg), 2 diacylglycerol (DAG) ester 3 diacylglycerol (DAG, 4 μg), 4 free fatty acids (palmitic acid 2 μg), 5 phosphaphatidylcholine (PtdCho, 20 μg), 6 monogalactosyldiacylglycerol (MGDG, 20 μg), 7 digalactosyldiacylglycerol (DGDG, 40 μg) 8 carotenoids extracted from C. reinhardtii. b TAG contents of 21 gr and CC503 under TAP and TAP-N conditions estimated by HPLC–ELSD. Error bar n = 3 technical replicates from one 3-L bioreactor culture. Ave. of % DW ± standard deviation
Fig. 5.
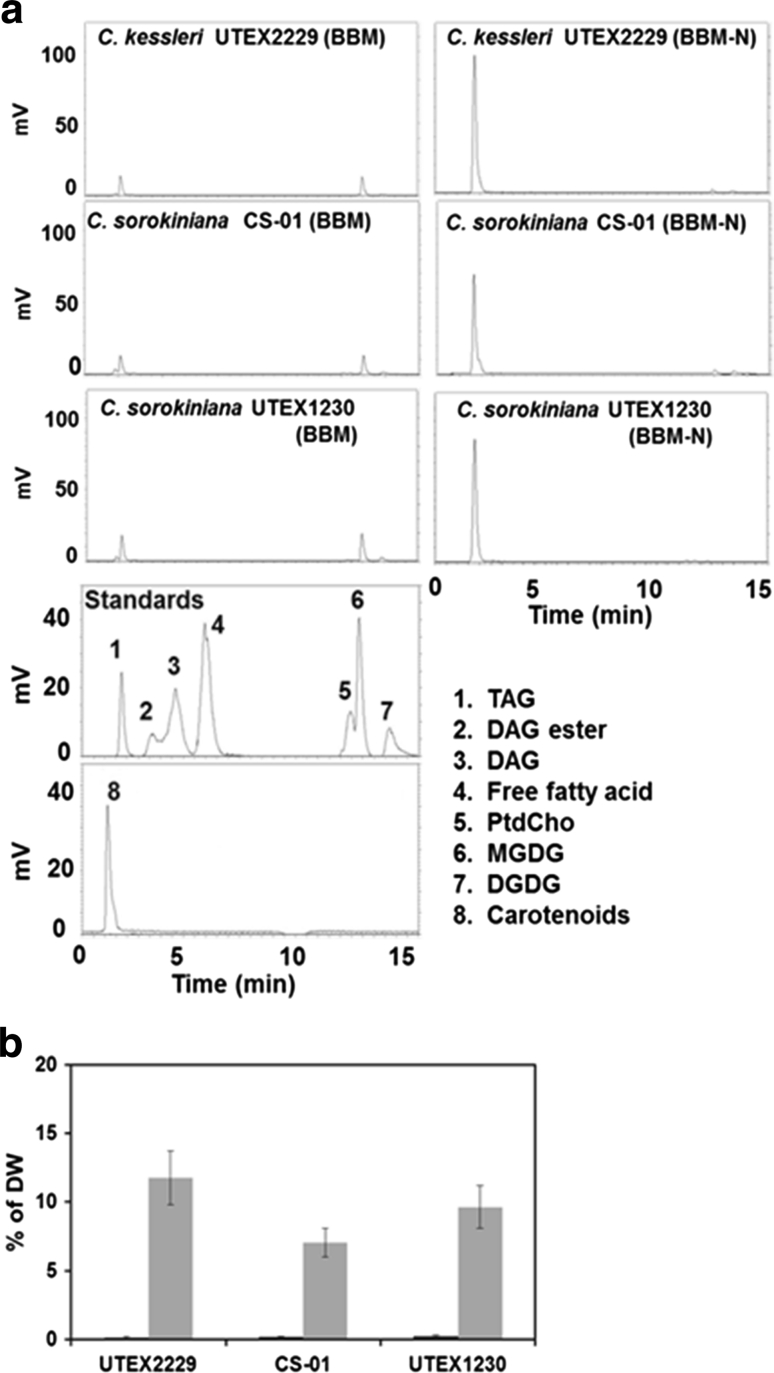
Comparison of TAG detection in C. kessleri UTEX 2229, C. sorokiniana CS-01, C. sorokiniana UTEX 1230 under BBM and BBM-N (a). Separation of neutral lipid from polar lipid and glycolipid, and authentic standards by HPLC–ELSD. 1 triacylglycerols (TAG, 1 μg), 2 diacylglycerol (DAG) ester 3 diacylglycerol (DAG, 4 μg), 4 free fatty acids (palmitic acid 2 μg), 5 phosphaphatidylcholine (PtdCho, 20 μg), 6 monogalactosyldiacylglycerol (MGDG, 20 μg), 7 digalactosyldiacylglycerol (DGDG, 40 μg), 8 carotenoids extracted from C. reinhardtii. (b) TAG contents of C. kessleri UTEX 2229, C. sorokiniana CS-01 and UTEX 1230 under BBM and BBM-N by HPLC–ELSD. Error bar n = 3 technical replicates from one 3-L bioreactor culture. Ave. of % DW ± standard deviation
Fig. 7.
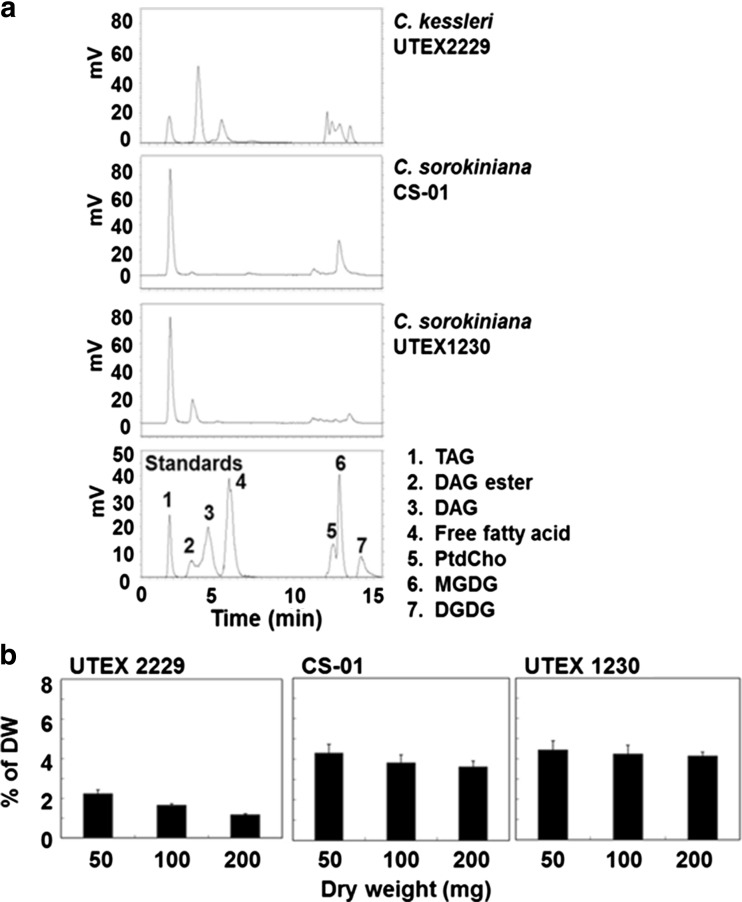
Comparison of TAG detection and contents in C. kessleri UTEX 2229, C. sorokiniana CS-01, C. sorokiniana UTEX 1230. a Separation of neutral lipid from polar lipid and glycolipid, and authentic standards by HPLC–ELSD. 1 triacylglycerols (TAG, 1 μg), 2 diacylglycerol (DAG) ester 3 diacylglycerol (DAG, 4 μg), 4 free fatty acids (palmitic acid 2 μg), 5 phosphaphatidylcholine (PtdCho, 20 μg), 6 monogalactosyldiacylglycerol (MGDG, 20 μg), 7 digalactosyldiacylglycerol (DGDG, 40 μg), 8 carotenoids extracted from C. reinhardtii. b TAG contents of C. kessleri UTEX 2229, C. sorokiniana CS-01 and UTEX 1230 at 50, 100 and 200 mg by HPLC–ELSD. Error bar n = 3 technical replicates from one 80-L aquarium culture. Ave. of % DW ± standard deviation
TLC for neutral lipid was separated by heptane/diethyl ether/acetic acid (70:30:1, v/v/v) and TAG were co-migrated at the same position as the standard TAG from soy beans stained by iodine stream and scraped for FAME (fatty acid methyl esters) analysis. Trans-methylation to FAME was performed as previously described [3]. FAME were analyzed using an Agilent 6890 Series GC System with Agilent 5973 Network Mass Selective Detector (Agilent Technologies, DE, USA). Chromatography was carried out using a 200 m × 250 μm × 0.25 μm Varian GC Capillary column (Varian Inc., CA, USA). The GC inlet was held at 270 °C and 1 μL of the sample was injected in splitless mode. The oven temperature was programmed from 130 °C (10 min hold) to 160 °C (7 min hold), from 160 to 190 °C (7 min hold), from 190 to 220 °C (22 min hold) and from 220 to 250 °C (17 min hold) at a rate of 10 °C/min for each step with helium as the carrier gas. The total analysis time was 75 min. The GC/MS was carried out using 70 eV EI and the data was evaluated with the total ion count. The total lipid content was calculated based on the 15:0 TAG internal standard and the ration of peak area of the total FAME composition sum and 15:0 FAME was applied to obtain the lipid content percentages in DW. All data include averages ± standard deviations of three technical replicates.
