Abstract
Accurate, fast, and affordable analysis of the cellular component of blood is of prime interest for medicine and research. Yet, most often sample preparation procedures for blood analysis involve handling steps prone to introducing artifacts, whereas analysis methods commonly require skilled technicians and well-equipped, expensive laboratories. Developing more gentle protocols and affordable instruments for specific blood analysis tasks is becoming possible through the recent progress in the area of microfluidics and lab-on-a-chip-type devices. Precise control over the cell microenvironment during separation procedures and the ability to scale down the analysis to very small volumes of blood are among the most attractive capabilities of the new approaches. Here we review some of the emerging principles for manipulating blood cells at microscale and promising high-throughput approaches to blood cell separation using microdevices. Examples of specific single-purpose devices are described together with integration strategies for blood cell separation and analysis modules.
Keywords: lab-on-a-chip, point-of-care diagnostic, cell separation, sample preparation, microfluidic
INTRODUCTION
Blood is a treasure of information about the functioning of the whole body. Every minute, the entire blood volume is recirculated throughout the body, delivering oxygen and nutrients to every cell and transporting products from and toward all different tissues. At the same time, cells of the immune system are transported quickly and efficiently through blood, to and from every place in the body where they perform specific immuno-surveillance functions. As a result, blood harbors a massive amount of information about the functioning of all tissues and organs in the body. Consequently, blood sampling and analysis are of prime interest for both medical and science applications, and hold a central role in the diagnosis of many physiologic and pathologic conditions, localized or systemic. However, tapping into this wealth of information, for clinical and scientific applications, requires not only the understanding of the biology involved but also adequate technologies.
Our knowledge about blood has always evolved in parallel with the general knowledge of biology, and several breakthroughs were facilitated by technological advances (1). The first use of microscopes in the seventeenth century allowed the observation of cells in blood. Later, in the nineteenth century, the development of tissue-staining techniques allowed the initial characterization the first blood cell populations. Even today, the examination of the peripheral blood film using Wright-Giemsa staining procedures and the full blood count are two of the most basic and yet the most informative investigations performed in hematology (2). Only flow cytometry techniques, available since the 1960s, can rival these in terms of finer details and higher throughput, representing today the golden standard in cell identification and separation. However, the sophistication of these latest techniques requires increasingly higher levels of skill and specialization from their users. Additionally, the high-quality standards of accuracy, reliability, and timeliness imposed since 1988 by the Clinical Laboratory Improvement Act (CLIA) reshaped the field of blood analysis (3). The analysis of the cellular component in particular has been affected by restricting it to highly specialized and strictly regulated laboratories, to the detriment of point-of-care diagnostic tools. However, despite considerable automation, even in advanced laboratories, a significant portion of blood handling is still performed manually or in conditions that may significantly alter the results of subsequent analysis. Reducing the chances for such errors to occur, reducing the time from blood collection to analysis, and increasing the availability of analysis techniques at the place where blood is collected are recognized challenges that would require faster, cheaper, and more comprehensive approaches.
Among the new technologies with an increasingly broader impact in biology, microfluidics and miniaturized lab-on-a-chip-type devices are extremely attractive for blood analysis. For clinical applications, bringing complete labs for blood analysis to the bedside through point-of-care analyzers capable of comprehensive diagnostic is poised to reshape the delivery of health care. New devices for convenient use at home or in doctors’ offices would allow for rapid and accurate diagnostic and prognostic, based on blood cells, of infectious diseases, cancers, and inflammatory responses. These may also allow better matching between drugs and patient pathophysiology, reducing side effects and improving efficiency of therapy. In drug discovery, microfluidic devices may redefine the entrance criteria for clinical trials and test for these criteria in a time- and cost-effective way. Nonetheless, in small-animal studies, microfabricated devices would only use minute amounts of blood for analysis, allowing for repetitive sampling at multiple time points and minimizing the adverse effects of blood drawing. Even more ambitious, in the discovery mode research, microfabricated devices for sample preparation would open new possibilities by allowing comprehensive genomic and proteomic analysis from small homogenous subpopulations down to single cells. On the whole, on-chip blood sample preparation would lead to more gentle, fast, and consistent manipulation of the living cells, and therefore more accurate and better quality of extracted information.
This review summarizes recent progress in the fast-paced field of microfluidics, assessing their impact on sample preparation techniques for blood cell analysis. We first present a brief overview of the challenges for blood sample preparation. We then emphasize the physical principles employed by new technologies and underline some of the enabling aspects of these compared to current approaches. Finally, we present some of the attempts to integrate different technologies and protocols into more complex lab-on-a-chip devices, addressing specific point-of-care diagnostic needs.
BRIEF OVERVIEW OF THE CHALLENGES FOR BLOOD SAMPLE PREPARATION
Blood has two main components, plasma and cells. Blood cells represent approximately 45% of the blood volume, and different cells perform different functions in different conditions. Thus, depending on the diagnostic needs, only some of the blood cells are of interest, and these cells need to be isolated from those that are irrelevant or may even alter the analysis (of the desired cells). A number of steps are common for any blood sample preparation techniques, although they are not always distinct and some overlap between steps is common (Figure 1). After a decision is made on what the target subpopulation should be, on the basis of clinical or research needs, the cells of interest are identified and separated from the blood sample and made available for the subsequent analysis steps.
Figure 1.
Schematics of the main preparation steps for blood samples and potential impact of on-chip blood sample preparation in medicine and sciences.
Cells in blood form a heterogenous and living mixture that continuously responds and adapts to the smallest changes in physiology. Each of these characteristics poses extreme challenges for extracting information from blood cells. The massive numbers and wide diversity of cell types present complicate precise identification of the target cells from a whole blood sample. There are roughly 5 × 106 cells in each microliter of blood, including red blood cells (RBCs), reticulocytes, platelets, and white blood cells (WBCs), which are most often the target of analysis. For every leukocyte in blood there are a thousand more RBCs with it. If WBCs are the target, all RBCs need to be removed, as they do not carry useful information and may even interfere with the subsequent analysis of the leukocytes. Leukocytes themselves are also very diverse. The classical separation of WBCs into five classes, neutrophils, lymphocytes, monocytes, eosinophils, and basophils (Table 1), is already too general for many applications. Recent advances in immunology have revealed a considerable number of subpopulations and sub-subpopulations that only can be identified by the presence of specific proteins on the surface of cells, also described as cluster of differentiation (CD) markers (4). Usually, not one but a combination of markers is used to classify cells, identify their function, or reveal specific cellular responses to pathological situations in the body. This extreme diversity poses serious challenges for achieving the desired purity of the final sample.
TABLE 1.
Absolute and relative number of cell populations and subpopulations in normal blood (adapted from Reference 11)
| Cell type | Average/µL | Percent of WBC |
|---|---|---|
| Erythrocytes (RBC) | 5,000,000 | |
| Reticulocytes | 30,000–70,000 | |
| Platelets | 200,000–500,000 | |
| All leukocytes (total) | 5000–10,000 | 100% |
| Neutrophils | 4000–8000 | 40%–66% |
| Monocytes | 200–800 | 4%–8% |
| Eosinophils | 50–300 | 1%–3% |
| Basophils | 0–100 | 0%–1% |
| Lymphocytes (total) | 1000–4000 | 20%–40% |
| CD4 + T Cell | 400–1600 | 15%–20% |
| CD8 + T Cell | 200–800 | 7%–10% |
| B-Cell | 200–800 | 8%–12% |
| NK | 100–500 | 4%–6% |
Gentle extraction of the identified target cells from the complex blood sample is an even greater challenge. Because most of the leukocytes are designed to respond quickly to changes in their environment, they can easily be altered by the handling procedures during the separation steps. Several studies document that the exposure of cells to improper stimuli during the blood processing steps can alter the original immuno-phenotype of the separated cells (5–7). If these alterations occur, subsequent analysis may not be able to discriminate between the primary state of target cells in the blood and the sample preparation artifacts.
Addressing the challenges of blood cell diversity and susceptibility to alterations can benefit from lab-on-a-chip technologies. The new separation and analysis methods can be more than just a miniaturization of current bulk assays. Building on demonstrated capabilities for microenvironment alteration and handling of individual cells (8, 9), more sophisticated techniques can be imagined to undertake the particular challenges of blood analysis. Gentle manipulation and precise control of the microenvironment of each individual cell in blood can be efficiently accomplished at microscale and would reduce the risks of altering the original state of the cells of interest. New separation principles and unique capabilities of integrating these with cell analysis are essential elements for accomplishing the goal of unaltered blood cell analysis.
ON-CHIP SEPARATION OF CELLS IN MICROSCALE FIELDS THROUGH DIFFERENTIAL ACTION ON DIFFERENT TYPES/SUBTYPES OF CELLS
The use of physical fields for the separation of cells takes advantage of the heterogeneity of physical properties of the blood cells. After exposure to fields of different nature, differences in size, density, electrical permittivity, dielectric characteristics, or adhesiveness among cells can be revealed in the form of forces differentially acting on cells of a particular type. Populations of cells that share similar behavior upon field exposure can then be differentially manipulated and eventually partitioned from the whole sample. It is important to emphasize that for most of the applications, separation using uniform fields can be achieved in the absence of additional cell-labeling or cell-modification steps. Devices using these principles may also perform the steps of identification and separation, as presented in Figure 1, simultaneously, resulting in a simpler design and higher reliability.
Small-scale implementation greatly favors on-chip field separation. Very strong fields can be created over small distances and small displacements on the scale of cell size can be used for effective separation. Even more important for cell separation, regardless of the nature of these fields, is microscale control, which allows for the manipulation of single cells and particles in suspension, with the potential for extreme purity or efficiency of the separation. Since the principles of separation of particles from solutions by using fields were outlined by Giddings in 1991 (10), a variety of fields have been employed in small-scale applications. Those relevant to blood separation are presented in the following sections.
Microfabricated Devices for Cell Separation Using Mechanical Forces
Differences in cell size are the easiest to observe when examining a blood smear under the microscope (11). Also, common centrifugation methods rely on differences in size and density among cells for blood separation. Geometrical differences between blood cells can also be exploited in microfabricated devices that achieve cell separation based on the mechanical restriction of cell displacement. Such devices are relatively easy to build using common microfabrication technologies; however, they are not very accommodating to changes in properties of the samples being separated. Separating target cells of new size usually requires a new design or at least a new device with modified dimensions. Also, despite simplicity, microfabricated devices working solely on the principle of size separation still have to overcome drawbacks such as limited efficiency or poor purity of the final sample.
