Abstract
Human herpesvirus-6 (HHV-6) A and 6B are ubiquitous betaherpesviruses viruses with lymphotropic and neurotropic potential. As reported earlier, these viruses establish latency by integration into the telomeres of host chromosomes. Chromosomally integrated HHV-6 (CIHHV-6) can be transmitted vertically from parent to child. Some CIHHV-6 patients are suffering from neurological symptoms, while others remain asymptomatic. Four patients with CIHHV-6 and CNS dysfunction were treated with valganciclovir or foscarnet. HHV-6 replication was detected by reverse transcriptase polymerase chain reaction amplification of a late envelope glycoprotein. In this study we also compared the inherited and persistent HHV-6 viruses by DNA sequencing. The prevalence of CIHHV-6 in this cohort of adult patients from the USA suffering from a wide range of neurological symptoms including long term fatigue were found significantly greater than the reported 0.8% in the general population. Long-term antiviral therapy inhibited HHV-6 replication as documented by loss of viral mRNA production. Sequence comparison of the mRNA and the inherited viral genome revealed that the transcript is produced by an exogenous virus. In conclusion, the data presented here document that some individuals with CIHHV-6 are infected persistently with exogenous HHV-6 strains that lead to a wide range of neurological symptoms; the proposed name for this condition is inherited herpesvirus 6 syndrome or IHS.
INTRODUCTION
Human herpesvirus-6 A and 6B (HHV-6A and HHV-6B) are ubiquitous betaherpesviruses viruses known for their lymphotropic and neurotropic potential [Hall et al. 1998, Santoro et al. 1999]. Primary HHV-6B infection results in exanthema subitum and typically occurs during the first two years of life [Yamanishi et al. 1988]. These viruses persist after primary infection; viral reactivation is associated with a variety of adult conditions and complications including encephalitis, drug-induced hypersensitivity or drug rash with eosinophilia and systemic symptoms, and transplant rejection [Tohyama et al. 2007, Watanabe et al. 2008].
Unlike other human herpesviruses, HHV-6 viruses establish latency by integration into the telomeres of host chromosomes and may be inherited [Arbuckle et al. 2010]. In a screen of blood donors in the United States and the United Kingdom, the prevalence of inherited HHV-6 in the general population is approximately 0.8% [Hall et al. 2004, Hall et al. 2008, Hudnall et al. 2008, Leong et al. 2007]. The prevalence of CIHHV-6 is increased in children referred for encephalitis, solid organ and stem cell transplant recipients as well individuals with lymphoproliferative disorders [Griffiths et al. 1999, Hubacek et al. 2012, Kidd et al. 2000, Lee et al. 2011, Lee et al. 2012, Pellett et al. 2012, Potenza et al. 2009, Torelli et al. 1995, Ward et al. 2007, Zerr et al. 2011]. In these populations, the prevalence of CIHHV-6 averages 2%.
In vitro studies and clinical reports indicate that HHV-6 may be reactivated from its integrated form [Arbuckle et al. 2010]. However, detection of reactivation is difficult because patients with inherited HHV-6 consistently present with viral DNA copy number above 0.5 million/ml in whole blood [Pellett et al. 2012]. There have been suggestions that elevated anti-HHV6 IgG levels are indicative of reactivation [Ablashi et al. 2000]; however, antibody response is variable among patients and may yield inconclusive results. Detection of mRNA, however, seems to be a promising marker for viral reactivation and several research groups have detected active HHV-6 replication using reverse-transcriptase PCR [Caserta et al. 2007, Ihira et al. 2012, Norton et al. 1999, Van et al. 2001].
A recent publication described the successful treatment of two CIHHV-6 patients with detectable HHV-6 glycoprotein mRNA in whole blood [Montoya et al. 2012]. Prior to treatment, both patients exhibited neurological symptoms including cognitive impairment and depression with concomitant abnormal quantitative EEG readings. Six weeks of foscarnet treatment resulted in the resolution of neurological symptoms and normalization of brain waves; however, symptoms returned after cessation of antiviral treatment.
Previous studies reported reduced antibody titers to HHV-6 glycoprotein B in the serum of individuals with CIHHV-6, when compared to those of healthy controls [Tanaka-Taya et al. 2004]. This suggests that there is an immune tolerance in these individuals, and the central hypothesis of this study is that repeated reactivation or exogenous infection may contribute to illness in symptomatic patients with CIHHV-6.