Results and Discussion
Development of Triacylglycerol Detection and Quantification by HPLC–ELSD Methods
Gas chromatographic analysis combined with purification of lipid classes by silica TLC or column chromatography is commonly used for TAG analyses. However, this approach is time-consuming and labor intensive and cannot be used to analyze large numbers of samples efficiently. In order to develop a rapid method for measuring TAG accumulation in C. reinhardtii 21 gr (wild type) and CC503 (wall-less) under conditions of nitrogen depletion, total neutral lipids were extracted and separated by HPLC–ELSD. The separation was performed by a silica column (Luna 3 μm silica 100 × 4.6 mm) with a solvent system consisting of a non-polar solvent A (hexane/isopropanol/acetic acid, 98.9:1:0.1 v/v/v,) and a more polar solvent B (isopropanol/acetic acid, 99.9:0.1 v/v,), allowing for resolution of the major lipid classes (carotenoids, triacylglycerides, diacylglycerides, free fatty acids, phospholipids and glycolipids). Seven TAG including tripentadecanoin, trihexadecanoin (palmitin), triheptadecanoin (margarin), trioctadecanoin (stearin), trioctadecenoin (vaccenin), triocadecadienoin (linolein) and trioctadecatrienoin (alpha-linolenin) (15:0, 16:0, 17:0, 18:0, 18:1, 18:2 and 18:3 TAG) were selected as standards and reproducibility of retention times and peak areas are compared in Table 2. Two different concentrations (1.0 and 5.0 μg) of each standard were injected 12 times. All of the retention times were averaged at 1.9 min. The percentage of the ratio of standard deviation and the average (%RSD) retention times were 0.4–1.1 in all standards at both concentrations. The averages of peak areas were 0.7–0.9 × 106 at 1.0 μg and 8.2–9.6 × 106 at 5.0 μg with outliers 7.5 × 106 and 4.8 × 106 as 17:0 and 18:0 TAG, respectively. The % RSD of peak areas at 1 and 5 μg were at 4.9–7.1 and 3.0–5.8 respectively that indicated the stability of TAG in this HPLC system (Table 2).
Comparison of Calibration Curves Measured by HPLC–ELSD and GC/MS
GC analyses in Chlamydomonas in addition to other algal species confirmed FAME 16:0 and 18:2 as primary fatty acids in lipid profiles [3, 10], therefore trihexadecanoin (16:0 TAG) and trioctadecenoin (18:2 TAG) were selected as authentic standards and their calibration curves are shown in Fig. 1. The concentration ranges of the calibration curves for both TAG by HPLC–ELSD were from 0.5 to 15 μg. Following the 15 μg peaks, saturation of the authentic standard created a plateau in the standard curve. The equations from the calibration curves were
| 1 |
for trihexadecanoin and
| 2 |
for trioctadienoin based on the peak area points at 1, 2.5, 5 and 10 μg by HPLC–ELSD. In contrast, both TAG were measured at the mass ranges from 5 to 250 ng by GC/MS. The calibration curves exhibited a linear trend at concentrations of 5, 10, 25, and 50 ng and presented a plateau fashion following 100 ng. The equations from the linear ranges of the calibration curves by GC/MS were
| 3 |
for trihexadecanoin and
| 4 |
for trioctadienoin. The equations show no significant difference between trihexadecanoin and trioctadienoin by both HPLC–ELSD (Eqs. 1, 2) and GC/MS (Eqs. 3, 4), however the detection level of 1–10 μg TAG by HPLC–ELSD is higher than that of 5–50 ng by GC/MS. This indicates the sensitivity of GC is 1,000 times higher than using HPLC–ELSD.
FAME Analysis by TLC/GC and Separation of Different Lipid Classes by HPLC–ELSD in C. reinhardtii
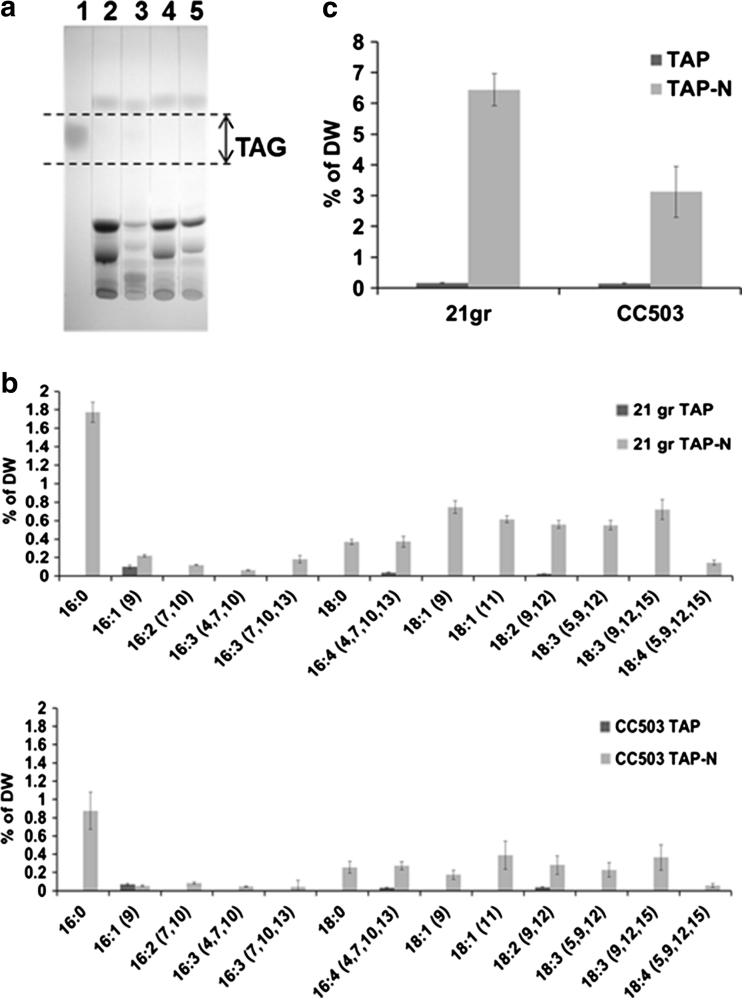
Two C. reinhardtii strains, 21 gr (wild type) and CC503 (wall-less), were grown in TAP and TAP media without a nitrogen source. TAG levels were compared by TLC/GC and HPLC–ELSD. TAG contents were first quantified by the conventional TLC/GC method. After lipid extraction, TAG in the crude lipid extract was separated by TLC and TAG on the TLC was scraped at the same position as the co-migrated soy TAG (Fig. 2a). The scraped TAG was eluted and trans-esterified to FAME and the FAME were analyzed as lipid compositions by GC/MS (Fig. 2b). The total lipid and TAG compositions of 21 gr and CC503 under nitrogen depletion were analyzed as the same types of compositions; 16:0 16:1–16:4 and 18:0–18:4 (carbon number: double bond number, the major fatty acid). The fatty acid profile shown in Fig. 2b is consistent with that reported previously in other C. reinhardtii strains [3, 10]. After FAME separation by GC/MS, the concentration of FAME was calculated based on the internal standard and total TAG contents were estimated as the sum of the FAME and expressed as the percentage of the DW (Fig. 2b, c). The TAG contents of 21 gr and CC503 under nitrogen deprivation were 6 and 3 % respectively but below 1 % in regular TAP media (Fig. 2c). The 3 % of CC503 TAG content under nitrogen depletion (biomass 5.28 mg/L) at day 7 and was lower than the 6 % TAG content of 21 gr (biomass 43.0 mg/L) due to biomass differences, which might indicate that the wall-less strain CC503 produces a lower oil production in comparison to the wild type 21 gr.