Dense arrays of posts spaced 5 µm or 7 µm apart, and weir-type structures of 3.5 µm height were employed for the separation of leukocytes from whole blood (12). Successive arrays of narrow channels with sizes decreasing from 20 × 10 µm to 5 × 2.5 µm were also tested for separation of tumor cells from blood samples spiked with cultured tumor cells (Figure 2) (13). The common principle was that the leukocytes and other larger cells would be trapped at the entrance of the array, while, at the same time, erythrocytes would easily pass through the narrow spaces because of their smaller size and their particular biconcave discoid shape. However, experiments showed that WBCs, although bigger in size, could also “squeeze” through the same narrow spaces, limiting the overall efficiency of capture to values between 7% for the weir and 35% for the 5 µmspaced posts (12). Additionally, only a limited number of cells could be separated before the captured cells altered the flow through the device, an important issue for blood samples containing millions of cells. In a slightly different device featuring a 5 µm lattice of posts with a polar urethane coating, one interesting phenomenon was observed during the passage of cells (14). Among the leukocytes trapped at the entrance of the array, granulocytes and T lymphocytes were differentially trapped at different distances into the array. Trapping occurred in the absence of adhesive proteins on the posts, and it has been speculated that differences in viscosity between the two cell types or differences in nonspecific stickiness following the interaction between granulocytes and T lymphocytes during the separation process may be responsible (14). The limited success of these early approaches only underlines the challenges in understanding and exploiting the interaction between mammalian cells and mechanical structures for separation purposes.
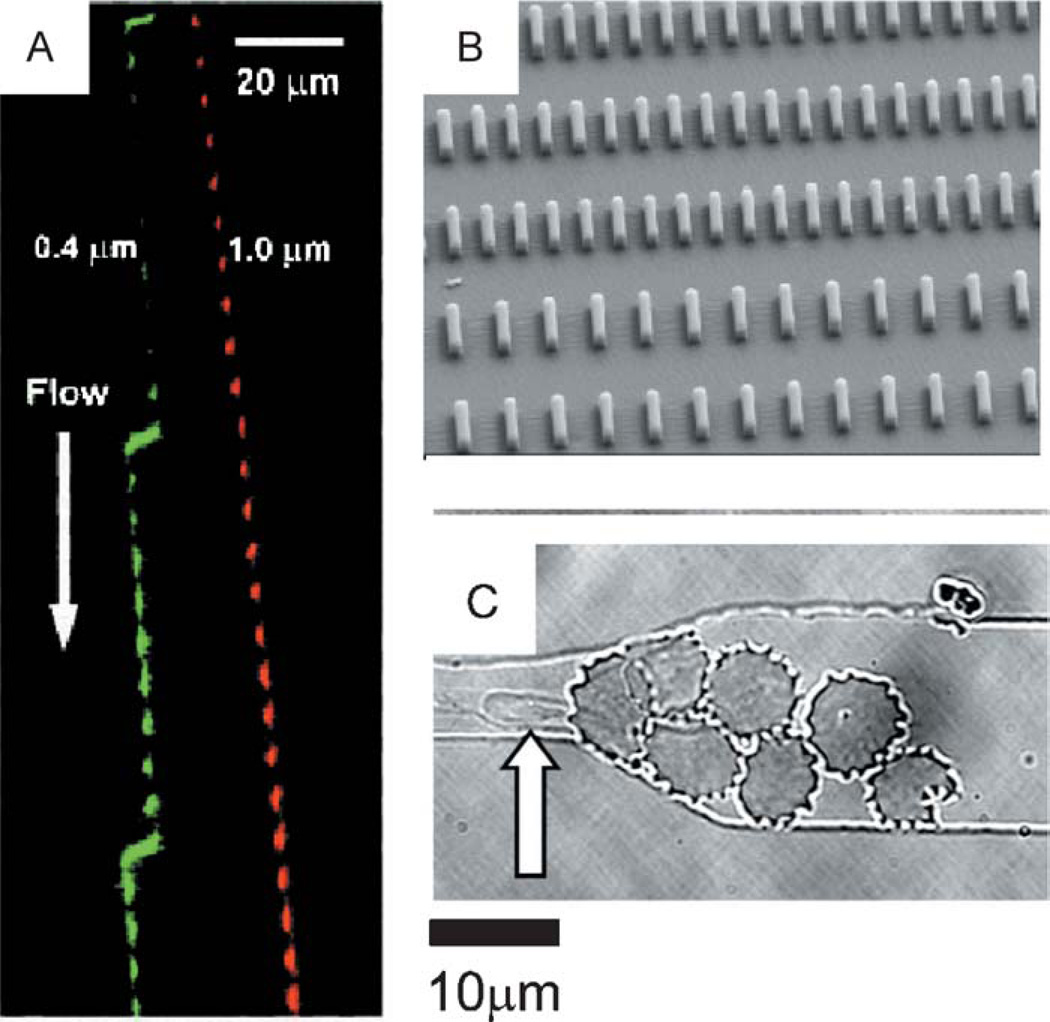
Figure 2.
Separation of cells using mechanical interactions. (A) Deterministic lateral displacement of fluorescent particles flown through an array of posts. Larger particles are deviated more from the axial flow stream (reprinted with permission from Reference 15; Copyright 2004 AAAS). (B) Two-dimensional array of posts for the isolation of cells based on size characteristics. Posts are spaced from 5 to 20 µm and allow the passage of erythrocytes while capturing the larger cells (13; Copyright 2004 IEEE). (C) A small group of infected RBCs in the schizont malaria stage blocking the entrance to a 6 µm constricted capillary. A normal RBC (arrow) passes through the labyrinth of infected cells and flows easily through the channel constriction (18; Copyright 2003 National Academy of Sciences, U.S.A.).
Using a completely different approach, the particularities of particle movement in laminar flow have been exploited for the separation of particles. Although not yet demonstrated for the separation of cells in blood, some microfabricated devices have been shown to discriminate few-micrometers differences in particle size, of the order observed between cells in blood. In one example, particles are separated through differential lateral displacement owing to asymmetric bifurcation of laminar flow around obstacles with sizes comparable to the size of the particles (Figure 2) (15). Alternatively, particles suspended in liquid were continuously separated upon passage through a narrow segment of width comparable to particle size. Two streams, one with particles and one without, were merged in the pinched microchannel, resulting in the alignment of particles to the sidewall. With all cells touching the same wall on the side of the cell stream, smaller cells had their geometrical center in the stream lines closer to the sidewall, whereas larger cells had their center closer to the stream lines in the middle of the channel. Consequently, after passage through the narrow section, cells that were initially randomly distributed across stream lines were segregated in different stream lines depending on their size. Segregation was increased even further when the flow stream spread again in a larger channel, with larger particles aligned toward the center and smaller particles pushed toward the walls (16). Finally, particularities of flow under a microstep structure in a translation movement over a flat surface were used in separating micron-sized particles and bacteria. Whereas the small particles were trapped under the step, larger particles went in the opposite direction, with the countercurrent generated by the motion of the step structure (17). Although several steps remain before these devices can be tested with mammalian cells, it is becoming evident that control of the microscale flow in the vicinity of cells is one critical element for the success of separation.
Although the deformability of cells of different sizes is reducing the efficiency of separation based on size, other devices have been designed to separate cells based on their mechanical rigidity in addition to size parameters. Malaria is one example of a disease where the increased rigidity of the RBCs leads to medical complications. Microfabricated devices, simulating capillary vessels as microchannels with sizes from 2 µm up to 6 µm, have been used to effectively separate more rigid, infected RBCs from normal, uninfected erythrocytes (18). Increased rigidity of the infected RBCs blocks their passage, even through large capillaries, and infected cells accumulate at the entrance to the channel (Figure 2). Interestingly, when more infected cells are trapped at the entrance of the capillaries, normal cells can still pass through the blockage formed by infected cells (18). Efforts to develop cheap and reliable diagnostic devices for malaria are not limited to mechanical separation of infected cells. Other principles, presented in the next section, have emerged, underlying the flexibility of microfabricated approaches.
Effective separation of plasma and cellular fractions from whole blood has also been accomplished using mechanical forces in microdevices. In general, the purpose of such a procedure is to prepare plasma for subsequent analysis by eliminating the potential confounding effects of cellular components on plasma biochemical assays. On-chip plasma separation is targeted to replace the classical benchtop centrifugation and allow integration with an increasing number of small, single-use, lab-on-a-chip-type devices for plasma analysis (19, 20). Plasma has been separated by flowing whole blood through microfilter-like parallel arrays of shallow channels (21), microchannel bent structures (22), or by centrifugation of 5 µL of blood in lab-on-a-disk-type devices (23, 24). Gentle handling of cells in many of these devices helps reduce the potential for the release of cellular factors in the plasma samples during the separation procedure. Conversely, target cells may be washed from plasma proteins, which may interfere with subsequent cell analysis. Overall, better understanding of the micromechanical interactions between cells and microstructures is poised to lead to very simple devices for cell separation, with a great impact on reducing the costs and increasing the accessibility of diagnostic and monitoring tests.
Dielectrophoresis in Microfabricated Devices for Cell Separation
Dielectrophoresis (DEP) uses the electrical polarization of cells in nonuniform electric fields to induce translational motion or reorientation of cells, affecting their dynamic behavior. Mammalian cells readily polarize when exposed to electric fields; however, their function is not affected by electric fields in quite a large range of intensities. The induced polarization depends on a multitude of factors related to cellular physiologic conditions, such as the bilipid membrane characteristics, internal structure, or size of the nucleus (25). In addition, depending on the frequency of the alternative electric field they are exposed to, the cells may polarize in the direction of the field vector (positive DEP) or in the opposite direction (negative DEP). The variability in cell response to electric fields is selective enough to differentiate not only between cell types but even between activation states of similar cells (26). On the other hand, the multitude of variables dictating the outcome of DEP makes the comparison among different results from different experimental systems rather difficult.
Because electrical fields scale down favorably at microscale, low voltages are enough to produce intense electrical fields. This and other technological advantages work in favor of the dielectrophoretic principles for separation in lab-on-a-chip devices. For example, alternate or direct electric fields are relatively easy to implement and control in microdevices. Thermal and hydrolysis effects that are detrimental for living cells, and that plagued early large-scale DEP devices, can be avoided. Exquisite control over the characteristics of the electric fields at length scales comparable to cell size can be achieved by the use of microfabricated electrodes and features. Nonetheless, DEP devices may be easier to reconfigure for different cell types than any other type of microdevices. Because cells respond differently depending on the frequency of the applied alternate electric fields, it may be possible to adjust DEP devices for separating different cell subtypes just by changing the field frequency or amplitude. As a consequence of this display of favorable factors, a large number of microscale devices have been proposed for different applications involving the manipulation of cells and particles in suspensions.
Some common principles for cell separation from mixtures using dielectric fields in lab-on-a-chip devices have already been outlined in several reviews (25, 27, 28) and many of them can be extended to cell separation from blood. Depending on the size of the sample, the strategies recommended for dielectrophoretic separation are different. For small samples containing few cells, the cells of interest may be separated by trapping them in predefined locations. For larger samples, when the number of cell traps would be prohibitively high, cells are usually diverted from the mixture into distinct flow streams based on their dielectric properties and thus separated in time or space.
Cell trapping using DEP is based on the creation of energy traps of sizes comparable to the cells to be captured. Typically, traps created by positive DEP are used for separation of cells from flowing mixtures. Cells that are trapped are in stable equilibrium and can be released by simply turning off the electric field (Figure 3,) (29). Alternatively, perturbations of the electric field can be induced through arrays of insulators (30). The selectivity of cell capture from a mixture can be enhanced when the frequency of the electric field is chosen such that the unwanted cells are driven away by negative DEP (31). However, if the crossover frequency for the two types of cells to be separated is close, separation may not be very effective. Cell separation can be further perturbed by uneven fluid flow conditions or imperfections in the electric field in large electrode arrays (28).
