Abstract
Computational analysis of human adenovirus type 4 (HAdV-E4), a pathogen that is the only HAdV member of species E, provides insights into its zoonotic origin and molecular adaptation. Its genome encodes a domain of the major capsid protein, hexon, from HAdV-B16 recombined into the genome chassis of a simian adenovirus. Genomes of two recent field strains provide a clue to its adaptation to the new host: recombination of a NF-I binding site motif, which is required for efficient viral replication, from another HAdV genome. This motif is absent in the chimpanzee adenoviruses and the HAdV-E4 prototype, but is conserved amongst other HAdVs. This is the first report of an interspecies recombination event for HAdVs, and the first documentation of a lateral partial gene transfer from a chimpanzee AdV. The potential for such recombination events are important when considering chimpanzee adenoviruses as candidate gene delivery vectors for human patients.
Keywords: Adenovirus, Molecular evolution, Recombination, Zoonosis
Introduction
Adenoviruses are found in many vertebrate hosts, spanning fish to human (Benkö et al., 2005; Davison et al., 2003). Human adenoviruses (HAdVs) may be pathogens, causing symptoms ranging from mild to severe, including death. They may also infect asymptomatically and persistently (Garnett et al., 2009), and are found to coinfect with other HAdVs, with up to four viruses characterized in some patients (Barrero et al., 2012; Metzgar et al., 2005; Vora et al., 2006). A wide spectrum of diseases are reported, involving the respiratory, ocular, gastrointestinal and genitourinary systems, as well as a metabolic disorder (obesity) (Echavarria, 2008). Since the initial nearly simultaneous reports of HAdVs as respiratory pathogens in 1953 (Hilleman and Werner, 1954; Rowe et al., 1953), they have been examined extensively, leading to insights in cell biology, molecular biology, immunology and epidemiology, as noted in a report of the 10th International Adenovirus Meeting in 2012 (Umea, Sweden) (Greber et al., 2013). Reflecting their importance in health and biotechnology, new HAdVs continue to be identified and characterized at high-resolution using genomics, albeit with some disagreement on using whole genome data to characterize, type and classify candidate new types (Seto et al., 2011) rather than serology-based methods (Aoki et al., 2011).
All of the HAdV prototype genomes are now completely sequenced (manuscript submitted for publication), providing a reference data set for comparative analyses amongst these prototypes, other archived historically important HAdVs and newly emergent HAdV strains. Detailed understanding of how these new pathogens emerged has come from such analyses. For example, an emergent highly contagious keratoconjunctivitis (EKC) pathogen (Engelmann et al., 2006) was revealed as a recombinant with at least three parents (Walsh et al., 2009); an emergent acute respiratory disease (ARD) pathogen causing a fatality and subsequent transmission as a highly contagious ocular disease pathogen (Henquell et al., 2009) was revealed as a recombinant (Robinson et al., 2011); and a re-emergent ARD pathogen that has the serological profile of a renal tract pathogen (Yang et al., 2009; Zhu et al., 2009) was revealed as a recombinant with two parents (Walsh et al., 2010).
Adenoviruses are important biomedical tools as vectors for epitope and gene delivery (Darr et al., 2009; Fujishiro et al., 2005; Graham and Prevec, 1992; Stone et al., 2006). Serendipitously, the recent attention to non-human simian adenoviruses (SAdVs) in this capacity has brought additional genomes into the data set (Roy et al., 2004a, 2004b, 2012, 2009, 2006), complementing the HAdV genomes and allowing for higher resolution into origins of other HAdVs. One example is HAdV-E4, which has been shown to contain a genome with relatively high similarity to several chimpanzee adenoviruses (Purkayastha et al., 2005a). HAdV-E4 is an important respiratory and ocular pathogen, as one of two HAdVs warranting a vaccine in the 1960s and again recently (Gaydos and Gaydos, 1995; Kajon et al., 2007; Lyons et al., 2008; Russell et al., 2006). Its origins were probed in different eras with then-available and then-novel protocols and instruments, including serological cross-reactivity of both its hexon and fiber; protein chemistry measurements (i.e., molecular weights of “internal polypeptides”); limited peptide sequencing (N-terminal amino acids); and limited nucleotide sequence analysis, e.g., restriction enzyme digests (Adrian et al., 1986; Gruber et al., 1993; Li and Wadell, 1988; Norrby and Wadell, 1969; Wadell, 1984). Recently, limited nucleotide sequencing of the putative epsilon epitope that contributes to its serum neutralization has provided additional clues about its phylogenetic relationships and origins (Ebner et al., 2005; Madisch et al., 2005; Pring-Akerblom et al., 1995; Sarantis et al., 2004). Speculations that HAdV-E4 directly derived from a putative common ancestor of HAdVs, is a recombinant of species B and C, or is related to chimpanzee adenoviruses were based on these limited data sets (Gruber et al., 1993; Li and Wadell, 1988; Wadell, 1984). Later, with its genome sequenced and a newly available set of SAdVs (Purkayastha et al., 2005a, 2005b; Roy et al., 2004a) for comparison, a definitive zoonotic origin was revealed based on nucleotide and amino acid sequence identities (Purkayastha et al., 2005a). With the recent upload of additional SAdV genomes into the public database (Roy et al., 2009), a more complete and granular understanding emerges, as reported here. The HAdV-E4 genome is a recombinant that contains the hexon loops 1 and 2 (L1 and L2) of HAdV-B16, comprising approximately 2.5% of the whole genome, in a genome chassis of SAdV-E26, similar to the genome structure reported for the re-emergent recombinant respiratory pathogen HAdV-B55 (Walsh et al., 2010). Included in this report are the computational analyses of two more recent HAdV-E4 field strains that provide an additional view as to the mechanism of how this virus adapted to the human host: A recombination that provides the NF-I binding motif, which is conserved across all other HAdVs as part of the DNA replication motifs, from a species B2 HAdV distinguishes these two recent isolates. Notably, this motif is either absent amongst the chimpanzee adenoviruses and both the type 4 prototype (HAdV-E4p) and the then-contemporary field strain (HAdV-E4vac) genomes (Purkayastha et al., 2005a, 2005b) or is a version that is unique to the chimpanzee adenoviruses.
Results
Comparative genomic analysis
The genomes of HAdV-E4 field strains (FS) isolated recently from two U.S. military basic trainees at separate geographic locations and presenting with ARD were sequenced, analyzed, and compared with genomes from strains isolated approximately 60 years ago. Table 1 provides details of these strains.
Table 1.
HAdV-E4 strains. Details of the type 4 viruses are presented, including alternative names, year of isolation, location of isolation, GenBank accession number, and the genome size.
|
HA4V-E4 strains | |||||||
|---|---|---|---|---|---|---|---|
| Strain | Designation | Alt. name | Alt. name | Isol. yr. | Location isol. | Acc. no. | Genome size |
| Prototype | HAdV-E4p | RI-67 | 1952 | Ft. Wood, MO | AY594253 | 35,990 | |
| Vaccine | HAdV-E4vac | Wyeth | 1962 | Camp Lejune, NC | AY594254 | 35,994 | |
| Field strain 1 | HAdV-E4FSl | NHRC 42606 | HAdV-E4a2 | 2003 | Ft. Jackson, SC | AY599835 | 35,965 |
| Field strain 2 | HAdV-E4FS2 | NHRC3 | HAdV-E4al | 2002 | Brooks AFB, TX | AY599837 | 35,964 |
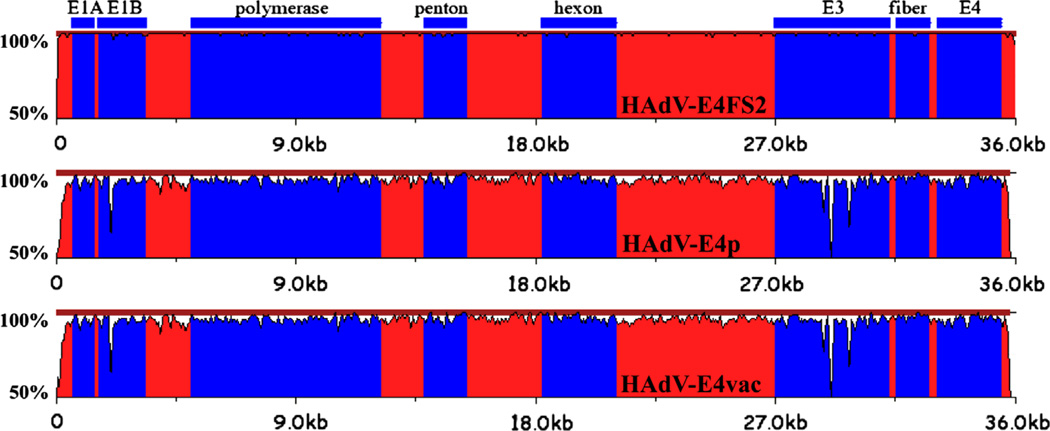
Computational analysis confirmed both field strain viruses as HAdV-E4 genomes and as being highly similar to each other. Alignments of the genomes using Sequencher and MEGA showed there are 46 base substitutions and seven indels, comprising five one-base indels and two three-base indels, between these two field strain genomes. Both are similar to, but divergent from, the two HAdV-E4 genomes isolated approximately fifty years earlier (prototype and vaccine). For reference, HAdV-E4vac differs from the prototype by 18 mismatches and two insertions (one one-base and one three-base). Pairwise nucleotide sequence alignments of the genomes, shown in Fig. 1, reflect the high levels of similarity and identity across the genomes. Global pairwise genome comparisons were made using zPicture (http://zpicture.dcode.org/), a sequence alignment tool that analyzes pairs of the HAdV-E4 genome sequences against each other for regions of similarity. As shown in Fig. 1, these results graphically complement the sequence data analysis demonstrating that the field strains are nearly identical to each other, as well as are highly similar to the prototype and vaccine strains, but with differences across the entire genome. Detailed protein percent identity analysis across the genome confirmed this (data not shown). Using the prototype (1952) and vaccine (1962) HAdV-E4 genomes as queries, both field strains share common divergent regions from the query genomes, specifically at the ITR, E1B, and E3 regions (Fig. 1). Calculations of the whole genome nucleotide sequence percent identities show that HAdV-E4FSl and HAdV-E4FS2 are nearly identical at 99.9%; both are similar to the prototype at 95.1% and to the vaccine at 95.1% (For reference, the prototype and vaccine genomes are 99.9% identical to each other). The HAdV-E4FSl genome is similar to the SAdV-E25 genome at 89.2% and to SAdV-E26 at 89.8%.
Fig. 1.