Fig. 2.
TAG analysis by TLC/GC method in C. reinhardtii 21 gr and CC503 under TAP and TAP-N conditions. a TAG separation by TLC. Lane 1, soy extraction stained by iodine stream; 2, 21 gr in TAP; 3, 21 gr on TAP-N; 4, CC503 in TAP; 5, CC503 in TAP-N. Arrow region shows the TAG part to be scraped. b TAG lipid compositions of 21 gr and CC503 under TAP and TAP-N conditions. c TAG contents of 21 gr and CC503 under TAP and TAP-N conditions quantified by GC/MS. Error bar n = 3 technical replicates from one 3-L bioreactor culture. Ave. of % DW ± standard deviation
The separation of TAG from carotenoids, DAG, phospholipids, galactolipids and phospholipid in C. reinhardtii 21 gr and CC503 under nitrogen depletion is shown in Fig. 3a. In the chromatograph of authentic standards and carotenoids extracted from C. reinhardtii, carotenoids, TAG, DAG and free fatty acids were eluted at the non-polar phase for 7 min, followed by a 1-min wash phase then PtdCho, MGDG and DGDG were eluted by isopropanol for another 7 min. The retention times of carotenoids (8), TAG (1), diacylglycerol ester (DAG ester, 2), DAG (3) and palmitic acid as free fatty acids (4) were eluted at 1.6, 1.9, 3.2, 4.1 and 5.5 min respectively and the retention times of phosphatidylcholine (PtdCho, 5), monogalactosyldiacylglycerol (MGDG, 6) and digalactosyldiacylglycerol (DGDG, 7) were at 12.0, 12.3 and 13.7 min respectively. Because the solvent system started from a non-polar solvent in this HPLC system and considering the solubility of the polar lipids in a non-polar solvent, detection of the phospholipid and glycolipid standards required 10 times higher concentrations than those of neutral lipids. Some phospholipids and glycolipids such as PE and DGTS could not be detected in this system. Sulfolipid SQDG has less than or similar solubility to DGTS and therefore also did not display a peak in this system [28, 29]. After the respective modification of [9, 30], detection of the different classes of lipids and separation of TAG were able to be improved in this system. The separation of lipids was characteristic in C. reinhardtii 21 gr and CC503 under nitrogen depletion. Considering the difference in total lipids between the gravimeter and FAME was more than 50 % in C. reinhardtii, the crude lipid extract was dissolved in non-polar hexane and filtered using a SPE column to remove the high amount of non-fatty acid types of lipids and served to protect the HPLC column. In both 21 gr and CC503 under nitrogen depletion, carotenoids as a minor lipid and TAG as a major lipid were detected at 1.6 and 1.9 min respectively. DAG ester, DAG and free fatty acids in the non-polar elution phase and PtdCho, MGDG and DGDG after isopropanol wash phase were not detected in this system (Fig. 3a). In contrast to the lipid profile by TLC (Fig. 2a), the profile by HPLC–ELSD was simpler (Fig. 3a). This is because the non-fatty acid types of lipids including glycolipids and phospholipids were removed by an SPE filter from the crude lipid extract and also the non-polar solvent was used for the running solvent in HPLC and polar lipid classes were less sensitive detection due to their reduced solubility compared to non-polar lipid classes, e.g. TAG, DAG and free fatty acids (1–4 μg) vs. PtdCho, MGDG and DGDG (20–40 μg). Traces of carotenoids and TAG were detected at 1.6 and 1.9 min respectively in both strains in TAP media. Considering 16:0 TAG demonstrated high reproducibility and a stable calibration curve (Table 2; Fig. 1) in combination with the major fatty acid in C. reinhardtii, palmitic acid (16:0), Eq. 1 from the calibration curves of trihexadecanoin (16:0 TAG) were applied to the TAG quantification of the samples from C. reinhardtii 21 gr and CC503 under nitrogen depletion. TAG content of 21 gr was 6 % and CC503 was 3 % under nitrogen depletion but below 1 % in TAP media. This indicates the values are as consistent as those estimated by the TLC/GC methods.
Comparison of Lipid Contents by TLC/GC Methods and HPLC–ELSD in Three Chlorella Strains Under BBM and BBM Without Nitrogen (BBM-N)
Using a gravimeter and other different quantification methods, total lipid content in C. sorokiniana was reported to be over a wide range from 3 to 52 %. Lipids can be extracted by various methods such as Bligh and Dyer [27], Soxhlet with chloroform/methanol [31], direct trans-methylation [12, 32] or using an automated extraction system [33]. After the extraction, extracted lipids were analyzed by either gravimeter, gas chromatography or spectrophotometer and then quantified by total FAME based on dry weight (%DW), relative percentages from total FAME estimation, lipid productivity (mg/L/day) and lipid per cell (g/cell) [18, 31–37] .
