Summary
PPAR δ was identified as a target of APC through the analysis of global gene expression profiles in human colorectal cancer (CRC) cells. PPARδ expression was elevated in CRCs and repressed by APC in CRC cells. This repression was mediated by β-catenin/Tcf-4-responsive elements in the PPARδ promotor. The ability of PPARs to bind eicosanoids suggested that PPARδ might be a target of chemopreventive non-steroidal anti-inflammatory drugs (NSAIDs). Reporters containing PPARδ-responsive elements were repressed by the NSAID sulindac. Furthermore, sulindac was able to disrupt the ability of PPARδ to bind its recognition sequences. These findings suggest that NSAIDs inhibit tumorigenesis through inhibition of PPARδ, the gene for which is normally regulated by APC.
Introduction
Colorectal cancer (CRC) is the second leading cause of cancer deaths in the United States. Recent advances in the understanding of CRC have raised expectations that this growing knowledge might lead to improved cancer prevention. In this regard, the identification of genetic alterations that underlie the initiation of colorectal tumors and of drugs which can prevent colorectal tumors show particular promise. Here, we suggest a molecular basis for the convergence of these two heretofore separate lines of investigation.
A growing body of evidence has shown that nonsteroidal anti-inflammatory drugs (NSAIDs) can suppress colorectal tumorigenesis in both humans and rodents (reviewed in Smalley and DuBois, 1997; Thun, 1997). In the general population, epidemiological studies have documented a decreased risk of CRC deaths associated with use of the NSAID aspirin. In individuals with familial adenomatous polyposis (FAP), an inherited predisposition to multiple colorectal polyps, the NSAID sulindac can reduce both the size and number of colorectal tumors. Likewise, sulindac and other NSAIDs have proven to be effective in prevention of intestinal tumorigenesis in mouse models of FAP. The molecular basis for these striking chemopreventive effects has been attributed to inhibition of cyclooxygenases (COX) and the resulting decrease in prostaglandin production (reviewed in Prescott and White, 1996). Consistent with this, expression of COX-2 is elevated in human colorectal tumors (Eberhart et al., 1994; Sano et al., 1995), and inactivation of the COX-2 gene in mice is associated with decreased intestinal tumorigenesis (Oshima et al., 1996).
However, other observations are difficult to reconcile with COX being the sole target of NSAIDs in the colon. For example, NSAID derivatives that lack the ability to inhibit COX have been shown to inhibit colon tumor growth in vivo and in vitro (Piazza et al., 1995, 1997; Mahmoud et al., 1998; Reddy et al., 1999). Conversely, colon cancer cells totally devoid of COX activity are growth inhibited as effectively as cells producing COX (Hanif et al., 1996; Elder et al., 1997). Likewise, COX-1 and COX-2 null mouse embryo fibroblast cells remain sensitive to the antiproliferative and antineoplastic effects of NSAIDs (Zhang et al., 1999). In those colon cancer cells producing COX, COX-produced prostaglandins cannot rescue cells from NSAID-associated growth arrest in vivo or in vitro (Narisawa et al., 1984; Hanif et al., 1996; Chan et al., 1998). The concentration of NSAIDs that inhibit growth is 10 to 100 times higher than that required to inhibit COX activity, suggesting the existence of additional cellular targets (Hanif et al., 1996; Ahnen, 1998; Charalambous et al., 1998; Simmons et al., 1999). Finally, many studies have demonstrated that the COX-2 protein is elevated in the neoplastic epithelial cells of human tumors (Eberhart et al., 1994; Sano et al., 1995), while COX-2 expression in mouse intestinal tumors is confined to nonneoplastic stromal cells (Oshima et al., 1996; Shattuck-Brandt et al., 1999). Thus, the mouse and human COX-2 proteins are located within disparate cellular populations of the colon, yet tumorigenesis in both species is prevented by the same NSAIDs. This suggests either that it is coincidental that the same agents can exert chemopreventive effects in mouse and human, or that other targets of NSAIDs, common to the neoplastic cells of both species, might exist and provide a link to the molecular pathogenesis described below.
Molecular genetic studies have identified a series of genetic alterations associated with the development of benign tumors (adenomas) and their progression to malignant disease (carcinomas) (reviewed in Kinzler and Vogelstein, 1996). In terms of prevention, the alterations that occur early in this process are of most interest. Inactivating mutations of the APC tumor suppressor pathway occur early and are found in most colorectal adenomas and carcinomas. Moreover, inherited mutations of APC cause FAP, characterized by the development of hundreds to thousands of colorectal adenomas. Several studies have suggested that APC’s association with β-catenin (Rubinfeld et al., 1993; Su et al., 1993) might be critical to its tumor-suppressive effects. In the colon, β-catenin binds to the Tcf-4 transcription factor, providing a domain that activates genes containing Tcf-4-binding sites in their regulatory regions (Behrens et al., 1996; Molenaar et al., 1996). Wild-type APC can promote the degradation of β-catenin (Munemitsu et al., 1995) and inhibit β-catenin/Tcf-4 regulated transcription (CRT), while disease-associated APC mutants are deficient in this ability (Korinek et al., 1997; Morin et al., 1997). In tumors lacking APC mutations, oncogenic mutations of β-catenin can lead to increased CRT (Morin et al., 1997; Rubinfeld et al., 1997).
The targets of this increased CRT are therefore likely to provide insights into APC’s suppressive effects. We have recently used SAGE technology to analyze changes in gene expression following inhibition of CRT by APC in human CRC cells and identified the c-myc oncogene as a direct target of CRT (He et al., 1998a). However, like many other critical regulators of cell growth, APC is likely to exert its effects through several effectors. Here, we report the identification of another target of the APC pathway, peroxisome proliferator-activated receptor δ (PPARδ; a.k.a. PPARβ, NUC1, and FAAR) (Schmidt et al., 1992; Amri et al., 1995; Jow and Mukherjee, 1995), which provides a link between NSAID-mediated chemoprevention and the genetic alterations identified in colorectal tumors.
PPARδ belongs to the nuclear receptor superfamily, which includes the steroid hormone, thyroid hormone, retinoid, and PPAR subfamilies as well as a growing number of orphan receptors (Kastner et al., 1995; Mangelsdorf et al., 1995; Lemberger et al., 1996). The PPAR subfamily comprises at least three distinct subtypes found in vertebrate species: PPARα, PPARδ, and PPARγ. The nuclear receptor family members function as ligand-dependent sequence-specific activators of transcription (Mangelsdorf et al., 1995; Lemberger et al., 1996). We found that the NSAIDs sulindac and indomethacin could mimic the effects of APC by downregulating the transcriptional activity of PPARδ. This inhibition appeared to be due to disruption of the DNA binding ability of PPARδ/RXR heterodimers. These observations demonstrate that APC and NSAIDs inhibit a mutual target, PPARδ, thereby providing a link between the genetic alterations underlying tumor development and cancer chemoprevention.
Results
APC Represses PPARδ Expression
The effects of APC on gene expression were explored using SAGE as previously reported (He et al., 1998a). In brief, gene expression was examined in a human CRC cell line with inducible APC (HT29-APC) and a control cell line with an inducible lacZ gene (HT29-β-gal) 9 hr after induction. SAGE analysis of 55,233 and 59,752 tags from APC-expressing and control cells, respectively, led to the identification of 14,346 different transcripts, the majority of which were not differentially expressed. Because biochemical studies have indicated that APC directly represses CRT, we focused on the repressed transcripts. One of the most highly repressed tags corresponded to PPARδ (24 tags in HT29-β-gal versus 5 tags in HT29-APC).
