Abstract
Development of non-viral particles for gene delivery requires a greater understanding of the properties that enable gene delivery particles to overcome the numerous barriers to intracellular DNA delivery. Linear poly(beta-amino) esters (PBAE) have shown substantial promise for gene delivery, but the mechanism behind their effectiveness is not well quantified with respect to these barriers. In this study, we synthesized, characterized, and evaluated for gene delivery an array of linear PBAEs that differed by small changes along the backbone, side chain, and end-group of the polymers. We examined particle size and surface charge, polymer molecular weight, polymer degradation rate, buffering capacity, cellular uptake, transfection, and cytotoxicity of nanoparticles formulated with these polymers. Significantly, this is the first study that has quantified how small differential structural changes to polymers of this class modulate buffering capacity and polymer degradation rate and relates these findings to gene delivery efficacy. All polymers formed positively charged (zeta potential 21–29 mV) nanosized articles (~ 150 nm). The polymers hydrolytically degraded quickly in physiological conditions, with half-lives ranging from 90 minutes to 6 hours depending on polymer structure. The PBAE buffering capacities in the relevant pH range (pH 5.1 – 7.4) varied from 34% to 95% protonable amines, and on a per mass basis, PBAEs buffered 1.4–4.6 mmol H+/g. When compared to 25 kDa branched polyethyleneimine (PEI), PBAEs buffer significantly fewer protons/mass, as PEI buffers 6.2 mmol H+/g over the same range. However, due to the relatively low cytotoxicity of PBAEs, higher polymer mass can be used to form particles than with PEI and total buffering capacity of PBAE-based particles significantly exceeds that of PEI. Uptake into COS-7 cells ranged from 0% to 95% of cells and transfection ranged from 0% to 93% of cells, depending on the base polymer structure and the end-modifications examined. Five polymers achieved higher uptake and transfection efficacy with less toxicity than branched-PEI control. Surprisingly, acrylate-terminated base polymers were dramatically less efficacious than their end-capped versions, both in terms of uptake (1–3% for acrylate, 75–94% for end-capped) and transfection efficacy (0–1% vs. 20–89%), even though there are minimal differences between acrylate and end-capped polymers in terms of DNA retardation in gel electrophoresis, particle size, zeta potential, and cytotoxicity. These studies further elucidate the role of polymer structure for gene delivery and highlight that small molecule end-group modification of a linear polymer can be critical for cellular uptake in a manner that is largely independent of polymer/DNA binding, particle size, and particle surface charge.
Keywords: Gene delivery, Poly(β-amino) ester, Buffering capacity, Polymer degradation
Introduction
Gene therapy is the treatment of disease through insertion or modification of DNA in cells. This treatment has tremendous implications for improving human health because almost all human diseases have a genetic component, including cancer. The fundamental challenge for successful gene therapy is finding both a safe and effective delivery system.1 The traditional method for gene therapy has been viral gene delivery. Viruses have evolved to transduce cells with high efficacy but are limited by low cargo capacity, resistance to repeated infection, difficulty in production and quality control, and safety concerns.2
All of these challenges can be overcome with non-viral methods that utilize biomaterials, which can be designed to deliver genes similar to a synthetic virus. Biodegradable cationic polymers such as poly(ester amines) and poly(amido amines) are promising for non-viral gene delivery due to their ability to condense plasmid DNA into small and stable nanoparticles, their ability to promote cellular uptake, facilitate escape from the endosome, and allow for DNA release in the cytoplasm.3 Studying these properties is integral to understanding how to design biomaterials for optimal transfection efficacy. In order to deliver its plasmid cargo to the nucleus of the target cell, a particle must be able to cross the cell membrane, escape endocytosis, and release the plasmid intracellularly to allow for trafficking to the nucleus. Each of these steps is essential, and the contribution of small changes to the chemical structure of the polymers to these mechanistic steps will be examined in this manuscript.
There are multiple necessary components for effective gene delivery using cationic polymers. First, the polymers must bind strongly to the DNA, encapsulating or condensing it to prevent its degradation. A cationic polymer, through positively charged amine groups, allows for electrostatic interactions with anionic DNA. Cationic polymers such as poly-L-lysine (PLL) have been demonstrated to form stable polymer/DNA complexes.4 The next step is cellular uptake, where the polymer/DNA nanoparticles must penetrate the lipid bilayer plasma membrane. These polyplexes or nanoparticles are generally taken into cells through endocytosis. Positively charged particles are important for attraction to anionic proteoglycans on the cell surface. Both particle size and surface charge play key roles in this step. Other potential uptake methods include ligand-specific / receptor-specific mediated endocytosis through various particle coatings 5, 6 or covalent attachment.7, 8
Once in the endosome, the particles are then subjected to the endosomal-lysosomal pathway, where the complexes need to avoid being enzymatically degraded by lysosomes or recycled out of the membrane. Bypassing lysosomal degradation has been a bottleneck in improving intracellular gene delivery. It has been shown that polymer/DNA particles can escape the endosome into the cytoplasm through the “proton sponge effect.” 9 Inside of the endosome, the pH drops from 7.4 to around 5.1, where a polymers’ secondary and tertiary amine groups can buffer the acidification.10 An influx of ions into the endosome can lead to osmotic swelling and eventual bursting to release the polyplexes into the cytoplasm.11 The buffer capacity of titratable amine groups can effectively facilitate endosomal rupture, inducing efficient gene expression.10, 12 An example of the importance of buffer capacity is that polyethylenimine (PEI) has an advantage over PLL in transfection due to its high buffering capacity. Studies have shown that polymers with secondary or tertiary amines are either able to provide more time to escape the endosome or mediate endosomal escape. Other strategies that have been utilized to promote endosomal escape include functionalizing polymers with endosomolytic peptides 13, 14, which utilize pH sensitive conformational changes that promote endosomal escape.
Once inside the cytoplasm, it is beneficial for the polymer to degrade to enhance release of the DNA to prevent polymer-mediated cytotoxicity. For effective plasmid release, polymers can be designed to degrade hydrolytically through ester linkages 15 or reducibly through disulfide linkages.16 Amine-containing polymers that can degrade hydrolytically have shown to have much higher transfection and lower cytoxicity than polymers such as PEI 17 and degradable versions of PEI have shown improved efficacy and lower cytotoxicity than non-degradable versions.18 The DNA plasmid must then overcome nuclear import. This is more easily achievable in dividing cells when the nuclear envelope breaks down during mitosis. Another strategy for nuclear import is appending nuclear localization signals (NLS) to DNA which may help carry it into the nucleus.19, 20 Measuring gene expression ensures that all of these intracellular barriers have been crossed including transcription and translation of the exogenous DNA.
Biodegradable cationic polymers such as end-modified poly(beta-amino ester)s have been demonstrated as promising biomaterials for non-viral gene delivery among various cell types.21–23 End-modification with diamine monomers has shown that some of these polymers can rival adenovirus for gene delivery in vitro.17 Additionally, PBAEs have been shown to have promise in the treatment of cancer in vitro and in vivo.24, 25 However, while previous studies have investigated certain physical and biological parameters, 26–28 they have not fully looked at the chemical properties and mechanistic details that may fully explain the advantages that the lead structures possess.29 In particular, differences in polymer buffering capacity, polymer degradation, and the mechanistic differences between the same linear polymers with acrylate end-groups compared to differing amine-containing small molecules as end-groups needed to be more fully evaluated. This study aims to elucidate the polymer properties and biological process most responsible for the high gene delivery efficacy of end-modified PBAEs.
Experimental Section
Cell Culture
COS-7 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) with Lglutamine and sodium pyruvate (DMEM 11995, Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Cells were grown at 37°C in a humid 5% CO2 atmosphere.