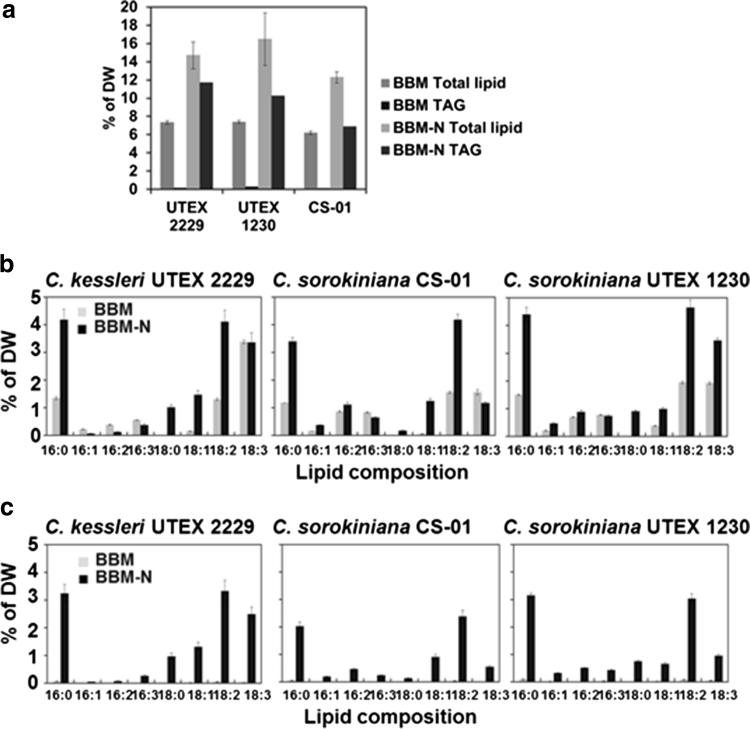
To obtain a rapid and accurate TAG content in Chlorella, the method using HPLC–ELSD was compared with the conventional TLC/GC method. Total lipid and TAG levels in C. kessleri UTEX 2229, C. sorokiniana CS-01 and UTEX 1230 were determined by the TLC/GC method under BBM and BBM-N growth conditions (Fig. 4). Figure. 4a shows that FAME total lipid content ranged from 12 to 17 % DW under BBM-N in all three Chlorella species, while 6–8 % DW under BBM. TAG accumulated 7–12 % DW under BBM-N, while below 0.3 % DW in BBM. Fatty acid composition of total lipids and TAG in the Chlorella species were compared in BBM and BBM-N and are shown in Fig. 4b, c. The fatty acids were detected at 16:0, 16:1, 16:2, 18:0, 18:1, 18:2 and 18:3 in both FAME total lipids and TAG, which are consistent with previous studies on the major fatty acids found in other Chlorella species such as C. zofingiensis [4], C. vulgaris [1, 38], C. sorokiniana [1, 18] and C. protothecoides [39].
Fig. 4.
Contents and compositions of FAME total lipids and FAME triacylglycerols (TAG) of dried Chlorella by GC/MS under BBM and BBM in absence of nitrogen source (BBM-N). a Contents of FAME total lipids and TAG in C. kessleri UTEX 2229, C. sorokiniana CS-01 and UTEX 1230. b Compositions of FAME total lipids in C. kessleri UTEX 2229, C. sorokiniana CS-01 and UTEX 1230. c FAME TAG compositions in C. kessleri UTEX 2229, C. sorokiniana CS-01 and UTEX 1230. Error bar n = 3 technical replicates from one 3 L bioreactor culture. Ave. of % DW ± standard deviation
The levels of each fatty acid differed under BBM and BBM-N. The ranges of FAME compositions in the total lipids were 0.2–4.5 % DW under BBM-N while 0.2–3 % under BBM in all Chlorella strains. The major fatty acids detected in the Chlorella strains were palmitic acid (16:0), linoleic acid (18:2) and linolenic acid (18:3). The respective ratio of fatty acids in C. kessleri UTEX 2229 measured 28, 28 and 23 % total lipids, C. sorokiniana CS-01 measured 28, 34 and 10 % total lipids and C. sorokiniana UTEX 1230 measured 27, 28 and 21 % total lipids under BBM-N conditions. The ratio of major fatty acids in C. kessleri UTEX 2229 measured 18, 18 and 46 % total lipids, C. sorokiniana CS-01 measured 19, 25 and 23 % total lipids and C. sorokiniana UTEX 1230 measured 20, 26 and 26 % total lipids under BBM. The levels of 16:0 and 18:2 of FAME total lipids under BBM-N in all Chlorella strains increased 2–3 times higher than those of BBM and the content of 18:3 of FAME total lipids under BBM-N in C. sorokiniana UTEX 1230 increased almost two times more than that in BBM condition. This might indicate the involvement of high palmitic acid production in plastid and also higher activity of fatty acid desaturase in the endoplasmic reticulum or plastid observed in plants [40, 41]. Figure 4c shows the fatty acid composition of TAG by the TLC/GC method. The compositions of all Chlorella strains under BBM-N showed the same pattern as the total lipid compositions and the TAG content was within the range of 0.01–4 % DW.
The separation of lipids was characteristic in the three Chlorella strains under BBM and BBM-N by HPLC–ELSD (Fig. 5a). Under BBM conditions TAG and a trace of carotenoids were detected at 1.9 and 1.7 min respectively in the non-polar elution phase while MGDG and traces of DGDG were measured at 13 and 13.8 min respectively after the isopropanol wash phase in all Chlorella strains. Under BBM-N conditions, the dominant TAG peak was detected at 1.9 min and traces of unknown peaks were detected, which might indicate that the flow toward the TAG biosynthesis pathway is dominant under BBM-N condition. The equations from the calibration curves of trihexadecanoin (16:0 TAG, Eq. 1) were applied to the TAG quantification of the samples from C. kessleri UTEX 2229, C. sorokiniana CS-01 and UTEX 1230 under BBM and BBM-N. The levels of Chlorella TAG detected by HPLC–ELSD under BBM and BBM-N are shown in Fig. 5b. The TAG contents in C. kessleri UTEX 2229, C. sorokiniana CS-01 and UTEX 1230 were 12, 7 and 10 % DW, respectively under BBM-N, while all the Chlorella strains under BBM was under 0.2 % DW.
