Abstract
Understanding how naïve virus-specific CD8+ T cells influence the type of immune response generated after virus infection is critical for the development of enhanced therapeutic and vaccination strategies to exploit CD8+ T cell-mediated immunity. Recent technological advances in T cell isolation and T receptor sequencing have allowed for greater understanding of the basic structure of immune T cell repertoires, the diversity of responses within and between individuals, and changes in repertoires over time and in response to infection conditions. In this review, we discuss the current understanding of how T cell repertoires contribute to potent antiviral responses. Additionally we compare the state of the art in receptor sequencing, highlighting the advantages and disadvantages of the three most common approaches: next-generation sequencing, template-switch anchored RT-PCR, and multiplex single cell PCR. Finally, we describe how TCR sequencing has delineated the relationship between naïve and immune T cell repertoires.
Introduction
T cell responses to virus infection, indeed T cell responses to many different antigens, are often characterized by reproducible biases in the responding repertoire [1–3]. The extent of the bias may be classified according to a system proposed by Turner et al (2006) [1], where Type 1 bias occurs when responding T cells express T cell receptors (TCRs) that show reproducibility only in Vβ or Vα usage; Type II bias reflects a preferred TCR Vα or Vβ in addition to a reproducible CDR3 motif; and Type III bias occurs when responding T cells use an identical Vα or Vβ region, CDR3 sequence and J region. These findings indicate that, for different antigens, the T cell pool available for recognition is vastly different, and multiple factors (including convergent recombination, prolonged antigen stimulation, and structural constraints) determine the extent of antigen-driven TCR bias both within and between individuals (reviewed elsewhere [1–4]). The extent to which an epitope-specific T cell response relies on a small subset of TCRs, or can utilize a broad range of TCRs is highly relevant, since utilization of a diverse range of TCRs in response to viral infection has been shown in many systems to promote effective viral control, prevent viral escape and enhance recognition of heterologous viruses [5–9]. This is typically thought to be due to the ability of a diverse range of TCRs to provide greater flexibility in the recognition of variant epitopes. In fact studies have shown a link between the extent of TCR diversity induced in response to viral epitopes and the likelihood of escape in HIV and SIV infection [5,7,10].
It appears however that the increased protection afforded by more diverse TCR repertoires may extend beyond their ability to prevent the emergence of escape mutants. This was elegantly demonstrated in an analysis of CD8+ T cell responses to the immunodominant Herpes Simplex Virus (HSV) glycoprotein B (gB)-derived peptide (495–502; SSIEFARL), presented by either Kb or Kbm8 MHC class I molecules [6]. Infection of mice with HSV resulted in a CD8+ T cell dependent survival in B6bm8 mice and death in wt B6 mice, an outcome that was directly correlated with the highly diverse gB-specific repertoire in Kbm8 mice and a much more restricted Kb-specific repertoire. Furthermore, the increased TCR diversity was correlated with enhanced avidity for peptide-MHC class I molecules (pMHCI) [6]. Because the incorporation of mutations by HSV is relatively uncommon, especially in structural proteins such as glycoprotein B, these data indicated that the protection afforded by increased TCR diversity arose instead as a consequence of a greater range of TCRs providing more scope for the selection of high avidity TCRs [6], which have been shown to clear virus more efficiently [11].This interpretation is supported by other studies that show that particular TCRs, rather than global diversity, are associated with improved cross-recognition and superior control of HIV infection [12–16]. Thus, the advantages conferred by highly diverse TCR repertoires noted in some studies may lie predominantly in the inclusion of TCRs with high avidity for pMHC.
Collectively, it is clear that the composition and complexity of TCR usage in the antiviral T cell response impacts substantially on an individual’s ability to deal with virus infection. In this review we discuss the various methods of TCR repertoire analysis and the impact of the naïve epitope-specific T cell repertoire on the immune response. These two areas have recently undergone great technological advances that directly affect our understanding of, and our ability to manipulate, TCR usage in response to viral infection.