Pairwise genome comparative analysis. zPicture (http://zpicture.dcode.org/) utilizes a local alignment algorithm, BlastZ, to display the regions of nucleotide sequence similarity between HAdV-E4FS1 and query genomes: HAdV-E4FS2 (top); HAdVE4p (middle); and HAdV-E4vac (bottom). Of note are the divergences at the ends of the genomes, corresponding to the semi-conserved inverted terminal repeat (ITR) sequences that contain critical functions, including DNA replication. The genome sequences are arrayed on the horizontal and the percent identity of genome pairs from 50% to 100% is noted along the y-axis. The colors are arbitrary and are used to provide contrast: blue regions highlight select genes or genome regions (E3), noted above the alignments. Red regions include both coding and noncoding sequences.
Protein homology analysis
Protein coding regions for HAdV-E4FSl were annotated and compared with representative data sets from members of the HAdV seven species (A–G), as was performed for HAdV-E4p (Purkayastha et al., 2005a). Reflecting the high level of similarity between the field strains and the two older genomes, similar values were found (data not presented). Of interest is the homology to the chimpanzee adenoviruses, particularly with the recently sequenced genome of SAdV-E26, providing a slightly higher level of similarity than SAdV-E25 across the genome. Shown in Table 2 are these data, including HAdV-B16, a human adenovirus that shows the highest percent similarity (93.5%) in the hexon sequence. As the hexon gene contains a variable region and a constant region, it was further dissected to examine these two domains individually: loop 1 (L1) and constant region (C). The constant regions of all show near identity; L1 reflects the discriminatory higher similarity of HAdV-B16 (94.8%) in contrast to the SAdVs (ca., 64%). In contrast, the fiber of SAdV-E26 shows the highest similarity (93.2%) as opposed to 29.5% for HAdV-B16.
Table 2.
Protein percent identities of select HAdV-E4FSl proteins to their homologs in SAdVs and HAdV-B16. Amino acid identities of proteins to their counterparts in species E SAdVs and species B1 HAdV-B16 were calculated using EMBOSS (http://emboss.sourceforge.net/), run with default parameters. Specifically, global alignments were performed using the Needle program of EMBOSS. A BLOSUM62 matrix was used for the amino acid sequence analysis. Reflecting the bipartite nature of the hexon gene, one of the two variable loop regions (L1) has been extracted and analyzed separately; the complementary constant (C) region is also analyzed.
| Protein | % Identity to homolog |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| L1 55kDa |
IIIa | Penton base |
VI | Hexon | Hexon (L1) |
Hexon (c) |
VIII | Fiber | |
| SAdV-E22 | 94.2 | 92.9 | 94.4 | 83.4 | 88.8 | 64.3 | 99.4 | 95.6 | 61.8 |
| SAdV-E23 | 93.9 | 92.6 | 94.4 | 84 | 87.1 | 62.9 | 99.4 | 96 | 59.8 |
| SAdV-E24 | 93.9 | 92.9 | 94.8 | 83.7 | 88.8 | 64.5 | 99.4 | 96 | 71.1 |
| SAdV-E25 | 94.9 | 97 | 95.3 | 86.5 | 88.9 | 64.7 | 100 | 97.4 | 90.8 |
| SAdV-E26 | 95.2 | 98 | 90 | 88.8 | 88.9 | 63.9 | 100 | 97.4 | 93.2 |
| HAdV-B16 | 83.2 | 88.2 | 80.2 | 65.2 | 93.5 | 94.8 | 97.5 | 90.7 | 29.5 |
Phylogenetic analysis
Whole genome
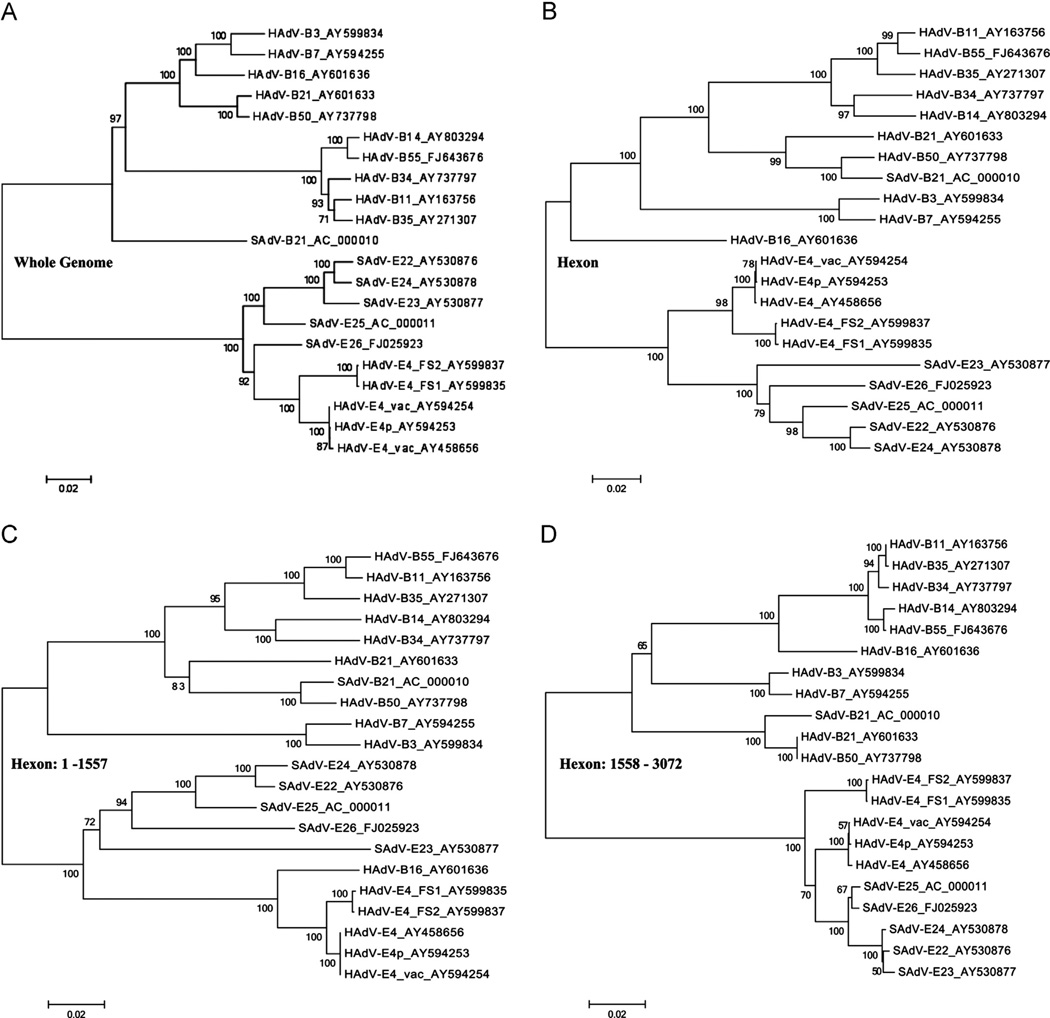
A representative of a much larger phylogenetic analysis of all HAdV and SAdV available genomes is shown in Fig. 2. Multiple sequence alignment and phylogenetic analysis of the whole genomes reflected the high levels of similarity between HAdV-E4 genomes and five of the six SAdV genomes, all of which were isolated from chimpanzees. As the HAdV-E4 genomes formed a subclade with a robust bootstrap value (1 0 0), they are phylogenetically distinct from the simian counterparts (Fig. 2A). SAdV-E26 is the closest genome from the simian set. The two type 4 field strains (2000s era collection) form a subclade within this, as do the two 1950–1960s era HAdV-E4p and -E4vac strains. This may reflect a common lineage given the time of isolation differences, rather than separate lineages resulting from independent zoonotic events. The vaccine strain genome originated from a live-vaccine tablet and was passaged minimally following recovery from the tablet (AY594254) (Purkayastha et al., 2005b). All of these species E genomes are contained in a clade distinguishing them from the other six species A–D and F–G. HAdV-B16 is in the clade that contained all of the subspecies B1 members; there was a clade with all of the subspecies B2 members, reflecting one criterion for the subspecies designation, i.e., a restriction enzyme (RE) digestion pattern of Smal that was noted as supporting the subspecies delineation which was also based on “apparent molecular weight of the internal polypeptides” (Wadell et al., 1980). Nota bene, SAdV-B21 is in the clade containing these species B HAdVs.
Fig. 2.
Phylogenomic analysis of HAdV genomes and hexons from species B and E, including select SAdVs. Bootstrap-confirmed neighbor-joining phylogenetic trees of the whole-genome (A) and hexon gene sequences are presented, with the hexon gene (B) further dissected for the detailed analyses of two domains: (C) nucleotides 1 to 1557, representing the proximal sequences and including the variable region and epitopic loopsL1 and L2, and (D) nucleotides 1558 to 3072, representing the remaining distal portion and including the conserved region. All phylogenetic trees were generated after multiple sequence alignments using the MAFFT software (http://www.ebi.ac.uk/tools/mafft) and default parameters. Bootstrapped, neighbor-joining trees with 1000 replicates were constructed using MEGA4.0.2 software with a maximum-composite-likelihood model. Bootstrap numbers shown at the nodes indicate the percentages of 1,000 replications producing the clade, with values above 80 considered robust. The scale bar is in units of nucleotide substitutions per site.
Hexon, and domains
As the above analyses of the hexon sequences suggested an identity with a human adenovirus, HAdV-B16, the hexon gene was examined using sequence alignment and phylogenetics. As shown in Fig. 2B, the hexons from type 4 HAdVs form a clade that is distinct from that of the non-human SAdVs (bootstrap 100); however, HAdV-B16 is excluded from this clade, despite sequence similarity. Therefore, the hexon was examined in greater detail. Previous reports noted the variable region as a candidate to recombine, generating new genome types and novel pathogens (Liu et al., 2012; Matsushima et al., 2011; Robinson et al., 2011; Walsh et al., 2009, 2010) (manuscripts in press; in preparation); therefore, the hexon sequence was divided into two halves for detailed analysis of each, one comprising nucleotides 1 to 1557 (Fig. 2C) and the other comprising nucleotides 1558 to 3072 (Fig. 2D), reflecting the variable region, including L1 and L2, and the constant or conserved region, respectively, as noted by Madisch et al. (2005).L1 and L2 putatively comprise the serum neutralization (SN) epitope. Phylogenetic analysis of nucleotides 1 to 1557 showed that HAdV-B16 formed a subclade with the HAdV-E4 sequences, with SAdV-E23 as the next closest simian virus (Fig. 2C). All of the species E SAdVs are contained in a subclade while the rest of the species B HAdV genomes formed a separate distinct clade, which included SAdV-B21. With one exception, that of SAdV-E23 forming its own subclade away from the other SAdVs (bootstrap value 72), the bootstrap values are all above 80, indicating high levels of confidence. In the larger data set trees, phylogenetic relationships forL1 and L2, separately, reflected the same pattern (data not shown). In contrast, sequences corresponding to nucleotides 1558 to 3072 cluster together, as was found for the whole genome phylogenetic tree, with HAdV-B16, subspecies B1, forming its own subclade off the branch for all subspecies B2 viruses (Fig. 2D). Bootstrap values are at 100. This unique bipartite observation may account for the 93.5% identity of the HAdV-B16 hexon to its counterpart in HAdV-E4FS1 (rather than a higher value). As noted earlier, separating the variable L1 from the constant region highlights the higher similarity of HAdV-E4FSl to HAdV-B16 (94.8%) in contrast to the SAdVs (ca., 64%).