Materials
Monomers were purchased from commercial vendors and used as received. 4-amino-1-butanol (S4), 5-amino-1-pentanol (S5), 1,4-butanediol diacrylate (B4), 1,6-hexanediol diacrylate (B6), 1-(3-aminopropyl)-4-methylpiperazine (E7) were purchased from Alfa-Aesar, Ward Hill, MA. 1,3-propanediol diacrylate (B3) and 1,5-pentanediol diacrylate (B5) were purchased from Monomer-Polymer and Dajac Laboratories (Trevose, PA). 2-methyl-1,5-diaminopentane (E4) was purchased from TCI America (Portland, OR). 2-(3-aminopropylamino)ethanol (E6) and branched 25 kDa poly(ethylene imine) (PEI) were purchased from Sigma Aldrich (St. Louis, MO). Enhanced green fluorescent protein plasmid driven by a CMV promoter (eGFP) was obtained from Aldevron (Fargo, ND). CellTiter 96 AQueous One MTS assay was purchased from Promega (Fitchburg, WI) and used according to manufacturer’s instructions.
Polymer Synthesis
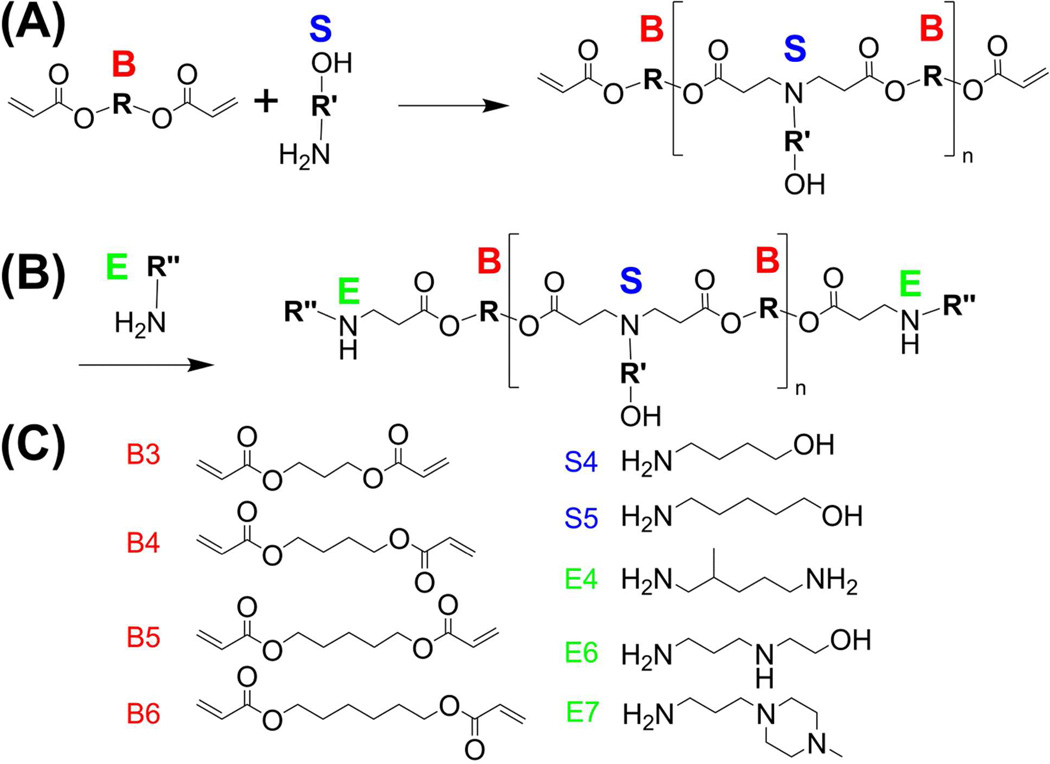
Polymers were synthesized using a two-step procedure (Figure 1). As an example, acrylate-terminated poly(1,4-butanediol diacrylate-co-4-amino-1-butanol), B4-S4, was first synthesized in a solvent-free fashion at different acrylate: amine monomer molar ratios (1.2:1, 1.1:1, 1.05:1). Reactions took place in glass vials in the dark under magnetic stirring for 24 h at 90°C. As a second step, amine-containing small molecules were individually conjugated to the ends of each polymer. Excess amine is used to fully end-modify the base polymer. 80 mg of polymer in 480 OL of DMSO was mixed with 320 µL of a 0.5M solution of the end capping amine in DMSO in 1.5 mL eppendorf tubes in a multi-tube vortexer with constant agitation for 1 h at room temperature. As an example, B4-S4 synthesized at a 1.1:1 ratio was end-modified by E7, and formed the B4-S4-E7 1.1:1 end-modified polymer. Polymers were stored at 100 mg/ml in anhydrous DMSO at −4°C with desiccant until use. Polymer nomenclature refers to the number of carbons between functional groups as we have previously described.29, 30 For example, polymer B4-S4 contains 4 carbons between acrylate groups in the polymer backbone, “B”, and 4 carbons between the amine and alcohol groups in the side chain, “S.” Polymer structure was characterized on a Bruker spectrometer by 1H NMR spectroscopy (400 MHz, d6-DMSO) and completion of end-modification was verified by elimination of the peaks corresponding to the acrylate termini of the polymer (at 5.9–6.4 ppm).30 Spectra for B3-S5-Ac (acrylate-terminated base polymer) and B3-S5-E7 can be found in Figure S1.
Figure 1.
(A) The base polymer is formed via Michael addition of diacrylates (“B”) and primary amines (“S”); the diacrylates are added in excess to form an acrylate terminated precursor. (B) In a second step, the diacrylate terminated base polymer is end-modified with an end capping amine (“E”), to form the end-modified polymer. (C) Monomers used in this study.
Particle Size and Charge
Particle size was determined both by dynamic light scattering (DLS) using a Mavern Zetasizer Nano ZS (Malvern Instruments, Malvern, UK, detection angle 173°, 633 nm laser) and by nanoparticle tracking analysis (NTA) using a Nanosight NS500 (Amesbury, UK, 532 nm laser). Particle charge was determined using a Mavern Zetasizer Nano ZS (Malvern Instruments, Malvern, UK). Polymer/DNA nanoparticles were made at a 60 w/w ratio in 25 mM sodium acetate buffer (pH = 5.0) at 30 ng/OL DNA and diluted into 150 mM PBS, pH 7.4. For the measurements on the Zetasizer, particles were diluted 5-fold into PBS, and particle size is reported as the intensity-weighted Z-averaged of the particle diameter in nm. Average electrophoretic mobilities were measured at 25°C and zeta potentials were analyzed using the Smoluchowsky model. For the NTA analysis, particles were diluted 50 to 100 fold into PBS such that particle number would be between 108 and 109 particles/mL, and particle size is determined from a 60s movie from which the Brownian motion of the particles was assessed as previously described.28
Gel Electrophoresis
The gel electrophoresis experiments were conducted in 1% agarose gels made with 1 µg/ml ethidium bromide in the gel. Particles were formed at 30 ng/µL DNA and at a 60 w/w ratio (polymer:DNA; 1.8 µg polymer/µL) and allowed to complex for 10 minutes before glycerol was added, with or without bromophenol blue (15 mg/mL), a negatively charged dye used to visualize the extent that the gel runs, and then immediately added to the gel. The gels were run for 40 min with 100 V applied. Gels were visualized with a Visi-Blue™ transilluminator (UVP, Upland, CA).
Buffering Capacity
The buffering capacities of the polymers were determined through acid-base titration. Ten micrograms of polymer in DMSO at 100 mg/mL was dissolved in 10 mL of 0.1 M NaCl solution. The pH of polymer solutions was set to pH 3 using 1 M HCl and titrated to pH 11 using 0.1 M NaOH. The pH of solutions was measured after each addition using a Mettler Toledo S20 pH meter. Buffer capacity was calculated in two ways: by taking the ratio of total protons buffered between pH 7.4 and 5.1 to the total amines of the polymer and by taking the ratio of protons buffered between pH 7.4 and 5.1 to total polymer mass. Titration of NaCl without the presence of polymer was used as background control. For end-modified PBAEs, the buffering contribution from excess free end-capping amine monomer was subtracted out to characterize the buffering of the polymers.