In this study, nested reverse transcriptase PCR assay was utilized to amplify U100 envelope glycoprotein mRNA as a means of detecting HHV-6 replication in blood samples isolated from symptomatic individuals with CIHHV-6 [Norton et al. 1999]. The goal of this study is to use mRNA detection as a means of determining the efficacy of antiviral treatment and optimal treatment time. Additionally, sequence analyses on HHV-6 late mRNA and inherited viral DNA were performed to investigate the persistence of exogenous HHV-6 in individuals with CIHHV-6.
METHODS
Study Subjects, Treatment, Statistical Analysis
Four patients diagnosed with inherited HHV-6 provided written informed consent prior to the start of this study, and the study was approved by the Institutional Review Board. All subjects presented with more than 0.5 million DNA copy numbers of HHV-6 in whole blood, with concomitant positive results of HHV-6 in hair follicles as detected by Viracor-IBT laboratories, Inc. Patients presented with fatigue and neurological symptoms including, but not limited to, depression, hypersomnia, memory and cognitive impairment. In this study, patients received therapeutic antiviral therapy consisting of either twice daily valganciclovir (patients 1,2, and 3) or 60 mg/kg foscarnet (patient 4) per day. Patients receiving short term treatment received 900 mg valganciclovir twice daily; patients receiving long term treatment received 900mg valganciclovir twice daily for three weeks and 450mg twice daily for three weeks or longer. Blood samples were collected in Paxgene DNA, RNA, and heparin tubes before and during treatment.
A two-tailed Fisher exact test was used to determine the significance of CIHHV-6 prevalence among this cohort of US adult patients suffering from neurological symptoms..
Detection of Viral mRNA by Nested Reverse Transcriptase PCR Assay
Nested reverse transcriptase PCR was used to amplify HHV-6 glycoprotein U100 mRNA in intracellular RNA from whole blood. Blood samples were collected in PAXgene blood RNA tubes and RNA was isolated using Trizol reagent. Total RNA was converted to cDNA using the GoScript Reverse Transcription System (Promega) and 5 ng of total RNA was used for PCR. Two rounds (30 cycles each) of PCR were performed using RedTaq polymerase (Sigma), as described previously [Norton et al. 1999]. Primers used for amplification are as follows: U100Round1F: CTAAATTTTCTACCTCCGAAATGT; U100Round1R: GAGTCCAT GAGTTAGAAGATT; U100Round2F: ACTACTACCTTAGAAGATATAG; U100Round2R: AAGC GCGTGCAGGTTTCCCAA [Norton et al. 1999]. RNA isolated from uninfected or infected Molt3 T lymphocytes was used as a negative or positive control, respectively. Reverse transcription and subsequent PCR amplification were conducted independently for each patient sample. Additionally, a reverse transcriptase null reaction was performed to ensure the absence of DNA in RNA samples. For a cellular control, beta actin was amplified with 30 cycles of PCR. Amplified products were separated by agarose gel electrophoresis, stained with ethidium bromide, and visualized using UV light in the Gel Doc Molecular Imaging System (Bio-Rad).
PCR Amplification, Cloning, Sequencing and Virus Isolation
A portion of the U100 gene was amplified by PCR amplification using RedTaq Polymerase (Sigma). DNA was isolated from agarose gel using Wizard SV Gel and PCR Clean-Up System (Promega) and TA cloned using the pGEMT-Easy cloning system. Cloned genes were sequenced using the ABI3130XL capillary sequencing instrument. Sequencing was performed at the Moffitt Cancer Center Molecular Genomics Core Facility.
Restriction Enzyme Digestion and Southern Blot Hybridization
Five micrograms of cellular DNA from patient PBMCs or 1ng of virion DNA was digested with SacI and separated using agarose gel electrophoresis. DNA fragments were transferred to nitrocellulose by vacuum blotting and hybridized with 32P-labeled HHV6A U1102 cloned into a BAC vector [Tang et al. 2010].
Alignment of nucleotide sequences
To obtain alignment scores, nucleotide sequences were aligned using ClustalW2 using default alignment parameters (DNA weight matrix: IUB, Gap open: 10, Gap extension: 0.2, Gap distance: 5, Numiter: 1).Alternately, multiple sequences were aligned using the Mauve algorithm and visualized using Jalview in the Virus Pathogen Database and Analysis Resource (ViPR).