FAME Analysis by TLC/GC and Separation of Different Lipid Classes by HPLC–ELSD in Chlorella Strains
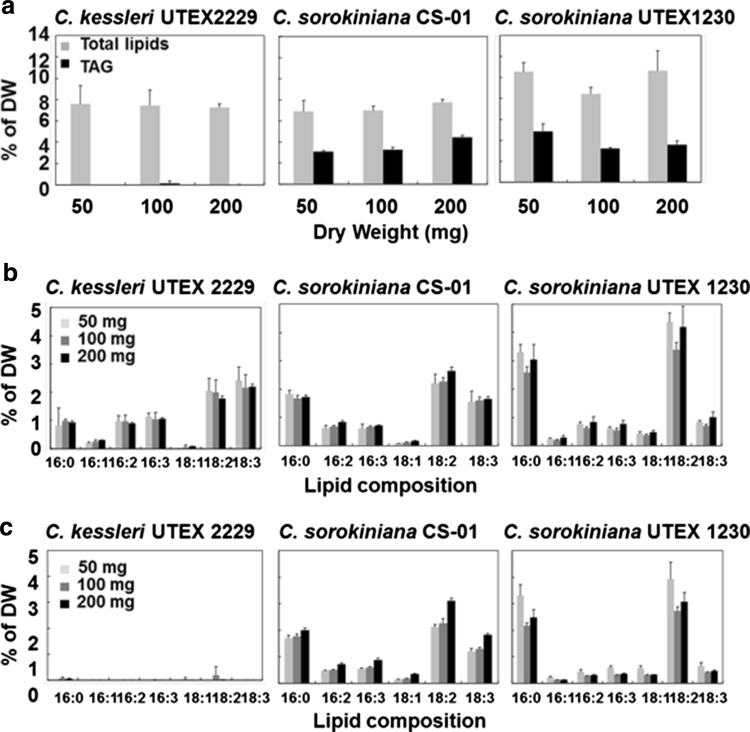
To assess accuracy of lipid analysis using the HPLC–ELSD method, the lipid contents were compared not only under nitrogen deprivation conditions but also in different amounts of DW. The comparison of the lipid analysis by the TLC/GC and HPLC–ELSD methods were performed with C. kessleri UTEX 2229, C. sorokiniana CS-01 and UTEX 1230. Total lipid and TAG levels in the three Chlorella strains were first determined by the TLC/GC method (Fig. 6). Figure 6a shows the range of FAME total lipid content from 7 to 11 % DW in all three Chlorella strains and TAG accumulation from 4 to 6 % DW in C. sorokiniana CS-01 and UTEX 1230 under late stationary phase. In contrast, TAG accumulation in C. kessleri UTEX 2229 was hardly detectable. The fatty acid composition of the total lipids and TAG in C. kessleri UTEX 2229, C. sorokiniana CS-01 and UTEX 1230 are shown in Fig. 6b, c. The fatty acids detected were 16:0, 16:1, 16:2, 18:0, 18:1, 18:2 and 18:3 both in the FAME total lipids and TAG. The levels of each fatty acid differed in the three Chlorella strains. The ranges of FAME compositions in total lipids were 0.2–3 % DW in both C. kessleri UTEX 2229 and C. sorokiniana CS-01 while slightly higher in C. sorokiniana UTEX 1230, 0.3–5 % DW. The major fatty acids detected in the Chlorella strains were palmitic acid (16:0), linoleic acid (18:2) and linolenic acid (18:3). Respective major fatty acids in C. kessleri UTEX 2229 were measured at 12, 26 and 30 % total lipids, C. sorokiniana CS-01 measured 24, 33 and 22 % total lipids, and C. sorokiniana UTEX 1230 measured 30, 40 and 9 % total lipids. Figure 6c shows the fatty acid composition and content of purified TAG after separation by TLC and analysis and quantification by GC analyses. The compositions of C. sorokiniana CS-01 and UTEX 1230 showed the same pattern as the total lipid compositions and the TAG content was within the range of 0.3–4 % DW, however in C. kessleri UTEX 2229 TAG was nearly undetectable. Although total lipids in C. kessleri UTEX 2229 accumulated to the same level as C. sorokiniana CS-01 and UTEX 1230, it is unclear whether the failure to detect TAG in C. kessleri UTEX 2229 was due to the absence of TAG synthesis and accumulation of TAG or rapid degradation of TAG. While the DW of Chlorella species differed from 50, 100 and 200 mg, the percentages of FAME total lipids and TAG per DW, excluding TAG of C. kessleri UTEX 2229, demonstrated consistency at 8–12 and 3–5 %, respectively using this method (Fig. 6a–c).
Fig. 6.
Contents and compositions of FAME total lipids and FAME triacylglycerols (TAG) at 50, 100 and 200 mg of dried Chlorella by GC/MS at the late stationary phase. a Contents of FAME total lipids and TAG in C. kessleri UTEX 2229, C. sorokiniana CS-01 and UTEX 1230. b Compositions of FAME total lipids in C. kessleri UTEX 2229, C. sorokiniana CS-01 and UTEX 1230. c FAME TAG compositions in C. kessleri UTEX 2229, C. sorokiniana CS-01 and UTEX 1230. Error bar n = 3 technical replicates from one 80 L aquarium culture. Ave. of % DW ± standard deviation
The separation of lipids was characteristic in Chlorella by HPLC–ELSD (Fig. 7a). In C. kessleri UTEX 2229 TAG, DAG and free fatty acids were detected after 1.8, 3.7 and 5.1 min, respectively in the non-polar elution phase while PtdCho, MGDG and DGDG were measured after 11.9, 12.4 and 13.2 min, respectively after the isopropanol wash phase. The CS-01 TAG peak was detected at 1.8 min and MGDG was detected at 12.3 min. In C. sorokiniana UTEX 1230 TAG and DAG were detected at 1.8 and 2.8 min, respectively with other unknown trace peaks. The equation from the calibration curves for trihexadecanoin (Eq. 1) was applied to the TAG quantification of the samples from C. kessleri UTEX 2229, C. sorokiniana CS-01 and UTEX 1230. The levels of Chlorella TAG detected by HPLC–ELSD are shown in Fig. 7b. The levels of TAG in C. kessleri UTEX 2229 were <2.5 % DW which is below the detection limit by GC/MS analysis. In C. sorokiniana CS-01 and UTEX 1230 levels of TAG were 5 % DW and were consistent between different amounts of DW (50, 100 and 200 mg).
The HPLC–ELSD system was able to detect TAG at a level which was below detection by GC analysis and also allowed us to separate not only TAG but also other lipids such as DAG, free fatty acids, PtdCho, MGDG and DGDG in a single 15 min HPLC run. Although the levels of total lipids in C. kessleri UTEX 2229 by GC/MS analysis were at the same levels as those of C. sorokiniana CS-01 and UTEX 1230, only the levels of TAG in C. kessleri UTEX 2229 were below detection (Fig. 6a). However, application of HPLC–ELSD to lipids of C. kessleri UTEX 2229 showed lower levels of TAG while a higher peak of DAG and free fatty acids in the same trail run (Fig. 7a). A previous study examining the lipid profile of C. minutissima grown under nitrogen starvation reported TAG and monoglyceride were the major lipid classes, and free fatty acid and DAG were reduced as assessed using HPLC–ELSD [23]. This might indicate the degradation of TAG or less activity of diacylglycerol acyl transferase that converts DAG to TAG [42, 43].
Comparison of Total Lipid Content by Gravimeter and FAME Total Lipid and TAG Contents by TLC/GC and HPLC–ELSD Methods
Total lipid contents of the wild type strain 21 gr by gravimeter were 24 and 23 % and those of CC503 were 21 and 18 % of DW, while FAME total lipid contents of 21 gr were 10 and 14 % and those of CC503 were 7 and 8 % DW under TAP and TAP without nitrogen conditions, respectively (Table 3). This difference between total lipids by gravimeter and FAME total lipids indicates that the non-fatty acid type of lipids such as chlorophyll, carotenoids, tocopherols and sterols might represent 50 % of the total lipids in C. reinhardtii 21 gr and CC503. The FAME TAG contents of 21 gr and CC503 were measured at <1 % in TAP media and 6 % (wild type 21 gr) and 3 % DW (CC503) under nitrogen depletion by both TLC/GC and HPLC–ELSD methods. Under conditions of nitrogen depletion, TAG accumulated in C. reinhardtii 21 gr and CC503, and that the prominent TAG peak that is used to quantitate TAG accumulation by HPLC–ELSD represents primarily TAG and is not a mixture of other cell lipids (for example carotenoids, which would result in an overestimation of TAG content by HPLC–ELSD if co-eluting with the TAG peak).
Table 3.