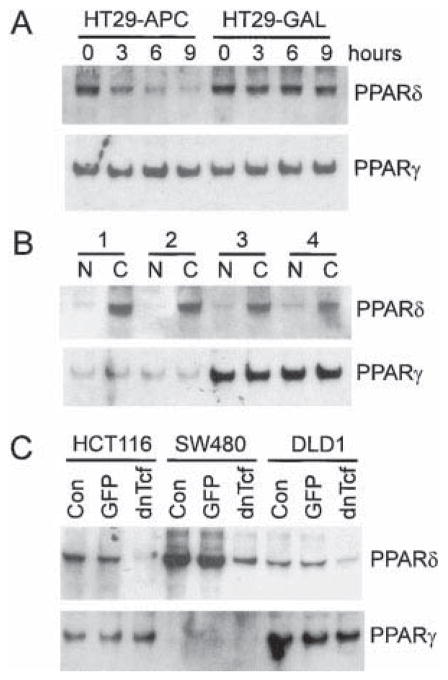
To confirm the SAGE data, we performed Northern blot analysis of RNA from HT29-APC and HT29β-gal cells using PPAR probes (Figure 1A). Repression of PPARδ was evident as early as 3 hr after APC induction, whereas no change was detectable in HT29 β-gal cells even 9 hr after induction. In contrast, expression of PPARγ was not affected by expression of APC, and the other known PPAR subfamily member, PPARα, was not expressed at detectable levels (Figure 1A and data not shown). The ability of APC to repress PPARδ expression suggested that expression of PPARδ should be elevated in primary CRCs, where CRT is often increased by mutations in APC or β-catenin. Northern blot analysis of PPAR expression in CRCs and normal colorectal mucosa revealed a marked increase in PPARδ expression in each of four cancers studied (Figure 1B). In contrast, there was no increase in PPARγ expression in the cancers of these patients (Figure 1B).
Figure 1. Expression of PPARδ in Human CRC Cells.
(A) Decreased expression of PPARδ following induction of APC in CRC cells. Expression of APC (HT29-APC) or β-galactosidase (HT29-GAL) was induced with (110 μM) ZnCl2 for the indicated times in HT29 CRC cells containing the respective genes under the control of a modified metallothionein promotor. Total RNA (10 μg) was isolated and analyzed by Northern blot analysis with probes specific for PPARδ and PPARγ.
(B) Increased expression of PPARδ in primary CRCs. Northern blot analyses with probes specific to PPARδ and PPARγ were performed on total RNA (10 μg) isolated from matched primary CRCs (C) and normal colon epithelium (N) removed from four different patients.
(C) Expression of PPARδ in CRCs is dependent on Tcf-4-mediated transcription. CRC cells with increased β-catenin/Tcf-4-mediated transcription due to either APC (SW480, DLD1) or β-catenin (HCT116) mutations were either mock infected (Con) or infected with adenovirus expressing GFP (GFP) or a dominant-negative mutant of Tcf-4 (dnTcf). Total RNA (10 μg) was isolated and analyzed by Northern blot analysis with probes specific for PPARδ and PPARγ.
APC Inhibits CRT of the PPARδ Gene
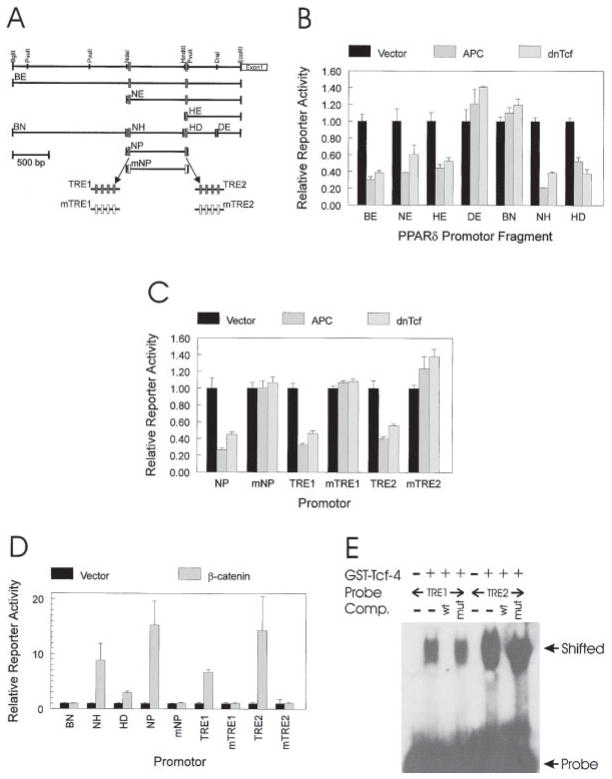
To explore the basis for the repression of PPARδ, we isolated and sequenced a 3.1 kb genomic region upstream of the PPARδ transcription start site (GenBank accession #AF187850) and used it to analyze APC responsiveness (Figure 2). A luciferase reporter construct containing this fragment (BE) upstream of a minimal promotor was markedly repressed by APC expression (Figures 2A and 2B). Analysis of nested deletions and pro-motor fragments identified two APC-responsive fragments (fragment NH and HD, Figures 2A and 2B). The sequence of these fragments revealed two putative Tcf-4-binding sites, one (TRE1) located 1543 bp upstream of the PPARδ transcription start site in fragment NH and the other (TRE2) located 759 bp upstream in fragment HD. A fragment spanning these two sites conferred marked APC repression, which was completely abrogated by disruption of the putative Tcf-4-binding sites (NP versus mNP, Figure 2C). Moreover, either of the putative Tcf-4-binding sites in isolation could confer APC responsiveness in a sequence-specific manner (TRE1 versus mTRE1, and TRE2 versus mTRE2 in Figure 2C).
Figure 2. APC Regulates PPARδ Expression through CRT.
(A) PPARδ promotor. A restriction map of the 3.1 kb region upstream of the first exon of PPARδ is shown. Restriction fragments BE, NE, HE, DE, BN, NH, HD, and NP were used to construct reporters for measuring APC and β-catenin responsiveness. Filled boxes represent potential Tcf-4-binding sites, and open boxes represent the same sites engineered to contain mutations that abolish Tcf-4 binding. mNP represents fragment NP with both potential Tcf-4-binding sites mutated. TRE1 and TRE2 contain four repeats of the two Tcf-4-binding sites, respectively. mTRE1 and mTRE2 are mutant forms of TRE1 and TRE2.
(B) The PPARδ promotor is repressed by APC and dnTcf. SW480 CRC cells were transfected with the indicated PPARδ promotor luciferase reporters (0.4 μg), with a β-galactosidase expression vector (0.2 μg pCMVβ), and with 1.0 μg of either a control (Vector), APC, or dnTcf expression vector. Luciferase activity is reported relative to the vector control after normalizing for transfection efficiency through β-galactosidase activity. Bars represent the means of three independent replicates with error bars representing the unbiased standard deviations.
(C) APC and dnTcf responsiveness is mediated by two putative Tcf-4-binding sites. PPARδ promotor fragments with intact and mutated Tcf-4-binding sites were tested for APC and dnTcf-4 responsiveness as described in (B). Bars represent the means of three independent replicates, with error bars representing the unbiased standard deviations.
(D) β-catenin transactivation maps to the same promotor regions mediating APC and dnTcf responsiveness. 293 cells were transfected with the indicated PPARδ promotor luciferase reporters (0.4 μg), with a β-galactosidase expression vector (0.2 μg pCMVβ), and with 1.0 μg of either a no insert control (Vector) or an oncogenic β-catenin (β-catenin) expression vector. Luciferase activity was reported as described for (B). Bars represent the means of three independent replicates, with error bars representing the unbiased standard deviations.