Degradation Studies
Two and a half milliliters of a 100 mg/ml solution of polymer in DMSO was added to 247.5 mL of phosphate buffered saline (PBS) solution at 37°C, and magnetically stirred to mix. At each time point, 25 mL of this solution was removed and frozen, then lyophilized to remove the water. This sample was dissolved in 1 mL of a solution of 94% THF, 5% DMSO and 1% piperidine, and organic phase permeation chromatography (GPC) was performed using the same solvent as an eluent at a flow rate of 1 mL/minute. The detector (Waters 2414 refractive index detector) and columns (three Waters Styragel columns, HR1, HR3, and HR4, in series) were maintained at 40°C throughout the runs. Polymer molecular weights presented are relative to monodisperse polystyrene standards (Shodex, Japan).
GFP transfections, with and without labeled plasmid
Fifteen thousand COS-7 cells were plated in 100 µL per well in clear 96-well tissue culture plates (Starstedt) to allow for overnight adhesion. For transfection experiments, eGFP pDNA was diluted into 25 mM NaAc buffer (pH 5.0) to form a final concentration of 60 ng/µL. Polymers stored at 100 mg/ml in DMSO were aliquoted out into 96-well plates and diluted to 3.6 µg/µL in 25 mM NaAc, and equal volumes of diluted polymers and diluted DNA were mixed by pipetting up and down in another 96-well plate. Ten minutes after mixing DNA and polymer solutions, 20 µL of nanoparticles were added to 100 µL of media (DMEM containing 10% FBS, 1% penicillin/streptomycin v/v) on the cells for a final pDNA dose of 600 ng/well. PEI/DNA complexes were formed at a 3:1 polymer to DNA weight ratio and formed in 150 mM NaCl as previously described 23, and PEI/DNA complexes were added to the cells for a final pDNA dose of 600 ng/well. Four hours after transfection, the cells were washed with PBS and 100 µL of fresh media was added to the cells. Forty-eight hours after transfection, gene expression was measured using flow cytometry (Accuri C6 with HyperCyt high-throughput adaptor); gating was performed on FlowJo software and GFP+ cells were gated as a subpopulation of cells by two-dimensional gating of FL1 vs FL2 separate increased autofluorescence signal from increased signal (for examples, see Figure S2).
For DNA uptake studies, eGFP pDNA was labeled with Cy3 using the Label IT® Tracker™ kit (Mirus Biopharma) following manufacturer’s instructions, and diluted into unlabeled pDNA resulting in a net ratio of 331 nucleotides / dye. Particles were formulated the same was as with the transfection experiment (but with labeled DNA), except that after washing the cells and changing the media 4 hours post transfection, the cells were washed again 2×, trypsin was added, and the cells were run on flow cytometry as above. Gating was performed on FlowJo 7.6.5 software and uptake was determined by two-dimensional gating (as a subpopulation of cells) of FL1 vs FL2 to separate increased autofluorescence signal from increased signal (for examples, see Figure S2).
Cell viability testing
For cell viability testing, transfection was performed as normal, but twenty-four hours after transfection, cell viability was measured by the AQueousOne CellTiter MTS assay; after addition of the CellTiter reagent (20 µL/well), cells were incubated at 37°C for 1 hour and then absorbance at 490 nm was measured on a plate reader (Synergy 2). Background absorbance from media and reagent were subtracted off, and the absorbance was normalized to untreated cells.
Statistical Analysis
Assays were performed in quadruplicate, and presented data are mean ± SD. All statistics were performed using the GraphPad Prism 5 software package. To examine multiple comparisons, such as differences between nanoparticle size with Acrylate (Ac) and Amine (Am) terminated polymers, we performed 1-way ANOVA with Bonferroni post tests.
Results and Discussion
Synthesis and Characterization of Polymer Array
PBAEs have been extensively investigated in a high-throughput fashion for their ability to mediate non-viral gene delivery in vitro, but significant characterization of the polymer properties that lead to overcoming the barriers to intracellular gene delivery have not been fully explored.3, 27, 31, 32 PBAEs have achieved transfection efficacies comparable to adenovirus for transfection of human endothelial cells,17 have been used systems for efficient siRNA mediated gene knockdown,33, 34 and have been used to target cancer in vitro and in vivo.24, 25 Hydrophobicity appears to play a significant role in the ability of PBAEs to mediate efficient gene delivery 30 and the number of plasmids per particle that PBAEs form can also play a role.28
To evaluate in greater detail why PBAEs are effective for non-viral gene delivery and to determine how subtle changes to structure affect efficacy, we synthesized an array of polymers with single carbon changes to the backbone, side chain, and small changes to the end-modifying amine (Figure 1). We synthesized 4 polymers with single carbon changes to the backbone (B3-S5-Ac, B4-S5-Ac, B5-S5-Ac, B6-S5-Ac) and end-modified each of those with E7 (B3-S5-E7, B4-S5-E7, B5-S5-E7, B6-S5-E7). We also synthesized two polymers with single carbon difference to the side chain (B4-S4 and B4-S5) and end-modified those polymers with 3 end-capping amines (E4, E6, E7), and finally we synthesized B4-S4 at three different amine:acrylate ratios (1.2, 1.1, 1.05) to generate different molecular weight versions of the base polymer, and end-capped those with a single end-capping amine (E7). We then studied polymer properties that we hypothesized would be related to the ability to overcome the barriers to intracellular gene delivery. We evaluated at nanoparticle size, zeta potential, and ability to retard DNA from moving on a gel to look at stable particle formation. We studied the PBAEs’ buffering capacity to investigate how these polymers might be able to escape the endosome, and the polymer degradation rate to assess the ability of the polymer to promote release of DNA as well as avoid cytotoxicity. Finally, we evaluated particle uptake, viability, and transfection efficacy as biological outcomes, and as a way to assess where in the process particular polymer structure were failing or succeeding in overcoming barriers to gene delivery.
Particle size and charge
Previous work with cationic polymers for DNA delivery has indicated that formation of small, positively charged nanoparticles is a prerequisite for efficient transfection.1, 35 However, too high of a charge density can lead to unwanted toxicity, and there may be some intermediate, optimal charge density depending on the polymer of interest. These competing effects can been seen in HPMA-oligolysine copolymers 36 and cationic glycopolymers,37 polymers synthesized by Reversible Addition-Fragmentation chain Transfer (RAFT) polymerization.