Figure 3.
Microfabricated DEP devices for the separation of cells. (A) Linear array of fluorescently tagged cells captured using DEP traps. (B, C) Pseudocolored scanning electron micrograph showing a single trap consisting of four electroplated gold electrodes arranged trapezoidally along with the substrate interconnects and an eight-trap array. Scale bars: (B) 20 µm, (C) 100 µm (reprinted with permission from Reference 29; Copyright 2002 American Chemical Society). (D, E) Selective trapping of malaria-infected RBCs from a heterogeneous mixture using DEP (32; reproduced with permission from The Royal Society of Chemistry). (F) Hyperlayer separation through the differential sedimentation of cells in a dielectrophoretic field (43; Copyright 2004 Biophysical Society).
In the absence of flow, discrimination may be improved by dielectric levitation and concentration of target cells on the surface of electrode arrays. For example, malaria-infected erythrocytes that have different electrical properties compared with normal RBCs owing to changes in their ionic content were concentrated at the center of a spiral array of electrodes (Figure 3).A sensitivity of 20 infected cells separated from 106 total cells has been reported by the use of simple, four-pole, planar devices (32). Separated cells can be examined further by different detection and imaging systems (fluorescence, cell morphometry, etc.), and some of them selectively released to improve the purity of the final sample.
Cells from larger samples, containing millions of cells, can be separated using hyperlayer techniques, by applying DEP forces to all cells flowing in microchannels from electrode arrays located at the bottom of the channel. Three forces acted simultaneously on every cell: sedimentation forces that were proportional to particle weight, DEP forces that fell exponentially with the distance between the cells and the electrodes, and drag forces that were proportional to fluid velocity in the channel (Figure 3). Equilibrium positions at different heights inside the channel for different cells were the result of the balance between changing levitation and constant sedimentation forces. For each cell, different velocities along the channel were the consequence of the parabolic velocity flow profile across the channel. As a result, cells from a heterogeneous mixture were separated along the channel and captured at different time intervals at the outlet (25). Differences between normal and cancer cells were exploited for hyperlayer separation of whole blood samples. Cells of the metastatic human breast cancer cell line MDA231 were selectively captured from whole blood by balancing the hydrodynamic and dielectrophoretic forces acting on cells upon their passage over a microelectrode array (33). Inter-digitated electrodes patterned on the bottom of thin flow chambers were used to levitate normal and cancerous cells at different levels, depending on their different dielectric properties. Separation of 50,000 tumor cells from a mixture of 10 µL of blood was achieved under the influence of a parabolic flow profile in less than 5 min (34). Although impressive, the efficacy of separation in these devices would need to be improved even further for applications such as the detection of cancerous cells in clinical samples of blood. Especially in the early stages of cancer, when only a few tumor cells are present in blood, larger volumes of blood, of the order of milliliters, would need to be processed before one or more cancer cells could be detected.
Physical separation of cells in distinct streams using DEP has also been demonstrated. A complex system where several steps of using DEP forces to funnel cells into narrow streams, break aggregates, and trap or separate cells along the stream has been used to separate individual cells (35). Separation of cells from the stream has been demonstrated for macrophages, T cells, and B cells. Upon passage over an array of interdigitated electrodes, at 400 µm from the edge of the array, the cell stream was separated into three distinct 150-µm-wide bands (36). More recent experiments from the same group, using a leukemia cell line, showed that further increases in selectivity could be achieved by focusing the cells in a narrow stream on top of the electrodes (37). Following a different approach, tumor cells from a Jurkat cell line were separated by deflection through negative DEP from erythrocytes in phosphate buffered solution (38). Other tumor cell lines mixed with whole blood were also separated by the use of dielectrophoretic forces in devices with arrays of microelectrodes (39).
When the contrast between the electrical properties of cells is less clear, cells can still be enriched in complex mixtures. Fivefold enrichment of hematopoietic cells (CD34+) in whole blood samples was observed in fractions collected after cell passage over a dielectrophoretic electrode array. Stem cells remained viable, were not altered by this enrichment method, and were capable of forming colonies in culture (40, 41). Thirty-fivefold enrichment of leukocytes from whole blood diluted 1:1000 in sucrose buffer has been reported by the use of the hyperlayer DEP separation method (42). Optimization of the separation conditions led to relatively high performance separation of T and B lymphocytes from monocytes, T (or B) lymphocytes from granulocytes, and monocytes from granulocytes. Because DEP devices are usually not complicated and do not involve cell-labeling or cell-modification steps, they are always a serious candidate for any on-chip hematological analysis (43).
The use of electric fields for cell separation is not limited to dielectrophoresis devices. Selective destruction of cells in a mixture has been shown by the use of pulsed electric fields of up to 2 kV/cm (44). Although not implemented in a microdevice, but nonetheless using microelectrodes spaced 400 µm apart, this method is based on the fact that the strength of the electric fields can be adjusted such that pores can be opened in larger cells before smaller cells are affected. Exposure of blood (mixture of peripheral blood mononuclear cells) to pulsed electric fields caused stepwise elimination of large monocytes without affecting smaller lymphocytes or stem cells. Through the same method, peripheral blood has been enriched in hematopoietic stem cells (CD34+/CD38−) and stem cell function preserved (44).
Microscale Optical Interactions for Cell Separation
Approaches using light to separate cells from complex mixtures are particularly attractive because they avoid mechanical contact between cells and surfaces and reduce the potential of activation. Optical trapping for manipulation and sorting of individual cells can be readily achieved by the use of laser tweezer-type devices. Although cell handling can be very precise, only one cell at a time can be manipulated (45). Scaling up the separation for millions of cells at a time, like those in small blood samples, would be very challenging using such systems. Thus, devices for parallel processing of a large number of cells in parallel have been tested. A microfluidic system using a diode laser bar has been demonstrated for the separation of cells based on their size. While larger cells are deviated when passing through the laser enlarged beam, smaller cells are not, and consequently, separation of large and small particles can be achieved (46). A dynamically reconfigurable optical lattice, produced as an interference pattern, was also used to separate particles cells from mixtures based on size and refractive index (47). More recently, the same principles of tunable optical lattices have been used to separate erythrocytes from leukocytes with more than 95% efficiency (48). This type of device is easily reconfigurable by adjusting the interference pattern, and because it has no narrow channels it is less prone to clogging during its use. The increased availability and continuously decreasing costs of solid-state coherent light sources are additional arguments for the use of microscale optical interactions for the separation of cells from blood samples.
Microfabricated Devices for Cell Separation Using Magnetic Interactions
Intuitively, the iron-bearing hemoglobin molecules contained only by erythrocytes would lead to differences in the magnetic properties between RBCs and other blood cells. However, practical measurements showed that, in spite of the higher iron content, RBCs have, at least in oxygenated blood, the same diamagnetic behavior as WBCs. A weak paramagnetic behavior has been observed for RBCs in deoxygenated blood. Only reduced forms of hemoglobin, deoxihemoglobin and methemoglobin, and pathologic hemozoin in RBCs infested with malaria are paramagnetic (49). In the presence of high magnetic fields and gradients, such differences can be used to separate leukocytes from RBCs. This was first demonstrated by Melville et al. (50) through the capture of erythrocytes from whole blood on a metal mesh in the vicinity of a powerful magnet. Although large magnets are still needed to produce the intense magnetic fields, microscale devices have the advantage of producing high magnetic field gradients in the proximity of microscopic magnetic features. Analytical magnetapheresis has been demonstrated in experimental settings involving microscopic features (51, 52). Particles and cells with various magnetic susceptibilities were selectively deposited and separated from suspensions flowing in a thin layer under a magnetic field applied perpendicular to the flow (52). The selectivity of magnetic separation of cells can be tremendously enhanced by coupling magnetic beads decorated with antibodies against proteins on the surface of the cells, a process generally known as magnetic cell sorting (MACS). Because in this process the identification of cells is based on protein recognition rather than intrinsic magnetic properties of the cells, devices using these principles are discussed separately in the next section.
Biochemical Interactions for the Separation of Cells in Microfabricated Devices
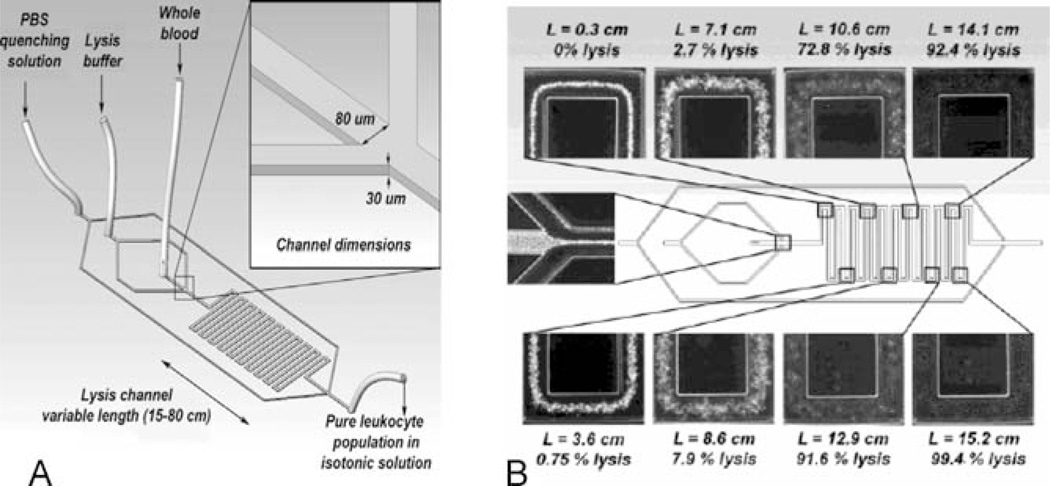
Biochemical differences among cells can be effectively used to selectively destroy some cell subpopulations after uniform exposure of the whole sample to a selectively toxic environment. It has been known, for example, for almost a century, that WBCs are more resistant to solutions of ammonium chloride than RBCs, which are readily lysed within tens of seconds after exposure (53). In regular separation procedures, bulk volumes of blood are mixed with bulk volumes of the lysing agent for periods of time long enough to account for diffusion and RBC lysis. At the same time, WBCs are also exposed and may be affected by the lysing solution; shortening the exposure time is of interest for minimizing the chance for WBC alteration (54). Although calculations and experiments show that lysing agent diffusion is usually the limiting factor for speeding up the reaction, it is evident that shortening the diffusion distance would provide significant advantages. This can be easily achieved in microfluidic devices, and complete lysis of erythrocytes and approximately 100% recovery of leukocytes were accomplished after exposure to an isotonic lysis buffer for less than 30 s (Figure 4). Alternatively, complete lysis of erythrocytes in hypotonic deionized water occurs two times faster, in only 15 s, whereas the leukocytes appeared to be intact (55). Following RBC lysis, leukocytes are immediately returned to isotonic physiological conditions by the addition of buffer. Microfluidic selective cell lysis not only results in improved yield and viability of the WBCs compared with bulk methods, but by assuring equal treatment for all cells in the sample it may also have a favorable effect on subsequent analysis by reducing yet another source of variability.
Figure 4.