(E) Putative Tcf-4-binding sites in the PPARδ promotor bind Tcf-4. GEMSA was performed using 32P-labeled probes containing either putative Tcf-4-binding sites TRE1 or TRE2. GEMSA was performed in the presence of a GST fusion protein containing the Tcf-4 DNA-binding domain as indicated. Wild-type (wt) or mutant (mut) competitors corresponding to the Tcf-4-binding sites were used as indicated.
As noted above, an obvious basis for the APC responsiveness of PPARδ would be inhibition of CRT. Consistent with this, there was a perfect concordance between the ability of APC to repress PPARδ promotor fragments (Figures 2B and 2C) and the ability of oncogenic β-catenin to induce transcriptional activity (Figure 2D). Likewise, there was a perfect concordance between APC responsiveness and the ability of a dominant-negative Tcf-4 (dnTcf) expression vector to inhibit transcriptional activity (Figures 2B and 2C). As with APC responsiveness, the β-catenin transactivation and the dnTcf repression were abrogated by mutation of the putative Tcf-4-binding sites (Figures 2C and 2D). The ability of Tcf-4 to directly bind to the PPARδ TRE sites was demonstrated by gel electrophoresis mobility shift assays (GEMSA). Both putative binding sites demonstrated significant Tcf-4 binding, which was inhibited by their cognate wild-type binding sequences but not by their mutant counterparts (Figure 2E).
The above results suggested that APC repressed PPARδ expression by interfering with CRT and that alterations in this pathway could lead to increased expression of PPARδ in CRCs. To further evaluate the generality of this pathway, we examined the ability of dnTcf to interfere with PPARδ expression in other human CRC cell lines. Like the HT29 cells in which PPARδ expression was first identified (Figure 1A), SW480 and DLD1 cells contain inactivating mutations of APC. HCT116 cells have an activating mutation of β-catenin. As expected from the study of primary tumors (Figure 1B), PPARδ expression was detected in all the lines (Figure 1C). Moreover, PPARδ expression was inhibited in each line by infection with an adenovirus containing a dnTcf expression cassette but not by a control adenovirus containing a GFP expression cassette (Figure 1C). In contrast, PPARγ expression was barely detectable in SW480 cells, and dnTcf had no effect on PPARγ expression in any of the lines tested.
Definition of PPARδ-Responsive Elements
To explore the functional significance of PPARδ repression, we developed reporters for PPARδ function. Although downstream targets of PPARδ are unknown, studies of other PPAR family members have defined a prototypic response element. Maximum DNA binding and activation are achieved through heterodimerization between a PPAR protein and RXR (Gearing et al., 1993). Accordingly, the prototypic PPAR response element ACO from the acyl-CoA oxidase gene promotor contains two copies of the core binding sequence AGGTCA separated by one base pair (Tugwood et al., 1992; Mangelsdorf et al., 1995; Lemberger et al., 1996; Juge-Aubry et al., 1997). PPARα and PPARγ bind this consensus efficiently, whereas PPARδ does not (see below). To define a PPARδ-responsive element, we performed in vitro binding site selection for both PPARδ and RXRα. Analysis of 28 binding sites selected with a RXRα GST fusion protein identified (A/G)GGTCA as the core consensus for RXR (Figure 3A). Analysis of 20 sites selected with a PPARδ GST fusion protein revealed a novel binding consensus (CGCTCAC), which was distinct from the previously defined PPARα/γ consensus (Figure 3B).
Figure 3. Development of a PPARδ-Specific Reporter.
(A) RXRα consensus binding site. PCR products of a randomized oligonucleotide template that bound a GST fusion protein containing the DNA-binding domain of RXRα were selected, cloned, and sequenced. The sequences of 28 clones are shown, manually aligned to derive the consensus binding sequence indicated at the bottom.
(B) PPARδ consensus binding site. PCR products of a randomized oligonucleotide template that bound a GST fusion protein containing the DNA-binding domain of PPARδ were selected, cloned, and sequenced. The sequences of 20 clones are shown, manually aligned to derive the consensus binding sequence indicated at the bottom.
(C) Binding specificity of PPARα, PPARδ, and PPARγ. Oligonucleotides containing the indicated binding elements (DRE or ACO) were 32P-labeled and incubated with GST fusion proteins containing either the PPARα, PPARδ, PPARγ, RXRα, or no DNA binding domain (−). DNA binding was assessed by GEMSA, where “Probe” indicates the unbound probe and “Shifted” indicates bound probe.
(D) Binding specificity of PPARδ/RXRα and PPARγ/RXRα heterodimers. Oligonucleotides containing the indicated binding elements (DRE or ACO) were 32P-labeled and incubated with in vitro translated PPARδ, PPARγ, and RXRα as indicated. Bindings were supplemented with PPARδ ligand cPGI (10 μM) or PPARγ ligand BRL 49653 (10 μM) as indicated. DNA binding was assessed by GEMSA, where “Probe” indicates the unbound probe and “Shifted” indicates bound probe.
(E) DRE confers PPARδ but not PPARγ responsiveness. 293 cells were transfected with DRE luciferase reporter (0.3 μg), a β-galactosidase expression vector (0.2 μg pCMVβ), and with 1.0 μg of either empty vector (Control), PPARδ, or PPARγ expression vectors. Where indicated, cells were treated with the PPARδ ligand cPGI (20 μM) or the PPARγ ligand BRL 49653 (20 μM). Luciferase activity was reported as relative luciferase activity after correction for transfection efficiency using β-galactosidase activity. Bars represent the means of three independent replicates with the error bars representing the unbiased standard deviations.
(F) ACO confers PPARγ but not PPARδ responsiveness. The same as in (E) except an ACO luciferase reporter was used.
The combination of the PPARδ and RXRα consensus sequences should form a PPARδ-binding element in vitro and a PPARδ-responsive element in vivo. We generated a putative PPARδ-responsive element (DRE, 5′-CGCTCACAGGTCA-3′) by joining the PPARδ- and RXRα-binding sites. GEMSA analysis of DRE revealed binding to PPARδ GST fusion protein but not to PPARα or PPARγ GST fusion proteins (Figure 3C). In contrast, the prototypic PPAR-responsive element ACO (5′-AGG ACAAAGGTCA-3′) bound PPARα and PPARγ but not PPARδ GST fusion proteins (Figure 3C). RXRα GST fusion protein demonstrated weaker binding to both responsive elements. To further test the specificity of these elements, we performed GEMSA analysis with in vitro translated PPARδ/RARα and PPARγ/RARα heterodimers in the presence or absence of ligand stimulation. Under the conditions used, binding of PPARδ/RXRα and PPARγ/RXRα heterodimers to their cognate elements could not be detected in the absence of ligand. However, PPARδ/RXRα binding to DRE was markedly induced by the PPARδ ligand cPGI, and PPARγ/RXRα binding to ACO was induced by the PPARγ ligand BRL49653 (Figure 3D). In contrast, PPARγ/RXRα heterodimers did not bind DRE in the presence of BRL49653 nor did PPARδ/RXRα bind ACO in the presence of cPGI. To test the specificity of these response elements in cells, we constructed luciferase reporters containing either the DRE or ACO elements. Transfection of 293 cells with PPARδ resulted in activation of the DRE reporter that was enhanced by cPGI (Figure 3E) but did not activate the ACO reporter even in the presence of cPGI (Figure 3F). In contrast, expression of PPARγ did not activate the DRE reporter (Figure 3E) but did activate the ACO reporter that was enhanced by BRL 49653 (Figure 3F).