RESULTS
Prevalence of CIHHV-6 in cohort of patients with neurological symptoms
To reveal the prevalence of CIHHV-6 in 337 US patients suffering from a wide range of neurological symptoms, including long term fatigue, peripheral blood samples were tested by quantitative PCR by Viracor-IBT laboratories. Seven patients had higher than 0.5 million HHV-6 copies per ml of blood. A two-tailed Fisher exact test was used to determine the significance of CIHHV-6 prevalence among these patients. The CIHHV-6 rate among this cohort of US adult patients suffering from neurological symptoms is 2.1% (7/337). This is significantly greater (p=.03) than the expected value of 0.8% reported in the general US population [Hall et al. 2004, Hall et al. 2008].
Long term antiviral treatment abrogates viral mRNA production
The four patients enrolled in this study harbored over 0.5 million copies of HHV-6 DNA copy numbers per ml of peripheral blood. Patients presented with a variety of neurological symptoms including but not limited to headache, blurred vision, and memory impairment, as well as other symptoms such as generalized pain and long term fatigue.
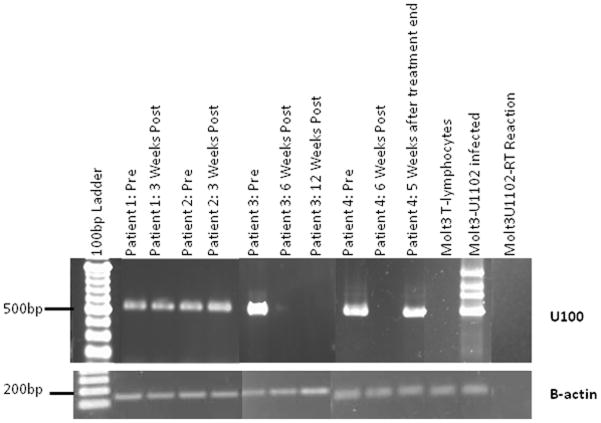
To determine if the symptoms were due to replicating HHV-6, nested RT-PCR was employed to detect the late envelope glycoprotein U100 mRNA in whole blood samples from patients with CIHHV-6 [Norton et al. 1999]. Initial RT-PCR assays were performed on blood samples isolated from symptomatic patients prior to the administration of antiviral medications; follow up samples were taken during or after treatment. For each RT-PCR reaction 5ng of total RNA isolated from peripheral blood was used. In all cases, U100 mRNA was detected in blood samples, in the absence of antiviral treatment (Figure 1). On the other hand, ten control single donor samples obtained from the local blood bank were negative for U100 mRNA (data not shown). In the absence of reverse transcriptase, PCR products were not detected (Figure 1). Long term (≥six weeks) administration of foscarnet (patient 4) or valganciclovir (patient 3) resulted in the abrogation of U100 mRNA expression (Figure 1), while short term (three weeks) administration of valganciclovir was not sufficient to eliminate viral gene expression in two unrelated patients (patients 1 and 2). Five weeks after the cessation of intravenous foscarnet treatment (patient 4), U100 mRNA was detected in the whole blood of patient 4. In all cases, resolution of symptoms was concurrent with the reduction of mRNA expression. This suggests that U100 mRNA detection is a reliable method of detecting HHV-6 persistence. However, treatment efficacy is variable and appears to be dependent on the length of treatment, with treatment length of greater than or equal to six weeks being optimal for valganciclovir and foscarnet.
Figure 1. Amplification of Late Envelope Glycoprotein U100 mRNA.
Nested reverse transcriptase PCR was used to amplify HHV-6 glycoprotein U100 mRNA from whole blood. Total RNA was converted to cDNA and two rounds of PCR were performed. Second round PCR products were separated by agarose gel electrophoresis, stained with ethidium bromide, and visualized using UV light. Top U100 RT-PCR; Bottom: Beta-actin RT-PCR
The late U100 viral mRNA originates from an exogenous HHV-6 strain
Amplified U100 cDNA was sequenced to confirm that the RT-PCR products were derived from mature mRNA that lacks introns rather than the inherited viral DNA. Sequencing was also performed to determine if the detected mRNA originated from the inherited virus, or from an exogenous virus. The U100 gene of the inherited viral genome was also amplified and both cDNA and inherited HHV-6 DNA sequences of U100 were cloned and sequenced. For sequences from the inherited viral DNA fragments, introns were removed using previously accepted splice sites prior to alignment using ClustalW2. Isolated cDNA sequences were also compared to the U100 sequences for HHV-6A U1102 and GS, or HHV-6B Z29. All patient cDNA sequences were more similar to HHV-6A U1102 than HHV-6A GS and differed from HHV-6A U1102 by only a CG inversion at bases 193/194 (Figure 2). On the other hand, the inherited genomic sequences were more similar to HHV-6A GS. The inherited HHV-6A sequences were greater than 99% identical to each other for three patients, as were all the reactivated viral sequences.