In a series of cationic glycopolymers with either a positively charged pendant group or a sugar, Ahmed et al. showed that increased carbohydrate content significantly reduces toxicity but also reduces transfection efficacy.37 Studies on a library of HPMA-oligolysine copolymers revealed that size of the charged moieties matters; 5- and 10-lysine long oligocations were more effective than 15-lysine long oligocations. Shorter lysine chains were more salt stable (5 > 10 > 15). However, polymers with 10 lysine long oligocations were the best at transfection, followed by 5, then 15, indicating that there was some medium optimum between even distribution of charge and larger charged groups.36 In addition, poly(glycoamidoamine) (PGAA) polymers, synthesized with repeating ethylenamines, were shown to be optimal transfection reagents when there were 4 ethylenamines; having 5 or 6 ethylenamines does not increase transfection but does increase toxicity.38
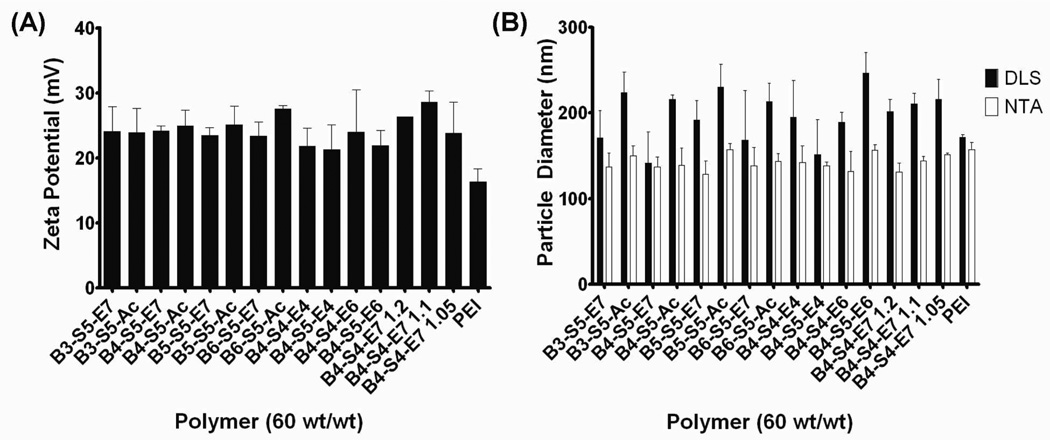
All polymers in this study spontaneously formed positively-charged (+21 to +29 mV zeta potential) nanoparticles in the 130–150 nm diameter range (Figure 2). Previous studies have indicated that a zeta potential of greater than +10 mV was required for PBAE nanoparticle transfection.27 Acrylate-terminated polymers were found to be slightly larger than their endmodified versions by dynamic light scattering measurements; on average, E7-modified B3/4/5/6-S5 polymers were 170±20 nm in diameter, versus 221±8 nm for acrylate-terminated versions. There is a statistically significant difference between B4-S5-E7 and B4-S5-Ac (p < 0.05), but no statistically significant difference between the other pairs. There was no statistical difference between any of the polymers when looking at size by nanoparticle tracking analysis. In this study, we measured particle size in two ways: dynamic light scattering (DLS) and nanoparticle tracking analysis (NTA). NTA directly measures number-averaged size, thus the average particle size by number-weighting is the same for all acrylate and amine pairs. DLS measurements are intrinsically intensity-weighted, where large infrequent particles can cause a disproportionate contribution to the average size. Thus, particle formulations where DLS and NTA measurements are the same, such as B4-S5-E7, are monodisperse and formulations such as B4-S5-E6 are more polydisperse and have a minority component of slightly larger particles. It is only in case of B4-S5-E7 and B4-S5-Ac that the presence of a minority component of larger particles is statistically significant by end-group, and in this case, the number-average size remains the same.
Figure 2.
(A) Zeta potential of selected polymers. (B) Nanoparticle diameters measured using dynamic light scattering and nanoparticle tracking analysis. (Note – Figure legend was changed in this draft)
Not surprisingly given the relatively narrow distribution of particle sizes and zeta potentials, particle size and zeta potential was not correlated with particle uptake or transfection efficacy to any significant degree (Figure S3a-c). However, particle size by dynamic light scattering appeared to be relatively negatively correlated with particle uptake (Figure S3d), indicating that smaller particles (as measured by DLS) tended to get taken up more efficiently than larger ones; this remains a weak trend. One potential explanation is that as DLS size is intensity-weighted, a relatively small number of larger particles would skew the DLS average much more than NTA number average. Thus, if these larger particles are particularly inefficient at being taken up by the cells, and they segregate DNA away from the smaller particles, these formulations overall would be less efficient in being taken up by cells. Generally, these nanoparticles were found to all be very similar in surface charge and particle size, yet they had substantial differences in uptake and transfection as will be further described below.
Gel electrophoresis
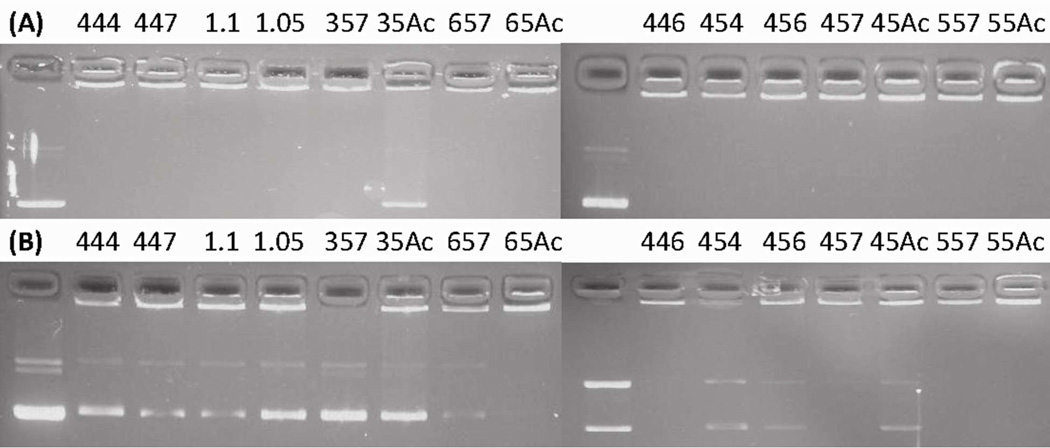
In addition to a basic requirement to form small, positively charged nanoparticles, for efficient DNA delivery, the particles must bind to and protect DNA effectively. At 60 w/w, all of the 14 polymers completely retarded the DNA except for B3-S5-Ac (Figure 3a). When bromophenol blue was added to the lanes containing particles, the most hydrophobic polymers containing B5 and B6 diacrylates in their backbone were able to retard the DNA (B5-S5-Ac, B5-S5-E7, B6-S5-Ac, B6-S5-E7) as well as B4-S4-E6 and B4-S5-E7. The remaining polymers were unable to completely retard the DNA migration (Figure 3b). Interestingly, acrylate terminated polymers and their amine-terminated counterparts bound DNA in similar patterns, with the largest discrepancy occurring with respect to the difference between B3-S5-Ac and B3-S5-E7. In this, even with no competition, B3-S5-Ac was unable to retard the DNA electrophoresis, up to a 150 w/w ratio, while the end-modified polymer was able to retain the DNA even at a low 15 w/w ratio (Figure S4). Hydrophobicity of the polymer seemed to play a large role in polymer binding affinity, as the four most hydrophobic polymers (B5-S5-Ac, B5-S5-E7, B6-S5-Ac, B6-S5-E7) were all able to retain the DNA even after addition of the bromophenol blue. This may indicate a significant hydrophobic effect for the binding of PBAEs with DNA. Previous studies have demonstrated that hydrophobicity plays a strong role in enhancing gene delivery with PBAEs, generally increasing transfection efficacy but also increasing cytotoxicity 30; these data may provide a mechanism for this effect.
Figure 3.
Gel electrophoresis of PBAE/DNA nanoparticles formed at 60 w:w (polymer:DNA ratio) (A) without bromophenol blue and (B) with bromophenol blue. For brevity, polymer names were shortened to remove the B-S-E designation, such that 444 is B4-S4-E4 and 357 is B3-S5-E7.