Comparison of total lipids, FAME total lipids and TAG quantification by gravimeter, HPLC–ELSD and GC/MS in C. reinhardtii
| C. reinhardtii | Gravimeter | GC/MS | HPLC–ELSD | |
|---|---|---|---|---|
| Total lipids | FAME total lipids | FAME TAG | TAG | |
| 21 gr | ||||
| TAP | 23.67 ± 0.37 | 9.96 ± 0.95 | 0.17 ± 0.02 | 0.11 ± 0.01 |
| TAP-N | 23.12 ± 0.38 | 13.88 ± 0.88 | 6.44a ± 0.53 | 6.44b ± 0.34 |
| CC503 | ||||
| TAP | 20.95 ± 1.21 | 7.42 ± 0.14 | 0.14 ± 0.02 | 0.10 ± 0.02 |
| TAP-N | 17.63 ± 1.10 | 7.85 ± 0.27 | 3.12c ± 0.08 | 3.15d ± 0.18 |
Error bar: n = 3 technical replicates from one 3 L bioreactor culture. Ave. of % DW ± standard deviation
a 54.59 (sum of peak area)/3.89 (15:0 peak area) × 500 μg (internal standard)/1,000 = 7.02 mg, 7.02 mg × 100/109 (mg DW) = 6.44 %
b {4.38 (peak area) + 1.67/1.94} × 2,250 (dilution factor)/1,000 = 7.02 mg, 7.02 mg × 100/109 (mg DW) = 6.44 %
c 24.03 (sum of peak area)/3.57 (15:0 peak area) × 500 μg (internal standard)/1,000 = 3.38 mg, 3.38 mg × 100/108 (mg DW) = 3.12 %
d {1.26 (peak area) + 1.67/1.94} × 2,250 (dilution factor)/1,000 = 3.40 mg, 3.40 mg × 100/108 (mg DW) = 3.15 %
The quantification of total lipids and TAG contents by gravimeter, GC/MS and HPLC–ELSD in three Chlorella strains under BBM and BBM-N are shown in Table 4. Total lipid content under BBM-N measured by gravimeter ranged from 17 to 26 % DW while only 16–19 % DW under BBM in all the Chlorella strains. The FAME total lipid content determined by GC/MS was 12–16 % DW under BBM-N, while 6–7 % DW under BBM. The levels of TAG measured by GC/MS with TLC or HPLC–ELSD were similar at 12, 7 and 10 % DW in C. kessleri UTEX 2229, C. sorokiniana CS-01 and UTEX 1230 respectively, however under BBM, TAG levels ranged from 0.2 to 0.3 % DW in all the Chlorella strains which further indicates the consistency of the TAG content measured by the TLC/GC and HPLC–ELSD methods. In comparison to BBM, BBM-N conditions accumulated twice the contents of FAME total lipids and 2–3 times higher TAG levels in the late stationary phase in C. sorokiniana (Figs. 4a vs. 6a; Tables 4 vs. 5). Additionally, Chlorella in BBM-N accumulated 2–4 times higher than TAG contents in C. reinhardtii under nitrogen deprivation (Figs. 2c vs. 4a; Tables 3 vs. 4).
Table 4.
Comparison of TAG quantification by HPLC–ELSD and GC/MS under BBM and BBM-N in Chlorella
| Chlorella | Weight | GC/MS | HPLC–ELSD–LT | |
|---|---|---|---|---|
| Total lipids by a gravimeter | FAME total lipids | FAME TAG | TAG | |
| C. kessleri UTEX 2229 | ||||
| BBM | 18.57 ± 0.35 | 7.35 ± 0.18 | 0.15 ± 0.02 | 0.17 ± 0.04 |
| BBM-N | 26.27 ± 2.02 | 14.70 ± 1.47 | 11.71 ± 1.30 | 11.77 ± 196 |
| C. sorokiniana CS-01 | ||||
| BBM | 15.53 ± 0.41 | 6.20 ± 0.17 | 0.12 ± 0.04 | 0.19 ± 0.01 |
| BBM-N | 16.53 ± 1.04 | 12.28 ± 0.63 | 6.89 ± 0.62 | 7.06 ± 1.04 |
| C. sorokiniana UTEX 1230 | ||||
| BBM | 18.78 ± 0.58 | 7.40 ± 0.16 | 0.28 ± 0.07 | 0.28 ± 0.08 |
| BBM-N | 23.72 ± 0.67 | 16.48 ± 2.89 | 10.24 ± 0.54 | 9.65 ± 1.53 |
Error bar: n = 3 technical replicates from one 3-L bioreactor culture). Ave. of % DW ± standard deviation
ND not detected. Samples were measured as % DW
Table 5.
Comparison of TAG quantification by HPLC–ELSD and GC/MS in Chlorella
| Chlorella | Weight | GC/MS | HPLC–ELSD–LT | |
|---|---|---|---|---|
| Total lipids by a gravimeter | FAME total lipids | FAME TAG | TAG | |
| C. kessleri UTEX 2229 | ||||
| 50 mg | 18.73 ± 1.10 | 7.60 ± 1.72 | ND | 2.25 ± 0.19 |
| 100 mg | 17.16 ± 0.54 | 7.47 ± 1.44 | 0.15 ± 0.22 | 1.66 ± 0.07 |
| 200 mg | 17.87 ± 0.38 | 7.25 ± 0.32 | 0.04 ± 0.01 | 1.18 ± 0.06 |
| C. sorokiniana CS-01 | ||||
| 50 mg | 13.71 ± 1.32 | 6.89 ± 1.06 | 3.08 ± 0.13 | 4.13 ± 0.43 |
| 100 mg | 14.29 ± 1.01 | 7.00 ± 0.20 | 3.29 ± 0.12 | 3.82 ± 0.38 |
| 200 mg | 13.46 ± 1.62 | 7.76 ± 0.28 | 4.44 ± 0.19 | 3.62 ± 0.29 |
| C. sorokiniana UTEX 1230 | ||||
| 50 mg | 18.88 ± 0.90 | 10.56 ± 0.88 | 4.85 ± 0.73 | 4.46 ± 0.42 |
| 100 mg | 14.05 ± 0.87 | 8.42 ± 0.67 | 3.20 ± 0.12 | 4.25 ± 0.41 |
| 200 mg | 14.11 ± 1.09 | 10.66 ± 0.88 | 3.58 ± 0.36 | 4.14 ± 0.19 |
Error bar: n = 3 technical replicates from one 80 L aquarium culture). Ave. of % DW ± standard deviation)
ND not detected. Samples were measured as % DW
Table 5 shows the comparison of quantification of total lipids and TAG by gravimeter, GC/MS and HPLC–ELSD in the three Chlorella strains. Total lipid content measured by gravimeter ranged from 14 to 19 % DW, while FAME total lipid content determined by GC/MS was 7–11 % DW. The levels of TAG measured by GC/MS with TLC or HPLC–ELSD were similar, ranging from 3 to 5 % DW in C. sorokiniana CS-01 and UTEX 1230 which indicated the measurement of TAG was consistent between the different spectrometric methods. The failure of TAG detection under 1 % DW by GC/MS in C. kessleri UTEX 2229 may be contributed to the limited detection by the GC/MS and TLC method in Chlorella while the profile of lipid classes can be detected at 1–2 % DW of TAG by the HPLC–ELSD methods. Furthermore C. sorokiniana UTEX 1230 and CS-01 provided stable TAG contents in the large-scale 80 L aquarium culture but not C. kessleri UTEX 2229. Therefore C. sorokiniana UTEX 1230 and CS-01 might be better strains than C. kessleri UTEX 2229 for biofuel application.
Comparison of the Time Courses of the TLC/GC and HPLC–ELSD Methods
TAG has been widely separated and quantified by the TLC and GC methods [3, 5, 44] and studied recently by using ESI–MS [45, 46] and LC/MS [47]. The conventional TLC/GC method was compared to the HPLC–ELSD method for TAG quantitation. Sample preparation using the TLC/GC method requires on average 4.8 h compared to 1.3 h using the HPLC–ELSD method. TAG amounts determined by HPLC–ELSD were comparable to those obtained by TLC/GC, provided that sufficient starting material was used so that the TAG measured by HPLC–ELSD is in the linear range of detection. After lipid extraction, HPLC–ELSD is a one-step procedure to detect and quantify the level of TAG and therefore efficient for high-throughput screening for lipid content in various and abundant algae under different conditions. Furthermore as the HPLC–ELSD method was established for separation and quantification of algal TAG which is chemically similar to most conventional fuels, it will take a great advantage for biofuel research and lipid analysis. For purposes of screening candidate algae and for determining optimal growth conditions for high oil yields, HPLC–ELSD provides a rapid and simple method for monitoring patterns of lipid accumulation in algae both of laboratory strains such as Chlamydomonas and industrially relevant strains such as Chlorella.