Figure 2. Nucleotide Alignment of cDNA and Genomic Sequences.
PCR amplified HHV-6 glycoprotein cDNA or corresponding genomic DNA fragments were TA cloned and sequenced. Obtained sequences were aligned using the Mauve algorithm and visualized in JalView in the ViPr database.
The cDNA and inherited genomic sequences differed from each other in all cases. U100 cDNA shared only an average of 96.3% nucleotide sequence identity with the inherited HHV-6A viral genome. U100 cDNA from patient 1 was more similar to HHV-6A U1102, while the inherited HHV-6 DNA sequence was 98% identical to HHV-6B Z29. In addition, theU100 mRNA sequence and inherited viral DNA sequences shared only a 90% nucleotide sequence identity. Differences in the genomic DNA and cDNA sequences demonstrate that these symptomatic patients with inherited HHV-6 harbor more than one HHV-6 virus, and the U100 mRNA detected by RT-PCR originates from an exogenous virus.
Inherited HHV-6 viral genome sequences are heterogeneous
The results clearly indicate that the reactivated virus from these patient samples did not emanate from the inherited virus; in all cases the reactivated viruses were more similar to HHV-6A U1102 than HHV-6A GS. Additionally, there was a high level of similarity between the U100 DNA sequences of the inherited viruses. In order to compare the genome of the inherited HHV-6 viruses to the HHV-6A GS virus, restriction fragment length analysis was performed on DNA from the PBMCs of four patients or GS virion DNA. Briefly, DNA was subjected to restriction enzyme digestion using SacI, fragments were separated by electrophoresis, blotted onto nitrocellulose and probed with a radiolabeled HHV-6A U1102 BAC clone (Figure 3). Despite similarities among the inherited HHV-6 banding patterns, Southern blot hybridization shows distinct banding patterns for each inherited virus and a significantly diverged banding patter from HHV-6A GS, indicating that all inherited viruses evaluated in this experiment markedly differ from HHV-6A GS and also uniquely distinct from each other.
Figure 3. Restriction Fragment Analysis of Inherited HHV-6 viruses.
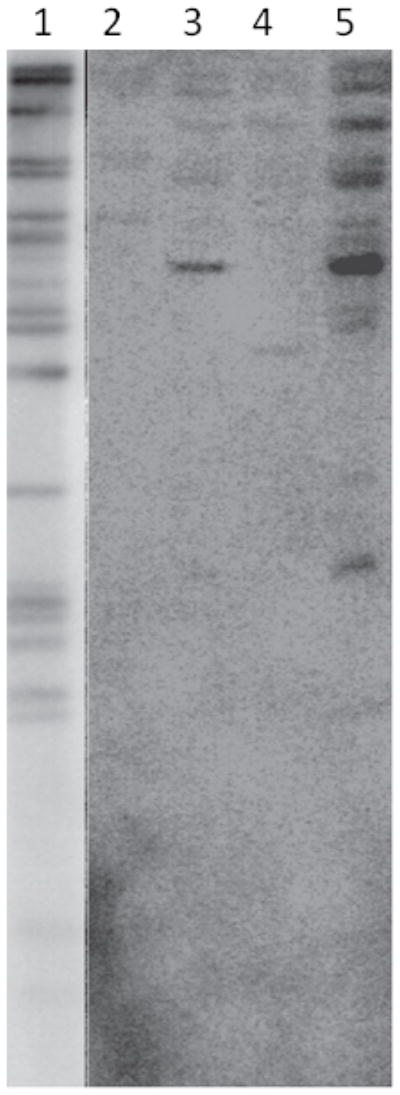
Cellular DNA from patient PBMCs or virion DNA from HHV-6 GS was digested using SacI. Fragments were separated by electrophoresis, blotted onto nitrocellulose and probed with a radiolabeled HHV-6A U1102 BAC clone. Lane 1: HHV-6A GS virion DNA, Lane 2: patient 5, Lane 3: patient 3, Lane 4: patient 4, Lane 5: patient 6; patient 6 is Sibling 1 from Family 1, diagnosed with IHS, (Arbuckle et al. 2010 and 2012).
DISCUSSION
Inheritance of HHV-6 infrequently occurs in the healthy, general US population and the reported prevalence is around 0.8% [Hall et al. 2004, Hall et al. 2008]. However, there is a noted increased prevalence in hospitalized individuals and individuals displaying neurological dysfunction [Pellett et al. 2012]. Importantly, the prevalence CIHHV-6 in this patient population suffering from a wide range of neurological symptoms is significantly higher (over 2%), suggesting a possible role of CIHHV-6 in pathology. In support of this hypothesis, late viral mRNA production was documented in all four unrelated patients with CIHHV-6 and neurological symptoms. In contrast, viral mRNA was reported in only 8% of asymptomatic CIHHV-6 individuals [Hall et al. 2008].