Sequence recombination analysis
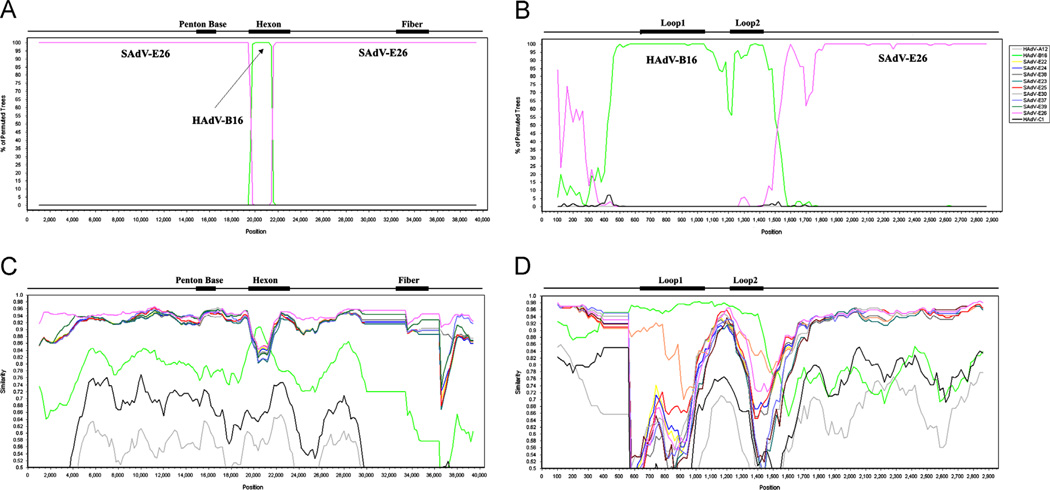
With the data from the protein percent identities and the phylogenomic analysis suggesting recombination, a detailed examination of the genome nucleotide sequence was completed using the Simplot software. One option of this software, Bootscan, revealed that the HAdV-E4FSl hexon contained near identity to its counterpart from HAdV-B16 in the first third or proximal portion of the gene (Fig. 3A and B). Sequences in the hexon recombination region appear to be less similar to the parental sequences (Fig. 3B) suggesting that the recombination is not recent and mutations have contributed to the divergence noted by the Bootscan analysis. The span of this sequence that aligns with HAdV-B16 is from nucleotides 360 to 1257, representing 897 bases of the hexon gene, which is 2811 nucleotides in length (31.9%) and encompasses L1 and L2, the putative epitopes for SN. As the HAdV-E4FSl genome is 35,965 bases in length, the recombinant HAdV-B16 portion comprises 2.5% of the HAdV-E4FSl genome. The Simplot analysis also supports this recombination observation (Fig. 3C and D).
Fig. 3.
Nucleotide sequence recombination analysis of the whole genome and the hexon gene of HAdV-E4FSl. The genome of HAdV-E4FSl (AY599837) was analyzed for sequence recombination events with HAdV and SAdV genomes using the software Simplot, which is available on-line (http://sray.med.som.jhmi.edu/SCRoftware/simplot/). Bootscan analysis of the genome (A) and hexon (B) shows a lateral transfer of a portion of the hexon gene from HAdV-B 16. These are supported by the Simplot analysis of the genome (C) and hexon (D). Unlike the Simplot panels (C and D), only the SAdV-E26 genome, rather than the entire set of SAdV-E22 through -E26 sequences, is presented in the Bootscan panels (A and B), as similar genomes compete out the signal. For Bootscan, removing four of the five genomes and repeating the analysis with each individually, gives a clearer representation, reflecting the Simplot result for the highest similarity match. MAFFT software was used to align the sequences prior to recombination analysis (http://mafft.cbrc.jp/alignment/server/). Default parameter settings for the Simplot software were used for analyzing the hexon sequences: window size (200 nucleotides [nt]), step size (20 nt), replicates used (n=100), gap stripping (on), distance model (Kimura) and tree model (neighbor-joining). Similarly, whole genomes were analyzed, beginning with an initial alignment using MAFFT and following with recombination analysis using Simplot and Bootscan. Only the window size and step size were altered for the genome analysis (1000 and 200, respectively), with the rest of the default parameters unchanged. Genome nucleotide positions are noted along the x-axis, and the percentages of permutated trees that supported grouping are marked along the y-axis. For reference, select genome and gene-specific landmarks are noted above each graph. Colors: pink, SAdV-E26 and green, HAdV-B16.
Inverted terminal repeat (ITR) sequence analysis
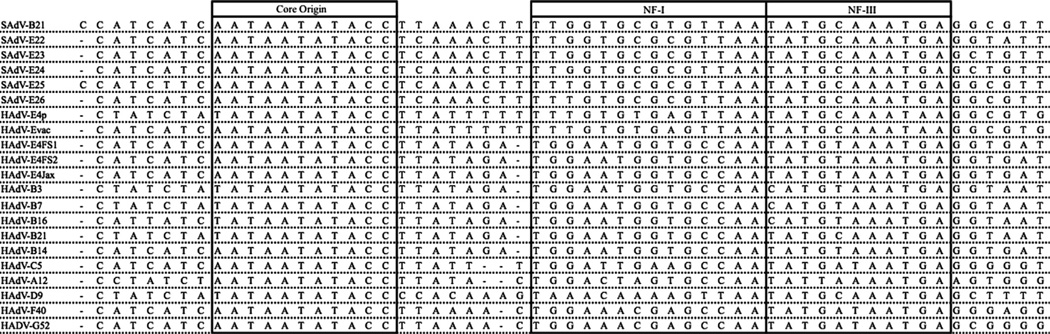
The ITRs from both HADV-E4 field strain genomes and a third, sequenced from another contemporary field strain (Jax78) (Houng et al., 2006), contained identical sequences that diverge at the terminal 5’ end from the HAdV-E4p and -E4vac genomes, as shown in Fig. 4. Sequences from the HAdV-E4p and -E4vac ITRs diverge from each other as well in the first eight nucleotides, with the HAdV-E4p sequence (CTATCTAT) identical to its counterparts in HAdV-B3, -B7, -B21, and -D9. This eight-base sequence of HAdV-E4vac (CATCATCA) was identical to the sequences found in the SAdV-B21, -E22, -E23, -E24, and -E26 genomes, with SAdV-B21 containing an extra C at the 5’-terminus. SAdV-E25 has the same extra C and a single base difference at nucleotide 6 as well (CCATCTTCA). The adjacent core origin sequence was absolutely canonical, and the sequences then diverged with clusters observable for the intervening sequence as well as for the NF-I and NF-III transcription factor binding motifs. Between the core origin and the NF-I motif, the intervening sequence of the fields strains (TTATAGA) was unlike the one found for both HAdV-E4p and -E4vac (TTATTTTT), but identical to the counterparts from species B (TTATAGA).
Fig. 4.
Multiple sequence alignment of nucleotide sequences from the inverted terminal repeat (ITR). Edited Clustal Omega-generated (www.ebi.ac.uk/Tools/msa/clustalo/) sequence alignments of ca. 58 bases from the 5’-termini of five HAdV-E4 genomes are compared with representative counterparts from other HAdV species and several SAdVs. For reference, the three sequence motifs implicated in adenovirus DNA replication are labeled and boxed: (1) “core origin”; (2) NF-I transcription factor binding motif; and (3) NF-III transcription factor binding motif. The Clustal Omega-generated alignment was manually edited to position gaps to generate an optimal alignment, using the SAdV-B21 as reference.
HAdVs utilize two host transcription factors, nuclear factors I and III (NF-I and NF-III), as part of the viral DNA replication complex (Hatfield and Hearing, 1991,1993; Mul et al., 1990; Pruijn et al., 1988). With the NF-I site reported for HAdV-C2 and -C5 as reference (Mul et al., 1990), permutations of putative NF-I motifs were located and the abutting putative NF-III motifs were identified. SAdVs contained a set of similar sequences that deviated from the NF-I motifs found for the HAdVs; both HAdV-E4p and -E4vac shared an identical sequence (TTTGTGTGAGTTAA) with resemblance to the SAdV motifs. However, the three recently circulating HAdV-E4 field strains had a NF-I motif (TGGAATGGTGCCAA) identical to the ones from HAdV species B (TGGAATGGTGCCAA). Examination of the NF-III motif showed the HAdV-E4p and -E4vac sequences to be nearly identical to the ones found in the SAdVs, whereas the same motif in the three field strains (TATGTTAAATGA) was identical to the NF-III motif found in the subspecies B2 virus, HAdV-B14; this differed slightly from the ones for subspecies B1 viruses (CATGTAAATGA).