Acknowledgments
The authors thank Dr. K. Horken and Dr. D. Weeks for providing the C. reinhardtii cultures, Dr. M Wan at Johns Hopkins University for providing C. sorokiniana CS-01, Cameron Murphy for technical assistance of HPLC–ELSD, Anji Reddy Konda for introducing the Bligh and Dyer method and TLC, and R. Cahoon and Dr. E. Cahoon for their critical revision of the manuscript. This work was supported by Department of Energy renewable biofuels.
Abbreviations
- Ave.
Average
- Conc.
Concentration
- DAG
Diacylglycerol(s)
- DGDG
Digalactosyldiacylglycerol
- DW
DW
- EI
Electron ionization
- ELSD
Evaporative light scattering detector
- ESI
Electrospray ionization
- FAME
Fatty acid methyl ester(s)
- HPLC
High performance liquid chromatography
- GC
Gas chromatography
- LC
Liquid chromatography
- MGDG
Monogalactosyldiacylglycerol
- MS
Mass spectrum
- PtdCho
Phosphatidylcholine
- RSD
Ratio of standard deviation
- SD
Standard deviation
- TAP
Tris acetate phosphate
- TAG
Triacylglycerol(s)
- TIC
Total ion count
- TLC
Thin layer chromatography
References
- 1.Hu Q, Sommerfeld M, Jarvis E, Ghirardi M, Posewitz M, Seibert M, Darzins A. Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J. 2008;54(4):621–639. doi: 10.1111/j.1365-313X.2008.03492.x. [DOI] [PubMed] [Google Scholar]
- 2.Durrett TP, Benning C, Ohlrogge J. Plant triacylglycerols as feedstocks for the production of biofuels. Plant J. 2008;54(4):593–607. doi: 10.1111/j.1365-313X.2008.03442.x. [DOI] [PubMed] [Google Scholar]
- 3.Msanne J, Xu D, Konda AR, Casas-Mollano JA, Awada T, Cahoon EB, Cerutti H. Metabolic and gene expression changes triggered by nitrogen deprivation in the photoautotrophically grown microalgae Chlamydomonas reinhardtii and Coccomyxa sp. C-169. Phytochemistry. 2012;75:50–59. doi: 10.1016/j.phytochem.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, Huang J, Sun Z, Zhong Y, Jiang Y, Chen F. Differential lipid and fatty acid profiles of photoautotrophic and heterotrophic Chlorella zofingiensis: assessment of algal oils for biodiesel production. Bioresour Technol. 2011;102(1):106–110. doi: 10.1016/j.biortech.2010.06.017. [DOI] [PubMed] [Google Scholar]
- 5.Moellering ER, Benning C. RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot Cell. 2010;9(1):97–106. doi: 10.1128/EC.00203-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramadan MF, Morsel JT. Determination of the lipid classes and fatty acid profile of Niger (Guizotia abyssinica Cass.) seed oil. Phytochem Anal. 2003;14(6):366–370. doi: 10.1002/pca.729. [DOI] [PubMed] [Google Scholar]
- 7.Amaral JS, Cunha SC, Alves MR, Pereira JA, Seabra RM, Oliveira BP. Triacylglycerol composition of walnut (Juglans regia L.) cultivars: characterization by HPLC–ELSD and chemometrics. J Agric Food Chem. 2004;52(26):7964–7969. doi: 10.1021/jf048918n. [DOI] [PubMed] [Google Scholar]
- 8.Yunoki K, Sato M, Seki K, Ohkubo T, Tanaka Y, Ohnishi M. Simultaneous quantification of plant glyceroglycolipids including sulfoquinovosyldiacylglycerol by HPLC–ELSD with binary gradient elution. Lipids. 2009;44(1):77–83. doi: 10.1007/s11745-008-3248-4. [DOI] [PubMed] [Google Scholar]
- 9.Graeve M, Janssen D. Improved separation and quantification of neutral and polar lipid classes by HPLC–ELSD using a monolithic silica phase: application to exceptional marine lipids. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877(20–21):1815–1819. doi: 10.1016/j.jchromb.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Siaut M, Cuine S, Cagnon C, Fessler B, Nguyen M, Carrier P, Beyly A, Beisson F, Triantaphylides C, Li-Beisson Y, et al. Oil accumulation in the model green alga Chlamydomonas reinhardtii: characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol. 2011;11:7. doi: 10.1186/1472-6750-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bumbak F, Cook S, Zachleder V, Hauser S, Kovar K. Best practices in heterotrophic high-cell-density microalgal processes: achievements, potential and possible limitations. Appl Microbiol Biotechnol. 2011;91(1):31–46. doi: 10.1007/s00253-011-3311-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodolfi L, Chini Zittelli G, Bassi N, Padovani G, Biondi N, Bonini G, Tredici MR. Microalgae for oil: strain selection, induction of lipid synthesis and outdoor mass cultivation in a low-cost photobioreactor. Biotechnol Bioeng. 2009;102(1):100–112. doi: 10.1002/bit.22033. [DOI] [PubMed] [Google Scholar]
- 13.Schenk PM, Thomas-Hall SR, Stephens E, Marx UC, Mussgnug JH, Posten C, Kruse O, Hankamer B. Second generation biofuel: high-efficiency microalgae for biodiesel production. Bioenerg Res. 2008;1:20–43. doi: 10.1007/s12155-008-9008-8. [DOI] [Google Scholar]
- 14.Heredia-Arroyo T, Wei W, Hu B. Oil accumulation via heterotrophic/mixotrophic Chlorella protothecoides. Appl Biochem Biotechnol. 2010;162(7):1978–1995. doi: 10.1007/s12010-010-8974-4. [DOI] [PubMed] [Google Scholar]
- 15.Feng P, Deng Z, Fan L, Hu Z. Lipid accumulation and growth characteristics of Chlorella zofingiensis under different nitrate and phosphate concentrations. J Biosci Bioeng. 2012;114(4):405–410. doi: 10.1016/j.jbiosc.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 16.Gao C, Xiong W, Zhang Y, Yuan W, Wu Q. Rapid quantitation of lipid in microalgae by time-domain nuclear magnetic resonance. J Microbiol Methods. 2008;75(3):437–440. doi: 10.1016/j.mimet.2008.07.019. [DOI] [PubMed] [Google Scholar]
- 17.Liu ZY, Wang GC, Zhou BC. Effect of iron on growth and lipid accumulation in Chlorella vulgaris. Bioresour Technol. 2008;99(11):4717–4722. doi: 10.1016/j.biortech.2007.09.073. [DOI] [PubMed] [Google Scholar]
- 18.Vigeolas H, Duby F, Kaymak E, Niessen G, Motte P, Franck F, Remacle C. Isolation and partial characterization of mutants with elevated lipid content in Chlorella sorokiniana and Scenedesmus obliquus. J Biotechnol. 2012;162(1):3–12. doi: 10.1016/j.jbiotec.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 19.Breuer G, Lamers PP, Martens DE, Draaisma RB, Wijffels RH. The impact of nitrogen starvation on the dynamics of triacylglycerol accumulation in nine microalgae strains. Bioresour Technol. 2012;124C:217–226. doi: 10.1016/j.biortech.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Pruvost J, Van Vooren G, Le Gouic B, Couzinet-Mossion A, Legrand J. Systematic investigation of biomass and lipid productivity by microalgae in photobioreactors for biodiesel application. Bioresour Technol. 2011;102(1):150–158. doi: 10.1016/j.biortech.2010.06.153. [DOI] [PubMed] [Google Scholar]
- 21.Huang YY, Beal CM, Cai WW, Ruoff RS, Terentjev EM. Micro-Raman spectroscopy of algae: composition analysis and fluorescence background behavior. Biotechnol Bioeng. 2009;105(5):889–898. doi: 10.1002/bit.22617. [DOI] [PubMed] [Google Scholar]