Unexpectedly, the sequence of late mRNAs and markedly differed from the inherited viral genome sequences. The presence of an exogenous persistently replicating HHV-6 virus suggests that patients with CIHHV-6 exhibit immune tolerance or a weakened immune response to the virus. It has been shown that the humoral response to inherited HHV-6 differs from primary infection. A previous study supporting the notion of immune tolerance in CIHHV-6 patients [Tanaka-Taya et al. 2004] reported that only 14% had detectable antibodies directed at glycoprotein B, the major neutralizing epitope. In contrast, there was 60% detection in healthy adult controls [Tanaka-Taya et al. 2004]. Reduced gB titers suggest that individuals with CIHHV-6 may have a reduced ability to fight a secondary HHV-6 infection. To date, there have been no studies on the rate of superinfection in patients with inherited HHV-6. This study suggests that, in symptomatic CIHHV-6 patients, infection with an exogenous HHV-6 virus may be a frequent occurrence. Additionally, one may propose that superinfection is the differentiating factor between symptomatic and asymptomatic individuals with CIHHV-6; however, this was not investigated in this study. Taken together, it may be proposed that some CIHHV-6 individuals acquire and are infected persistently with exogenous HHV-6 strains that lead to a wide range of neurological symptoms. A fitting name for this condition is inherited herpesvirus 6 syndrom or IHS.
The results indicate that patient response to antiviral therapy using oral valganciclovir or intravenous foscarnet was largely dependent on treatment length; a three week treatment with valganciclovir was ineffective in preventing virus reactivation, as indicated by the recurrent expression of U100 mRNA. Recent reports document the long-term benefit of antiviral drug therapy of two patients with IHS [Montoya et al. 2012]. Both of these patients have suffered debilitating neurological symptoms but antiviral therapy resulted in marked and long-lasting improvement also documented by quantitative EEG [Montoya et al. 2012]. Currently the two patients have no detectable U100 mRNA in their PBMCs and are free of neurological symptoms.
There have been previous reports of in vitro reactivation of integrated HHV-6 by chemical inducers, such as TPA and trichostatin A [Arbuckle et al. 2010]. However, it remains unclear as to whether or not inherited HHV-6 strains retain their ability to reactivate and cause persistent infection. In this study, sequencing of genomic DNA and cDNA indicated that the detected mRNA arose only from an exogenous HHV-6 virus in all the patient samples evaluated, rather than the inherited viruses. There was no evidence of reactivation of the CIHHV-6 virus. However it is possible that this occurs as well and that the copy number is too low to be detected.
To date, there have been no reports on HHV-6-specific CD4 or CD8 responses in CIHHV-6 patients. Two independent groups recently reported that the large structural phosphoprotein, U11, is an immunodominant CD4 and CD8 epitope for HHV-6B [Martin et al. 2012, Nastke et al. 2012]. Future studies could reveal whether patients with IHS fail to develop adequate T cell-mediated protective immunity.
Another finding of this study was that in all cases, the partial U100 sequence inherited by three of the patients was more similar to HHV-6A GS than HHV-6A U1102. This finding may be attributed to the fact that the GS strain was isolated in the United States, the location of all patients. To date, the GS strain is the only HHV-6A virus isolated in the United States and little is known as to the variation of HHV-6 viral sequences by geographic location.
Differences between the inherited and reactivated viruses may pose difficulties when designing PCR primers, and may lead to failure to identify individuals with CIHHV-6 in routine clinical assays.
The SacI restriction enzyme cleavage profiles of the inherited strains were highly heterogeneous as compared to strain GS. This may reflect significant sequence divergence; however, it is also possible that the inherited viruses suffered deletions and/or rearrangements resulting in a markedly different restriction enzyme cleavage profile.
Another question that remains unanswered is at what point during human evolution did HHV-6 enter the germline? Inheritance of CIHHV-6 follows Mendelian genetics and the sequence of inherited and current HHV-6 isolates are quite divergent suggesting that inheritance of the virus in some families dates back hundreds or thousands of years.