1. Methods of repertoire analysis
In the last few years, the study of TCR repertoire has significantly expanded with the introduction of new sequencing protocols. Currently, three types of technical approaches are employed for the study of repertoire: i) Next generation sequencing, ii) Template-switch anchored PCR and iii) Multiplex single-cell PCR. We will discuss each approach and their respective advantages and disadvantages.
i) Next-generation sequencing
Reported methods for next-generation sequencing of TCR repertoire rely on panels of multiplex primers targeting the V-regions and J-regions or C-regions of the TCR locus. These techniques can utilize genomic DNA (when targeting V- and J- regions) or cDNA (when targeting V- and J- or V- and C-regions) [17,18]. This method allows a comprehensive characterization of a TCR chain (α or β), with the primary limitation being the size of the sample itself. Approximately two million reads are produced in a single analysis, allowing exhaustive sampling from most sample sizes. This technique has been used to determine the overall level of TCRα and β sharing among different individuals, which occurs at a much higher level than expected if repertoire generation were truly random [4,19]. A combination of convergent recombination and preferential V-J recombination has been suggested to underlie the abundance of these public clonotypes.
By the nature of the protocol (bulk sequencing of single chain amplicons), pairing of the α and β chains is not currently feasible. However, a protocol utilizing nanowell single cell sorting followed by emulsion PCR and a next-generation sequencing protocol has been reported to allow paired heavy and light chain amplification and sequencing of the immunoglobulin locus. The emulsion PCR kept individual cells in isolation and the end product was a fusion [19] between heavy and light chains mediated by a primer that targeted both constant regions. Notably, this protocol relied on an initial single cell isolation in order to maintain heavy and light chain pairing [20].
This approach, or similar ones utilizing isolated cells and bar coding, could be adapted to TCR loci with significant promise for mass amplification of complete clonotypes. However, one complication that needs to be considered is the frequent (as high as 60%) co-expression of two TCRα alleles (though one allele is out of frame in the majority of cases). The fusion approach outlined above assumes only two TCR products in a given cell, when it is more common in T cells to have three [21].
ii) Template-switch anchored RT-PCR
Template-switch anchored RT-PCR exploits a reverse transcriptase-based method for adding a universal template at the 5′ end of an RNA molecule, allowing the unbiased amplification of RNA without prior knowledge of the 5′ sequence [22]. Instead, the added universal template is used in place of the panel of V-region primers used in the other methods discussed. This method had been widely applied to a number of experimental models to demonstrate repertoire variation among diverse pathological outcomes and to define key features of the convergent recombination hypothesis [7,23–26].
This protocol has largely been applied to bulk populations, with amplification followed by subcloning into a plasmid vector or deep sequencing of the amplicon product [22]. While most of the data reported with this method is TCRβ sequence, in principle it can be equally applied to TCRα. When utilized for bulk sequencing, this protocol faces the same limitations as current deep-sequencing based approaches in that determining the clonotype of an individual cell is not possible.
iii) Single cell PCR based methods
Multiple single cell methods have been used for repertoire analysis. Single cell methods have the distinct advantage of guaranteeing representative amplification of the underlying receptor distribution or reporting out the level of amplification failure. That is, with other methods it can be difficult to determine if a subset of the population is being missed, whereas the efficiency of single cell PCR is directly observed as the number of cells that do not amplify. Initially these methods used an RT step followed by nested PCR amplification on a single Vβ region, selected by flow cytometry sorting with Vβ specific antibodies [27,28]. More recently, multiplex methods allowing paired amplification of the Vβ and Vα regions have been developed [9,21,29,30].
The paired amplification of the TCRα and TCRβ loci allowed by single cell protocols are another distinct advantage of this approach. Determining the co-expression of dual TCRα chains (largely with one out of frame locus) is achieved by subcloning and sequencing of the mixed PCR product. This entire process is lower throughput than either of the other two methods discussed to a significant degree, which limits the application of this approach in addressing some questions. However, emulsion PCR and high-throughput barcoding technology in line with deep-sequencing platforms may overcome these limitations in the near future, allowing a high-efficiency, high-throughput single cell analysis.