Discussion
High-resolution data provides insight into emergent pathogens
Computational analysis of high-resolution genome data provides unprecedented views and insights to the understanding of microbial pathogens and infectious diseases (Relman, 2011), particularly in the context of newer additions to the archived, openly accessible and widely available genome database. In addition to providing information concerning phylogenetic relationships, which is conducive for typing adenoviruses (Seto et al., 2011), genome sequence data may be used as a tool for predicting emergent pathogens (manuscript in press) and for understanding the molecular evolution mechanisms involved in the genesis of novel viruses and emergent pathogens. Reported here, extending and enhancing a previous observation into the zoonotic origin of HdV-E4 (Purkayastha et al., 2005a), is another example of the insights revealed from whole genome analysis: molecular adaptation of a major adenovirus respiratory pathogen, HAdV-E4, to a new host. HAdV-E4 has been an intellectual challenge since its initial identification in 1954 as one of the first two HAdV isolated (Hilleman and Werner, 1954), particularly with its subsequent recognition as the only HAdV member in a species populated by five chimpanzee adenoviruses, SAdV-E22 through -E26. Speculations of HAdV-E4 as a virus that was “most closely related to a putative common archetype”; a novel virus resulting from recombination between two HAdVs from species B and C; and a highly divergent member of species B were supported by data using the then-most current and advanced methodologies (Bailey and Mautner, 1994; Gruber et al., 1993; Wadell, 1984). Two reports of newly isolated chimpanzee adenoviruses (Bailey and Mautner, 1994; Basnight et al., 1971; Rogers et al., 1967) led to speculation of a relationship with HAdV-E4, based on low-resolution genome data, e.g., restriction enzyme digestion patterns (Li and Wadell, 1988). Limited sequence data challenged its segregation into its own species (Bailey and Mautner, 1994). Additional low-resolution genome data, specifically GC% content, supported E as a species separate from B and C (Benkö et al., 2005). High-resolution and complete genome sequencing, coupled with a growing data set that included many simian adenovirus genomes, provided clearer evidence that HAdV-E4 arose through zoonosis, with a genome sequence that was more similar to SAdV-E25 than to other HAdVs, including members of species B and C (Purkayastha et al., 2005a). Data presented in this report shows a higher level of identity to SAdV-E26, a recently isolated chimpanzee adenovirus that is being screened as a candidate gene therapy vector (Roy et al., 2009) with presumably no seroprevalence in the human population. As both field strains reported here are nearly identical with respect to genome data, the same conclusions may be made for HAdV-E4FS2 as for HAdV-E4FSl, which is discussed in detail.
Interspecies recombination events
HAdV-E4 contains a recombinant genome, similar in structure to the genome of HAdV-B55, in which the hexonL1 and L2 of one genome are embedded in the genome chassis of another HAdV (Walsh et al., 2010), resulting in a novel pathogen. The reexamination of the two earlier published HAdV-E4 genomes and the analysis of two recently circulating field strains was enhanced by additional HAdV genomes in the database, completing the data set for original prototype genomes, and by more SAdV genomes. Recombination analysis revealed an interspecies event with HAdV-B16, or a yet-to-be-identified HAdV-B16-like virus, resulting in an emergent pathogen in which the hexonL1 and L2 of a human adenovirus genome are embedded in the genome chassis of a simian adenovirus. Nota bene, the “interspecies” recombination refers to the lateral partial gene transfer between HAdV species B and E, which is a novel observation. Additionally, it represents a lateral gene transfer between a human adenovirus and a chimpanzee adenovirus, presumably an ancestor to HAdV-E4, which is also a novel observation. It should also be noted that the prevailing a priori belief from 1980 should be updated, i.e., that the “established [species] serve as recombination barriers” based on observations then that “No intermediate strains with ‘parents’ from two different [species] have been reported and no evidence for recombination between Ad5 and Adl2 ts mutants could be found” (Wadell et al., 1980).
A second independent interspecies recombination event is revealed from comparing the NF-I motifs contained in the ITRs of three contemporary recent circulating HAdV-E4 field strains (TGGAATGGTGCCAA) with the counterparts from the two type 4 strains isolated nearly 60 years ago (TTTGTGTGAGTTAA). The older strains contain NF-I motifs that bear resemblance to those noted for the analyzed SAdVs (TTTGTGCGCGTTAA). This recombination event introduced a NF-I binding site from a species B HAdV into the ITR region housing the three viral DNA replication factor binding sites for the more recent type 4 viruses. This set of two host transcription factor binding motifs enhances DNA replication initiation up to 100 × both in vivo and in vitro (Mul et al., 1990). The field strain NF-I motif is characteristic of and similar to nearly all HAdVs, except HAdV-D9 as shown in Fig. 4, with its absence in the older type 4 strains characteristic of the examined SAdVs. The seven bases preceding this motif indicate this NF-I site may have resulted from a recombination with a species B virus as well. An inspection of the first 57 nucleotides of the ITRs from these two field strains shows they are identical to their counterparts from subspecies B2, represented in Fig. 4 by HAdV-B14, suggesting a recombination partner in the subspecies B2 in contrast to the findings of Houng et al. (2006).
NF-I and NF-III are reported to have viral growth enhancing effects independently and nonsynergistically (Rosenfeld et al., 1987). Stimulation of viral replication by the host transcription factor NF-I was noted to be strongly dependent on the concentration of the pTP-pol complex, with enhancement of up to 50 × at low pTP-pol concentrations but less than 2 × at high concentrations (Rosenfeld et al., 1987). Host transcription factor NF-III was noted to be much less dependent on this, but deletions and mutations in the NF-III binding site resulted in viruses that were defective for growth, including a high percentage of small plaques and yields that were 30–50 × below the wild-type levels (Hatfield and Hearing, 1993). The equivalent NF-I motif found for the older type 4 strains and the SAdVs, if they contain this motif, may either be optimized for the chimpanzee host, as species-specific motifs, or may bind alternative factors, as there are multiple NF-I genes in the genome (Giger et al., 2009). In addition, although the NF-I motif has been reported as necessary for HAdV DNA replication (Leegwater et al., 1985; Mul et al., 1990), it is also reported that the HAdV-E4 prototype does not absolutely require NF-I (Harris and Hay, 1988). This latter observation may reflect a less than optimal but viable pathogen in the human host.
Reconciliation of observations from serology and DNA sequencing
Serum neutralization (SN), centered on the epsilon epitope, which is presumably encoded by hexon loopsL1 and L2, has been used to differentiate and type HAdVs in the last century. In theory, since theL1 sequence of HAdV-E4FSl is highly similar to theL1 sequence of HAdV-B16 (94.8%), there should be a very close antigenic relationship. This is not the case, however, as two representative reports illustrate this contradiction. Rowe, et al. found that HAdV-E4 was neutralized by antiserum to HAdV-B16, but did not report a reciprocal with antiserum to HAdV-E4 (Rowe et al., 1958). In contrast, Rafajko (1964) reported that HAdV-E4 and HAdV-B16 cross-react at a “low level” in two-way crosses, with an 8 × titration in one direction but much a lower titration in the other direction. The “predominantly one-sided character” of the cross-neutralization between types 4 and 16 (Norrby and Wadell, 1969) was also reported by others, e.g., 2–4 × versus 16–32 × differences (Stevens et al., 1967). Additionally, SN data summaries from that era discussed similar conflicting results from several researchers (Hierholzer et al., 1991; Rowe et al., 1958; Stevens et al., 1967; Wadell, 1984; Wigand, 1975, 1987, 1965, 1985), with Wigand et al. (1965), reporting “[t]ypes 4 and 16 were neutralized by all heterologous sera tested with 5- to 20-fold lower heterologous than homologous titers” based on three antisera for each virus. Further, they note “[t]he neutralization method used cannot be regarded as either sensitive or accurate” (Wigand et al., 1965). Twenty-two years later, Wigand, in “Pitfalls in the identification of adenoviruses”, states HAdV-B16 “shows a variety of cross-reactions in SN. The prototype and some other strains show cross-SN with [HAdV-E4], others with [HAdV-B14] (citing Wigand, 1975), still others do not cross with another type ([Wigand] unpublished data)” (Wigand, 1987).
Another significant inconsistency with SN data regarding HAdV-E4 is the report that the virus was neutralized with antiserum to SAdV-E25 and vice-versa, sometimes. With the isolation of chimpanzee adenoviruses (SAdVs) in Basnight et al. (1971), Rogers et al. (1967), studies to probe adenoviral homologies were performed. SN experiments showed that Pan 9 (SAdV-E25) and HAdV-E4 resulted in “a definite reciprocal cross-neutralization with heterologous titers about 8-fold lower than homologous titers in either direction” (Willimzik et al., 1981). But, as noted in the same report, this “bilateral relationship” between SAdV-E25 and HAdV-E4 was not reported in the original studies on SAdV-E25 (Rogers et al., 1967). Seven reports were cited by Willimzik et al. (1981) as presenting negative results with respect to the same observation and, in most cases, involving HAdV antisera against SAdVs; on the other hand, publications were cited noting that SAdV antisera against HAdVs presented rare “concomitant neutralization found for two or three antisera of the same type” (Willimzik et al., 1981). With the genome data available, a similarity of 64.7% was calculated for the L1 sequences from HAdV-E4FSl and SAdV-E25. This provides a strong prediction that the antisera raised against the variable hexon domains will not cross-react if theL1 and L2 loops are the sole definitive epitopes.
Given high-resolution and complete genome data, it is clear that the SN epitope alone, recognized either by bench SN (Crawford-Miksza and Schnurr, 1996; Yuan et al., 2009) or PCR-based limited DNA sequence determination (Ebner et al., 2005; Madisch et al., 2005; Pring-Akerblom et al., 1995; Sarantis et al., 2004), is inadequate for understanding the relationships between HAdV-E4 and -B16. This is the first example in which the species profiles resulting from the whole genome phylogenetic tree are not identical to the profiles for the hexonL1 and L2 trees (manuscript in preparation):L1 and L2 for HAdV-B16 forms a clade with the species E counterparts, rather than with species B members, contrasting with genome phylogenetic trees and accepted taxo-nomic classification. Similarly, the PCR-based limited DNA sequence data (Ebner et al., 2005; Madisch et al., 2005; Pring-Akerblom et al., 1995; Sarantis et al., 2004) do not provide definitive resolution for typing and taxonomy, raising questions as to whether the particular one-dimensional linear string of nucleotide sequences captured by the primer sets actually represent the three-dimensional and perhaps non-linear cluster of amino acid sequences comprising antibody recognition. Assessing a single site along the genome also precludes recognition of genome recombination events that are relevant in defining a pathogen, tropism and disease.