Polymer buffering capacity
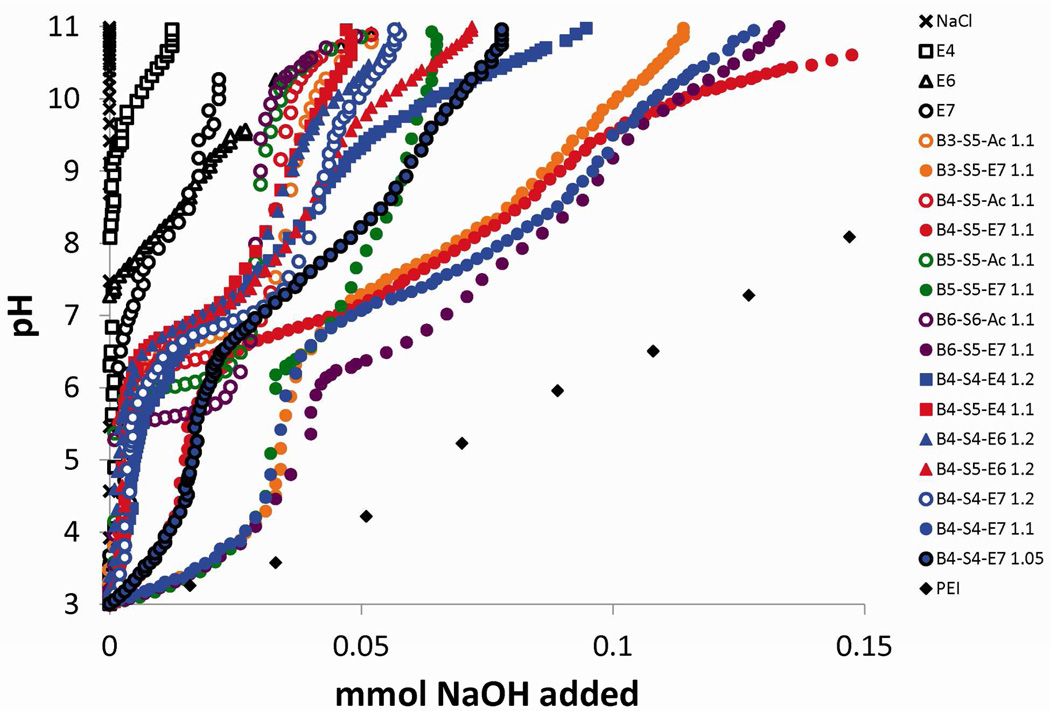
In order to escape the endosome, there needs to be a mechanism of endosomal escape. The ability of a polymer to buffer the endosome has been shown to be highly correlated with the amount of secondary and tertiary amine groups the polymers contain,10 as these amines tend to be protonatable across relevant pH ranges. To assess the ability of the polymers to buffer the endosome, titration curves were determined for polymers using acid-base titration (Figure 4). Using the titration curves, the buffering capacities of the polymers were calculated through the percentage of amine groups protonated between pH of 7.4 and 5.1. As references, sodium chloride (NaCl) showed a curve with no buffering indicated by its vertical slope, while polyethylenimine (PEI) displayed significant buffering indicated by a gradual slope between pH 5.1 and 7.4. Because PEI has an abundance of secondary and tertiary amine groups, it can buffer many protons in the endosome, where the pH drops from 7.4 to 5.1. The buffering capacities of gene delivery polymers affect their ability to escape the endosome via the proton sponge effect.9, 11
Figure 4.
Acid-base titration curves for selected polymers and normalized for 150 mM aqueous NaCl. Measurements were taken using a Mettler Toledo S20 pH meter. pH was adjusted to pH 3 with HCl, then titrated with NaOH. (Note – Figure labels were changed in this draft to reflect the general nomenclature)
Although the polymers differ only by small structural changes, their buffering capacities were found to have significant differences (Table 1). B4-S5-E7 1.05:1 was found to have the lowest buffering capacity on a per-amine basis and on a per-mass basis, while B6-S5-E7 1.1:1 was found to have the highest buffering on both measures, buffering 4.6 mmol H+/g and having a per-amine buffering capacity of 95%.
Table 1.
Buffer capacities of selected polymers were calculated using the titration curves found in Figure 2.
| Polymer | Buffer Capacity Per Amine: Protons buffered/ Total amines (%) |
Buffer Capacity Per Mass: Protons buffered/ Total mass (mmol H+/g) |
MW (kDa) |
Half Life (h) |
|---|---|---|---|---|
| B3-S5-E7 | 80 | 4.4 | 8.8 | 4.4 |
| B3-S5-Ac | 64 | 2.8 | ||
| B4-S5-E7 | 80 | 4.2 | 5.8 | 5.1 |
| B4-S5-Ac | 72 | 3.0 | ||
| B5-S5-E7 | 65 | 3.3 | 20.1 | 1.2 |
| B5-S5-Ac | 69 | 2.7 | ||
| B6-S5-E7 | 95 | 4.6 | 11.7 | 3.6 |
| B6-S5-Ac | 73 | 2.7 | ||
| B4-S4-E4 | 49 | 2.3 | 18.0 | 6.1 |
| B4-S5-E4 | 49 | 2.2 | 11.7 | 5.3 |
| B4-S4-E6 | 52 | 2.5 | 10.7 | 5.5 |
| B4-S5-E6 | 52 | 2.5 | 6.9 | 6.5 |
| B4-S4-E7 1.2 | 61 | 2.8 | 21.1 | 1.6 |
| B4-S4-E7 1.1 | 58 | 2.6 | 22.6 | 4.6 |
| B4-S4-E7 1.05 | 38 | 1.7 | 33.1 | 3.9 |
| PBAEs | 34–95 | 1.4–4.6 | 5.8–33.1 | 1.6–6.5 |
| PEI | 26 | 6.2 | 25.0 |
(Note – Polymer names were changed in this table in this draft to reflect the general nomenclature)
Additionally, when the buffering capacity of the PBAEs are compared to branched PEI on a per-mass basis, the comparison is initially not a favourable one. The PBAEs buffering capacity was concentrated in the relevant pH range (pH 5.1 – 7.4), with their per-amine buffering capacity varying from 34% to 95% as compared to 25 kDa polyethyleneimine (PEI), which only uses 25% of its amine content over that key range. However, due to the higher amine-density on PEI, on a per mass basis, all PBAEs buffered fewer protons than PEI (1.4–4.6 mmol H+/g for PBAEs vs 6.2 mmol H+/g for PEI). However, since the PBAEs are much less cytotoxic, and are typically formulated at 60 w/w compared to 1–3 w/w for PEI, the total buffering capacity of PBAE based particles significantly exceeds that of PEI. As an example, the PBAE with the lowest buffering capacity per mass can buffer 1.7 mmol H+/g, but on a formulation basis, since the PBAE formulation contain on average 20 times more polymer than PEI formulation, 60 w/w particles would be able to buffer 5.5-times as many protons as 3 w/w PEI.
The structure-function relationship for end-cap molecule and PBAE buffering extent per mass is clear (Table 1). Polymers end-capped with E7 generally have the highest buffer capacities, which is expected as the E7 group contributes two tertiary amines in its structure. Polymers end-capped with E6 also have relatively high buffer capacities as compared to E4, as each E6 group contributes an extra secondary amine group as opposed to an extra primary amine. Acrylate-terminated polymers showed only slightly lower buffering capacity compared to their end-modified counterparts (Table 1, Figure S5). The buffering for the acrylate-terminated polymers are not the lowest of the samples, indicating that the base polymers themselves, rather than their end-groups, drive buffering in the range of pH 5.1–7.4. Further, these results reveal that the modest differences in buffering capacity observed with these different polymer structures do not strongly correlate to the relatively large differences in particle uptake and transfection also observed with these structures (Fig. S3f,g).
In the context of intracellular gene delivery, endosomal buffering is required to mediate endosomal escape and facilitate transfection of the cell. We hypothesized that low buffering capacity would result in the stranding of nanoparticles in the endosomal/lysosomal system, resulting in high uptake not allowing for high transfection. In general, however, this trend is not perfect due to other confounding factors, and demonstrates that high buffering capacity is likely necessary but not sufficient for effective transfection with PBAEs. As predicted, there is no correlation between buffering capacity (per mass) and particle uptake, but there is a weak positive correlation between buffering capacity per mass and transfection (Fig. S3f). This makes intuitive sense, as improvements in buffering capacity should improve endosomal escape, thus enhancing the transfection of cells that have already taken up particles. This correlation is not that strong because of the generally very strong correlation between uptake and transfection seen in this cell type.