Diagnosis of IHS requires detection of CIHHV-6 status and detection of persistent viral infection. Several commercial companies offer quantitative PCR assays that may reveal the presence of CIHHV-6 but there are no clinical laboratories offering specific tests to detect persistent HHV-6 infection. The study presented here shows that the RT-PCR assay for the HHV-6 U100 mRNA offers a means of distinguishing latent infections from lytic/persistent infections. Since HHV-6 has been also recognized as an agent involved in organ rejection of transplant recipients the RT-PCR assay could guide a physician’s decision on whether administration of antiviral drugs is warranted.
Table 1.
Patient Information Summary
| HHV-6 Status | |||||||
|---|---|---|---|---|---|---|---|
| Patient | Integrated HHV-6Strain | Antiviral | Treatment Duration | Dose | Pre Treatment | 3 Weeks Post | 6 Weeks Post |
| 1 | B | Valganciclovir | 3 weeks | 1800 mg | Positive | Positive | N/A |
| 2 | A | Valganciclovir | 3 weeks | 1800 mg | Positive | Positive | N/A |
| 3 | A | Valganciclovir | 6+ weeks | 900–1800 mg | Positive | N/A | Negative |
| 4 | A | Foscarnet | 6+ weeks | 60mg/kg | Positive | N/A | Negative |
Acknowledgments
This work was supported by grants provided by the National Institute of Health 5R01CA111196 and the HHV-6 Foundation.
Contributor Information
Shara N. Pantry, Email: spantry@health.usf.edu.
Maria M. Medveczky, Email: mmedvecz@health.usf.edu.
Jesse H. Arbuckle, Email: jesse.arbuckle@nih.gov.
Janos Luka, Email: jluka@bioworldantibodies.com.
Jose G. Montoya, Email: gilberto@stanford.edu.
Jianhong Hu, Email: JHu@ufscc.ufl.edu.
Rolf Renne, Email: RRenne@ufscc.ufl.edu.
Daniel Peterson, Email: dan@ishere.com.
Joshua C. Pritchett, Email: joshuapritchett@hhv-6foundation.org.
Dharam V. Ablashi, Email: dharam_ablashi@hhv-6foundation.org.
Peter G. Medveczky, Email: pmedvecz@health.usf.edu.
References
- Ablashi DV, Eastman HB, Owen CB, Roman MM, Friedman J, Zabriskie JB, Peterson DL, Pearson GR, Whitman JE. Frequent HHV-6 reactivation in multiple sclerosis (MS) and chronic fatigue syndrome (CFS) patients. J Clin Virol. 2000;16:179–191. doi: 10.1016/s1386-6532(99)00079-7. [DOI] [PubMed] [Google Scholar]
- Arbuckle JH, Medveczky MM, Luka J, Hadley SH, Luegmayr A, Ablashi D, Lund TC, Tolar J, De Meirleir K, Montoya JG, Komaroff AL, Ambros PF, Medveczky PG. The latent human herpesvirus-6A genome specifically integrates in telomeres of human chromosomes in vivo and in vitro. Proc Natl Acad Sci U S A. 2010;107:5563–8. doi: 10.1073/pnas.0913586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caserta MT, Hall CB, Schnabel K, Lofthus G, McDermott MP. Human herpesvirus (HHV)-6 and HHV-7 infections in pregnant women. J Infect Dis. 2007;196:1296–303. doi: 10.1086/522430. [DOI] [PubMed] [Google Scholar]
- Griffiths PD, Ait-Khaled M, Bearcroft CP, Clark DA, Quaglia A, Davies SE, Burroughs AK, Rolles K, Kidd IM, Knight SN, Noibi SM, Cope AV, Phillips AN, Emery VC. Human herpesviruses 6 and 7 as potential pathogens after liver transplant: prospective comparison with the effect of cytomegalovirus. J Med Virol. 1999;59:496–501. doi: 10.1002/(sici)1096-9071(199912)59:4<496::aid-jmv12>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Hall CB, Caserta MT, Schnabel KC, Long C, Epstein LG, Insel RA, Dewhurst S. Persistence of human herpesvirus 6 according to site and variant: possible greater neurotropism of variant A. Clin Infect Dis. 1998;26:132–137. doi: 10.1086/516280. [DOI] [PubMed] [Google Scholar]