Limited and missing DNA sequence data sets provide misleading and puzzling conclusions, for example, in Madisch et al. (2005) reported the L1 and L2 sequences of HAdV-B16 and HAdV-E4 forming a clade, with SAdV-E25 branching from it distantly; but observed HAdV-E4 and SAdV-E25 forming a subclade within the branch for species C in the gamma (hemagglutination) epitope representing the fiber gene. They noted the inclusion of SAdV-E25 in their analysis was due to the report that HAdV-E4 was more similar to the SAdVs (Purkayastha et al., 2005a), and suggested a recombination, noting a limited analysis by the absence of the HAdV-B16 data (Madisch et al., 2005). Their hypothesis of “an interspecies recombination mechanism in the phylogeny of both HAdV-E4 and SAdV-E25, combining a species HAdV-C-like fiber gene with a species HAdV-B16 genetic backbone” (Madisch et al., 2005), is shown to be incorrect in this report, as data support a recombination scenario in which the hexon L1 and L2 domains were laterally transferred into the genome of a SAdV. Similarly, Sarantis et al. (2004) used a different primer set to amplify the hexon hypervariable 7 region of 49 serotypes; their data suggested HAdV-B16 and HAdV-E4 were of the same species E, which was distantly related to species B, comprising only HAdV-B3 and HAdV-B7. The rest of the species B members formed a cluster branching from the clusters of species A and F, and all three branch from species C. In contrast, Ebner et al. (2005), sequenced the hexon genes from 51 serotypes and, based on separate phylogenetic analysis of the variable region (765 aa) and the constant region (229 aa), suggested the hexon of HAdV-B16 was a product of recombination between species E (a clade with HAdV-E4) and species B (a clade with HAdV-B3 and HAdV-B7) in the variable and constant regions, respectively. This putative recombinant was suggested by Pring-Akerblom, et al. in 1995 from analyzing hexon sequences of HAdV-B3, -E4, and -B16, concluding “[if] nevertheless AV4 should have arisen by recombination, one parent most likely seems to be an AV16 strain” (Pring-Akerblom et al., 1995). Their goal was to reconcile SN data showing cross-reactions between HAdV-B16 and -E4 (Wigand, 1987). In support of this, they observed a “high degree of homology” between the HAdV-E4 and HAdV-B16 hexon genes, including, surprisingly, in the hypervariable L1 and L2 regions, as well as the expected conserved region. However, loop 4 distinguished HAdV-E4 from both HAdV-B3 and -B16, supporting “a separate subgenus” in contrast to sequence-based observations from Bailey and Mautner (1994) who observed a “close evolutionary relationship” between HAdV-E4 and species B viruses in several genome regions, challenged the species E assignment.
On the origin of species E, human adenovirus type 4
Sequencing archived viruses, including historically important or intriguing ones, and circulating descendants provides unique opportunities for understanding how they evolve and adapt. Although it is extremely difficult to envision the past with absolute certainty, high-resolution data provide more informed hypotheses and additional data. Using multiple approaches, the genomes of a prototype HAdV-E4 (1952), a prominent field strain “vaccine” (1962) and two current field strains (2002 and 2003) provide snapshots of the molecular evolution of this highly contagious human pathogen. The recent availability of all of the prototype HAdV genomes and the recent interest in simian adenoviruses and their genomes have provided the resources for refining the earlier view that HAdV-E4p contained genome sequences that had higher similarity to five SAdV genomes rather than earlier hypothesized relationships to HAdV species B and C partial sequences. The refined view is that HAdV-E4 viruses contain a genome comprising 97% SAdV-E26-like genome chassis, with a hexon containing the L1 and L2 of HAdV-B16-like virus. Presumably this hexon provides compatibility with the new host. This evolutionary change has been selected and is preserved in the two type 4 strains circulating nearly fifty years later. In turn, the newer field strains have an additional genome change, acquiring a DNA replication factor binding motif that is similar to ones found in other HAdVs. As NF-I is noted to be a virus growth enhancer that acts independently and nonsynergistically of its partner NF-III, and as its absence may be compensated by a higher concentration of the pTP-pol complex, its absence or “human-imperfect” sequence in HAdV-E4p and -E4vac may not have resulted in an evolution “cul-de-sac”. Rather, the pathogen may be less fit in competition but still viable and infectious. Upon the second recombination event, providing a human-complementary NF-I, HAdV-E4 may be more robust and better equipped to survive selection during host passages.
Pathoepidemiology of HAdV-E4
The proposed two stage evolutionary path allowing HAdV-E4 to establish itself in a new host may explain the reports of HAdV-E4 outbreaks and infections predominantly in the U.S. military population in the 1970s and earlier. With the caveat that it is impossible to know when exactly a mutant appears in the population and given that records and virus collection have been less than optimal, and that HAdVs, in general, are underreported, HAdV type surveys show a lower number of HAdV-E4 infections in the civilian population prior to the 1970s, despite it being one of the two first HAdVs isolated and identified (Hilleman and Werner, 1954). A summary from The Seattle Virus Watch presented HAdV typing data from Seattle (1965–1969) and New York (1961–1965) noting only three HAdV-E4 were identified out of 520 HAdVs, and two out of 564, respectively (Fox et al., 1977), in civilian populations. In a 1984 review, Wadell notes and cites a report that “Ad4 is rarely isolated from children in Europe and the United States; it was isolated from four children out of 1800” (Wadell, 1984). A 15-year study (1967–1981) of a children Group Day Care Center found no HAdV-E4 isolates amongst 298 HAdV isolates, even though the researchers were aware of its circulation in the U.S. military (Pacini et al., 1987). In contrast, HAdV-E4 was first isolated during an outbreak at Ft. Leonard Wood (MO; U.S.A.) (Hilleman and Werner, 1954) and one of the first respiratory pathogen surveys, spanning 1966–1971, noted it and HAdV-B7 as causal factors in 60% of basic military trainees hospitalized with ARD (Dudding et al., 1973). HAdV-E4 presented enough of a problem that by 1965, an initial live human diploid cell vaccine was developed and tested (Chanock et al., 1966). Subsequently, the predominance of HAdV-E4 and HAdV-B7 in the military drove the development and deployment of vaccines against both in the past and currently (Binn et al., 2007; Couch et al., 1963; Gaydos and Gaydos, 1995; Top et al., 1971). After successfully preventing adenovirus-caused ARD, the original vaccines were discontinued and the subsequent rise of HAdV respiratory pathogens to pre-vaccine levels of HAdV-E4 and -B7, as causative agents of ARD, has been well documented (Blasiole et al., 2004; Kajon et al., 2007; Metzgar et al., 2007). This provided a view of the robustness of HAdV-E4. In a retrospective surveillance study spanning 1996, the last year of routine vaccine use, to 2002, the displacement of other HAdV respiratory pathogens by HAdV-E4 is detailed, with both HAdV-E4 and B7 initially constituting a small proportion of the ARD-causing HAdV pathogens identified in 1996, at 4% each. A follow-up study showed HAdV-B7 “virtually disappearing” in 1998 with HAdV-E4 numbers increasing to 73% and then displacing the previously predominant strain HAdV-B21 (58% in 1996). HAdV-B21 was noted as the dominant circulating strain prior to the discontinuation of the vaccines (Blasiole et al., 2004), i.e., during the suppression of HAdV-E4. From 1999–2002, without the vaccine, the proportion of HAdV-E4 isolated from military recruit camps rose from 98% to 99.9% (Kajon et al., 2007). Another study of the recruit training camps, continuing this surveillance, documented the predominance of HAdV-E4, with a few cases of HAdV-B7, B3, and species C viruses reported in 2003 to April 2006 (Metzgar et al., 2007). This study also noted abrupt and unexpected outbreak of HAdV-B14, displacing HAdV-E4 and other HAdVs in 2006; but when the study ended in August 2006, it appeared HAdV-E4 was again displacing all other HAdVs (Metzgar et al., 2007). Currently, HAdV-E4 circulates in the civilian population as well as the military, for example, a report surveying hospital patients in Southern Taiwan (2001–2002) and typing 317 HAdV isolates revealed HAdV-E4 as the major respiratory HAdV pathogen at 57% in 2001.
A limited examination of ITRs sequenced from 61 HAdV-E4 strains archived in the U.S. military from 1953 to 1996 suggests the “first Ad4 strain carrying the new 208 bp ITR was initially isolated in 1976 from Georgia, USA” and a “second new Ad4 genotype was isolated from France two years later” (Houng et al., 2006). The caveat is that the analysis focused on the differences in the first eight nucleotides at the terminus and divergence after the 22nd nucleotide, rather than the replication/transcription factor binding motifs, and the full sequence data set has not been released to the public. In addition, their hypothesis that the 5’-ITR was recombined from a subspecies B1 virus into the current strains of HAdV-E4 (Houng et al., 2006) is incorrect, as closer inspection shows the initial seven nucleotides CATCATC are identical for the type 4 field strains and subspecies B2, differing from CTATCTA observed for subspecies B1 HAdV-B3 and -B7. Additionally, following the NF-III motif, GGTGAT is found for the recent type 4 field strains and subspecies B2 HAdVs, differing again from the GGTAAT found for subspecies B1 HAdVs. Nevertheless, the survey of archived type 4 strains provides a putative time-line for the second recombination event and may account for the spread of this virus into the general population.
Conclusions
The two independent time-separated interspecies recombination events found in the genomes of two recently circulating HAdV-E4 field strains, coupled with the overall high similarity of the genome to SAdV genomes, provide a unique view of the emergence of a human viral pathogen in very high resolution. Optimization of human adenovirus type 4 to a new host is explained by the capture of a HAdV partial hexon gene, as well as the integration of a binding site, from another HAdV, for a human transcription factor that expedites adenoviral DNA replication. As HAdV-E4 is an important highly contagious human respiratory and ocular pathogen, these results emphasize the value of genomics and bioinformatic approaches to the study of viruses and infectious disease agents, in general, and adenovirus, specifically. Adenoviruses are important biomedical tools also, as candidate gene and epitope delivery vectors. Simian non-human adenoviruses are increasingly being considered as alternatives to HAdVs due to concerns of seroprevalence. This report of lateral DNA and epitope transfers between HAdVs and SAdVs, specifically, and observations of recombinants, in general, serve as caveats when considering chimpanzee adenoviruses as candidate gene delivery vectors for human patients.
Materials and methods
Stocks, virus growth and DNA purification
HAdV-E4FSl, also known as NHRC 42606 (Ft. Jackson, SC; 2003), was obtained from Kevin Russell (Naval Health Research Center; San Diego, CA) and HAdV-E4FS2, also known as NHRC 3 (Brooks AFB; San Antonio, TX; 2002), was obtained from Linda Canas (Brooks AFB; San Antonio, TX). These strains were confirmed as type 4 by molecular typing, including PCR and micro-array analysis (Lin et al., 2006). They are further distinguished as HAdV-E4a2 and HAdV-E4al, respectively, by RE analysis (Kajon et al., 2007). Viruses were grown in A-549 cells (ATCC CCL-185), with the virus growth and DNA purification by Virapur,L1C. (San Diego, CA), using protocols as reported earlier (Lauer et al., 2004; Purkayastha et al., 2005a, 2005b).