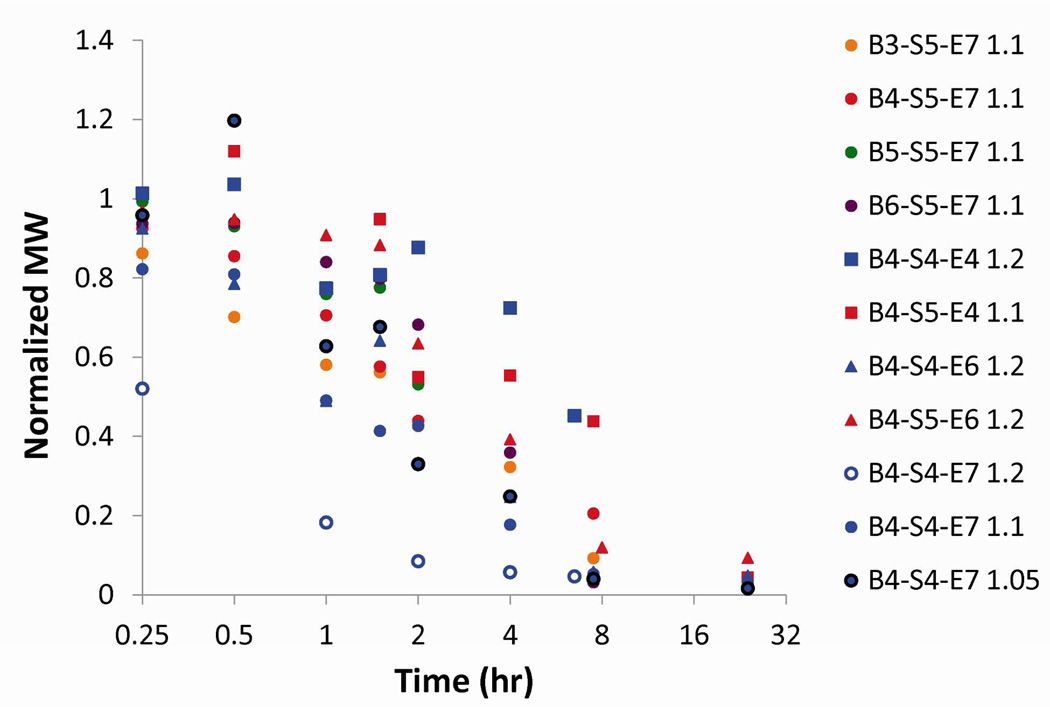
Polymer degradation
Intracellular DNA delivery requires that the polymer forms stable complexes that can bind DNA, protect it from enzymatic degradation, and enter cells. However, successful transfection also requires that the DNA be released for efficient transcription of mRNA.39 Polymer degradation rate is an important chemical parameter as it determines the time scale that the polymers have to escape the endosome and enter the cytoplasm for effective transfection. It is also important to characterize the polymer degradation mechanisms to evaluate the basis of possible reduced cytotoxicity compared to other cationic polymers. Cationic polymers that cannot degrade effectively will likely not be as biocompatible with cells. An example of this is PEI, which typically mediates high uptake and has a very high total buffer capacity (Table 1), but has a lower transfection efficacy and higher toxicity than other polymers.40 Polymer degradation can be helpful in terms of enhancing the release of DNA from the polymer and reducing cytotoxicity; but if degradation is too quick, it could decrease particle stability, DNA protection, and cellular uptake.
Generally, the PBAEs degrade very rapidly in aqueous conditions, with half-lives in PBS at 37°C ranging from 90 minutes to just over 6 hours (Figure 5, Table 1). We hypothesized that due to the trade off between wanting to increase particle stability extracellularly, but also promote DNA release intracellularly, we may find a biphasic response between polymer half-life and transfection efficacy. We found that a modest biphasic trend is demonstrated when we compare transfection to half-life (Figure S3e). In addition, the two polymers with the shortest half-lives (B5-S5-E7 at 1.2 h and B4-S4-S7 1.2:1 at 1.6 h) showed the largest discrepancy between uptake and transfection, which may suggest that modestly long (> 2 h) half lives are required to protect the DNA all the way to the nucleus. Overall, these degradation rates are surprisingly rapid. Faster than anticipated degradation of polymers is observed in other related systems such as poly(glycoamidoamine) (PGAA) polymers, which contain hydroxyl groups alpha to the amide bonds and secondary amines in the backbone of the polymer, and show rapid hydrolysis at pH 7.4 (half-lives of around 20 hr), even though amide bonds should hydrolyze much more slowly than ester bonds.41 Interestingly, there is faster degradation in pH 7.4 than in pH 5; this is attributed to the effect of the proximal –OH group.41 In particular, the secondary amines in the PGAAs are located a similar distance away from the amide bonds as the tertiary amines in the PBAEs are to the ester bonds, supporting the theory that they could likely be responsible for the rapid degradation of the PBAEs seen here. This very rapid degradation rate could be another reason for the general effectiveness and low cytotoxicity seen with this class of polymers, but it does pose potential challenges for eventual in vivo translation of this technology.
Figure 5.
Degradation of polymers over time by GPC. MW is normalized (set = 1) to initial MW at t = 0. The majority of polymers showed extensive degradation by 4 hours.
Particle Uptake, Transfection Efficacy, and Cytotoxicity
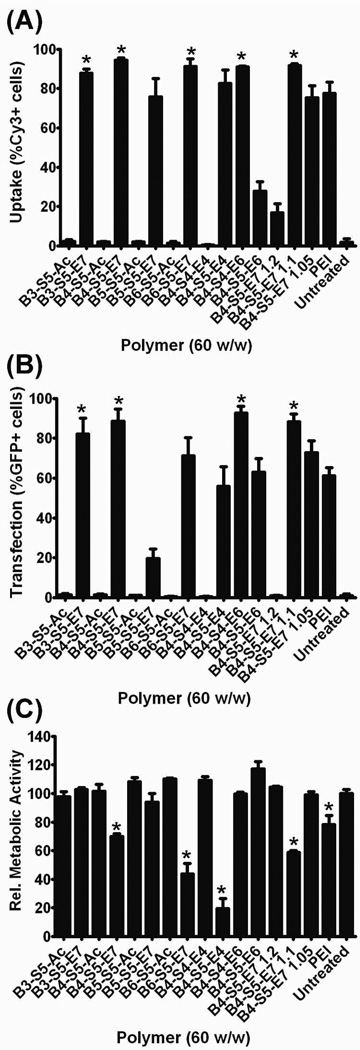
In order to compare polymer properties to biological outcomes, we investigated the cellular uptake, transfection efficacy, and cytotoxicity of our nanoparticle formulations (Figure 6). Polymers B4-S5-E6, B4-S4-E7 1.1:1, B3-S5-E7, and B4-S5-E7 were all found to have superior uptake and transfection to PEI, and B6-S5-E7 was found to have superior uptake but comparable transfection to PEI (Figure 6a,b; p < 0.05 for all comparisons). Most of the tested polymers were non-cytotoxic at the formulation ratio and dose tested with the exceptions being B4-S5-E4, B4-S4-E7 1.1, B4-S4-E7, B4-S4-E7, and PEI, which showed increased cytotoxicity relative to untreated controls (Figure 6c; p < 0.05 for all comparisons).
Figure 6.