Microfluidic lysis device for selective lysis of RBCs in whole blood. (A) Device design and construction. (B) Snapshots illustrating lysis of erythrocytes in microfluidic channels. L denotes the distance traveled in the microchannels and % lysis denotes the percentage of lysed erythrocytes. Complete erythrocyte lysis is achieved in less than 30 s (reprinted with permission from Reference 55). (Copyright 2004 American Chemical Society.)
ON-CHIP CELL MICROARRAYS FOR BLOOD SAMPLE PREPARATION
In the previous section, we described the use of microscale fields for the separation of target cells from whole blood. In all devices chosen as examples, separation was accomplished under the selective action of uniform fields, which would differentially act on target cells, separating them from the mixture without the need for additional means for cell identification. In the following section, the focus is on devices in which the processes of identification and of separation of the target cells from mixtures are dissociated into distinct steps. Although the implementation of two distinct steps leads to more complex devices, it also appears to be beneficial in terms of efficiency and accuracy of separation. More precise identification, involving potentially more than one criterion, can be combined with more effective means of cell handling, resulting in more pure populations.
There are two distinct approaches for the identification and separation of cells based on the number of cells simultaneously processed. In the serial approach, comparable to flow cytometry, cells are aligned in a single row, identified as they pass one by one in front of a detector module, and immediately separated using a distinct module. In the parallel approach, cells are first positioned in a regular array and then simultaneously identified and separated in pure subpopulations. At microscale, the first approach would result in cell-sorting devices that are smaller, cheaper, and portable compared with present flow cytometers. The second approach, using the array format that allows simultaneous processing of a larger number of cells, would lead to significant improvement in throughput and productivity of cell separation compared with the current fluorescence activated cells sorting (FACS) systems. The impact of microfabrication techniques on serial cell sorting is limited to incremental improvements by miniaturizing existing systems or by expanding the identification criteria. By contrast, in the case of cell arrays, microfabrication plays a fundamental role because only technologies working at the same scale as the cell size can achieve both the precise positioning of the cells in the array format and gentle extraction of target cells without altering them in the process.
Microfluidic Flow Cell Sorters
The current state of the art for cell identification and separation is FACS technology. Cells labeled with fluorescent dyes are identified as they pass through an array of excitation laser beams and optical detectors. Target cells, according to their fluorescence characteristics, are then isolated into droplets that are separated using switching electric fields. Although effective in rapidly separating cells with high accuracy, FACS machines are large, expensive, and require skilled technicians to operate them. Although being able to sort up to 106 cells/s, when dealing with whole blood direct analysis, FACS becomes very time consuming owing to the large number of RBCs, reticulocytes, and platelets in the samples. Preparation steps to eliminate the unwanted cells and increase the proportion of target WBCs are usually required and, as discussed in Brief Overview of the Challenges for Blood Sample Preparation, above, may easily alter the target cells.
Recent developments in microfluidics, through the microscale implementation of optics and flow control structures, have led to the development of different microsystems for cell separation comparable to the FACS sorting machines. In a direct extension of the FACS optical systems, several different optical elements (waveguides, lens, and fiber-to-waveguide couplers) were integrated with microfluidic channels to form complete cell identification modules (56). Standard photolithography and regular thick photoresists (SU8) were used to create the microfluidic network and the optical components of the microsystem. Polystyrene beads were measured in the microchip flow cytometer, and three signals (forward scattering, large-angle scattering, and extinction) were measured simultaneously for each bead (56). In a different approach, optical identification has been combined with hydrodynamic liquid pumping and electrokinetic switching to demonstrate the separation of fluorescent-tagged 1-µm-sized bacteria cells. As in FACS, the sample is focused hydrodynamically; however, the selection of particles is achieved not by breaking the stream into droplets, but rather through electroosmotic current flow by switching the fluid stream to one or another channel at the downstream junction (57). Similar systems using optical identification of the target cells, such as electrokinetic focusing and switching (58), monolithic soft microvalves (59–61), or even optical tweezers (62), for the separation of cells have also been reported. By the isolation of cells directly from continuous streams, these last two approaches claim to avoid the technical problems of cell alterations associated with droplet formation and isolation of cells into droplets characteristic to FACS machines.
Other parameters, such as cell deformability and cell adhesivity, can be assessed in microfluidic devices by measuring the time for cell passage through micron-sized channels. The presence of an optical trap inside the narrow channel may eventually enhance the differences between normal and less deformable cells, allowing for the differentiation among RBCs from normal and cancer patients (63). The spectral impedance of individual erythrocytes was measured in a glass-polyimide microfluidic chip that uses integrated channels and electrodes (64). A laminar liquid flow carries the suspended particles through the measurement area and each particle’s impedance signal is measured at multiple frequencies, ranging from 100 kHz to 15 MHz. Although the spectral impedance of cells is an integrative measurement of membrane capacitance, cell size, cytoplasm resistance, and ionic channels, the device allows for differentiation between erythrocytes and other cell types (64). Capacitance measurement of cells has been shown to differentiate cells on the basis of their DNA content (65). Although in the original application cells were fixed and suspended in ethanol, it has been suggested that microchip sorters for live cells could be developed based on capacitance measurement principles (66).
When identification of specific protein markers on the surface of cells is accomplished using magnetic beads decorated with antibodies directed against these proteins, the subsequent separation steps are significantly simpler. Immunomagnetic separations from bulk mixtures have been extensively studied and are the principle behind numerous commercial products (67). Chaotic trajectories of the magnetic particles, generated through appropriate temporal variations of electric currents in microconductors imbedded in microchannel walls, have been employed to enhance the mixing of particles and cells (68). Ferrofluid-based capture reagents consisting of nanoparticles with a magnetic core surrounded by a polymeric layer coated with antibodies have also been reported for cell-separation applications (69). Limitations in the purity of the final product and efficiency of the separation steps can be largely eliminated by the use of microscale devices.
In the most simple form, cells tagged with magnetic beads are flown through a microchannel and captured in precise locations (70). In a more elaborate design, cells marked with magnetic beads are separated on an array of magnetic wires (71). Cells with magnetic beads attached through specific antibodies were introduced in a narrow stream at one side of a parallel array of magnetic wires. Large magnetic field gradients that formed at the edges of microfabricated cobalt-chrome-tantalum wires arranged diagonally were employed to deviate tagged cells away from the main stream and into separate channels. More recently, Inglis et al. proposed the use of magnetic beads with different magnetic susceptibility and different antibody specificity to simultaneously isolate cells of different types from complex mixtures, such as blood (72). RBCs coupled with magnetic microspheres were separated from whole blood after flowing the sample over an array of permanent magnets in alternating spatial arrangements. The effectiveness of the RBC magnetic separation and the purity of plasma solution was significantly better using the device compared with conventional centrifugal methods (73). The flexibility of magnetic separation systems can potentially be enhanced by the use of magnetic particles of different sizes in a device based on free-flow magnetophoresis principles. In one such example, small (2.8 µm) and large (4.5 µm) magnetic particles were deflected toward different exits upon passage through a horizontal magnetic channel in the presence of magnetic and gravitational fields (74). Overall, technical advantages such as the ability to create large magnetic field gradients using only modest magnetic sources and the capability for precise handling of the separated cells make the use of microscale magnetic sorters very attractive for point-of-care diagnostic devices.
Two-Dimensional Cell Microarrays for Cell Sorting
Compared with flow cytometry-type devices where cells have to pass in front of a detector one by one before they are separated, two-dimensional cell arrays allow for identification and separation of multiple cells at the same time. Microarray-based cell sorting has the potential for higher throughput and productivity, yet other advantages may become even more important. Whereas FACS systems allow one cell to be measured only once, arrayed devices would permit multiple measurements at multiple time points from the same cell. These devices would enable the separation of cells based on their individual dynamic responses to stimuli or drugs, or permit long-term observation of individual cells. Microarray devices allow the use of cell morphology, location of intracellular molecules, and cellular structures as criteria for cell separation. Such advantages make the development of microarray format cell sorters very attractive for clinical and research applications, and microarray format cell sorters have a great potential in blood cell separation.
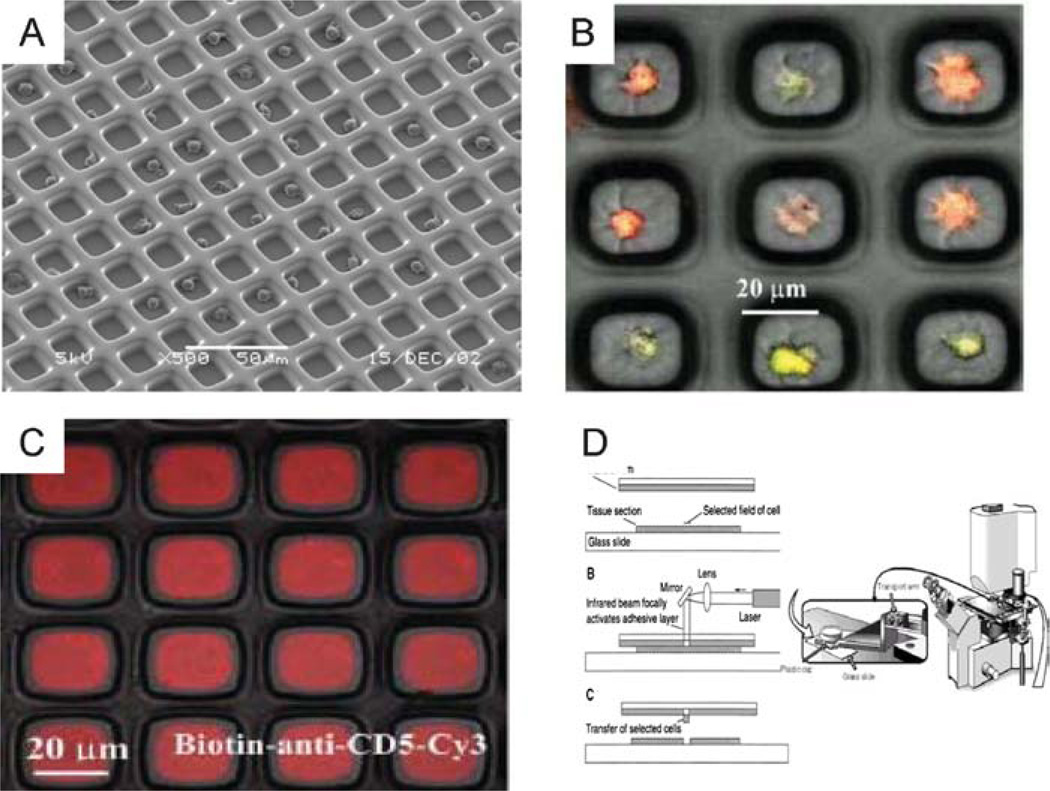
Several approaches have been explored for the creation of the regular arrangement of cells on the surface of flat platforms. Passive cell positioning through sedimentation from a cell suspension has been investigated through the creation of microscale wells or chambers in materials such as glass (75), silicon (76), cellulose ester membranes (77), or polyethylene glycol (78). The precision of cell localization in an array can be negatively affected by the nonspecific adhesion of cells in between the wells. Micropatterned PEG gels were proven to be very effective in this regard (Figure 5) (79). In addition, patterning the bottom of the PEG wells with adhesive proteins or monoclonal antibodies can alter the cell-substrate interaction, further enhancing the selectivity of cell capture in the array (79).