- 22.Na JG, Lee HS, Oh YK, Park JY, Ko CH, Lee SH, Yi KB, Chung SH, Jeon SG. Rapid estimation of triacylglycerol content of Chlorella sp. by thermogravimetric analysis. Biotechnol Lett. 2011;33(5):957–960. doi: 10.1007/s10529-011-0522-y. [DOI] [PubMed] [Google Scholar]
- 23. Tang H, Chen M, Garcia ME, Abunasser N, Ng KY, Salley SO (2011) Culture of microalgae Chlorella minutissima for biodiesel feedstock production. Biotechnol Bioeng 108(10):2280–2287 [DOI] [PubMed]
- 24.Blanc G, Duncan G, Agarkova I, Borodovsky M, Gurnon J, Kuo A, Lindquist E, Lucas S, Pangilinan J, Polle J, et al. The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell. 2010;22(9):2943–2955. doi: 10.1105/tpc.110.076406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris EH, editor. The Chlamydomonas sourcebook. A comprehensive guide to biology and laboratory use. San Diego: Academic Press; 1989. [DOI] [PubMed] [Google Scholar]
- 26.Bold HC. The morphology of Chlamydomonas chlamydogama sp. nov. Bull Torrey Bot Club. 1949;76:101–108. doi: 10.2307/2482218. [DOI] [Google Scholar]
- 27.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37(8):911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 28.Riekhof WR, Ruckle ME, Lydic TA, Sears BB, Benning C. The sulfolipids 2′-O-acyl-sulfoquinovosyldiacylglycerol and sulfoquinovosyldiacylglycerol are absent from a Chlamydomonas reinhardtii mutant deleted in SQD1. Plant Physiol. 2003;133(2):864–874. doi: 10.1104/pp.103.029249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riekhof WR, Sears BB, Benning C. Annotation of genes involved in glycerolipid biosynthesis in Chlamydomonas reinhardtii: discovery of the betaine lipid synthase BTA1Cr. Eukaryot Cell. 2005;4(2):242–252. doi: 10.1128/EC.4.2.242-252.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy EJ, Rosenberger TA, Horrocks LA. Separation of neutral lipids by high-performance liquid chromatography: quantification by ultraviolet, light scattering and fluorescence detection. J Chromatogr B. 1996;685:9–14. doi: 10.1016/0378-4347(96)00138-7. [DOI] [PubMed] [Google Scholar]
- 31.Maccarthy JJ, Patterson GW. Effects of cation levels of the nutrient medium on the biochemistry of Chlorella: I. concentration series. Plant Physiol. 1974;54(2):129–132. doi: 10.1104/pp.54.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cha TS, Chen JW, Goh EG, Aziz A, Loh SH. Differential regulation of fatty acid biosynthesis in two Chlorella species in response to nitrate treatments and the potential of binary blending microalgae oils for biodiesel application. Bioresour Technol. 2011;102(22):10633–10640. doi: 10.1016/j.biortech.2011.09.042. [DOI] [PubMed] [Google Scholar]
- 33.Singh M, Reynolds DL, Das KC. Microalgal system for treatment of effluent from poultry litter anaerobic digestion. Bioresour Technol. 2011;102(23):10841–10848. doi: 10.1016/j.biortech.2011.09.037. [DOI] [PubMed] [Google Scholar]
- 34.Zhou W, Li Y, Min M, Hu B, Chen P, Ruan R. Local bioprospecting for high-lipid producing microalgal strains to be grown on concentrated municipal wastewater for biofuel production. Bioresour Technol. 2011;102(13):6909–6919. doi: 10.1016/j.biortech.2011.04.038. [DOI] [PubMed] [Google Scholar]
- 35.Lu S, Wang J, Niu Y, Yang J, Zhou J, Yuan Y. Metabolic profiling reveals growth related FAME productivity and quality of Chlorella sorokiniana with different inoculum sizes. Biotechnol Bioeng. 2012;109(7):1651–1662. doi: 10.1002/bit.24447. [DOI] [PubMed] [Google Scholar]
- 36.Miao C, Chakraborty M, Chen S. Impact of reaction conditions on the simultaneous production of polysaccharides and bio-oil from heterotrophically grown Chlorella sorokiniana by a unique sequential hydrothermal liquefaction process. Bioresour Technol. 2012;110:617–627. doi: 10.1016/j.biortech.2012.01.047. [DOI] [PubMed] [Google Scholar]
- 37.Ratha SK, Babu S, Renuka N, Prasanna R, Prasad RB, Saxena AK. Exploring nutritional modes of cultivation for enhancing lipid accumulation in microalgae. J Basic Microbiol. 2012;52:1–10. doi: 10.1002/jobm.201290001. [DOI] [PubMed] [Google Scholar]
- 38.Gouveia L, Oliveira AC. Microalgae as a raw material for biofuels production. J Ind Microbiol Biotechnol. 2009;36(2):269–274. doi: 10.1007/s10295-008-0495-6. [DOI] [PubMed] [Google Scholar]
- 39.Santos CA, Ferreira ME, da Silva TL, Gouveia L, Novais JM, Reis A. A symbiotic gas exchange between bioreactors enhances microalgal biomass and lipid productivities: taking advantage of complementary nutritional modes. J Ind Microbiol Biotechnol. 2011;38(8):909–917. doi: 10.1007/s10295-010-0860-0. [DOI] [PubMed] [Google Scholar]
- 40.Chapman KD, Ohlrogge JB. Compartmentation of triacylglycerol accumulation in plants. J Biol Chem. 2012;287(4):2288–2294. doi: 10.1074/jbc.R111.290072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wallis JG, Browse J. Mutants of Arabidopsis reveal many roles for membrane lipids. Prog Lipid Res. 2002;41(3):254–278. doi: 10.1016/S0163-7827(01)00027-3. [DOI] [PubMed] [Google Scholar]
- 42.Wagner M, Hoppe K, Czabany T, Heilmann M, Daum G, Feussner I, Fulda M. Identification and characterization of an acyl-CoA: diacylglycerol acyltransferase 2 (DGAT2) gene from the microalga O. tauri. Plant Physiol Biochem. 2010;48(6):407–416. doi: 10.1016/j.plaphy.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 43.Khozin-Goldenbeg I, Cohen Z. Unraveling algal lipid metabolism: recent advance in gene identification. Biochimie. 2011;93:91–100. doi: 10.1016/j.biochi.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 44.Zhang M, Fan J, Taylor DC, Ohlrogge JB. DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell. 2009;21(12):3885–3901. doi: 10.1105/tpc.109.071795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bates PD, Durrett TP, Ohlrogge JB, Pollard M. Analysis of acyl fluxes through multiple pathways of triacylglycerol synthesis in developing soybean embryos. Plant Physiol. 2009;150(1):55–72. doi: 10.1104/pp.109.137737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Durrett TP, McClosky DD, Tumaney AW, Elzinga DA, Ohlrogge J, Pollard M. A distinct DGAT with sn-3 acetyltransferase activity that synthesizes unusual, reduced-viscosity oils in Euonymus and transgenic seeds. Proc Natl Acad Sci USA. 2010;107(20):9464–9469. doi: 10.1073/pnas.1001707107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Danielewicz MA, Anderson LA, Franz AK. Triacylglycerol profiling of marine microalgae by mass spectrometry. J Lipid Res. 2011;52(11):2101–2108. doi: 10.1194/jlr.D018408. [DOI] [PMC free article] [PubMed] [Google Scholar]