2. Influence of the naïve T cell repertoire on the antiviral immune response
The advantages of the single cell method, along with its throughput limitations, make it especially suitable for analyzing relatively restricted populations, including antigen-specific responses, where sampling of ~10s–100s of cells provides a representative sample. In the case of the naïve antigen-specific repertoire, where an entire animal may only contain a few hundred reactive cells, single-cell PCR allows for an exhaustive, representative characterization of the entire repertoire. Here we discuss the application of these methods to understanding the relationship between the naïve and immune repertoires.
There is great interest in how immune T cell populations are generated from their naïve counterparts, and thus how the naïve pool determines the nature of the immune response. Studies analyzing the course of the response of either single CD4+ TCRβ chain transgenic T cells to a nominal antigen [31] or CD8+ TCRαβ transgenic cells to a range of altered peptide ligands for which the TCR exhibited varying affinities [32], have broadly concluded that T cell recruitment is virtually complete after antigen challenge, suggesting that there is minimal loss of TCR diversity at the early phase of the T cell response. However, both of these studies suggested that high affinity cells were enriched over the course of the response, due to loss of extremely low affinity clones [31] and/or sustained proliferation of high affinity cells over time [32]. This is supported by another study in which CD8+ TCRαβ transgenic and single TCRβ chain transgenic cells were ‘barcoded’ with unique genetic tags, and transferred into mice before infection with recombinant LCMV expressing the cognate peptide [33]. Detection of barcode-labeled CD8+ T cells was used as an indication of the extent to which the naïve repertoire was recruited. In accordance with the previous studies, naïve CD8+ T cell recruitment was found to be virtually complete. While the latter study provides no information on differential expansion of T cell clones after recruitment, collectively these studies suggest that through comprehensive recruitment, the full extent of naïve TCR repertoire usage is represented in the immune response. However the fitness (or lack thereof) of the responding T cells is likely to influence their representation, and thus alter TCR diversity, in immune populations [31,32].
More recently, the development of a tetramer-based magnetic enrichment strategy, to directly detect and isolate endogenous antigen-specific T cell precursors from naïve individuals [34], has enabled an unprecedented analysis of how the naïve precursor pool shapes immune T cell populations. The influence of the naïve pool on the magnitude of T cell responses after antigen challenge has been, and continues to be, intensively studied (reviewed in [35]). However, only a few studies, including our own, have determined how TCR usage in naïve, endogenous (non-transgenic), epitope-specific populations influences that observed in the expanded immune population [34,36,37]. Moon and colleagues, in the original description of the enrichment technique, found that CD4+ T cells specific for I-Ab in complex with the variant 2W1S epitope from the I-E alpha chain, showed similar Vβ gene biases in the naïve and immune repertoires, suggesting that the immune population largely reflects the naïve population from which it is derived [34].
In our recent study, TCR CDR3β regions from naïve CTLps specific for a range of influenza-derived epitopes identified in the B6 model of virus infection were sequenced [36]. Analysis was restricted to TCRs expressing a TRBV chain that was previously defined as dominant in the immune response. Using a range of statistical measures to compare the naïve TCR usage to an extensive database of corresponding immune CDR3β sequences, several key findings emerged. Firstly, the broad characteristics of the epitope-specific populations were conserved between naive and immune sets, including modal CDR3β length, and preferred J region usage, supporting the original contention [34] that the immune pool reflects TCR usage in the naive population. Despite this, there was clear evidence of selective clonotype expansion from the naïve to the immune pool, as evidenced by exacerbated TRBV biases and more skewed clonotype distribution. Critically, and principally as a result of the skewed clonotype distribution, the extent of TCR diversity was significantly reduced in the immune (compared to naïve) populations for all four epitope-specificities analyzed. Moreover, it was found that, as a measure of TCR diversity at the population level, the extent to which CTL repertoires were ‘public’ (i.e. shared between multiple individuals) or ‘private’ (found only within individuals) tended to be encoded within the naïve repertoires [37]. Importantly, these findings were true for all four of the epitope specific CTLp populations analyzed, despite the fact that they showed marked differences in the extent of CTLp recruitment and expansion [36].