Genome sequencing and annotation
As reported for a similar project (Purkayastha et al., 2005b), genome sequencing of the two field strains was outsourced to Commonwealth Biotechnologies, Inc. (Richmond, VA), which used the Sanger chemistry method, utilizing DYEnamic ET Terminator Cycle Sequencing kits (Amersham Biosciences; Piscataway, NJ), and resolved the sequence ladders on an ABI Prism 377 DNA Sequencer (Applied Biosystems; Foster City, CA). These sequences were assembled with DNA Sequencher (GeneCodes, Inc.; Ann Arbor, MI), with an average five-fold coverage across the genomes and a minimum of three-fold including both orientations. This sequence was supplemented with PCR amplification and re-sequencing of areas that were found to be questionable upon sequence assembly and genome annotation. Quality control included genome annotation, and comparisons with earlier type 4 genome data, including the prototype and vaccine (Purkayastha et al., 2005a, 2005b). Annotation was performed using the Genome Annotation Transfer Utility (GATU) software tool (Tcherepanov et al., 2006), and recorded and visualized using Artemis, a genome viewer (http://www.sanger.ac.uk/resources/software/artemis/) (Berriman and Rutherford, 2003). Open reading frames were compared against GenBank entries for confirmation and protein similarity. Splice sites were predicted using the GenScan web server at MIT (http://www.genes.mit.edu/GENSCAN.html), and confirmed by comparisons to previously annotated type 4 genomes (Purkayastha et al., 2005a, 2005b).
Computational genome analyses
Sequence analyses were performed using publicly accessible software tools as described earlier (Walsh et al., 2009). Multiple whole-genome alignments were performed using multiple alignment, which utilizes fast Fourier transforms (MAFFT) (Katoh and Toh, 2008). This software is available online (http://www.ebi.ac.uk/Tools/mafft/.) and was applied using the default gap parameters in all alignments. Pairwise alignments, comparisons and visualization of genomes were performed using the zPicture software (http://zpicture.dcode.org) (Ovcharenko et al., 2004). ITRs were initially aligned with Clustal Omega (www.ebi.ac.uk/Tools/msa/clustalo/) (Larkin et al., 2007); several gaps and mismatches were edited by eye for an optimal alignment.
Sequence percent identity analysis
Protein and noncoding sequence annotations were completed as described previously (Purkayastha et al., 2005a, 2005b; Walsh et al., 2009). Global alignments were performed using the Needle program of EMBOSS (Olson, 2002). A BLOSUM62 matrix was used for the amino acid sequence analysis, and a DNA full matrix was used for nucleotide sequence analysis. To analyze the hexon L1 and L2 regions corresponding to the putative SN epsilon epitope (Crawford-Miksza and Schnurr, 1996; Yuan et al., 2009), the flanking sequences and protocols used were as described by Madisch et al. (2005).
Genome and gene recombination analysis
The recombination analysis software tool Simplot, including Bootscan, was used to complete a nucleotide sequence recombination assessment of the MAFFT aligned genes (Lole et al., 1999). Default parameter settings were used for the window size (200 nucleotides [nt]), step size (20 nt), replicates used (n = 100), gap stripping (on), distance model (Kimura) and tree model (neighbor joining). Similarly, whole genomes were analyzed, starting with an initial alignment using MAFFT and following with recombination analysis using Simplot and Bootscan. Only the window size and step size were altered (1000 and 200, respectively), with the rest of the default parameters unchanged.
Phylogenetic analysis
Whole genome and gene sequence alignments for phylogenetic trees were constructed using MAFFT, a web-accessible software tool. Select regions of the hexon sequences were extracted for detailed analysis, borders as noted by Madisch et al. (2005). Boot-strapped, neighbor-joining trees with 1000 replicates were constructed using MEGA4 software with the maximum-composite-likelihood method (Tamura et al., 2007). All of the other parameters were set by default.
GenBank accession numbers
Genome data and annotation files for HAdV-E4FSl (NHRC 42606) and HAdV-E4FS2 (NHRC 3), AY599835 and AY599837, respectively, were deposited in Genbank. Other HAdV genomes are as follows: HAdV-E4 prototype (AY594253) and vaccine (AY594254); HAdV-C2 (AC_000007); HAdV-B3 (DQ0-86466); HAdV-C5 (AC_000008); HAdV-B7 (AY594255); HAdV-D9 (AJ854486); HAdV-B11 (AF532578); HAdV-A12 (AC000005); HAdV-B14 (AY803294); HAdV-B16 (AY601636); HAdV-B21 (AY601633); HAdV-B34 (AY737797); HAdV-B35 (AY271307); and HAdV-F41 (DQ315364). Chimpanzee “simian” adenoviruses and genome accession numbers are as follows: SAdV-E21 (AC_000010); SAdV-E22 (AY530876); SAdV-E23 (AY530877); SAdV-E24 (AY530878); SAdV-E25 (AC_000011); and SAdV-E26 (FJ025923).
Acknowledgments
Genome sequencing was completed with support from the Department of Defense through which one of the authors (D.S.: 2002 to 2004) was affiliated with the U.S. Air Force Surgeon General Office, Directorate of Modernization (SGR) and the U.S. Air Force Epidemic Outbreak Surveillance (EOS) Program (Falls Church, VA), specifically by a grant from the U.S. Army Medical Research and Material Command (DAMD17-03-2-0089). Additional support was provided through the EOS Project, which was funded by HQ USAF Surgeon General Office, Directorate of Modernization (SGR), and the Defense Threat Reduction Agency (DTRA). One of the HAdV-E4 field strains was obtained from and collected with the support of the Global Emerging Infections Surveillance and Response System (GEIS), a Division of the Armed Forces Health Surveillance Center, Silver Spring, MD (www.afhsc.mil). D.S. thanks Clark Tibbetts (EOS; 2001 to 2005) and David Metzgar for earlier discussions on type 4 HAdVs, as well as the Office of the Provost at George Mason University for funding a Sabbatical Study Leave; Lilian Coralia Lopez Rojas Desjardins for providing a unique writing environment in Antigua (Guatemala); and Professor James Chodosh, his research group, and the Massachusetts Eye and Ear Infirmary, Harvard Medical School (Boston, MA) for providing a stimulating intellectual environment.
JC was supported by NIH grant EY013124 and a Research to Prevent Blindness Senior Scientific Investigator Award.
References
- Adrian T, Wadell G, Hierholzer JC, Wigand R. DNA restriction analysis ol adenovirus prototypes 1 to 41. Arch. Virol. 1986;91:277–290. doi: 10.1007/BF01314287. [DOI] [PubMed] [Google Scholar]
- Aoki K, Benko M, Davison AJ, Echavarria M, Erdman DD, Harrach B, Kajon AE, Schnurr D, Wadell G. Toward an integrated human adenovirus designation system that utilizes molecular and serological data and serves both clinical and fundamental virology. J. Virol. 2011;85:5703–5704. doi: 10.1128/JVI.00491-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey A, Mautner V. Phylogenetic relationships among adenovirus serotypes. Virology. 1994;205:438–52. doi: 10.1006/viro.1994.1664. [DOI] [PubMed] [Google Scholar]
- Barrero PR, Valinotto LE, Tittarelli E, Mistchenko AS. Molecular typing of adenoviruses in pediatric respiratory infections in Buenos Aires, Argentina (1999–2010) J. Clin. Virol. 2012;53:145–150. doi: 10.1016/j.jcv.2011.11.001. [DOI] [PubMed] [Google Scholar]
- Basnight M, Jr., Rogers NG, Gibbs CJ, Jr., Gajdusek DC. Characterization of four new adenovirus serotypes isolated from chimpanzee tissue explants. Am. J. Epidemiol. 1971;94:166–171. doi: 10.1093/oxfordjournals.aje.a121308. [DOI] [PubMed] [Google Scholar]
- Benkö MHB, Both GW, Russell WC, Adair BM, Ádám É, de Jong JC, Hess M, Johnson M, Kajon A, Kidd AHDLHQ-GL, Mautner V, Pring-Akerblom P, Wadell G. Family Adenoviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. The Eighth Report of the International Committee on Taxonomy of Viruses. San Diego, CA: Academic Press; 2005. pp. 213–228. [Google Scholar]
- Berriman M, Rutherford K. Viewing and annotating sequence data with Artemis. Brief. Bioinform. 2003;4:124–132. doi: 10.1093/bib/4.2.124. [DOI] [PubMed] [Google Scholar]
- Binn LN, Sanchez JL, Gaydos JC. Emergence of adenovirus type 14 in US military recruits—a new challenge. J. Infect. Dis. 2007;196:1436–1437. doi: 10.1086/522969. [DOI] [PubMed] [Google Scholar]
- Blasiole DA, Metzgar D, Daum LT, Ryan MA, Wu J, Wills C, Le CT, Freed NE, Hansen CJ, Gray GC, Russell KL. Molecular analysis of adenovirus isolates from vaccinated and unvaccinated young adults. J. Clin. Microbiol. 2004:1686–1693. doi: 10.1128/JCM.42.4.1686-1693.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanock RM, Ludwig W, Heubner RJ, Cate TR, Chu LW. Immunization by selective infection with type 4 adenovirus grown in human diploid tissue cultures. I. Safety and lack of oncogenicity and tests for potency in volunteers. 195. JAMA. 1966:445–52. [PubMed] [Google Scholar]
- Couch RB, Chanock RM, Cate TR, Lang DJ, Knight V, Huebner RJ. Immunization with types 4 and 7 adenovirus by selective infection of the intestinal tract. Am. Rev. Respir. Dis. 1963;88:394–03. doi: 10.1164/arrd.1963.88.3P2.394. SUPPL. [DOI] [PubMed] [Google Scholar]
- rawford-Miksza L, Schnurr DP. Analysis of 15 adenovirus hexon proteins reveals the location and structure of seven hypervariable regions containing serotype-specific residues. J. Virol. 1996;70:1836–1844. doi: 10.1128/jvi.70.3.1836-1844.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darr S, Madisch I, Hofmayer S, Rehren F, Heim A. Phylogeny and primary structure analysis of fiber shafts of all human adenovirus types for rational design of adenoviral gene-therapy vectors. J. Gen. Virol. 2009;90:2849–2854. doi: 10.1099/vir.0.014514-0. [DOI] [PubMed] [Google Scholar]