PPARδ Function Is Regulated by the APC/β-Catenin/Tcf-4 Pathway
The above findings suggested that PPARδ activity was regulated by APC/β-catenin/Tcf-4 pathway at the transcriptional level. To address the functional consequences of this transcriptional regulation in CRC cells, we used the PPARδ-specific reporters described above. Transfection of APC into a human CRC cell line resulted in downregulation of the PPARδ reporter DRE but had no effect on the PPARα/γ-responsive reporter ACO (Figure 4A). The lack of any effect on PPARα/γ demonstrated the specificity of this inhibition and made it unlikely that the effects were due to nonspecific toxicity of a tumor suppressor. Transfection of a dnTcf-4 expression vector also specifically repressed the PPARδ reporter but not the PPARα/γ reporter. To further eliminate the possibility of nonspecific toxic effects, we tested β-catenin’s ability to positively regulate PPARδ activity. Expression of oncogenic β-catenin mutants activated the PPARδ reporter but did not activate the PPARα/γ reporter (Figure 4B).
Figure 4. PPARδ Activity Is Regulated by APC, β-Catenin, and Sulindac.
(A) APC and dnTcf specifically repress PPARδ activity. PPARδ and PPARγ activity was assessed with the DRE and ACO luciferase reporters, respectively. SW480 CRC cells were transfected with the indicated luciferase reporters (0.4 μg of DRE or ACO), with a β-galactosidase expression vector (0.2 μg pCMVβ), and with 1.0 μg of either a control (Vector), APC, or dnTcf expression vector. Luciferase activity was calculated as described in Figure 2. Bars represent the means of three independent replicates with the error bars being the unbiased standard deviations.
(B) β-catenin expression increases PPARδ activity. The 293 human cell line was transfected with the indicated luciferase reporters (0.4 μg of DRE or ACO), with a β-galactosidase expression vector (0.2 μg pCMVβ), and with 0.8 μg of either a no insert control (Vector) or an oncogenic β-catenin expression vector.
(C) Sulindac and indomethacin specifically repress PPARδ activity. PPARδ and PPARγ activity was assessed as transcriptional activity of the DRE and ACO luciferase reporters, respectively. HCT116 and SW480 CRC cells were transfected with the indicated luciferase reporters (1.0 μg of DRE or ACO) and with a β-galactosidase expression vector (0.2 μg of pCMVβ). Cells were allowed to recover for 20 hr after transfection and then treated for 10 hr with the indicated concentrations (μM) of sulindac sulfide or indomethacin. Luciferase activity was reported relative to the control (0) after normalizing for transfection efficiency.
NSAIDs Suppress PPARδ Activity
Suppression of colorectal tumorigenesis by NSAIDs suggested that these compounds may be linked to the genetic alterations that drive tumorigenesis. The identification of PPARδ as a target of the APC tumor suppressor pathway suggested a specific link. Both precursors and products involved in eicosanoid metabolism have been shown to be ligands for PPARs (Keller et al., 1993; Yu et al., 1995; Forman et al., 1997; Kliewer et al., 1997). The ability of NSAIDs to perturb eicosanoid metabolism suggested that PPARs may be an ultimate target of NSAIDs in suppressing tumorigenesis (Prescott and White, 1996), and the above findings suggest that PPARδ could be a specific target. To explore this possibility, we tested the effects of the NSAID sulindac on PPARδ function. As noted in the Introduction, sulindac has been shown to suppress intestinal tumorigenesis in humans and mice. In culture, sulindac sulfide, the active metabolite of sulindac, has been shown to induce apoptosis in CRC cells (Piazza et al., 1995; Shiff et al., 1995; Hanif et al., 1996; Chan et al., 1998). Sulindac sulfide treatment resulted in a dose-dependent repression of PPARδ activity in CRC cells, as assessed with the DRE reporter (Figure 4C). A greater than 2-fold repression was observed at low concentrations of sulindac sulfide, and a greater than 10-fold reduction was noted at levels of sulindac sulfide that induced substantial degrees of apoptosis in these cells (Figures 4C and 5D). In contrast, sulindac sulfide had only a modest effect (less than 25% repression at the highest concentration tested) on PPARα/γ activity, assessed with the ACO reporter (Figure 4C). A similar dose-dependent suppression of PPARδ was observed with indomethacin, another NSAID (Figure 4C).
Figure 5. PPARδ Can Protect Colon Cancer Cells from Sulindac-Induced Apoptosis.
(A–D) PPARδ inhibits sulindac sulfide–induced apoptosis. HCT116 and SW480 cells were either mock infected (Uninfected) or infected with adenovirus expressing GFP (AdGFP) or PPARδ (AdPPARδ). Twenty hours after infection, cells were treated for 42 hr with sulindac sulfide. Apoptosis was assessed by the presence of apoptotic nuclei (condensation and fragmentation) after Hoechst 33258 staining. (A–C) Fluorescence microscopy of uninfected (A), AdGFP (B), or AdPPARδ (C) infected HCT116 cells treated with 125 μM of sulindac sulfide. (D) Bars represent the fraction of apoptotic nuclei after treatment with the indicated adenoviruses and concentration of sulindac sulfide (μM).
(E) PPARδ rescues sulindac sulfide inhibition of clonal growth. Cells were infected with the indicated adenovirus, treated with the indicated concentrations of sulindac sulfide, and plated. Clonal growth was scored as colony formation after 6 days. Colonies were visualized by staining with Crystal Violet (upper panel) and enumerated (lower panel).
Expression of PPARδ Partially Rescues Sulindac Sulfide–Induced Apoptosis
If suppression of PPARδ activity were contributing to sulindac-induced apoptosis, overexpression of PPARδ might be expected to protect against sulindac sulfide–induced apoptosis. We constructed an adenovirus (Ad-PPARδ) expressing PPARδ and a green fluorescent protein (GFP) marker using AdEasy technology (He et al., 1998b). The ability of AdPPARδ to suppress sulindac sulfide–induced apoptosis was compared to that of AdGFP, which contained only the GFP marker gene. AdPPARδ produced nearly a 5-fold decrease in apoptosis in HCT116 cells treated with 100 or 125 μM sulindac sulfide (Figures 5A–5D). Similar results were obtained with the SW480 cell line (Figure 5D). However, the suppression of apoptosis could be overridden at higher concentrations of sulindac sulfide (150 μM, Figure 5D). The results were further confirmed and extended by the ability of AdPPARδ to rescue inhibition of clonal cell growth by sulindac sulfide. Treatment of cells with sulindac sulfide resulted in a greater than 4-fold decrease in the number of colonies (Figure 5E). At 100 or 125 μM sulindac sulfide but not higher doses, this decrease could be completely rescued by infection with Ad-PPARδ, which actually resulted in a slightly increased number (~15%) of colonies. In contrast, the APC target and prototypic oncogene c-myc could not rescue the sulindac-mediated inhibition of clonal growth.