In summary, the application of single cell-based TCR repertoire analysis has demonstrated a similar relationship between endogenous naïve and immune T cell populations across multiple specificities, as has been observed in model transgenic systems and in endogenous CD4+ T cell populations. Specifically, while the complexity of TCR usage in the naïve pool has a substantial influence on the immune response after antigen challenge, the strength of the TCR-pMHC interaction can also influence the representation of clones within the response. In this way, the immune system safeguards TCR diversity and its associated advantages [1,5,6,8,9,24,38], while allowing high quality T cells to dominate [39]. Collectively, given the accordance of the data acquired to date, it seems plausible that this relationship between naïve and immune virus-specific populations may apply generally to all T cell responses.
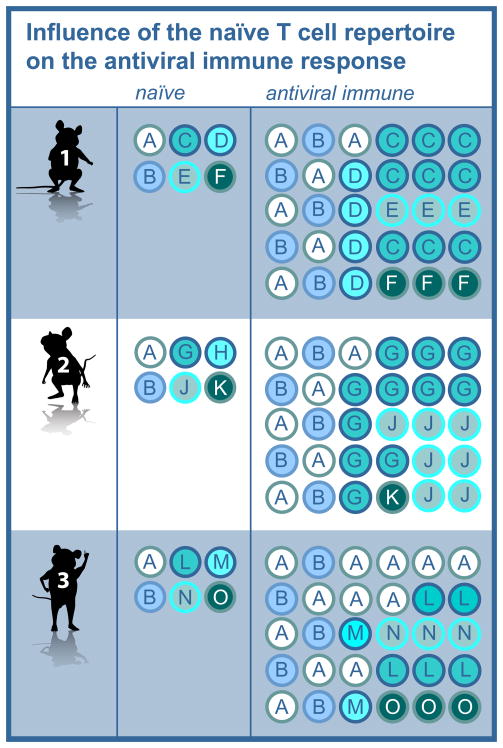
Figure 1. Influence of the naïve T cell repertoire on the antiviral immune response.
A schematic depicting the clonotypes present in the naïve and antiviral immune repertoires of three mice. Each letter corresponds to a unique clonotype. The antiviral immune repertoire broadly reflects the characteristics of the naïve repertoire, but for all epitopes shows significantly greater inequality in the distribution of clonotypes relative to the naïve repertoire, with some responses expand to a greater extent than others (e.g. clonotypes C, G, J and O). The extent of sharing is largely determined in the naïve repertoire (e.g. clonotypes A and B) and reflects sharing of both minor and dominant clones.
Highlights.
the complexity of TCR usage in antiviral T cell responses impacts on viral clearance
Single cell PCR based methods are optimal for analysis of antigen-specific T cell repertoires
Antiviral immune CTL repertoires broadly reflect the naïve CTL precursors from which they are derived
Selective clonal expansion is routinely observed from naïve into immune antiviral CTL populations
This selective expansion is likely driven by TCR avidity
Acknowledgments
This work was supported by National Health and Medical Research (NHMRC) Project grant AI1046333 and a Sylvia and Charles Viertel Senior Medical Research Fellowship awarded to N.L.G., and National Institutes of Health Grants AI077714 and AI091938, NIH/NIAID Contract HHSN266200700005C (St. Jude Center of Excellence for Influenza Research and Surveillance) and funds from ALSAC awarded to P.G.T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Nicole L. La Gruta, Email: nllg@unimelb.edu.au.
Paul G. Thomas, Email: paul.thomas@stjude.org.