Figure 5.
Leukocyte arrays on a PEG grid. (A) Scanning electron micrograph of the PEG grid and trapped lymphocytes (reprinted with permission from Reference 78; Copyright 2003 American Chemical Society). (B) Composite image of differentially labeled T and B cells occupying the same PEG microwells array. T lymphocytes are fluorescently labeled green and Raji B lymphocytes are labeled orange (79; reproduced with permission of The Royal Society of Chemistry). (C) Biotinylated anti-CD5-Cy3 antibody bound to the bottom of avidin-modified PEG microwells for specific cell capture (79; reproduced with permission from The Royal Society of Chemistry). (D) Laser capture microdissection principle for retrieving cells from arrays (92).
Regular methods for patterning cells on surfaces (80) are not directly useful for cell sorting in array format because they are either too slow or not selective enough to work with complex mixtures of cells. However, some patterning principles can be extrapolated for selective cell capturing in precise locations. Selective attachment of cells on patterned antibodies allows immunophenotyping and simultaneous sorting of specific leukocyte populations into multiple subpopulations (81, 82). The versatility of the concept is confirmed in the selection of antigen-specific B lymphocytes from a lymphocyte spleen extract using a microarray of antigens in a chip format. Following stimulation of B cells using IgM antibodies and monitoring of calcium flux, B cells responsive to antigen stimulation were identified (83). Micropatterned surfaces presenting specific proteins were created on negatively charged polyelectrolyte surfaces to arrange B cells in large arrays, for sensing or sorting purposes (84). Selective capturing of cells from flow on uniform surfaces covered with antibodies has been demonstrated in microfluidic devices (85). In microchannels of size comparable to the cell diameter, cells can be effectively pushed closer to the antibody-covered surfaces, facilitating interaction and capture (86). Also, a combination of PEG and antibodies can improve the selectivity of capture (85). Selective extraction of leukocytes from blood has also been reported in microfluidic devices using a biomimetic approach, involving adhesive rolling of leukocytes from whole blood on selectin-coated surfaces. Although RBCs do not interact with selectins and pass through the channels at higher speeds, leukocytes are slowed by the interaction with selectins, and they are consequently enriched inside the channel (87). The efficiency of selectin-based cell capturing can be enhanced to some degree by the presence of microstructure inside channels (88).
Alternatively, laser printing (89) and laser traps (90) can be used to precisely position cells on surfaces. Vertical-cavity surface emitting laser-driven infrared optical tweezers were used for remote manipulation of individual cells arrayed using direct current electrophoresis (91). The laser can also remove selected cells from their locations on the array (79), either by capture microdissection (92) or laser catapulting (93, 94). Through the high photonic pressure force from the focused laser beam, selected cells are ejected from the object plane and catapulted directly into a collection tube for further analysis. Optical cell arrays employ cells immobilized on optical fibers or waveguides, and absorbance, luminescence, or fluorescence signals are measured (95). The use of fiber optic arrays instead of automated digital microscopy can improve the overall throughput by 500-fold (96).
More complex sorting of cells can be achieved by creating an arrangement of cells that can be actively controlled to hold or release cells. To date there have been few examples of devices in which cells were captured in an array format, interrogated or studied at those locations, and then released. One example of a microarray for cell sorting is the micro dynamic flow cytometer (µDAC) developed by Maxwell et al. (97). At each cell-trapping location across the array, suction pressure can be applied to capture one cell per well from a cell suspension. Once cells are captured, they can be analyzed and the desired fractions released from their suction traps through a bubble-based actuator. Another approach relied on the use of an array of dielectrophoretic traps that can capture and hold cells as desired (29, 98). Cells in the array were used in parallel luminescent single-cell assays and then sorted based on their dynamic functional responses to stimuli. DEP traps were used to stably confine cells and hold them against disrupting fluid flows (99). These traps can be physically arrayed and electrically addressed, enabling dynamic array cytometry (29). Combinations of electrical and optical methods for cell manipulation have also been demonstrated. Direct current electrophoresis has been used to position negatively charged mammalian cells in array formats, and optical tweezers subsequently used to move cells (91).
Cells decorated with magnetic beads were captured on a microelectromagnet matrix formed by perpendicularly arrayed, 10-µm-spaced, conducting wires on a Si/SiO2 substrate. The insulating layer on top of the wires allowed the creation of maxima in the magnetic field between two adjacent wires. A force of maximum 40pN was induced in each bead, therefore trapping the cells attached to the magnetic beads in that location. By adjusting the currents in the array of wires, cells/particles can be trapped and transported across the matrix, at single-cell resolution, allowing for sorting/mixing capabilities. Although the functioning of the device has been shown only for bacteria cells, through the choice of the antibodies on beads, the authors suggest that bacteria from whole blood or even cells could possibly be trapped this way (100).
The variety of approaches to cell separation, using different principles and a wide range of selection criteria, underlines the growing interest and opportunity in the area of cell separation using microscale techniques. However, taking full advantage of the emerging technologies would require the integration of separation and analysis into unitary systems. After capturing the cells of interest, these systems would extract information from them, in the form of gene and/or protein expression, which can be used further for clinical diagnostics and medical research.
INTEGRATED LAB-ON-A-CHIP DEVICES FOR CELL SAMPLE ANALYSIS
The ultimate goal for blood sample preparation is to select homogenous target cell populations and make them available for analysis. Various lab-on-a-chip devices for a wide range of assays in small volumes, incorporating reaction chambers, separation columns, micropumps and valves, as well as other components, have been developed (9, 101). Proteins can be directly captured, isolated, and identified on chip using techniques such as electrophoresis, isoelectric focusing, and mass spectrometry (102). Lab-on-a-chip devices already exist for a range of nucleic acid analysis, including, but not limited to, PCR, gel electrophoresis, and hybridization assays (103). Nonetheless, cells can be explored using functional assays, by exposing them, on the chip, to precisely controlled microenvironments, revealing the particularities of cell response to stimuli or allowing one to probe into the intracellular mechanisms (104, 105). However, most often these devices start with a cell sample that has already been partially processed off the chip. Only a small number of lab-on-a-chip devices have been proposed that integrate steps starting from blood processing and separation of target cells to cell lysis and analysis.
The first example of an integrated device was demonstrated by Burns et al. in 1997 (106).A device that integrated components such as channels, heaters, temperature sensors, and fluorescence detectors was developed to analyze nanoliter-sized DNA samples. The device metered and measured out aqueous reagents and DNA-containing solutions, mixed the solutions together, amplified and digested the DNA to form discrete products, and achieved separation and detection of the products. No external lenses, heaters, or mechanical pumps were necessary to complete the sample processing and analysis (106). Following this demonstration, there have been several attempts to integrate multiple functions onto a single chip (Figure 6). Several reports have shown integration of simple functions on microfabricated devices that can perform lysis of cells (107), quantitative biochemical analysis of soluble components at the single-cell level (108), analysis of nucleic acids (109–111), or proteomic analysis (102). Gene transfection in cells sorted in a highly parallel format (112) has been used to alter specific cells for cell therapy purposes. Detection of pathogenic bacteria and genotyping can be accomplished in lab-on-a-chip devices (113–117). Single-cell injection, lysis, separation, and detection of intracellular constituents have been reported (107–108, 118–120). Complete on-the-chip processing of small samples for extraction of DNA and RNA has been demonstrated in nanoliter volumes, opening new opportunities for research (121).
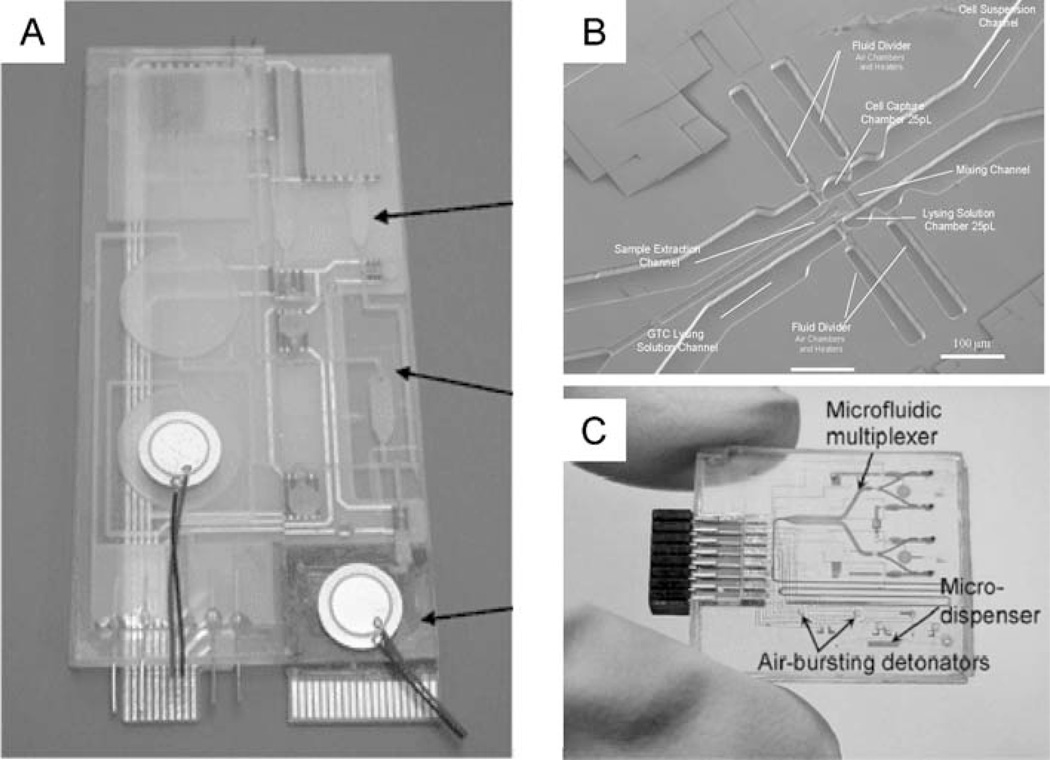
Figure 6.
Integrated devices for sample preparation and analysis. (A) Schematic of a plastic fluidic chip incorporating microfluidic networks, electrochemical and thermopneumatic pumps, valves, and a microarray chip (reproduced with permission from Reference 114; Copyright 2004 American Chemical Society). (B) Scanning electron micrograph of a microfluidic device for single-cell handling, lysis, and quantitative analysis (108; Copyright 2004 American Chemical Society). (C) Assembled biochip with microfluidic control system, air-bursting detonators for fluid driving, and blood-gases analyzers (20; Copyright 2004 IEEE).
Complete analysis of blood cells is of major interest for medical and science applications. In dealing with the complexity of the blood analysis process, modular approaches have an advantage because each step can be optimized individually. One such example, a microdevice that combines blood lysis, ultrasonic mixing for sample preparation, and on-chip PCR, showed promising results in the genetic analysis of bacteria from whole blood samples (121). Potential problems may arise, however, from the incompatibility between successive steps of analysis. For example, when WBCs were extracted on the chip from 3.5 µL of whole blood and PCR performed (12), clustering of RBCs and subsequent RBC lysis can release hemoglobin that inhibits PCR reactions. Overall, complex microdevices to address specific problems in blood cell analysis can be built using modular schemes and microfabrication technologies, allowing for utilization of a wider range of cell separation and analysis principles than at macroscale and for complex integration of modules working on distinct principles.