Sulindac Sulfide Disrupts the DNA Binding Ability of PPARδ/RXRα Heterodimers
To determine whether sulindac could inhibit the transcription of PPARδ like APC, we examined expression of PPARδ following sulindac sulfide treatment. Concentrations of sulindac sulfide that resulted in suppression of PPARδ activity and apoptosis had no effect on the level of PPARδ transcripts, excluding this possibility (Figure 6A). Because PPARs can bind eicosanoids (Keller et al., 1993; Yu et al., 1995; Forman et al., 1997; Kliewer et al., 1997), the effects of NSAIDs on PPARδ could be due to their ability to perturb eicosanoid metabolism. However, several studies (see Discussion) have suggested that the chemopreventive effects of NSAIDs are not simply related to their ability to suppress prostaglandin synthesis. We therefore considered the possibility that NSAIDs directly inhibit PPARδ activity. This notion was supported by the ability of some NSAIDs to activate PPARα/γ (Lehmann et al., 1997) and of indomethacin to alter the ligand responsiveness of TetR/PPARδ fusion proteins (Yu et al., 1995). We tested the ability of sulindac to inhibit cPGI-stimulated PPARδ/RXRα heterodimer DNA binding activity in vitro. Sulindac sulfide was able to inhibit binding of PPARδ/RXRα heterodimers to the DRE element (Figure 6B). Binding to DRE was also inhibited by the NSAID indomethacin and, at higher concentrations, the sulindac sulfide–related compound sulindac sulfone (Figure 6B). The relative concentrations of sulindac sulfide, indomethacin, and sulindac sulfone required to inhibit binding to DRE were roughly concordant with the concentrations required to induce apoptosis in CRC cells, with sulindac sulfide being the most potent and sulindac sulfone the least (Figures 5D and 6B; data not shown). None of these drugs had any effect on BRL49653-stimulated binding of PPARγ/RXRα heterodimers in an analogous assay performed with the ACO element (Figure 6C).
Figure 6. Mechanism of Suppression of PPARδ by NSAIDs.

(A) NSAIDs do not affect PPARδ, PPARγ, or RXRα expression. HCT116 and SW480 cells were treated with the indicated concentration (μM) of sulindac sulfide for 36 hr, and RNA was isolated. Northern blot analysis was performed on 10 μg of total RNA with a probe specific for PPARδ, PPARγ, or RXRα.
(B) NSAIDs suppress PPARδ DNA binding. The DRE-binding element was 32P-labeled and incubated with no lysate (Probe only) or in vitro translated PPARδ (δ), RXRα (RXRα), or both (δ + RXRα). PPARδ + RXRα + cPGI (10 μM) was included in all lysates treated with the indicated NSAIDs. DNA binding was assessed by GEMSA, where “Probe” indicates the unbound probe and “Shifted” indicates bound probe.
(C) NSAIDs do not suppress PPARγ DNA binding. The ACO-binding element was 32P-labeled and incubated with no lysate (Probe only) or in vitro translated PPARγ (γ), RXRα (RXRα), or both (γ + RXRα). PPARγ + RXRα + BRL 49653 (10 μM) was included in all lysates treated with the indicated NSAIDs. DNA binding was assessed by GEMSA, where “Probe” indicates the unbound probe and “Shifted” indicates bound probe.
Discussion
The above results demonstrate that PPARδ is a target of both APC and NSAIDs and suggest a model of how APC and NSAIDs operate to suppress intestinal tumorigenesis (Figure 7). In most CRCs, inactivating mutations of APC lead to elevated levels of CRT (Korinek et al., 1997; Morin et al., 1997). In rare CRCs without APC mutations, β-catenin mutations that render it resistant to APC-mediated degradation result in elevated β-catenin/Tcf-4-mediated transcription (Morin et al., 1997). In either case, this increased β-catenin/Tcf-4 activity leads to increased transcription of growth-promoting genes. Accordingly, restoration of APC function to CRC cells with defective APC function results in growth suppression and apoptosis (Morin et al., 1996). The genes that have been postulated to mediate the growth-promoting effects of β-catenin/Tcf-4 activity include those encoded by the c-myc oncogene (He et al., 1998a), the cyclin D1 gene (Tetsu and McCormick, 1999), and others (WISP, c-jun, and fra-1) (Pennica et al., 1998; Mann et al., 1999). The present findings suggest that PPARδ represents a β-catenin/Tcf-4 target with particular importance for chemoprevention. Whereas APC or β-catenin mutations can result in increased PPARδ activity, NSAIDs can compensate for this defect by suppressing PPARδ activity and promoting apoptosis. This suppression of PPARδ is mediated in part by the ability of some NSAIDs to directly inhibit the DNA binding activity of PPARδ. In addition, because fatty acids and eicosanoids can act as ligands and modifiers of PPAR activity (Keller et al., 1993; Yu et al., 1995; Prescott and White, 1996; Forman et al., 1997; Kliewer et al., 1997), PPARδ activity might be repressed by the NSAID-mediated changes in eicosanoid metabolism.
Figure 7. Unified Model for APC- and NSAID-Mediated Suppression of CRC.

Elements indicated in blue have been shown to have a tumor-suppressive effect, whereas elements in red have been shown to promote tumor formation. The effects of items in boxes have been demonstrated by genetic alterations. LOX, 5-lipoxygenase; sPLA2, secretory phospholipase 2; and COX, cyclooxygenase.
The above model can help explain several features of NSAID-mediated chemoprevention. First, the effectiveness of some NSAIDs in the prevention of colorectal adenomas can now be linked to genetic defects that underlie the initiation of these tumors and to the ability of NSAIDs to counterbalance the consequences of these genetic defects. Second, although NSAID functions have been linked to their inhibition of COX activity and the resulting inhibition of prostaglandin synthesis, several studies have suggested that the chemopreventive and apoptosis-inducing activities of NSAIDs are not entirely related to the inhibition of COX or to the decreased levels of prostaglandins (see Introduction). These results may be explained by the ability of NSAIDs to directly inhibit PPARδ. Indeed, the sulindac derivative sulindac sulfone, which is devoid of COX inhibitory activity, has apoptotic activity in vitro and chemopreventive activity in vivo and has been proposed as a chemopreventive agent that lacks the toxicity associated with traditional NSAIDs (Piazza et al., 1995, 1997; Mahmoud et al., 1998). Sulindac sulfone inhibited PPARδ activity, albeit at higher concentrations than that required for sulindac sulfide, consistent with its reduced chemopreventive and apoptosis-promoting activity. Third, recent studies have demonstrated that PPARγ agonists promote intestinal tumorigenesis in the Min mouse (Lefebvre et al., 1998; Saez et al., 1998), while the same agonists inhibit the growth of human CRC cells (Brockman et al., 1998; Sarraf et al., 1998). Although the conclusions of these studies were contradictory, they clearly demonstrated the ability of PPAR ligands to modify intestinal tumor growth. A role for PPARs in intestinal tumorigenesis is further suggested by the recent identification of loss-of-function mutations in one allele of PPARγ in 4 of 55 sporadic CRCs (Sarraf et al., 1999). However, unlike PPARδ, neither PPARγ nor PPARα is a target of the APC/β-catenin pathway. Whereas PPARδ is increased in expression in cancers, downregulated by APC, and upregulated by β-catenin, PPARγ and PPARα do not display these properties. Fourth, the ability of COX2 expression to modulate apoptosis (Tsujii and Dubois, 1995) and intestinal tumorigenesis (Oshima et al., 1996) may be partially related to its ability to alter the spectrum of ligands for PPARδ and other PPARs. In this regard, it is interesting to note that the PPARδ ligand cPGI can partially rescue infertility resulting from COX-2 deficiency (Lim et al., 1999). Finally, the ability of dietary fatty acids and secreted phospholipases to modify the spectrum of PPARδ ligands and thus alter PPARδ activity could account for their ability to affect CRC risk (Willett et al., 1990; Dietrich et al., 1993; MacPhee et al., 1995; Vanden Heuvel, 1999).