References
- **1.Turner SJ, Doherty PC, McCluskey J, Rossjohn J. Structural determinants of T-cell receptor bias in immunity. Nature Reviews Immunology. 2006;6:883–94. doi: 10.1038/nri1977. [DOI] [PubMed] [Google Scholar]
- 2.Gras S, Kjer-Nielsen L, Burrows SR, McCluskey J, Rossjohn J. T-cell receptor bias and immunity. Current opinion in immunology. 2008;20:119–25. doi: 10.1016/j.coi.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Miles JJ, Douek DC, Price DA. Bias in the alphabeta T-cell repertoire: implications for disease pathogenesis and vaccination. Immunology and cell biology. 2011;89:375–87. doi: 10.1038/icb.2010.139. [DOI] [PubMed] [Google Scholar]
- 4.Li H, Ye C, Ji G, Han J. Determinants of public T cell responses. Cell Res. 2012;22 :33–42. doi: 10.1038/cr.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *5.Charini WA, Kuroda MJ, Schmitz JE, Beaudry KR, Lin W, Lifton MA, Krivulka GR, Necker A, Letvin NL. Clonally diverse CTL response to a dominant viral epitope recognizes potential epitope variants. J Immunol. 2001;167:4996–5003. doi: 10.4049/jimmunol.167.9.4996. [DOI] [PubMed] [Google Scholar]
- *6.Messaoudi I, Guevara Patino JA, Dyall R, LeMaoult J, Nikolich-Zugich J. Direct link between mhc polymorphism, T cell avidity, and diversity in immune defense. Science. 2002;298:1797–800. doi: 10.1126/science.1076064. [DOI] [PubMed] [Google Scholar]
- 7.Price DA, West SM, Betts MR, Ruff LE, Brenchley JM, Ambrozak DR, Edghill-Smith Y, Kuroda MJ, Bogdan D, Kunstman K, et al. T cell receptor recognition motifs govern immune escape patterns in acute SIV infection. Immunity. 2004;21 :793–803. doi: 10.1016/j.immuni.2004.10.010. [DOI] [PubMed] [Google Scholar]
- *8.Cornberg M, Chen AT, Wilkinson LA, Brehm MA, Kim SK, Calcagno C, Ghersi D, Puzone R, Celada F, Welsh RM, et al. Narrowed TCR repertoire and viral escape as a consequence of heterologous immunity. J Clin Invest. 2006;116:1443–56. doi: 10.1172/JCI27804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *9.Wang GC, Dash P, McCullers JA, Doherty PC, Thomas PG. T cell receptor αβ diversity inversely correlates with pathogen-specific antibody levels in human cytomegalovirus infection. Sci Transl Med. 2012;4:128ra42. doi: 10.1126/scitranslmed.3003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **10.Kosmrlj A, Read EL, Qi Y, Allen TM, Altfeld M, Deeks SG, Pereyra F, Carrington M, Walker BD, Chakraborty AK. Effects of thymic selection of the T-cell repertoire on HLA class I-associated control of HIV infection. Nature. 2010;465:350–354. doi: 10.1038/nature08997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derby M, Alexander-Miller M, Tse R, Berzofsky J. High-avidity CTL exploit two complementary mechanisms to provide better protection against viral infection than low-avidity CTL. J Immunol. 2001;166:1690–7. doi: 10.4049/jimmunol.166.3.1690. [DOI] [PubMed] [Google Scholar]
- 12.Dong T, Stewart-Jones G, Chen N, Easterbrook P, Xu X, Papagno L, Appay V, Weekes M, Conlon C, Spina C, et al. HIV-specific cytotoxic T cells from long-term survivors select a unique T cell receptor. The Journal of experimental medicine. 2004;200:1547–57. doi: 10.1084/jem.20032044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gillespie GM, Stewart-Jones G, Rengasamy J, Beattie T, Bwayo JJ, Plummer FA, Kaul R, McMichael AJ, Easterbrook P, Dong T, et al. Strong TCR conservation and altered T cell cross-reactivity characterize a B*57-restricted immune response in HIV-1 infection. Journal of immunology. 2006;177:3893–902. doi: 10.4049/jimmunol.177.6.3893. [DOI] [PubMed] [Google Scholar]