- Hall CB, Caserta MT, Schnabel KC, Boettrich C, McDermott MP, Lofthus GK, Carnahan JA, Dewhurst S. Congenital infections with human herpesvirus 6 (HHV6) and human herpesvirus 7 (HHV7) J Pediatr. 2004;145:472–7. doi: 10.1016/j.jpeds.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Hall CB, Caserta MT, Schnabel K, Shelley LM, Marino AS, Carnahan JA, Yoo C, Lofthus GK, McDermott MP. Chromosomal integration of human herpesvirus 6 is the major mode of congenital human herpesvirus 6 infection. Pediatrics. 2008;122:513–20. doi: 10.1542/peds.2007-2838. [DOI] [PubMed] [Google Scholar]
- Hubacek P, Hrdlickova A, Spacek M, Zajac M, Muzikova K, Sedlacek P, Cetkovsky P. Prevalence of chromosomally integrated HHV-6 in patients with malignant disease and healthy donors in the Czech Republic. Folia Microbiol (Praha) 2012 doi: 10.1007/s12223-012-0180-z. [DOI] [PubMed] [Google Scholar]
- Hudnall SD, Chen T, Allison P, Tyring SK, Heath A. Herpesvirus prevalence and viral load in healthy blood donors by quantitative real-time polymerase chain reaction. Transfusion. 2008;48:1180–7. doi: 10.1111/j.1537-2995.2008.01685.x. [DOI] [PubMed] [Google Scholar]
- Ihira M, Enomoto Y, Kawamura Y, Nakai H, Sugata K, Asano Y, Tsuzuki M, Emi N, Goto T, Miyamura K, Matsumoto K, Kato K, Takahashi Y, Kojima S, Yoshikawa T. Development of quantitative RT-PCR assays for detection of three classes of HHV-6B gene transcripts. J Med Virol. 2012;84:1388–95. doi: 10.1002/jmv.23350. [DOI] [PubMed] [Google Scholar]
- Kidd IM, Clark DA, Sabin CA, Andrew D, Hassan-Walker A, Sweny P, Griffiths PD, Emery VC. Prospective study of human betaherpesviruses after renal transplantation: association of human herpesvirus 7 and cytomegalovirus co-infection with cytomegalovirus disease and increased rejection. Transplantation. 2000;69:2400–4. doi: 10.1097/00007890-200006150-00032. [DOI] [PubMed] [Google Scholar]
- Lee S, Brown Ra, Eid AJ, Razonable RR. Chromosomally integrated human herpesvirus-6 in kidney transplant recipients. Nephrology, dialysis, transplantation : official publication of the European Dialysis and Transplant Association - European Renal Association. 2011;26:2391–3. doi: 10.1093/ndt/gfr259. [DOI] [PubMed] [Google Scholar]
- Lee S, Brown RA, Razonable RR. Chromosomally integrated human herpesvirus-6 in transplant recipients. Transplant infectious disease : an official journal of the Transplantation Society. 2012;14:346–54. doi: 10.1111/j.1399-3062.2011.00715.x. [DOI] [PubMed] [Google Scholar]
- Leong HN, Tuke PW, Tedder RS, Khanom AB, Eglin RP, Atkinson CE, Ward KN, Griffiths PD, Clark DA. The prevalence of chromosomally integrated human herpesvirus 6 genomes in the blood of UK blood donors. J Med Virol. 2007;79:45–51. doi: 10.1002/jmv.20760. [DOI] [PubMed] [Google Scholar]
- Martin LK, Schub A, Dillinger S, Moosmann A. Specific CD8(+) T cells recognize human herpesvirus 6B. Eur J Immunol. 2012;42:2901–12. doi: 10.1002/eji.201242439. [DOI] [PubMed] [Google Scholar]
- Montoya JG, Neely MN, Gupta S, Lunn MR, Loomis KS, Pritchett JC, Polsky B, Medveczky PG. Antiviral therapy of two patients with chromosomally-integrated human herpesvirus-6A presenting with cognitive dysfunction. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology. 2012;55:40–5. doi: 10.1016/j.jcv.2012.05.016. [DOI] [PubMed] [Google Scholar]
- Nastke M, Becerra A, Yin L, Dominguez-Amorocho O, Gibson L, Stern LJ, Calvo-Calle J. Human CD4+ T cell response to human herpesvirus 6. J Virol. 2012;86:4776–92. doi: 10.1128/JVI.06573-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton Ra, Caserta MT, Hall CB, Schnabel K, Hocknell P, Dewhurst S. Detection of human herpesvirus 6 by reverse transcription-PCR. J Clin Microbiol. 1999;37:3672–5. doi: 10.1128/jcm.37.11.3672-3675.