DNA nanoparticle uptake (A), cellular transfection (B), and cellular viability (C) after application of nanoparticles to cells. Data are presented as mean ± SEM. * Statistically significant improvement (p < 0.05) vs. 25 kDa PEI control for uptake and transfection plots; statistically significantly increased cytotoxicity (p < 0.05) vs. untreated control. (Note – Figure labels were changed in this draft to reflect the general nomenclature)
Increasing the number of carbons along the polymer backbone from 3 through 6 and leaving the side chain length at 5 carbons tended to increase cytotoxicity without increasing transfection efficacy. This result mirrors previous findings in COS-7 cells and RPE cells, where there is a limit at which increasing hydrophobicity of the polymer backbone does not improve transfection efficacy and only increases toxicity.22, 30 Increasing the side-chain length from 4 carbons to 5 carbons resulted in mixed effects; end-modification of B4-S4 with E6 leaded to superior cell uptake and transfection compared to end-modification of B4-S5 with E6, but end-modification of B4-S5 with E4 had superior cell uptake and transfection compared to end-modification of B4-S4 with E4.
Increasing initial polymer molecular weight also had mixed effects. We synthesized B4-S4-E7 at 3 different molar ratios of acrylate to amine, resulting in 3 different molecular weights for the same base polymer. B4-S4-E7 at 1.1:1 had the highest transfection and uptake, but it also was the most toxic. These results are evocative of the recent work by Eltoukhy, et al, where they showed that intermediate length PBAEs mediated optimal transfection.42
Acrylate-terminated PBAEs were found to have significantly lower uptake and transfection than their amine-terminated counterparts (p < 0.001 for all comparisons). Previous studies had indicated that acrylate-terminated polymers showed significantly reduced transfection efficacies,26, 27, 43 but were not able to determine the specific cause. Our investigation shows that this difference in transfection efficacy is due to differential uptake of the acrylateterminated polymer nanoparticles as compared to the amine end-capped polymers. This is particularly striking given that there were no significant differences in particle formation as measured by gel electrophoresis or in nanoparticle properties with respect to particle sizes and zeta potentials. Furthermore, the acrylate-terminated polymers were non-cytotoxic. Therefore, end-capping linear PBAEs with small molecules containing amines is necessary for sufficient cell uptake and transfection in a manner largely independent from nanoparticle biophysical properties. Further studies on specific mechanisms of gene delivery uptake will help improve our understanding of how differential polymer structure affects transfection efficacy and we are currently undertaking these studies.
Conclusions
Development of non-viral nanoparticles for gene delivery requires a greater understanding of the properties that enable gene delivery nanoparticles to overcome the numerous barriers to intracellular DNA delivery. Here we evaluated the effects of small structural perturbations within an array of linear poly(beta-amino ester)s (PBAEs) on polymer properties which are related to the barriers to intracellular gene delivery. Previous work has not investigated PBAE buffering capacity or examined the degradation rate of PBAEs formed from Michael addition of a primary amine containing side chain and a diacrylate. Interestingly, the PBAE polymers generally showed very rapid degradation in physiological conditions (t½ = 90 min – 6 hours). On a per mass basis, PBAEs buffered 1.4–4.6 mmol H+/g. When compared to 25 kDa polyethyleneimine (PEI), PBAEs buffer significantly fewer protons/mass. However, since the PBAEs are much less cytotoxic and degrade so rapidly, they can be formulated at significantly higher weight ratios without substantial toxicity, and thus total buffering capacity of PBAE based particles significantly exceeds that of PEI. This may explain the requirement for higher w/w ratios to achieve optimal efficacy using PBAEs compared to other polymer systems, and the rapid degradation rate may explain the low toxicity observed with large amounts of polymer.
Acrylate-terminated base polymers were much less efficacious than corresponding small molecule amine-containing end-capped versions, both in terms of uptake and transfection, even though there are minimal differences between acrylate and amine-terminated polymers in terms of DNA retardation in gel electrophoresis, nanoparticle size, nanoparticle zeta potential, polymer buffering capacity and cytotoxicity. This is a very interesting finding, and further investigation into the source of the considerable difference in efficacy seen here would be important. These studies further elucidate the role of polymer structure for gene delivery and highlight that small molecule end-group modification of a linear polymer can be critical for cellular uptake in a manner that is largely independent of polymer/DNA binding, particle size, and particle surface charge.
Supplementary Material
Acknowledgements
The authors acknowledge support in part by the NIH (R21CA152473). JCS thanks the MSTP program for support.
References
- 1.Putnam D. Polymers for gene delivery across length scales. Nat Mater. 2006;5(6):439–451. doi: 10.1038/nmat1645. [DOI] [PubMed] [Google Scholar]
- 2.Check E. Gene therapy put on hold as third child develops cancer. Nature. 2005;433:561–561. doi: 10.1038/433561a. [DOI] [PubMed] [Google Scholar]
- 3.Green JJ, Langer R, Anderson DG. A combinatorial polymer library approach yields insight into nonviral gene delivery. Acc Chem Res. 2008;41(6):749–759. doi: 10.1021/ar7002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park T. Current status of polymeric gene delivery systems. Advanced Drug Delivery Reviews. 2006;58:467–486. doi: 10.1016/j.addr.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Shmueli R. Electrostatic Surface Modifications to Improve Gene Delivery. Expert Opinion Drug Delivery. 2010;7:535–550. doi: 10.1517/17425241003603653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou J, Liu J, Cheng CJ, Patel TR, Weller CE, Piepmeier JM, Jiang Z, Saltzman WM. Biodegradable poly(amine-co-ester) terpolymers for targeted gene delivery. Nat Mater. 2012;11(1):82–90. doi: 10.1038/nmat3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogris M, Steinlein P, Carotta S, Brunner S, Wagner E. DNA/polyethylenimine transfection particles: Influence of ligands, polymer size, and PEGylation on internalization and gene expression. Aaps Pharmsci. 2001;3(3) doi: 10.1208/ps030321. art. no. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kagaya H, Oba M, Miura Y, Koyama H, Ishii T, Shimada T, Takato T, Kataoka K, Miyata T. Impact of polyplex micelles installed with cyclic RGD peptide as ligand on gene delivery to vascular lesions. Gene Ther. 2012;19(1):61–69. doi: 10.1038/gt.2011.74. [DOI] [PubMed] [Google Scholar]
- 9.Boussif O. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl. Acad. Sci. U.S.A. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ou M. A family of bioreducible poly(disulfide amine)s for gene delivery. Biomaterials. 2009;30:5804–5814. doi: 10.1016/j.biomaterials.2009.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sonawane ND, Szoka FC, Verkman AS. Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. J Biol Chem. 2003;278(45):44826–44831. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- 12.Akinc A, Thomas M, Klibanov AM, Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J Gene Med. 2005;7(5):657–663. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- 13.Ogris M, Carlisle RC, Bettinger T, Seymour LW. Melittin enables efficient vesicular escape and enhanced nuclear access of nonviral gene delivery vectors. The Journal of biological chemistry. 2001;276(50):47550–47555. doi: 10.1074/jbc.M108331200. [DOI] [PubMed] [Google Scholar]