- 14.Yu XG, Lichterfeld M, Chetty S, Williams KL, Mui SK, Miura T, Frahm N, Feeney ME, Tang Y, Pereyra F, et al. Mutually exclusive T-cell receptor induction and differential susceptibility to human immunodeficiency virus type 1 mutational escape associated with a two-amino-acid difference between HLA class I subtypes. Journal of virology. 2007;81:1619–31. doi: 10.1128/JVI.01580-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Bockel DJ, Price DA, Munier ML, Venturi V, Asher TE, Ladell K, Greenaway HY, Zaunders J, Douek DC, Cooper DA, et al. Persistent survival of prevalent clonotypes within an immunodominant HIV gag-specific CD8+ T cell response. Journal of immunology. 2011;186:359–71. doi: 10.4049/jimmunol.1001807. [DOI] [PubMed] [Google Scholar]
- **16.Chen H, Ndhlovu ZM, Liu D, Porter LC, Fang JW, Darko S, Brockman MA, Miura T, Brumme ZL, Schneidewind A, et al. TCR clonotypes modulate the protective effect of HLA class I molecules in HIV-1 infection. Nature immunology. 2012;13:691–700. doi: 10.1038/ni.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *17.Robins HS, Campregher PV, Srivastava SK, Wacher A, Turtle CJ, Kahsai O, Riddell SR, Warren EH, Carlson CS. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009;114:4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang C, Sanders CM, Yang Q, Schroeder HW, Jr, Wang E, Babrzadeh F, Gharizadeh B, Myers RM, Hudson JR, Jr, Davis RW, et al. High throughput sequencing reveals a complex pattern of dynamic interrelationships among human T cell subsets. Proc Natl Acad Sci US A. 2010;107:1518–1523. doi: 10.1073/pnas.0913939107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robins HS, Srivastava SK, Campregher PV, Turtle CJ, Andriesen J, Riddell SR, Carlson CS, Warren EH. Overlap and effective size of the human CD8+ T cell receptor repertoire. Sci Transl Med. 2010;2:47ra64. doi: 10.1126/scitranslmed.3001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeKosky BJ, Ippolito GC, Deschner RP, Lavinder JJ, Wine Y, Rawlings BM, Varadarajan N, Giesecke C, Dörner T, Andrews SF, et al. High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire [Internet] Nature Biotechnology. 2013 doi: 10.1038/nbt.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *21.Dash P, McClaren JL, Oguin TH, Rothwell W, Todd B, Morris MY, Becksfort J, Reynolds C, Brown SA, Doherty PC, et al. Paired analysis of TCRα and TCRβ chains at the single-cell level in mice. J Clin Invest. 2011;121:288–295. doi: 10.1172/JCI44752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quigley MF, Almeida JR, Price DA, Douek DC. Unbiased molecular analysis of T cell receptor expression using template-switch anchored RT-PCR. Curr Protoc Immunol. 2011;(Unit10.33) doi: 10.1002/0471142735.im1033s94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Ye C, Ji G, Wu X, Xiang Z, Li Y, Cao Y, Liu X, Douek DC, Price DA, et al. Recombinatorial biases and convergent recombination determine interindividual TCRβ sharing in murine thymocytes. J Immunol. 2012;189:2404–2413. doi: 10.4049/jimmunol.1102087. [DOI] [PubMed] [Google Scholar]
- *24.Price DA, West SM, Betts MR, Ruff LE, Brenchley JM, Ambrozak DR, Edghill-Smith Y, Kuroda MJ, Bogdan D, Kunstman K, et al. T cell receptor recognition motifs govern immune escape patterns in acute SIV infection. Immunity. 2004;21:793–803. doi: 10.1016/j.immuni.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 25.Price DA, Brenchley JM, Ruff LE, Betts MR, Hill BJ, Roederer M, Koup RA, Migueles SA, Gostick E, Wooldridge L, et al. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J Exp Med. 2005;202:1349–61. doi: 10.1084/jem.20051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price DA, Asher TE, Wilson NA, Nason MC, Brenchley JM, Metzler IS, Venturi V, Gostick E, Chattopadhyay PK, Roederer M, et al. Public clonotype usage identifies protective Gag-specific CD8+ T cell responses in SIV infection. J Exp Med. 2009;206:923–36. doi: 10.1084/jem.20081127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malherbe L, Mark L, Fazilleau N, McHeyzer-Williams LJ, McHeyzer-Williams MG. Vaccine adjuvants alter TCR-based selection thresholds. Immunity. 2008;28:698–709. doi: 10.1016/j.immuni.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maryanski JL, Jongeneel CV, Bucher P, Casanova JL, Walker PR. Single-cell PCR analysis of TCR repertoires selected by antigen in vivo: a high magnitude CD8 response is comprised of very few clones. Immunity. 1996;4:47–55. doi: 10.1016/s1074-7613(00)80297-6. [DOI] [PubMed] [Google Scholar]
- 29.Ozawa T, Tajiri K, Kishi H, Muraguchi A. Comprehensive analysis of the functional TCR repertoire at the single-cell level. Biochem Biophys Res Commun. 2008;367:820–825. doi: 10.1016/j.bbrc.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 30.Sun X, Saito M, Sato Y, Chikata T, Naruto T, Ozawa T, Kobayashi E, Kishi H, Muraguchi A, Takiguchi M. Unbiased analysis of TCRα/β chains at the single-cell level in human CD8+ T-cell subsets. PLoS ONE. 2012;7:e40386. doi: 10.1371/journal.pone.0040386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malherbe L, Hausl C, Teyton L, McHeyzer-Williams MG. Clonal selection of helper T cells is determined by an affinity threshold with no further skewing of TCR binding properties. Immunity. 2004;21:669–79. doi: 10.1016/j.immuni.2004.09.008. [DOI] [PubMed] [Google Scholar]
- **32.Zehn D, Lee SY, Bevan MJ. Complete but curtailed T-cell response to very low-affinity antigen. Nature. 2009;458:211–4. doi: 10.1038/nature07657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Heijst JW, Gerlach C, Swart E, Sie D, Nunes-Alves C, Kerkhoven RM, Arens R, Correia-Neves M, Schepers K, Schumacher TN. Recruitment of antigen-specific CD8+ T cells in response to infection is markedly efficient. Science. 2009;325:1265–9. doi: 10.1126/science.1175455. [DOI] [PubMed] [Google Scholar]
- **34.Moon JJ, Chu HH, Pepper M, McSorley SJ, Jameson SC, Kedl RM, Jenkins MK. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 2007;27:203–13. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jenkins MK, Moon JJ. The role of naive T cell precursor frequency and recruitment in dictating immune response magnitude. Journal of immunology. 2012;188:4135–40. doi: 10.4049/jimmunol.1102661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *36.La Gruta NL, Rothwell WT, Cukalac T, Swan NG, Valkenburg SA, Kedzierska K, Thomas PG, Doherty PC, Turner SJ. Primary CTL response magnitude in mice is determined by the extent of naive T cell recruitment and subsequent clonal expansion. J Clin Invest. 2010;120:1885–1894. doi: 10.1172/JCI41538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas PG, Handel A, Doherty PC, La Gruta NL. Ecological analysis of antigen-specific CTL repertoires defines the relationship between naive and immune T-cell populations. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1839–44. doi: 10.1073/pnas.1222149110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turner SJ, La Gruta NL, Kedzierska K, Thomas PG, Doherty PC. Functional implications of T cell receptor diversity. Current opinion in immunology. 2009;21:286–90. doi: 10.1016/j.coi.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malherbe L, Filippi C, Julia V, Foucras G, Moro M, Appel H, Wucherpfennig K, Guery JC, Glaichenhaus N. Selective activation and expansion of high-affinity CD4+ T cells in resistant mice upon infection with Leishmania major. Immunity. 2000;13:771–82. doi: 10.1016/s1074-7613(00)00075-3. [DOI] [PubMed] [Google Scholar]