- Davison AJ, Benko M, Harrach B. Genetic content and evolution of adenoviruses. J. Gen. Virol. 2003;84:2895–2908. doi: 10.1099/vir.0.19497-0. [DOI] [PubMed] [Google Scholar]
- Dudding BA, Top FH, Jr., Winter RE, Buescher EL, Lamson TH, Leibovitz A. Acute respiratory disease in military trainees: the adenovirus surveillance program, 1966–1971. Am. J. Epidemiol. 1973;97:187–198. doi: 10.1093/oxfordjournals.aje.a121499. [DOI] [PubMed] [Google Scholar]
- Ebner K, Pinsker W, Lion T. Comparative sequence analysis of the hexon gene in the entire spectrum of human adenovirus serotypes: phylogenetic, taxonomic, and clinical implications. J. Virol. 2005;79:12635–12642. doi: 10.1128/JVI.79.20.12635-12642.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echavarria M. Adenoviruses in immunocompromised hosts. Clin. Microbiol. Rev. 2008;21:704–715. doi: 10.1128/CMR.00052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelmann I, Madisch I, Pommer H, Heim A. An outbreak of epidemic keratoconjunctivitis caused by a new intermediate adenovirus 22/H8 identified by molecular typing. Clin. Infect. Dis. 2006;43:e64–e66. doi: 10.1086/507533. [DOI] [PubMed] [Google Scholar]
- Fox JP, Hall CE, Cooney MK. The Seattle virus watch. VII. Observations of adenovirus infections. Am. J. Epidemiol. 1977;105:362–386. doi: 10.1093/oxfordjournals.aje.a112394. [DOI] [PubMed] [Google Scholar]
- Fujishiro J, Takeda S, Takeno Y, Takeuchi K, Ogata Y, Takahashi M, Hakamata Y, Kaneko T, Murakami T, Okada T, Ozawa K, Hashizume K, Kobayashi E. Gene transfer to the rat kidney in vivo and ex vivo using an adenovirus vector: factors influencing transgene expression. Nephrol. Dial. Transplant. 2005;20:1385–1391. doi: 10.1093/ndt/gfh783. [DOI] [PubMed] [Google Scholar]
- Garnett CT, Talekar G, Mahr JA, Huang W, Zhang Y, Ornelles DA, Gooding LR. Latent species C adenoviruses in human tonsil tissues. J. Virol. 2009;83:2417–2428. doi: 10.1128/JVI.02392-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaydos CA, Gaydos JC. Adenovirus vaccines in the U.S. military. Mil. Med. 1995;160:300–304. [PubMed] [Google Scholar]
- Giger JM, Bodell PW, Baldwin KM, Haddad F. The CAAT-binding transcription factor 1/nuclear factor 1 binding site is important in beta- myosin heavy chain antisense promoter regulation in rats. Exp. Physiol. 2009;94:1163–1173. doi: 10.1113/expphysiol.2009.049692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham FL, Prevec L. Adenovirus-based expression vectors and recombi nant vaccines. Vol. 20. Reading, Mass: Biotechnology; 1992. pp. 363–390. [DOI] [PubMed] [Google Scholar]
- Greber UF, Arnberg N, Wadell G, Benko M, Kremer EJ. Adenoviruses— from pathogens to therapeutics: a report on the 10th International Adenovirus Meeting. Cell. Microbiol. 2013;15:16–23. doi: 10.1111/cmi.12031. [DOI] [PubMed] [Google Scholar]
- Gruber WC, Russell DJ, Tibbetts C. Fiber gene and genomic origin of human adenovirus type 4. Virology. 1993;196:603–611. doi: 10.1006/viro.1993.1516. [DOI] [PubMed] [Google Scholar]
- Harris MP, Hay RT. DNA sequences required for the initiation of adenovirus type 4 DNA replication in vitro. J. Mol. Biol. 1988;201:57–67. doi: 10.1016/0022-2836(88)90438-x. [DOI] [PubMed] [Google Scholar]
- Hatfield L, Hearing P. Redundant elements in the adenovirus type 5 inverted terminal repeat promote bidirectional transcription in vitro and are important for virus growth in vivo. Virology. 1991;184:265–276. doi: 10.1016/0042-6822(91)90843-z. [DOI] [PubMed] [Google Scholar]
- Hatfield L, Hearing P. The NFIII/OCT-1 binding site stimulates adenovirus DNA replication in vivo and is functionally redundant with adjacent sequences. J. Virol. 1993;67:3931–3939. doi: 10.1128/jvi.67.7.3931-3939.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henquell C, Boeuf B, Mirand A, Bacher C, Traore O, Dechelotte P, Labbe A, Bailly JL, Peigue-Lafeuille H. Fatal adenovirus infection in a neonate and transmission to health-care workers. J. Clin. Virol. 2009;45:345–348. doi: 10.1016/j.jcv.2009.04.019. [DOI] [PubMed] [Google Scholar]
- Hierholzer JC, Stone YO, Broderson JR. Antigenic relationships among the 47 human adenoviruses determined in reference horse antisera. Arch. Virol. 1991;121:179–197. doi: 10.1007/BF01316753. [DOI] [PubMed] [Google Scholar]
- Hilleman MR, Werner JH. Proceedings of the Society for Experimental Biology and Medicine. Vol. 85. New York, N.Y: Society for Experimental Biology and Medicine; 1954. Recovery of new agent from patients with acute respiratory illness; pp. 183–188. [DOI] [PubMed] [Google Scholar]
- Houng HS, Clavio S, Graham K, Kuschner R, Sun W, Russell KL, Binn LN. Emergence of a new human adenovirus type 4 (Ad4) genotype: identification of a novel inverted terminal repeated (ITR) sequence from majority of Ad4 isolates from US military recruits. J. Clin. Virol. 2006;35:381–387. doi: 10.1016/j.jcv.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Kajon AE, Moseley JM, Metzgar D, Huong HS, Wadleigh A, Ryan MA, Russell KL. Molecular epidemiology of adenovirus type 4 infections in US military recruits in the postvaccination era (1997–2003) J. Infect. Dis. 2007;196:67–75. doi: 10.1086/518442. [DOI] [PubMed] [Google Scholar]
- Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Vol. 23. Oxford, England: Bioinformatics; 2007. pp. 2947–2948. [DOI] [PubMed] [Google Scholar]
- Lauer KP, L1orente I, Blair E, Seto J, Krasnov V, Purkayastha A, Ditty SE, Hadfield TL, Buck C, Tibbetts C, Seto D. Natural variation among human adenoviruses: genome sequence and annotation of human adenovirus serotype 1. J. Gen. Virol. 2004;85:2615–2625. doi: 10.1099/vir.0.80118-0. [DOI] [PubMed] [Google Scholar]
- Leegwater PA, van Driel W, van der Vliet PC. Recognition site of nuclear factor I, a sequence-specific DNA-binding protein from HeLa cells that stimulates adenovirus DNA replication. EMBO J. 1985;4:1515–1521. doi: 10.1002/j.1460-2075.1985.tb03811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QG, Wadell G. The degree of genetic variability among adenovirus type 4 strains isolated from man and chimpanzee. Arch. Virol. 1988;101:65–77. doi: 10.1007/BF01314652. [DOI] [PubMed] [Google Scholar]
- Lin B, Wang Z, Vora GJ, Thornton JA, Schnur JM, Thach DC, Blaney KM, Ligler AG, Malanoski AP, Santiago J, Walter EA, Agan BK, Metzgar D, Seto D, Daum LT, Kruzelock R, Rowley RK, Hanson EH, Tibbetts C, Stenger DA. Broad-spectrum respiratory tract pathogen identification using resequencing DNA microarrays. Genome Res. 2006;16:527–535. doi: 10.1101/gr.4337206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu EB, Wadford DA, Seto J, Vu M, Hudson NR, Thrasher L, Torres S, Dyer DW, Chodosh J, Seto D, Jones MS. Computational and serologic analysis of novel and known viruses in species human adenovirus D in which serology and genomics do not correlate. PLoS One. 2012;7:e33212. doi: 10.1371/journal.pone.0033212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons A, Longfield J, Kuschner R, Straight T, Binn L, Seriwatana J, Reitstetter R, Froh IB, Craft D, McNabb K, Russell K, Metzgar D, Liss A, Sun X, Towle A, Sun W. A double-blind, placebo-controlled study of the safety and immunogenicity of live, oral type 4 and type 7 adenovirus vaccines in adults. Vaccine. 2008;26:2890–2898. doi: 10.1016/j.vaccine.2008.03.037. [DOI] [PubMed] [Google Scholar]
- Madisch I, Harste G, Pommer H, Heim A. Phylogenetic analysis of the main neutralization and hemagglutination determinants of all human adenovirus prototypes as a basis for molecular classification and taxonomy. J. Virol. 2005;79:15265–15276. doi: 10.1128/JVI.79.24.15265-15276.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima Y, Shimizu H, Phan TG, Ushijima H. Genomic characterization of a novel human adenovirus type 31 recombinant in the hexon gene. J. Gen. Virol. 2011;92:2770–2775. doi: 10.1099/vir.0.034744-0. [DOI] [PubMed] [Google Scholar]
- Metzgar D, Osuna M, Kajon AE, Hawksworth AW, Irvine M, Russell KL. Abrupt emergence of diverse species B adenoviruses at US military recruit training centers. J. Infect. Dis. 2007;196:1465–1473. doi: 10.1086/522970. [DOI] [PubMed] [Google Scholar]
- Metzgar D, Osuna M, Yingst S, Rakha M, Earhart K, Elyan D, Esmat H, Saad MD, Kajon A, Wu J, Gray GC, Ryan MA, Russell KL. PCR analysis of egyptian respiratory adenovirus isolates, including identification of species, serotypes, and coinfections. J. Clin. Microbiol. 2005;43:5743–5752. doi: 10.1128/JCM.43.11.5743-5752.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mul YM, Verrijzer CP, van der Vliet PC. Transcription factors NFI and NFIII/ oct-1 function independently, employing different mechanisms to enhance adenovirus DNA replication. J. Virol. 1990;64:5510–5518. doi: 10.1128/jvi.64.11.5510-5518.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrby E, Wadell G. Immunological relationships between hexons of certain human adenoviruses. J Virol. 1969;4:663–670. doi: 10.1128/jvi.4.5.663-670.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson SA. EMBOSS opens up sequence analysis. European Molecular Biology Open Software Suite. Brief. Bioinform. 2002;3:87–91. doi: 10.1093/bib/3.1.87. [DOI] [PubMed] [Google Scholar]
- Ovcharenko I, Loots GG, Hardison RC, Miller W, Stubbs L. zPicture: dynamic alignment and visualization tool for analyzing conservation profiles. Genome Res. 2004;14:472–77. doi: 10.1101/gr.2129504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacini DL, Collier AM, Henderson FW. Adenovirus infections and respiratory illnesses in children in group day care. J. Inf. Dis. 1987;156:920–927. doi: 10.1093/infdis/156.6.920. [DOI] [PubMed] [Google Scholar]