In addition to explaining several features of NSAID-mediated chemoprevention, our observations may have important ramifications for the development of chemopreventive agents. In particular, it is conceivable that the development of drugs that specifically target PPARδ might lead to more efficacious and less toxic means for CRC chemoprevention.
Experimental Procedures
Cell Lines and Chemicals
Human CRC cells (HT29, HCT116, SW480, and DLD1) and embryonic kidney cells (293) were obtained from ATCC (Manassas, VA). BRL49653 and cPGI were purchased from American Radiolabeled Chemicals and Cayman Chemical Company, respectively. Sulindac derivatives and indomethacin were purchased from BIOMOL. Unless otherwise indicated, all chemicals were purchased from Sigma (St. Louis, MO).
In Vitro DNA-Binding Site Selection for PPARδ and RXRα
GST fusion proteins containing the N-terminal DNA-binding domains of human PPARδ and RXRα were constructed by PCR amplifying codons 1–249 of PPARδ and 1–224 of RXRα and cloning them into pGEX-2TK vector. As controls, GST fusion proteins containing the DNA-binding domains of human PPARα (aa 1–249) and PPARγ (aa 1–248) were also constructed. To identify the potential consensus DNA sequence motifs recognized by PPARδ and RXRα, a previously described in vitro site selection procedure was utilized (Zawel et al., 1998). Selected PCR products were cloned in to pZero2.1, tested for binding, and sequenced as described.
Gel Electrophoretic Mobility Shift Assays
DNA-binding assays supplemented with Poly dIdC (6 μg/ml) were performed essentially as described (Zawel et al., 1998). For binding to PCR products derived from in vitro site selections, 1.0–1.5 μg of protein and 50 ng of DNA were used. For competitions, a 100-fold excess of unlabeled probe was used. For GEMSA with GST fusion proteins, 0.3–0.5 μg of fusion protein and 0.5 ng of 32P kinase labeled (~106 dpm) DNA were used. The probes for Tcf-4 binding were as previously reported (Korinek et al., 1997). For GEMSA with in vitro translated proteins, 0.1 to 0.2 μl of programmed lysate and 32P-labeled probe (~106 dpm) was used. The DRE probe was formed by annealing 5′-GCGTGAGCGCTCACAGGTCAATTCG-3′ and 5′-CCGAATT GACCTGTGAGCGCTCACG-3′. The ACO probe was formed by annealing 5′-GCGGACCAGGACAAAGGTCACGTTC-3′ and 5′-CGA ACGTGACCTTTGTCCTGGTCCG-3′.
Construction of a PPARδ-Responsive Reporter
The following oligonucleotides containing PPARδ and RXR recognition motifs that were identified from in vitro site selection approach were synthesized: 5′-CTAGCGTGAGCGCTCACAGGTCAATTCGGTGAGCGCTCACAGGTCAATTCG-3′ and 5′-CTAGCGAATTGACCTGTGAGCGCTCACCGAATTGACCTGTGAGCGCTCACG-3′. As a control, the following oligonucleotides containing a PPARα and PPARγ-responsive element from the acyl-CoA oxidase promotor were also synthesized: 5′-CTAGCGGACCAGGACAAAGGTCACGTTCGGACCAGGACAAAGGTCACGTTCG-3′ and 5′-CTAGCGAACGTGACCTTTGTCCTGGTCCGAACGTGACCTTTGTCCTGGTCCG-3′. The oligonucleotide cassettes were dimerized and cloned into pBV-Luc, a luciferase reporter plasmid with very low basal activity (He et al., 1998a). All constructs were verified by DNA sequencing.
Constructions of PPARδ Promotor Reporters
Three independent BAC clones containing the PPARδ promotor sequence were obtained by screening a BAC library (Research Genetics). For the construction of PPARδ promotor reporters, corresponding restriction fragments (illustrated in Figure 2A) were sub-cloned into pBV-Luc. The mNP reporter was constructed by cloning a PCR product into pBV-Luc, whereas p4XTRE1-Luc, p4XmTRE1-Luc, p4XTRE2-Luc, and p4XmTRE2-Luc were constructed from oligonucleotides. Details of construction and oligonucleotide sequences are available upon request or at www.coloncancer.org/ppar.htm.
Reporter Assays
Reporter plasmid, effector plasmid, and β-gal control plasmid were transfected into cells using LipofectAmine (Life Technologies). Twenty-four hours after transfection, cells were lysed and collected for assays of luciferase activity using Promega’s Luciferase Assay System.
In Vitro Transcription and Translation Assays
The full-length proteins of PPARδ, PPARγ, and RXRα were generated by in vitro transcription-coupled translation of PCR products using the TNT T7 Quick-Coupled Transcription/Translation System (Promega). PCR primer sequences are available upon request or at www.coloncancer.org/ppar.htm.
Generation of Recombinant Adenovirus Expressing PPARδ or c-Myc
A PCR product of human PPARδ was cloned into pCMV-HAHA and verified by sequencing. This expression cassette was used to generate recombinant adenovirus using the AdEasy system as previously described (He et al., 1998b). Additional details of the construction are available upon request or at www.coloncancer.org/ppar.htm. AdMyc was generated in a similar fashion and was a gift of H. Hermeking.
Acknowledgments
We thank Carlo Rago, Christopher Torrance, Victor Velculescu, Leigh Zawel, Lin Zhang, and Wei Zhou for their help and advice and Heiko Hermeking for providing the AdMyc. This work is supported by National Institutes of Health grants CA57345 and CA62924. B. V. is an investigator of the Howard Hughes Medical Institute. K. W. K. received research funding from Genzyme Molecular Oncology (Genzyme). Under a licensing agreement between the Johns Hopkins University and Genzyme, the SAGE technology was licensed to Genzyme, and K. W. K. and B. V. are entitled to a share of royalties received by the University from sales of the licensed technology. The SAGE technology is freely available to academia for research purposes. K. W. K. and B. V. are consultants to Genzyme. The University and researchers (K. W. K. and B. V.) own Genzyme stock, which is subject to certain restrictions under University policy. The terms of this arrangement are being managed by the University in accordance with its conflict of interest policies.