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellett PE, Ablashi DV, Ambros PF, Agut H, Caserta MT, Descamps V, Flamand L, Gautheret-Dejean A, Hall CB, Kamble RT, Kuehl U, Lassner D, Lautenschlager I, Loomis KS, Luppi M, Lusso P, Medveczky PG, Montoya JG, Mori Y, Ogata M, Pritchett JC, Rogez S, Seto E, Ward KN, Yoshikawa T, Razonable RR. Chromosomally integrated human herpesvirus 6: questions and answers. Rev Med Virol. 2012;22:144–155. doi: 10.1002/rmv.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potenza L, Barozzi P, Masetti M, Pecorari M, Bresciani P, Gautheret-Dejean a, Riva G, Vallerini D, Tagliazucchi S, Codeluppi M, Di Benedetto F, Gerunda GE, Narni F, Torelli G, Luppi M. Prevalence of human herpesvirus-6 chromosomal integration (CIHHV-6) in Italian solid organ and allogeneic stem cell transplant patients. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2009;9:1690–7. doi: 10.1111/j.1600-6143.2009.02685.x. [DOI] [PubMed] [Google Scholar]
- Santoro F, Kennedy PE, Locatelli G, Malnati MS, Berger EA, Lusso P. CD46 Is a Cellular Receptor for Human Herpesvirus 6. Cell. 1999;99:817–827. doi: 10.1016/s0092-8674(00)81678-5. [DOI] [PubMed] [Google Scholar]
- Tanaka-Taya K, Sashihara J, Kurahashi H, Amo K, Miyagawa H, Kondo K, Okada S, Yamanishi K. Human herpesvirus 6 (HHV-6) is transmitted from parent to child in an integrated form and characterization of cases with chromosomally integrated HHV-6 DNA. J Med Virol. 2004;73:465–73. doi: 10.1002/jmv.20113. [DOI] [PubMed] [Google Scholar]
- Tang H, Kawabata A, Yoshida M, Oyaizu H, Maeki T, Yamanishi K, Mori Y. Human herpesvirus 6 encoded glycoprotein Q1 gene is essential for virus growth. Virology. 2010;407:360–367. doi: 10.1016/j.virol.2010.08.018. [DOI] [PubMed] [Google Scholar]
- Tohyama M, Hashimoto K, Yasukawa M, Kimura H, Horikawa T, Nakajima K, Urano Y, Matsumoto K, Iijima M, Shear NH. Association of human herpesvirus 6 reactivation with the flaring and severity of drug-induced hypersensitivity syndrome. Br J Dermatol. 2007;157:934–40. doi: 10.1111/j.1365-2133.2007.08167.x. [DOI] [PubMed] [Google Scholar]
- Torelli G, Barozzi P, Marasca R, Cocconcelli P, Merelli E, Ceccherini-Nelli L, Ferrari S, Luppi M. Targeted integration of human herpesvirus 6 in the p arm of chromosome 17 of human peripheral blood mononuclear cells in vivo. J Med Virol. 1995;46:178–88. doi: 10.1002/jmv.1890460303. [DOI] [PubMed] [Google Scholar]
- Van dB, Locatelli G, Geerts L, Fagà G, Ieven M, Goossens H, Bottiger D, Oberg B, Lusso P, Berneman ZN. Development of reverse transcriptase PCR assays for detection of active human herpesvirus 6 infection. J Clin Microbiol. 2001;39:2308–10. doi: 10.1128/JCM.39.6.2308-2310.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward KN, Leong HN, Thiruchelvam AD, Atkinson CE, Clark DA. Human herpesvirus 6 DNA levels in cerebrospinal fluid due to primary infection differ from those due to chromosomal viral integration and have implications for diagnosis of encephalitis. J Clin Microbiol. 2007;45:1298–1304. doi: 10.1128/JCM.02115-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H, Daibata M, Tohyama M, Batchelor J, Hashimoto K, Iijima M. Chromosomal integration of human herpesvirus 6 DNA in anticonvulsant hypersensitivity syndrome. Br J Dermatol. 2008;158:640–2. doi: 10.1111/j.1365-2133.2007.08382.x. [DOI] [PubMed] [Google Scholar]
- Yamanishi K, Okuno T, Shiraki K, Takahashi M, Kondo T, Asano Y, Kurata T. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet. 1988;1:1065–7. doi: 10.1016/s0140-6736(88)91893-4. [DOI] [PubMed] [Google Scholar]
- Zerr DM, Fann JR, Breiger D, Boeckh M, Adler AL, Xie H, Delaney C, Huang M, Corey L, Leisenring WM. HHV-6 reactivation and its effect on delirium and cognitive functioning in hematopoietic cell transplantation recipients. Blood. 2011;117:5243–9. doi: 10.1182/blood-2010-10-316083. [DOI] [PMC free article] [PubMed] [Google Scholar]