- Pring-Akerblom P, Trijssenaar FE, Adrian T. Sequence characterization and comparison of human adenovirus subgenus B and E hexons. Virology. 1995;212:232–236. doi: 10.1006/viro.1995.1474. [DOI] [PubMed] [Google Scholar]
- Pruijn GJ, van Miltenburg RT, Claessens JA, van der Vliet PC. Interaction between the octamer-binding protein nuclear factor III and the adenovirus origin of DNA replication. J. Virol. 1988;62:3092–3102. doi: 10.1128/jvi.62.9.3092-3102.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purkayastha A, Ditty SE, Su J, McGraw J, Hadfield TL, Tibbetts C, Seto D. Genomic and bioinformatics analysis of HAdV-4, a human adenovirus causing acute respiratory disease: implications for gene therapy and vaccine vector development. J. Virol. 2005a;79:2559–2572. doi: 10.1128/JVI.79.4.2559-2572.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purkayastha A, Su J, McGraw J, Ditty SE, Hadfield TL, Seto J, Russell KL, Tibbetts C, Seto D. Genomic and bioinformatics analyses of HAdV-4vac and HAdV-7vac, two human adenovirus (HAdV) strains that constituted original prophylaxis against HAdV-related acute respiratory disease, a reemer-ging epidemic disease. J. Clin. Microbiol. 2005b;43:3083–3094. doi: 10.1128/JCM.43.7.3083-3094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafajko RR. Production and standardization of adenovirus types 1 to 18 reference antisera. Am. J. Hyg. 1964;79:310–319. doi: 10.1093/oxfordjournals.aje.a120385. [DOI] [PubMed] [Google Scholar]
- Relman DA. Microbial genomics and infectious diseases. N. Engl. J. Med. 2011;365:347–357. doi: 10.1056/NEJMra1003071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson CM, Singh G, Henquell C, Walsh MP, Peigue-Lafeuille H, Seto D, Jones MS, Dyer DW, Chodosh J. Computational analysis and identification of an emergent human adenovirus pathogen implicated in a respiratory fatality. Virology. 2011;409:141–147. doi: 10.1016/j.virol.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers NG, Basnight M, Gibbs CJ, Gajdusek DC. Sequence-specific interactions between cellular DNA-binding proteins and the adenovirus origin of DNA replication. Mol. Cell. Biol. 1967;216:446–449. doi: 10.1128/mcb.7.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld PJ, O'Neill EA, Wides RJ, Kelly TJ. Sequence-specific interactions between cellular DNA-binding proteins and the adenovirus origin of DNA replication. Mol. Cell. Biol. 1987;7:875–886. doi: 10.1128/mcb.7.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe WP, Hartley JW, Huebner RJ. Proceedings of the Society for Experimental Biology and Medicine. Vol. 97. Society for Experimental Biology and Medicine; 1958. Serotype composition of the adenovirus group; pp. 465–170. [DOI] [PubMed] [Google Scholar]
- Rowe WP, Huebner RJ, Gilmore LK, Parrott RH, Ward TG. Proceedings of the Society for Experimental Biology and Medicine. Vol. 84. New York, N.Y.: Society for Experimental Biology and Medicine; 1953. Isolation of a cytopathogenic agent from human adenoids undergoing spontaneous degeneration in tissue culture; pp. 570–573. [DOI] [PubMed] [Google Scholar]
- Roy S, Gao G, Clawson DS, Vandenberghe LH, Farina SF, Wilson JM. Complete nucleotide sequences and genome organization of four chimpanzee adenoviruses. Virology. 2004a;324:361–372. doi: 10.1016/j.virol.2004.03.047. [DOI] [PubMed] [Google Scholar]
- Roy S, Gao G, Lu Y, Zhou X, Lock M, Calcedo R, Wilson JM. Characterization of a family of chimpanzee adenoviruses and development of molecular clones for gene transfer vectors. Hum. Gene Ther. 2004b;15:519–530. doi: 10.1089/10430340460745838. [DOI] [PubMed] [Google Scholar]
- Roy S, Sandhu A, Medina A, Clawson DS, Wilson JM. Adenoviruses in fecal samples from asymptomatic rhesus macaques, United States. Emerg Infect. Dis. 2012;18:1081–1088. doi: 10.3201/eid1807.111665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Vandenberghe LH, Kryazhimskiy S, Grant R, Calcedo R, Yuan X, Keough M, Sandhu A, Wang Q, Medina-Jaszek CA, Plotkin JB, Wilson JM. Isolation and characterization of adenoviruses persistently shed from the gastrointestinal tract of non-human primates. PLoS Pathog. 2009;5:e1000503. doi: 10.1371/journal.ppat.1000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Zhi Y, Kobinger GP, Figueredo J, Calcedo R, Miller JR, Feldmann H, Wilson JM. Generation of an adenoviral vaccine vector based on simian adenovirus 21. J. Gen. Virol. 2006;87:2477–2485. doi: 10.1099/vir.0.81989-0. [DOI] [PubMed] [Google Scholar]
- Russell KL, Hawksworth AW, Ryan MA, Strickler J, Irvine M, Hansen CJ, Gray GC, Gaydos JC. Vaccine-preventable adenoviral respiratory illness in US military recruits, 1999–2004. Vaccine. 2006;24:2835–2842. doi: 10.1016/j.vaccine.2005.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarantis H, Johnson G, Brown M, Petric M, Tellier R. Comprehensive detection and serotyping of human adenoviruses by PCR and sequencing. J. Clin. Microbiol. 2004;42:3963–3969. doi: 10.1128/JCM.42.9.3963-3969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto D, Chodosh J, Brister JR, Jones MS. Using the whole-genome sequence to characterize and name human adenoviruses. J. Virol. 2011;85:5701–5702. doi: 10.1128/JVI.00354-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens DA, Schaeffer M, Fox JP, Brandt CD, Romano M. Standardization and certification of reference antigens and antisera for 30 human adenovirus serotypes. Am. J. Epidemiol. 1967;86:617–633. doi: 10.1093/oxfordjournals.aje.a120771. [DOI] [PubMed] [Google Scholar]
- Stone D, di Paolo NC, Lieber A. Development of group B adenoviruses as gene transfer vectors. Biotechnol. Genet. Eng. Rev. 2006;22:101–123. doi: 10.1080/02648725.2006.10648067. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tcherepanov V, Ehlers A, Upton C. Genome Annotation Transfer Utility (GATU): rapid annotation of viral genomes using a closely related reference genome. BMC Genomics. 2006;7:150. doi: 10.1186/1471-2164-7-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Top FH, Jr., Grossman RA, Bartelloni PJ, Segal HE, Dudding BA, Russell PK, Buescher EL. Immunization with live types 7 and 4 adenovirus vaccines. I. Safety, infectivity, antigenicity, and potency of adenovirus type 7 vaccine in humans. J. Infect. Dis. 1971;124:148–154. doi: 10.1093/infdis/124.2.148. [DOI] [PubMed] [Google Scholar]
- Vora GJ, Lin B, Gratwick K, Meador C, Hansen C, Tibbetts C, Stenger DA, Irvine M, Seto D, Purkayastha A, Freed NE, Gibson MG, Russell K, Metzgar D. Co-infections of adenovirus species in previously vaccinated patients. Emerg. Infect. Dis. 2006;12:921–930. doi: 10.3201/eid1206.050245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadell G. Molecular epidemiology of human adenoviruses. Curr. Top. Microbiol. Immunol. 1984;110:191–220. doi: 10.1007/978-3-642-46494-2_7. [DOI] [PubMed] [Google Scholar]
- Wadell G, Hammarskjold ML, Winberg G, Varsanyi TM, Sundell G. Genetic variability of adenoviruses. Ann. N.Y. Acad. Sci. 1980;354:16–12. doi: 10.1111/j.1749-6632.1980.tb27955.x. [DOI] [PubMed] [Google Scholar]
- Walsh MP, Chintakuntlawar A, Robinson CM, Madisch I, Harrach B, Hudson NR, Schnurr D, Heim A, Chodosh J, Seto D, Jones MS. Evidence of molecular evolution driven by recombination events influencing tropism in a novel human adenovirus that causes epidemic keratoconjunctivitis. PLoS One. 2009;4:e5635. doi: 10.1371/journal.pone.0005635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh MP, Seto J, Jones MS, Chodosh J, Xu W, Seto D. Computational analysis identifies human adenovirus type 55as a re-emergent acute respira tory disease pathogen. J. Clin. Microbiol. 2010;48:991–993. doi: 10.1128/JCM.01694-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigand R. Terminal dilution purification of adenovirus prototypes, and the antigenic relationship between types 4, 16 and 14. Arch. Virol. 1975;49:323–328. doi: 10.1007/BF01318241. [DOI] [PubMed] [Google Scholar]
- Wigand R. Pitfalls in the identification of adenoviruses. J. Virol. Methods. 1987;16:161–169. doi: 10.1016/0166-0934(87)90001-2. [DOI] [PubMed] [Google Scholar]
- Wigand R, Bauer H, Lang F, Adam W. Neutralization of the adenoviruses types 1 to 28: specificity and antigenic relationships. Arch. Gesamte Virusforsch. 1965;15:188–199. doi: 10.1007/BF01257730. [DOI] [PubMed] [Google Scholar]
- Wigand R, Sehn N, Hierholzer JC, De Jong JC, Adrian T. Immunological and biochemical characterization of human adenoviruses from subgenus B. I. Antigenic relationships. Arch. Virol. 1985;84:63–78. doi: 10.1007/BF01310554. [DOI] [PubMed] [Google Scholar]
- Willimzik HF, Kalter SS, Lester TL, Wigand R. Immunological relation ship among adenoviruses of humans, simians, and nonprimates as determined by the neutralization test. InterVirology. 1981;15:28–36. doi: 10.1159/000149211. [DOI] [PubMed] [Google Scholar]
- Yang Z, Zhu Z, Tang L, Wang L, Tan X, Yu P, Zhang Y, Tian X, Wang J, Zhang Y, Li D, Xu W. Genomic analyses of recombinant adenovirus type 11a in China. J. Clin. Microbiol. 2009 doi: 10.1128/JCM.00282-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Qu Z, Wu X, Wang Y, Liu L, Wei F, Gao H, Shang L, Zhang H, Cui H, Zhao Y, Wu N, Tang Y, OJn L. Molecular modeling and epitopes mapping of human adenovirus type 3 hexon protein. Vaccine. 2009;27:5103–5110. doi: 10.1016/j.vaccine.2009.06.041. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Zhang Y, Xu S, Yu P, Tian X, Wang L, Liu Z, Tang L, Mao N, Ji Y, Li C, Yang Z, Wang S, Wang J, Li D, Xu W. Outbreak of acute respiratory disease in China caused by B2 species of adenovirus type 11. J. Clin. Microbiol. 2009;47:697–703. doi: 10.1128/JCM.01769-08. [DOI] [PMC free article] [PubMed] [Google Scholar]