References
- Ahnen DJ. Colon cancer prevention by NSAIDs: what is the mechanism of action? Eur J Surg Suppl. 1998;582:111–114. doi: 10.1080/11024159850191544. [DOI] [PubMed] [Google Scholar]
- Amri EZ, Bonino F, Ailhaud G, Abumrad NA, Grimaldi PA. Cloning of a protein that mediates transcriptional effects of fatty acids in preadipocytes. Homology to peroxisome proliferator-activated receptors. J Biol Chem. 1995;270:2367–2371. doi: 10.1074/jbc.270.5.2367. [DOI] [PubMed] [Google Scholar]
- Behrens J, von Kries JP, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of β-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- Brockman JA, Gupta RA, Dubois RN. Activation of PPARγ leads to inhibition of anchorage-independent growth of human CRC cells. Gastroenterology. 1998;115:1049–1055. doi: 10.1016/s0016-5085(98)70072-1. [DOI] [PubMed] [Google Scholar]
- Chan TA, Morin PJ, Vogelstein B, Kinzler KW. Mechanisms underlying nonsteroidal antiinflammatory drug-mediated apoptosis. Proc Natl Acad Sci USA. 1998;95:681–686. doi: 10.1073/pnas.95.2.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charalambous D, Skinner SA, O’Brien PE. Sulindac inhibits colorectal tumour growth, but not prostaglandin synthesis in the rat. J Gastroenterol Hepatol. 1998;13:1195–1200. [PubMed] [Google Scholar]
- Dietrich WF, Lander ES, Smith JS, Moser AR, Gould KA, Luongo C, Borenstein N, Dove W. Genetic identification of Mom-1, a major modifier locus affecting Min-induced intestinal neoplasia in the mouse. Cell. 1993;75:631–639. doi: 10.1016/0092-8674(93)90484-8. [DOI] [PubMed] [Google Scholar]
- Eberhart CE, Coffey RJ, Radhika A, Giardiello FM, Ferrenbach S, DuBois RN. Up-regulation of cyclooxygenase 2 gene expression in human colorectal adenomas and adenocarcinomas. Gastroenterology. 1994;107:1183–1188. doi: 10.1016/0016-5085(94)90246-1. [DOI] [PubMed] [Google Scholar]
- Elder DJ, Halton DE, Hague A, Paraskeva C. Induction of apoptotic cell death in human colorectal carcinoma cell lines by a cyclooxygenase-2 (COX-2)-selective nonsteroidal anti-inflammatory drug: independence from COX-2 protein expression. Clin Cancer Res. 1997;3:1679–1683. [PubMed] [Google Scholar]
- Forman BM, Chen J, Evans RM. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors α and δ. Proc Natl Acad Sci USA. 1997;94:4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gearing KL, Gottlicher M, Teboul M, Widmark E, Gustafsson JA. Interaction of the peroxisome-proliferator-activated receptor and retinoid X receptor. Proc Natl Acad Sci USA. 1993;90:1440–1444. doi: 10.1073/pnas.90.4.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanif R, Pittas A, Feng Y, Koutsos MI, Qiao L, Staiano-Coico L, Shiff SI, Rigas B. Effects of nonsteroidal anti-inflammatory drugs on proliferation and on induction of apoptosis in colon cancer cells by a prostaglandin-independent pathway. Biochem Pharmacol. 1996;52:237–245. doi: 10.1016/0006-2952(96)00181-5. [DOI] [PubMed] [Google Scholar]
- He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998a;281:1509– 1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci USA. 1998b;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jow L, Mukherjee R. The human peroxisome proliferator-activated receptor (PPAR) subtype NUC1 represses the activation of hPPAR a and thyroid hormone receptors. J Biol Chem. 1995;270:3836–3840. doi: 10.1074/jbc.270.8.3836. [DOI] [PubMed] [Google Scholar]
- Juge-Aubry C, Pernin A, Favez T, Burger AG, Wahli W, Meier CA, Desvergne B. DNA binding properties of peroxisome proliferator-activated receptor subtypes on various natural peroxisome proliferator response elements. Importance of the 5′-flanking region. J Biol Chem. 1997;272:25252–25259. doi: 10.1074/jbc.272.40.25252. [DOI] [PubMed] [Google Scholar]
- Kastner P, Mark M, Chambon P. Nonsteroid nuclear receptors: what are genetic studies telling us about their role in real life? Cell. 1995;83:859–869. doi: 10.1016/0092-8674(95)90202-3. [DOI] [PubMed] [Google Scholar]
- Keller H, Dreyer C, Medin J, Mahfoudi A, Ozato K, Wahli W. Fatty acids and retinoids control lipid metabolism through activation of peroxisome proliferator-activated receptor-retinoid X receptor heterodimers. Proc Natl Acad Sci USA. 1993;90:2160–2164. doi: 10.1073/pnas.90.6.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B. Lessons from hereditary colon cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Sundseth SS, Jones SA, Brown PJ, Wisely GB, Koble CS, Devchand P, Wahli W, Willson TM, Lenhard JM, Lehmann JM. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors α and γ. Proc Natl Acad Sci USA. 1997;94:4318–4323. doi: 10.1073/pnas.94.9.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a β-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- Lefebvre AM, Chen I, Desreumaux P, Najib J, Fruchart JC, Geboes K, Briggs M, Heyman R, Auwerx J. Activation of the peroxisome proliferator-activated receptor γ promotes the development of colon tumors in C57BL/6J-APCMin/+ mice. Nat Med. 1998;4:1053–1057. doi: 10.1038/2036. [DOI] [PubMed] [Google Scholar]
- Lehmann JM, Lenhard JM, Oliver BB, Ringold GM, Kliewer SA. Peroxisome proliferator-activated receptors α and γ are activated by indomethacin and other non-steroidal anti-inflammatory drugs. J Biol Chem. 1997;272:3406–3410. doi: 10.1074/jbc.272.6.3406. [DOI] [PubMed] [Google Scholar]
- Lemberger T, Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: a nuclear receptor signaling pathway in lipid physiology. Annu Rev Cell Dev Biol. 1996;12:335–363. doi: 10.1146/annurev.cellbio.12.1.335. [DOI] [PubMed] [Google Scholar]
- Lim H, Gupta RA, Ma W, Paria BC, Moller DE, Morrow JD, DuBois RN, Trzaskos JM, Dey SK. Cyclooxygenase-2-derived prostacyclin mediates embryo implantation in the mouse via PPARδ. Genes Dev. 1999;13:1561–1574. doi: 10.1101/gad.13.12.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPhee M, Chepenik K, Liddell R, Nelson K, Siracusa L, Buchberg A. The secretory phospholipase A2 gene is a candidate for the Mom1 locus, a major modifier of ApcMin-induced intestinal neoplasia. Cell. 1995;81:957–966. doi: 10.1016/0092-8674(95)90015-2. [DOI] [PubMed] [Google Scholar]
- Mahmoud NN, Boolbol SK, Dannenberg AJ, Mestre JR, Bilinski RT, Martucci C, Newmark HL, Chadburn A, Bertagnolli MM. The sulfide metabolite of sulindac prevents tumors and restores enterocyte apoptosis in a murine model of familial adenomatous polyposis. Carcinogenesis. 1998;19:87–91. doi: 10.1093/carcin/19.1.87. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann B, Gelos M, Siedow A, Hanski ML, Gratchev A, Ilyas M, Bodmer WF, Moyer MP, Riecken EO, Buhr HJ, Hanski C. Target genes of β-catenin-T cell-factor/lymphoid-enhancer-factor signaling in human colorectal carcinomas. Proc Natl Acad Sci USA. 1999;96:1603–1608. doi: 10.1073/pnas.96.4.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. XTcf-3 transcription factor mediates β-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- Morin PJ, Vogelstein B, Kinzler KW. Apoptosis and APC in colorectal tumorigenesis. Proc Natl Acad Sci USA. 1996;93:7950–7954. doi: 10.1073/pnas.93.15.7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- Munemitsu S, Albert I, Souza B, Rubinfeld B, Polakis P. Regulation of intracellular β-catenin levels by the adenomatous polyposis coli (APC) tumor-suppressor protein. Proc Natl Acad Sci USA. 1995;92:3046–3050. doi: 10.1073/pnas.92.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narisawa T, Hermanek P, Habs M, Schmahl D. Reduction of carcinogenicity of N-nitrosomethylurea by indomethacin and failure of resuming effect of prostaglandin E2 (PGE2) against indomethacin. J Cancer Res Clin Oncol. 1984;108:239–242. doi: 10.1007/BF00402475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima M, Dinchuk JE, Kargman SL, Oshima H, Hancock B, Kwong E, Trzaskos JM, Evans JF, Taketo MM. Suppression of intestinal polyposis in APC (716) knockout mice by inhibition of cyclooxygenase 2 (COX-2) Cell. 1996;87:803–809. doi: 10.1016/s0092-8674(00)81988-1. [DOI] [PubMed] [Google Scholar]