- 14.Meyer M, Philipp A, Oskuee R, Schmidt C, Wagner E. Breathing life into polycations: functionalization with pH-responsive endosomolytic peptides and polyethylene glycol enables siRNA delivery. J Am Chem Soc. 2008;130(11):3272–3273. doi: 10.1021/ja710344v. [DOI] [PubMed] [Google Scholar]
- 15.Lynn DM, Langer R. Degradable poly(beta-amino esters): Synthesis, characterization, and self-assembly with plasmid DNA. J Am Chem Soc. 2000;122:10761–10768. [Google Scholar]
- 16.Lin C, Engbersen JF. Effect of chemical functionalities in poly(amido amine)s for non-viral gene transfection. Journal of controlled release : official journal of the Controlled Release Society. 2008;132(3):267–272. doi: 10.1016/j.jconrel.2008.06.022. [DOI] [PubMed] [Google Scholar]
- 17.Green JJ, Zugates GT, Tedford NC, Huang Y, Griffith LG, Lauffenburger DA, Sawicki JA, Langer R, Anderson DG. Combinatorial modification of degradable polymers enables transfection of human cells comparable to adenovirus. Adv Mater. 2007;19(19):2836–2842. [Google Scholar]
- 18.Forrest ML, Koerber JT, Pack DW. A degradable polyethylenimine derivative with low toxicity for highly efficient gene delivery. Bioconjug Chem. 2003;14(5):934–940. doi: 10.1021/bc034014g. [DOI] [PubMed] [Google Scholar]
- 19.Zanta MA. Gene delivery: A single nuclear localization signal peptide is sufficient to carry DNA to the cell nucleus. Proc. Natl. Acad. Sci. U.S.A. 1999;96:91–96. doi: 10.1073/pnas.96.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dean DA, Strong DD, Zimmer WE. Nuclear entry of nonviral vectors. Gene Ther. 2005;12(11):881–890. doi: 10.1038/sj.gt.3302534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sunshine J, Green JJ, Mahon K, Yang F, Eltoukhy A, Nguyen DN, Langer R, Anderson DG. Small molecule end groups of linear polymer determine cell-type gene delivery efficacy. Adv Mater. 2009;21(48):4947–4951. doi: 10.1002/adma.200901718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sunshine JC, Sunshine SB, Bhutto I, Handa JT, Green JJ. Poly(beta-Amino Ester)- Nanoparticle Mediated Transfection of Retinal Pigment Epithelial Cells In Vitro and In Vivo. PloS one. 2012;7(5):e37543. doi: 10.1371/journal.pone.0037543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhise NS, Gray RS, Sunshine JC, Htet S, Ewald AJ, Green JJ. The relationship between terminal functionalization and molecular weight of a gene delivery polymer and transfection efficacy in mammary epithelial 2-D cultures and 3-D organotypic cultures. Biomaterials. 2010;31(31):8088–8096. doi: 10.1016/j.biomaterials.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Showalter SL, Huang YH, Witkiewicz A, Costantino CL, Yeo CJ, Green JJ, Langer R, Anderson DG, Sawicki JA, Brody JR. Nanoparticulate delivery of diphtheria toxin DNA effectively kills Mesothelin expressing pancreatic cancer cells. Cancer Biol Ther. 2008;7(10):1584–1590. doi: 10.4161/cbt.7.10.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tzeng SY, Guerrero-Cazares H, Martinez EE, Sunshine JC, Quinones-Hinojosa A, Green JJ. Non-viral gene delivery nanoparticles based on poly(beta-amino esters) for treatment of glioblastoma. Biomaterials. 2011;32(23):5402–5410. doi: 10.1016/j.biomaterials.2011.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zugates GT, Tedford NC, Zumbuehl A, Jhunjhunwala S, Kang CS, Griffith LG, Lauffenburger DA, Langer R, Anderson DG. Gene delivery properties of end-modified poly(betaamino ester)s. Bioconjug Chem. 2007;18(6):1887–1896. doi: 10.1021/bc7002082. [DOI] [PubMed] [Google Scholar]
- 27.Anderson DG, Akinc A, Hossain N, Langer R. Structure/property studies of polymeric gene delivery using a library of poly(beta-amino esters) Mol Ther. 2005;11(3):426–434. doi: 10.1016/j.ymthe.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 28.Bhise NS, Shmueli RB, Gonzalez J, Green JJ. A novel assay for quantifying the number of plasmids encapsulated by polymer nanoparticles. Small. 2012;8(3):367–373. doi: 10.1002/smll.201101718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Green JJ. 2011 Rita Schaffer Lecture: Nanoparticles for Intracellular Nucleic Acid Delivery. Ann Biomed Eng. 2012;40(7):1408–1418. doi: 10.1007/s10439-012-0550-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sunshine JC, Akanda MI, Li D, Kozielski KL, Green JJ. Effects of base polymer hydrophobicity and end-group modification on polymeric gene delivery. Biomacromolecules. 2011;12(10):3592–3600. doi: 10.1021/bm200807s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akinc A, Lynn DM, Anderson DG, Langer R. Parallel synthesis and biophysical characterization of a degradable polymer library for gene delivery. J Am Chem Soc. 2003;125(18):5316–5323. doi: 10.1021/ja034429c. [DOI] [PubMed] [Google Scholar]
- 32.Anderson DG, Lynn DM, Langer R. Semi-automated synthesis and screening of a large library of degradable cationic polymers for gene delivery. Angew Chem Int Ed Engl. 2003;42(27):3153–3158. doi: 10.1002/anie.200351244. [DOI] [PubMed] [Google Scholar]
- 33.Lee JS, Green JJ, Love KT, Sunshine J, Langer R, Anderson DG. Gold, poly(beta-amino ester) nanoparticles for small interfering RNA delivery. Nano Lett. 2009;9(6):2402–2406. doi: 10.1021/nl9009793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tzeng SY, Yang PH, Grayson WL, Green JJ. Synthetic poly(ester amine) and poly(amido amine) nanoparticles for efficient DNA and siRNA delivery to human endothelial cells. International journal of nanomedicine. 2011;6:3309–3322. doi: 10.2147/IJN.S27269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sunshine JC, Bishop CJ, Green JJ. Advances in polymeric and inorganic vectors for nonviral nucleic acid delivery. Therapeutic Delivery. 2011;2(4):493–521. doi: 10.4155/tde.11.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson RN, Chu DS, Shi J, Schellinger JG, Carlson PM, Pun SH. HPMA-oligolysine copolymers for gene delivery: optimization of peptide length and polymer molecular weight. J Control Release. 2011;155(2):303–311. doi: 10.1016/j.jconrel.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahmed M, Narain R. The effect of polymer architecture, composition, and molecular weight on the properties of glycopolymer-based non-viral gene delivery systems. Biomaterials. 2011;32(22):5279–5290. doi: 10.1016/j.biomaterials.2011.03.082. [DOI] [PubMed] [Google Scholar]
- 38.Ingle NP, Malone B, Reineke TM. Poly(glycoamidoamine)s: a broad class of carbohydratecontaining polycations for nucleic acid delivery. Trends Biotechnol. 2011;29(9):443–453. doi: 10.1016/j.tibtech.2011.04.012. [DOI] [PubMed] [Google Scholar]
- 39.Schaffer DV, Fidelman NA, Dan N, Lauffenburger DA. Vector unpacking as a potential barrier for receptor-mediated polyplex gene delivery. Biotechnol Bioeng. 2000;67(5):598–606. doi: 10.1002/(sici)1097-0290(20000305)67:5<598::aid-bit10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 40.Moghimi SM, Symonds P, Murray JC, Hunter AC, Debska G, Szewczyk A. A two-stage poly(ethylenimine)-mediated cytotoxicity: implications for gene transfer/therapy. Mol Ther. 2005;11(6):990–995. doi: 10.1016/j.ymthe.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y, Reineke TM. Degradation of poly(glycoamidoamine) DNA delivery vehicles: polyamide hydrolysis at physiological conditions promotes DNA release. Biomacromolecules. 2010;11(2):316–325. doi: 10.1021/bm9008233. [DOI] [PubMed] [Google Scholar]
- 42.Eltoukhy AA, Siegwart DJ, Alabi CA, Rajan JS, Langer R, Anderson DG. Effect of molecular weight of amine end-modified poly(beta-amino ester)s on gene delivery efficiency and toxicity. Biomaterials. 2012;33(13):3594–3603. doi: 10.1016/j.biomaterials.2012.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Akinc A, Anderson DG, Lynn DM, Langer R. Synthesis of poly(beta-amino ester)s optimized for highly effective gene delivery. Bioconjug Chem. 2003;14(5):979–988. doi: 10.1021/bc034067y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.