CONCLUSIONS
Rapid advances in understanding the particulates of blood sample preparation and technological developments in microfabrication are converging into new capabilities for blood analysis. As lab-on-a-chip devices become enabling tools for separation and analysis of small and homogenous subpopulations of cells precisely and without altering their state, new possibilities open for diagnostic applications and treatment. In parallel, microfluidic devices are new tools for discovery given their potential to perform comprehensive analysis from small samples, down to a single cell, which would have not been accessible otherwise.
ACKNOWLEDGMENTS
This work was supported in part by the National Institute of Biomedical Imaging and Bioengineering (BioMEMS Resource Center, P41 EB002503).
Contributor Information
Mehmet Toner, Email: mtoner@hms.harvard.edu.
Daniel Irimia, Email: dirimia@hms.harvard.edu.
LITERATURE CITED
- 1.Wintrobe MM. In: Blood, Pure and Eloquent: A Story of Discovery, of People, and of Ideas. Wintrobe MM, editor. New York: McGraw-Hill; 1980. p. 771. [Google Scholar]
- 2.Bauer J. Advances in cell separation: recent developments in counter-flow centrifugal elutriation and continuous flow cell separation. J. Chromatogr. B. 1999;722(1–2):55–69. doi: 10.1016/s0378-4347(98)00308-9. [DOI] [PubMed] [Google Scholar]
- 3.Kost GJ, Ehrmeyer SS, Chernow B, Winkelman JW, Zaloga GP, et al. The laboratory-clinical interface—point-of-care testing. Chest. 1999;115(4):1140–1154. doi: 10.1378/chest.115.4.1140. [DOI] [PubMed] [Google Scholar]
- 4.Goldsby RA, Kindt TJ, Osborne BA, Kuby J. Immunology. 5th ed. New York: W.H. Freeman; 2003. pp. A-1–A-12. [Google Scholar]
- 5.Fukuda S, Schmid-Schonbein GW. Centrifugation attenuates the fluid shear response of circulating leukocytes. J. Leukocyte Biol. 2002;72(1):133–139. [PubMed] [Google Scholar]
- 6.Hallden G, Nopp A, Ihre E, Peterson C, Lundahl J. Conditions in blood sampling procedures that extend the ex vivo stability of eosinophil activity markers in peripheral blood from allergic patients and healthy controls. Ann. Allergy Asthma Immunol. 1999;83(5):413–421. doi: 10.1016/S1081-1206(10)62839-6. [DOI] [PubMed] [Google Scholar]
- 7.Lundahl J, Hallden G, Hallgren M, Skold CM, Hed J. Altered expression of Cd11b/Cd18 and Cd62l on human mono-cytes after cell preparation procedures. J. Immunol. Methods. 1995;180(1):93–100. doi: 10.1016/0022-1759(94)00303-e. [DOI] [PubMed] [Google Scholar]
- 8.Vilkner T, Janasek D, Manz A. Micro total analysis systems. Recent developments. Anal. Chem. 2004;76(12):3373–3385. doi: 10.1021/ac040063q. [DOI] [PubMed] [Google Scholar]
- 9.Voldman J, Gray ML, Schmidt MA. Microfabrication in biology and medicine. Annu. Rev. Biomed. Eng. 1999;1:401–425. doi: 10.1146/annurev.bioeng.1.1.401. [DOI] [PubMed] [Google Scholar]
- 10.Giddings JC, Barman BN, Liu MK. Separation of cells by field-flow fractionation. ACS Symp. Ser. 1991;464:128–144. [Google Scholar]
- 11.Wintrobe MM. In: Wintrobe’s Clinical Hematology. 10th ed. Richard Lee EG, Foerster J, Lukens J, Paraskevas F, Greer JP, Rodgers GM, Wintrobe MM, editors. Philadelphia: Lippincott Williams & Wilkins; 1999. [Google Scholar]
- 12.Wilding P, Kricka LJ, Cheng J, Hvichia G, Shoffner MA, Fortina P. Integrated cell isolation and polymerase chain reaction analysis using silicon microfilter chambers. Anal. Biochem. 1998;257(2):95–100. doi: 10.1006/abio.1997.2530. [DOI] [PubMed] [Google Scholar]
- 13.Mohamed H, McCurdy LD, Szarowski DH, Duva S, Turner JN, Caggana M. Development of a rare cell fractionation device: application for cancer detection. IEEE Trans. Nanobiosci. 2004;3(4):251–256. doi: 10.1109/tnb.2004.837903. [DOI] [PubMed] [Google Scholar]
- 14.Carlson RH, Gabel CV, Chan SS, Austin RH, Brody JP, Winkelman JW. Self-sorting of white blood cells in a lattice. Phys. Rev. Lett. 1997;79(11):2149–2152. [Google Scholar]
- 15.Huang LR, Cox EC, Austin RH, Sturm JC. Continuous particle separation through deterministic lateral displacement. Science. 2004;304(5673):987–990. doi: 10.1126/science.1094567. [DOI] [PubMed] [Google Scholar]
- 16.Yamada M, Nakashima M, Seki M. Pinched flow fractionation: continuous size separation of particles utilizing a laminar flow profile in a pinched microchannel. Anal. Chem. 2004;76(18):5465–5471. doi: 10.1021/ac049863r. [DOI] [PubMed] [Google Scholar]
- 17.Vankrunkelsven S, Clicq D, Pappaert K, Ranson W, Det Tand C, et al. A novel microstep device for the size separation of cells. Electrophoresis. 2004;25(10–11):1714–1722. doi: 10.1002/elps.200405900. [DOI] [PubMed] [Google Scholar]
- 18.Shelby JP, White J, Ganesan K, Rathod PK, Chiu DT. A microfluidic model for single-cell capillary obstruction by Plasmodium falciparum infected erythrocytes. Proc. Natl. Acad. Sci. USA. 2003;100(25):14618–14622. doi: 10.1073/pnas.2433968100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lauks IR. Microfabricated biosensors and microanalytical systems for blood analysis. Acc. Chem. Res. 1998;31(5):317–324. [Google Scholar]
- 20.Ahn CH, Choi JW, Beaucage G, Nevin JH, Lee JB, et al. Disposable smart lab on a chip for point-of-care clinical diagnostics. Proc. IEEE. 2004;92(1):154–173. [Google Scholar]
- 21.Yang X, Hibara A, Tokeshi M, Morishima K, Kikutani Y, et al. Int. Conf. Miniat. Syst. Chem. Life Sci., 8th, Malmo, Sweden. Cambridge, UK: R. Soc. Chem; 2004. Interchannel microstructure for separation and analysis of plasma from whole blood; pp. 120–122. [Google Scholar]
- 22.Blattert C, Jurischka R, Tahhan I, Schoth A, Kerth P, Menz W. Int. Conf. Miniat. Syst. Chem. Life Sci., 8th, Malmo, Sweden. Cambridge, UK: R. Soc. Chem; 2004. Separation of blood cells and plasma in microchannel bend structures; pp. 483–485. [DOI] [PubMed] [Google Scholar]
- 23.Brenner T, Haeberie S, Zengerie R, Ducree J. Int. Conf. Miniat. Syst. Chem. Life Sci., 8th, Malmo, Sweden. Cambridge, UK: R. Soc. Chem; 2004. Continous centrifugal separation of whole blood on a disk; pp. 566–568. [Google Scholar]
- 24.Kang J, Cho H, Kwak S, Yoon D, Kim T. Int. Conf. Miniat. Syst. Chem. Life Sci., 8th, Malmo, Sweden. Cambridge, UK: R. Soc. Chem; 2004. Novel particle separation using spiral channel and centrifugal force for plasma separation from whole blood; pp. 614–616. [Google Scholar]
- 25.Gascoyne PRC, Vykoukal JV. Dielectrophoresis-based sample handling in general-purpose programmable diagnostic instruments. Proc. IEEE. 2004;92(1):22–42. doi: 10.1109/JPROC.2003.820535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffith AW, Cooper JM. Single-cell measurements of human neutrophil activation using electrorotation. Anal. Chem. 1998;70(13):2607–2612. doi: 10.1021/ac980070c. [DOI] [PubMed] [Google Scholar]
- 27.Hughes MP. Strategies for dielectrophoretic separation in laboratory-on-a-chip systems. Electrophoresis. 2002;23(16):2569–2582. doi: 10.1002/1522-2683(200208)23:16<2569::AID-ELPS2569>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 28.Gascoyne PRC, Vykoukal J. Particle separation by dielectrophoresis. Electrophoresis. 2002;23(13):1973–1983. doi: 10.1002/1522-2683(200207)23:13<1973::AID-ELPS1973>3.0.CO;2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voldman J, Gray ML, Toner M, Schmidt MA. A microfabrication-based dynamic array cytometer. Anal. Chem. 2002;74(16):3984–3990. doi: 10.1021/ac0256235. [DOI] [PubMed] [Google Scholar]
- 30.Lapizco-Encinas BH, Simmons BA, Cummings EB, Fintschenko Y. Dielectrophoretic concentration and separation of live and dead bacteria in an array of insulators. Anal. Chem. 2004;76(6):1571–1579. doi: 10.1021/ac034804j. [DOI] [PubMed] [Google Scholar]
- 31.Huang Y, Joo S, Duhon M, Heller M, Wallace B, Xu X. Dielectrophoretic cell separation and gene expression profiling on microelectronic chip arrays. Anal. Chem. 2002;74(14):3362–3371. doi: 10.1021/ac011273v. [DOI] [PubMed] [Google Scholar]
- 32.Gascoyne P, Mahidol C, Ruchirawat M, Satayavivad J, Watcharasit P, Becker FF. Microsample preparation by dielectrophoresis: isolation of malaria. Lab Chip. 2002;2(2):70–75. doi: 10.1039/b110990c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Becker FF, Wang XB, Huang Y, Pethig R, Vykoukal J, Gascoyne PRC. Separation of human breast-cancer cells from blood by differential dielectric affinity. Proc. Natl. Acad. Sci. USA. 1995;92(3):860–864. doi: 10.1073/pnas.92.3.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang J, Huang Y, Wang XB, Becker FF, Gascoyne PRC. Cell separation on microfabricated electrodes using dielectrophoretic/gravitational field flow fractionation. Anal. Chem. 1999;71(5):911–918. doi: 10.1021/ac981250p. [DOI] [PubMed] [Google Scholar]
- 35.Fiedler S, Shirley SG, Schnelle T, Fuhr G. Dielectrophoretic sorting of particles and cells in a microsystem. Anal. Chem. 1998;70(9):1909–1915. doi: 10.1021/ac971063b. [DOI] [PubMed] [Google Scholar]
- 36.Holmes D, Green NG, Morgan H. Microdevices for dielectrophoretic flow-through cell separation. IEEE Eng. Med. Biol. Mag. 2003;22(6):85–90. doi: 10.1109/memb.2003.1266051. [DOI] [PubMed] [Google Scholar]
- 37.Holmes D, Morgan H. Cell sorting and separation using dielectrophoresis. In: Morgan H, editor. Electrostatics 2003; Inst. Phys. Conf. Ser.no. 178; Bristol, UK: Inst. Phys. Publ; 2004. pp. 107–112. [Google Scholar]