- Pennica D, Swanson TA, Welsh JW, Roy MA, Lawrence DA, Lee J, Brush J, Taneyhill LA, Deuel B, Lew M, et al. WISP genes are members of the connective tissue growth factor family that are up-regulated in wnt-1-transformed cells and aberrantly expressed in human colon tumors. Proc Natl Acad Sci USA. 1998;95:14717–14722. doi: 10.1073/pnas.95.25.14717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza GA, Rahm ALK, Krutzsch M, Sperl G, Paranka NS, Gross PH, Brendel K, Burt RW, Alberts DS, Pamukcu R, Ahnen DJ. Antineoplastic drugs sulindac sulfide and sulfone inhibit cell growth by inducing apoptosis. Cancer Res. 1995;55:3110–3116. [PubMed] [Google Scholar]
- Piazza GA, Alberts DS, Hixson LJ, Paranka NS, Li H, Finn T, Bogert C, Guillen JM, Brendel K, Gross PH, et al. Sulindac sulfone inhibits azoxymethane-induced colon carcinogenesis in rats without reducing prostaglandin levels. Cancer Res. 1997;57:2909–2915. [PubMed] [Google Scholar]
- Prescott SM, White RL. Self-promotion? Intimate connections between APC and prostaglandin H synthase-2. Cell. 1996;87:783–786. doi: 10.1016/s0092-8674(00)81983-2. [DOI] [PubMed] [Google Scholar]
- Reddy BS, Kawamori T, Lubet RA, Steele VE, Kelloff GJ, Rao CV. Chemopreventive efficacy of sulindac sulfone against colon cancer depends on time of administration during carcinogenic process. Cancer Res. 1999;59:3387–3391. [PubMed] [Google Scholar]
- Rubinfeld B, Souza B, Albert I, Muller O, Chamberlain SH, Masiarz FR, Munemitsu S, Polakis P. Association of the APC gene product with β-catenin. Science. 1993;262:1731–1734. doi: 10.1126/science.8259518. [DOI] [PubMed] [Google Scholar]
- Rubinfeld B, Robbins P, El-Gamil M, Albert I, Porfiri E, Polakis P. Stabilization of β-catenin by genetic defects in melanoma cell lines. Science. 1997;275:1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- Saez E, Tontonoz P, Nelson MC, Alvarez JG, Ming UT, Baird SM, Thomazy VA, Evans RM. Activators of the nuclear receptor PPARγ enhance colon polyp formation. Nat Med. 1998;4:1058–1061. doi: 10.1038/2042. [DOI] [PubMed] [Google Scholar]
- Sano H, Kawahito Y, Wilder RL, Hashiramoto A, Mukai S, Asai K, Kimura S, Kato H, Kondo M, Hla T. Expression of cyclooxygenase-1 and -2 in human CRC. Cancer Res. 1995;55:3785–3789. [PubMed] [Google Scholar]
- Sarraf P, Mueller E, Jones D, King FJ, DeAngelo DJ, Partridge JB, Holden SA, Chen LB, Singer S, Fletcher C, Spiegelman BM. Differentiation and reversal of malignant changes in colon cancer through PPARγ. Nat Med. 1998;4:1046–1052. doi: 10.1038/2030. [DOI] [PubMed] [Google Scholar]
- Sarraf P, Mueller E, Smith WM, Wright HM, Kum JB, Aaltonen LA, de la Chapelle A, Spiegelman BM, Eng C. Loss-of-function mutations in PPARγ associated with human colon cancer. Mol Cell. 1999;3:799–804. doi: 10.1016/s1097-2765(01)80012-5. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Endo N, Rutledge SJ, Vogel R, Shinar D, Rodan GA. Identification of a new member of the steroid hormone receptor superfamily that is activated by a peroxisome proliferator and fatty acids. Mol Endocrinol. 1992;6:1634–1641. doi: 10.1210/mend.6.10.1333051. [DOI] [PubMed] [Google Scholar]
- Shattuck-Brandt RL, Lamps LW, Heppner Goss KJ, DuBois RN, Matrisian LM. Differential expression of matrilysin and cyclooxygenase-2 in intestinal and colorectal neoplasms. Mol Carcinog. 1999;24:177–187. [PubMed] [Google Scholar]
- Shiff SJ, Qiao L, Tsai LL, Rigas B. Sulindac sulfide, an aspirin-like compound, inhibits proliferation, causes cell cycle quiescence, and induces apoptosis in HT-29 colon adenocarcinoma cells. J Clin Invest. 1995;96:491–503. doi: 10.1172/JCI118060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons DL, Botting RM, Robertson PM, Madsen ML, Vane JR. Induction of an acetaminophen-sensitive cyclooxygenase with reduced sensitivity to nonsteroid antiinflammatory drugs. Proc Natl Acad Sci USA. 1999;96:3275–3280. doi: 10.1073/pnas.96.6.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley WE, DuBois RN. CRC and nonsteroidal anti-inflammatory drugs. Adv Pharmacol. 1997;39:1–20. doi: 10.1016/s1054-3589(08)60067-8. [DOI] [PubMed] [Google Scholar]
- Su LK, Vogelstein B, Kinzler KW. Association of the APC tumor suppressor protein with catenins. Science. 1993;262:1734–1737. doi: 10.1126/science.8259519. [DOI] [PubMed] [Google Scholar]
- Tetsu O, McCormick F. β-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- Thun MJ. Aspirin and gastrointestinal cancer. Adv Exp Med Biol. 1997;400A:395–402. doi: 10.1007/978-1-4615-5325-0_53. [DOI] [PubMed] [Google Scholar]
- Tsujii M, Dubois RN. Alterations in cellular adhesion and apoptosis in epithelial cells overexpressing prostaglandin endo-peroxide synthase 2. Cell. 1995;83:493–501. doi: 10.1016/0092-8674(95)90127-2. [DOI] [PubMed] [Google Scholar]
- Tugwood JD, Issemann I, Anderson RG, Bundell KR, McPheat WL, Green S. The mouse peroxisome proliferator activated receptor recognizes a response element in the 5′ flanking sequence of the rat acyl CoA oxidase gene. EMBO J. 1992;11:433–439. doi: 10.1002/j.1460-2075.1992.tb05072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Heuvel JP. Peroxisome proliferator-activated receptors: a critical link among fatty acids, gene expression and carcinogenesis. J Nutr. 1999;129:575–580. doi: 10.1093/jn/129.2.575S. [DOI] [PubMed] [Google Scholar]
- Willett WC, Stampfer MJ, Colditz GA, Rosner BA, Speizer FE. Relation of meat, fat, and fiber intake to the risk of colon cancer in a prospective study among women. N Engl J Med. 1990;323:1664–1672. doi: 10.1056/NEJM199012133232404. [DOI] [PubMed] [Google Scholar]
- Yu K, Bayona W, Kallen CB, Harding HP, Ravera CP, McMahon G, Brown M, Lazar MA. Differential activation of peroxisome proliferator-activated receptors by eicosanoids. J Biol Chem. 1995;270:23975–23983. doi: 10.1074/jbc.270.41.23975. [DOI] [PubMed] [Google Scholar]
- Zawel L, Dai JL, Buckhaults P, Zhou S, Kiinzler KW, Vogelstein B, Kern SE. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol Cell. 1998;1:611–617. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- Zhang X, Morham SG, Langenbach R, Young DA. Malignant transformation and antineoplastic actions of nonsteroidal anti-inflammatory drugs (NSAIDs) on cyclooxygenase-null embryo fibroblasts. J Exp Med. 1999;190:451–460. doi: 10.1084/jem.190.4.451. [DOI] [PMC free article] [PubMed] [Google Scholar]