- 38.Muller T, Schnelle T, Gradl G, Shirley SG, Fuhr G. Microdevice for cell and particle separation using dielectrophoretic field-flow fractionation. J. Liquid Chromatogr. Relat. Technol. 2000;23(1):47–59. [Google Scholar]
- 39.Pethig R. Dielectrophoresis: using inhomogeneous AC electrical fields to separate and manipulate cells. Crit. Rev. Biotechnol. 1996;16(4):331–348. [Google Scholar]
- 40.Stephens M, Talary MS, Pethig R, Burnett AK, Mills KI. The dielectrophoresis enrichment of CD34(+) cells from peripheral blood stem cell harvests. Bone Marrow Transplant. 1996;18(4):777–782. [PubMed] [Google Scholar]
- 41.Talary MS, Mills KI, Hoy T, Burnett AK, Pethig R. Dielectrophoretic separation and enrichment of Cd34+ cell subpopulation from bone-marrow and peripheral-blood stem-cells. Med. Biol. Eng. Comput. 1995;33(2):235–237. doi: 10.1007/BF02523050. [DOI] [PubMed] [Google Scholar]
- 42.Wang XB, Yang J, Huang Y, Vykoukal J, Becker FF, Gascoyne PRC. Cell separation by dielectrophoretic field-flow-fractionation. Anal. Chem. 2000;72(4):832–839. doi: 10.1021/ac990922o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang J, Huang Y, Wang XB, Becker FF, Gascoyne PRC. Differential analysis of human leukocytes by dielectrophoretic field-flow-fractionation. Biophys. J. 2000;78(5):2680–2689. doi: 10.1016/S0006-3495(00)76812-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eppich HM, Foxall R, Gaynor K, Dombkowski D, Miura N, et al. Pulsed electric fields for selection of hematopoietic cells and depletion of tumor cell contaminants. Nat. Biotechnol. 2000;18(8):882–887. doi: 10.1038/78504. [DOI] [PubMed] [Google Scholar]
- 45.Ozkan M, Wang M, Ozkan C, Flynn R, Birkbeck A, Esener S. Optical manipulation of objects and biological cells in microfluidic devices. Biomed. Microdevices. 2003;5(1):61–67. [Google Scholar]
- 46.Applegate RW, Squier J, Vestad T, Oakey J, Marr DWM. Optical trapping, manipulation, and sorting of cells and colloids in microfluidic systems with diode laser bars. Optics Express. 2004;12(19):4390–4398. doi: 10.1364/opex.12.004390. [DOI] [PubMed] [Google Scholar]
- 47.MacDonald MP, Spalding GC, Dholakia K. Microfluidic sorting in an optical lattice. Nature. 2003;426(6965):421–424. doi: 10.1038/nature02144. [DOI] [PubMed] [Google Scholar]
- 48.MacDonald MP, Neale S, Paterson L, Richies A, Dholakia K, Spalding GC. Cell cytometry with a light touch: sorting microscopic matter with an optical lattice. J. Biol. Regul. Homeost. Agents. 2004;18(2):200–205. [PubMed] [Google Scholar]
- 49.Paul F, Roath S, Melville D, Warhurst DC, Osisanya JOS. Separation of Malaria-infected erythrocytes from whole-blood—use of a selective high-gradient magnetic separation technique. Lancet. 1981;2(8237):70–71. doi: 10.1016/s0140-6736(81)90414-1. [DOI] [PubMed] [Google Scholar]
- 50.Melville D, Paul F, Roath S. Direct magnetic separation of red-cells from whole-blood. Nature. 1975;255(5511):706. doi: 10.1038/255706a0. [DOI] [PubMed] [Google Scholar]
- 51.Zborowski M, Ostera GR, Moore LR, Milliron S, Chalmers JJ, Schechter AN. Red blood cell magnetophoresis. Biophys. J. 2003;84(4):2638–2645. doi: 10.1016/S0006-3495(03)75069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fuh CB, Su YS, Tsai HY. Determination of magnetic susceptibility of various ion-labeled red blood cells by means of analytical magnetapheresis. J. Chromatogr. A. 2004;1027(1–2):289–296. doi: 10.1016/j.chroma.2003.08.103. [DOI] [PubMed] [Google Scholar]
- 53.Szilard P. A method for the isolation of white blood cells. J. Lab. Clin. Med. 1923;8:661–683. [Google Scholar]
- 54.Kouoh F, Levert H, Gressier B, Luyckx M, Brunet C, et al. Reduced ammonium chloride haemolysis time enhances the number of isolated functional rabbit polymorphonuclear neutrophils. APMIS. 2000;108(6):417–421. doi: 10.1034/j.1600-0463.2000.d01-77.x. [DOI] [PubMed] [Google Scholar]
- 55.Sethu P, Anahtar M, Moldawer LL, Tompkins RG, Toner M. Continuous flow microfluidic device for rapid erythrocyte lysis. Anal. Chem. 2004;76(21):6247–6253. doi: 10.1021/ac049429p. [DOI] [PubMed] [Google Scholar]
- 56.Wang Z, El-Ali J, Engelund M, Gotsaed T, Perch-Nielsen IR, et al. Measurements of scattered light on a microchip flow cytometer with integrated polymer based optical elements. Lab Chip. 2004;4(4):372–377. doi: 10.1039/b400663a. [DOI] [PubMed] [Google Scholar]
- 57.Dittrich PS, Schwille P. An integrated microfluidic system for reaction, high-sensitivity detection, and sorting of fluorescent cells and particles. Anal. Chem. 2003;75(21):5767–5774. doi: 10.1021/ac034568c. [DOI] [PubMed] [Google Scholar]
- 58.Fu LM, Yang RJ, Lin CH, Pan YJ, Lee GB. Electrokinetically driven micro flow cytometers with integrated fiber optics for on-line cell/particle detection. Anal. Chim. Acta. 2004;507(1):163–169. [Google Scholar]
- 59.Fu AY, Spence C, Scherer A, Arnold FH, Quake SR. A microfabricated fluorescence-activated cell sorter. Nat. Biotechnol. 1999;17(11):1109–1111. doi: 10.1038/15095. [DOI] [PubMed] [Google Scholar]
- 60.Fu AY, Chou HP, Spence C, Arnold FH, Quake SR. An integrated microfabricated cell sorter. Anal. Chem. 2002;74(11):2451–2457. doi: 10.1021/ac0255330. [DOI] [PubMed] [Google Scholar]
- 61.Studer V, Jameson R, Pellereau E, Pepin A, Chen Y. A microfluidic mammalian cell sorter based on fluorescence detection. Microelectron. Eng. 2004;73–74:852–857. [Google Scholar]
- 62.Wang MM, Tu E, Raymond DE, Yang JM, Zhang H, et al. Microfluidic sorting of mammalian cells by optical force switching. Nat. Biotechnol. 2005;23(1):83–87. doi: 10.1038/nbt1050. [DOI] [PubMed] [Google Scholar]
- 63.Lee W, Park J, Chung C, Han D, Chang J. Int. Conf. Miniat. Syst. Chem. Life Sci., 8th, Malmo, Sweden. Cambridge, UK: R. Soc. Chem; 2004. Combined Erythrocyte deformability test by single cell marching microstructure and optical trapping; pp. 213–215. [Google Scholar]
- 64.Gawad S, Schild L, Renaud P. Micromachined impedance spectroscopy flow cytometer for cell analysis and particle sizing. Lab Chip. 2001;1(1):76–82. doi: 10.1039/b103933b. [DOI] [PubMed] [Google Scholar]
- 65.Sohn LL, Saleh OA, Facer GR, Beavis AJ, Allan RS, Notterman DA. Capacitance cytometry: measuring biological cells one by one. Proc. Natl. Acad. Sci. USA. 2000;97(20):10687–10690. doi: 10.1073/pnas.200361297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prodan C, Mayo F, Claycomb JR, Miller JH, Benedik MJ. Low-frequency, low-field dielectric spectroscopy of living cell suspensions. J. Appl. Phys. 2004;95(7):3754–3756. [Google Scholar]
- 67.Safarik I, Safarikova M. Use of magnetic techniques for the isolation of cells. J. Chromatogr. B. 1999;722(1–2):33–53. [PubMed] [Google Scholar]
- 68.Suzuki H, Ho CM, Kasagi N. A chaotic mixer for magnetic bead-based micro cell sorter. J. Microelectromech. Syst. 2004;13(5):779–790. [Google Scholar]
- 69.Collarini EJ, Cain CA, Gammon D, Harriman B, Magee K, et al. Comparison of methods for erythroblast selection: application to selecting fetal erythroblasts from maternal blood. Cytometry. 2001;45(4):267–276. doi: 10.1002/1097-0320(20011201)45:4<267::aid-cyto10023>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 70.Furdui VI, Harrison DJ. Immuno-magnetic T cell capture from blood for PCR analysis using microfluidic systems. Lab Chip. 2004;4(6):614–618. doi: 10.1039/b409366f. [DOI] [PubMed] [Google Scholar]
- 71.Berger M, Castelino J, Huang R, Shah M, Austin RH. Design of a microfabricated magnetic cell separator. Electrophoresis. 2001;22(18):3883–3892. doi: 10.1002/1522-2683(200110)22:18<3883::AID-ELPS3883>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 72.Inglis DW, Riehn R, Austin RH, Sturm JC. Continuous microfluidic immunomagnetic cell separation. Appl. Phys. Lett. 2004;85(21):5093–5095. [Google Scholar]
- 73.Haik Y, Pai V, Chen CJ. Development of magnetic device for cell separation. J. Magn. Magn. Mater. 1999;194(1–3):254–261. [Google Scholar]
- 74.Pamme N, Manz A. Int. Conf. Miniat. Syst. Chem. Life Sci., 8th, Malmo, Sweden. Cambridge, UK: R. Soc. Chem; 2004. On-chip free-flow magnetophoresis—separation and detection of mixtures of magnetic particles in continuous flow; pp. 49–51. [Google Scholar]
- 75.Delehanty J, Shaffer K, Lin BL. A comparison of microscope slide substrates for use in transfected cell microarrays. Biosens. Bioelectron. 2004;20(4):773–779. doi: 10.1016/j.bios.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 76.Richter K, Orfert M, Howitz S, Thierbach S. Deep plasma silicon etch for microfluidic applications. Surf. Coat. Technol. 1999;116–19:461–467. [Google Scholar]
- 77.Xu CW. High-density cell microarrays for parallel functional determinations. Genome Res. 2002;12(3):482–486. doi: 10.1101/gr.213002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Revzin A, Tompkins RG, Toner M. Surface engineering with poly(ethylene glycol) photolithography to create high-density cell arrays on glass. Langmuir. 2003;19(23):9855–9862. [Google Scholar]
- 79.Revzin A, Sekine K, Sin A, Tompkins RG, Toner M. Development of a microfabricated cytometry platform for characterization and sorting of individual leukocytes. Lab Chip. 2005;5(1):30–37. doi: 10.1039/b405557h. [DOI] [PubMed] [Google Scholar]
- 80.Folch A, Toner M. Microengineering of cellular interactions. Annu. Rev. Biomed. Eng. 2000;2:227–256. doi: 10.1146/annurev.bioeng.2.1.227. [DOI] [PubMed] [Google Scholar]
- 81.Ko IK, Kato K, Iwata H. Antibody microarray for correlating cell phenotype with surface marker. Biomaterials. 2005;26(6):687–696. doi: 10.1016/j.biomaterials.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 82.Belov L, Huang P, Barber N, Mulligan SP, Christopherson RI. Identification of repertoires of surface antigens on leukemias using an antibody microarray. Proteomics. 2003;3(11):2147–2154. doi: 10.1002/pmic.200300599. [DOI] [PubMed] [Google Scholar]
- 83.Yamamura S, Rao S, Omori M, Tokimitsu Y, Kondo S. Int. Conf. Miniat. Syst. Chem. Life Sci., 8th, Malmo, Sweden. Cambridge, UK: R. Soc. Chem; 2004. High-throughput screening and analysis for antigen specific single-cell using microarray; pp. 78–80. [Google Scholar]
- 84.Kim H, Doh J, Irvine DJ, Cohen RE, Hammond PT. Large area two-dimensional B cell arrays for sensing and cell-sorting applications. Biomacromolecules. 2004;5(3):822–827. doi: 10.1021/bm034341r. [DOI] [PubMed] [Google Scholar]
- 85.Murthy SK, Sin A, Tompkins RG, Toner M. Effect of flow and surface conditions on human lymphocyte isolation using microfluidic chambers. Langmuir. 2004;20(26):11649–11655. doi: 10.1021/la048047b. [DOI] [PubMed] [Google Scholar]
- 86.Chang K-C, Hammer DA. The forward rate of binding of surface-tethered reactants: effect of relative motion between two surfaces. Biophys. J. 1999;76(3):1280–1292. doi: 10.1016/S0006-3495(99)77291-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chang WC, Liepmann D, Lee LP. A biomimetic method for extracting leukocytes from blood in microfluidic devices. Proc. Annu. Int. IEEE-EMBS Spec. Topic Conf. Microtechnol. Med. Biol. (Cat. No.02EX578), 2nd, Madison, 2–4. 2002 May;:184–188. [Google Scholar]
- 88.Chang WC, Lee LP, Liepmann D. Biomimetic technique for adhesion-based collection and separation of cells in a microfluidic channel. Lab Chip. 2005;5(1):64–73. doi: 10.1039/b400455h. [DOI] [PubMed] [Google Scholar]
- 89.Barron JWP, Ladouceur H, Ringeisen B. Biological laser printing: a novel technique for creating heterogeneous 3- dimensional cell patterns. Biomed. Microdevices. 2004;6(2):139–147. doi: 10.1023/b:bmmd.0000031751.67267.9f. [DOI] [PubMed] [Google Scholar]
- 90.Odde D, Renn M. Laser-guided direct writing of living cells. Biotechnol. Bioeng. 2000;67(3):312–318. doi: 10.1002/(sici)1097-0290(20000205)67:3<312::aid-bit7>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 91.Ozkan M, Pisanic T, Scheel J, Barlow C, Esener S, Bhatia SN. Electro-optical platform for the manipulation of live cells. Langmuir. 2003;19(5):1532–1538. [Google Scholar]
- 92.EmmertBuck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang ZP, et al. Laser capture microdissection. Science. 1996;274(5289):998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- 93.Thalhammer S, Lahr G, Clement-Sengewald A, Heckl WM, Burgemeister R, Schutze K. Laser microtools in cell biology and molecular medicine. Laser Phys. 2003;13(5):681–691. [Google Scholar]
- 94.Westphal G, Burgemeister R, Friedemann G, Wellmann A, Wernert N, et al. Noncontact laser catapulting: a basic procedure for functional genomics and proteomics. Methods Enzymol. 2002;356:80–99. doi: 10.1016/s0076-6879(02)56925-1. [DOI] [PubMed] [Google Scholar]
- 95.Taylor LC, Walt DR. Application of high-density optical microwell arrays in a live-cell biosensing system. Anal. Biochem. 2000;278(2):132–142. doi: 10.1006/abio.1999.4440. [DOI] [PubMed] [Google Scholar]
- 96.Krivacic RT, Ladanyi A, Curry DN, Hsieh HB, Kuhn P, et al. A rare-cell detector for cancer. Proc. Natl. Acad. Sci. USA. 2004;101(29):10501–10504. doi: 10.1073/pnas.0404036101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Maxwell RB, Gerhardt AL, Toner M, Gray ML, Schmidt MA. A microbubble-powered bioparticle actuator. J. Microelectromech. Syst. 2003;12(5):630–640. [Google Scholar]
- 98.Gray DS, Tan JL, Voldman J, Chen CS. Dielectrophoretic registration of living cells to a microelectrode array. Biosens. Bioelectron. 2004;19(7):771–780. doi: 10.1016/j.bios.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 99.Frenea M, Faure SP, Le Pioufle B, Coquet P, Fujita H. Positioning living cells on a high-density electrode array by negative dielectrophoresis. Mater. Sci. Eng. C-Biomim. Supramol. Syst. 2003;23(5):597–603. [Google Scholar]
- 100.Lee H, Purdon AM, Westervelt RM. Manipulation of biological cells using a microelectromagnet matrix. Appl. Phys. Lett. 2004;85(6):1063–1065. [Google Scholar]
- 101.Huang Y, Mather EL, Bell JL, Madou M. MEMS-based sample preparation for molecular diagnostics. Anal. BioAnal. Chem. 2002;372(1):49–65. doi: 10.1007/s00216-001-1191-9. [DOI] [PubMed] [Google Scholar]
- 102.Lion N, Rohner TC, Dayon L, Arnaud IL, Damoc E, et al. Microfluidic systems in proteomics. Electrophoresis. 2003;24(21):3533–3562. doi: 10.1002/elps.200305629. [DOI] [PubMed] [Google Scholar]
- 103.Auroux PA, Koc Y, deMello A, Manz A, Day PJR. Miniaturised nucleic acid analysis. Lab Chip. 2004;4(6):534–546. doi: 10.1039/b408850f. [DOI] [PubMed] [Google Scholar]
- 104.Tourovskaia A, Figueroa-Masot X, Folch A. Differentiation-on-a-chip: a microfluidic platform for long-term cell culture studies. Lab Chip. 2005;5(1):14–19. doi: 10.1039/b405719h. [DOI] [PubMed] [Google Scholar]
- 105.Andersson H, van den Berg A. Microfluidic devices for cellomics: a review. Sens. Actuators B: Chem. 2003;92(3):315–325. [Google Scholar]
- 106.Burns MA, Johnson BN, Brahmasandra SN, Handique K, Webster JR, et al. An integrated nanoliter DNA analysis device. Science. 1998;282(5388):484–487. doi: 10.1126/science.282.5388.484. [DOI] [PubMed] [Google Scholar]
- 107.Kim J, Hee JG, Jang S, Zoval JV, Da Silva NA, Madou MJ. Cell lysis on a microfluidic CD (compact disc) Lab Chip. 2004;4(5):516–522. doi: 10.1039/b401106f. [DOI] [PubMed] [Google Scholar]
- 108.Irimia D, Tompkins RG, Toner M. Single-cell chemical lysis in picoliter-scale closed volumes using a microfabricated device. Anal. Chem. 2004;76(20):6137–6143. doi: 10.1021/ac0497508. [DOI] [PubMed] [Google Scholar]
- 109.Yang J, Liu Y, Rauch C, Stevens R, Liu R, et al. High sensitivity PCR assay in plastic micro reactors. Lab Chip. 2002;2(4):179–187. doi: 10.1039/b208405h. [DOI] [PubMed] [Google Scholar]
- 110.Sethu P, Mastrangelo CH. Cast epoxy-based microfluidic systems and their application in biotechnology. Sens. Actuators B: Chem. 2004;98(2–3):337–346. [Google Scholar]
- 111.Chung Y, LY Jan MS, Lin JH, Cheng WC, Fan CY. Microfluidic chip for high efficiency DNA extraction. Lab Chip. 2004;4(2):141–147. doi: 10.1039/b310849j. [DOI] [PubMed] [Google Scholar]
- 112.Tixier-Mita A, Jun J, Ostrovidov S, Chiral M, Frenea M, et al. Int. Conf. Miniat. Syst. Chem. Life Sci., 8th, Malmo, Sweden. Cambridge, UK: R. Soc. Chem; 2004. A silicon micro-system for parallel gene transfection into arrayed cells; pp. 180–182. [Google Scholar]
- 113.Liu RH, Grodzinski P. Development of integrated microfluidic system for genetic analysis. J. Microlithogr. Microfabr. Microsyst. 2003;2(4):340–355. [Google Scholar]
- 114.Liu RH, Yang JN, Lenigk R, Bonanno J, Grodzinski P. Self-contained, fully integrated biochip for sample preparation, polymerase chain reaction amplification, and DNA microarray detection. Anal. Chem. 2004;76(7):1824–1831. doi: 10.1021/ac0353029. [DOI] [PubMed] [Google Scholar]
- 115.Liu Y, Garcia CD, Henry CS. Recent progress in the development of mu TAS for clinical analysis. Analyst. 2003;128(8):1002–1008. doi: 10.1039/b306278n. [DOI] [PubMed] [Google Scholar]
- 116.Lagally ET, Scherer JR, Blazej RG, Toriello NM, Diep BA, et al. Integrated portable genetic analysis microsystem for pathogen/infectious disease detection. Anal. Chem. 2004;76(11):3162–3170. doi: 10.1021/ac035310p. [DOI] [PubMed] [Google Scholar]
- 117.Lagally ET, Emrich CA, Mathies RA. Fully integrated PCR-capillary electrophoresis microsystem for DNA analysis. Lab Chip. 2001;1(2):102–107. doi: 10.1039/b109031n. [DOI] [PubMed] [Google Scholar]
- 118.Gao J, Yin X, Fang Z. Integration of single cell injection, cell lysis, separation and detection of intracellular constituents on a microfluidic chip. Lab Chip. 2004;4(1):47–52. doi: 10.1039/b310552k. [DOI] [PubMed] [Google Scholar]
- 119.Wu HK, Wheeler A, Zare RN. Chemical cytometry on a picoliter-scale integrated microfluidic chip. Proc. Natl. Acad. Sci. USA. 2004;101(35):12809–12813. doi: 10.1073/pnas.0405299101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.McClain MA, Culbertson CT, Jacobson SC, Allbritton NL, Sims CE, Ramsey JM. Microfluidic devices for the high-throughput chemical analysis of cells. Anal. Chem. 2003;75(21):5646–5655. doi: 10.1021/ac0346510. [DOI] [PubMed] [Google Scholar]
- 121.Hong JW, Studer V, Hang G, Anderson WF, Quake SR. A nanoliter-scale nucleic acid processor with parallel architecture. Nat. Biotech. 2004;22(4):435–439. doi: 10.1038/nbt951. [DOI] [PubMed] [Google Scholar]
- 122.Grodzinski P, Yang J, Liu RH, Ward MD. A modular microfluidic system for cell pre-concentration and genetic sample preparation. Biomed. Microdevices. 2003;5(4):303–310. [Google Scholar]