Abstract
Nematodes represent a diverse phylum of both free living and parasitic species. While the species Caenorhabditis elegans (C. elegans) is a valuable model organism, parasitic nematodes or helminths pose a serious threat to human health. Indeed, helminths cause many neglected tropical diseases that afflict humans. Nematode glycoconjugates have been implicated in evasive immunomodulation, a hallmark of nematode infections. One monosaccharide residue present in the glycoconjugates of several human pathogens is galactofuranose (Galf). This five-membered ring isomer of galactose has not been detected in mammals, making Galf metabolic enzymes attractive therapeutic targets. The only known pathway for biosynthetic incorporation of Galf into glycoconjugates depends upon generation of the glycosyl donor UDP-Galf by the flavoenzyme uridine 5’-diphosphate (UDP) galactopyranose mutase (UGM or Glf). A putative UGM encoding gene (glf-1) was recently identified in C. elegans. Because Galf has yet to be identified in any nematode glycan, we sought to assess the catalytic activity of the C. elegans glf-1 gene product (CeUGM). We found that CeUGM catalyzes the isomerization of UDP-Galf and UDP-galactopyranose (UDP-Galp). In the presence of enzyme, substrate, and a hydride source, a galactose–N5-FAD adduct was isolated, suggesting the CeUGM flavin adenine dinucleotide (FAD) cofactor serves as a nucleophile in covalent catalysis. Homology modeling and protein variants indicate that CeUGM possesses an active site similar to that of prokaryotic enzymes, despite the low sequence identity (~15%) between eukaryotic and prokaryotic UGM proteins. Even with the primary sequence differences, heterocyclic UGM inhibitors developed against prokaryotic proteins also inhibit CeUGM activity. We postulate that these inhibitors can serve as chemical probes of Galf in nematodes and as anthelmintic leads. Together, our data suggest that CeUGM facilitates the biosynthetic incorporation of Galf into nematode glycoconjugates through generation of the glycosyl donor UDP-Galf.
Keywords: glycobiology, UDP-galactopyranose mutase (UGM), UDP-galactofuranose, flavoprotein, nematode, C. elegans, parasitic, helminth, neglected tropical diseases, small molecule inhibition, anthelmintic
INTRODUCTION
Nematodes are a serious threat to agriculture1, livestock2, and human health.3 Plant parasitic nematodes cause estimated crop losses of US$100 billion annually4, and some 4 billion people worldwide are infected or at risk of nematode infection.5 Helminth infection and modulation of the host immune response are areas of intense research.6,7 Glycoconjugates displayed on the surface of helminths are thought to be major contributors to the observed immunomodulation.8-11 Indeed, several nematode glycoconjugates have been structurally characterized12 and shown to alter human immune responses.13-16 Thus, a more thorough understanding of nematode glycoconjugate biosynthesis can lead to new strategies for combating human helminth infections. The value of such studies is mounting as many helminth strains are becoming increasingly resistant to current chemotheraputics.2,17,18
A recently described gene in C. elegans, glf-1, is intriguing as it appears to encode a uridine 5’–diphosphate (UDP) galactopyranose mutase (UGM).19,20 UGM flavoproteins catalyze the production of the glycosyl donor UDP-Galf from UDP-Galp (Figure 1).21 The monosaccharide d-galactofuranose (Galf) is the thermodynamically disfavored, five-membered ring isomer of galactose. Galf residues are absent in mammals, yet they are a prominent component of glycoconjugates from several bacterial, fungal, and protozoan pathogens.22-24 A heterologous lipopolysaccharide (LPS) synthesis assay suggests that the glf-1 gene product mediates UDP-Galf biosynthesis.19 Intriguingly, C. elegans glf-1 deletion mutant experiments indicate that the enzyme is essential.25 Still, Galf residues have yet to be identified in a nematode glycan, and the catalytic activity of CeUGM has not been evaluated.24,26
Figure 1.
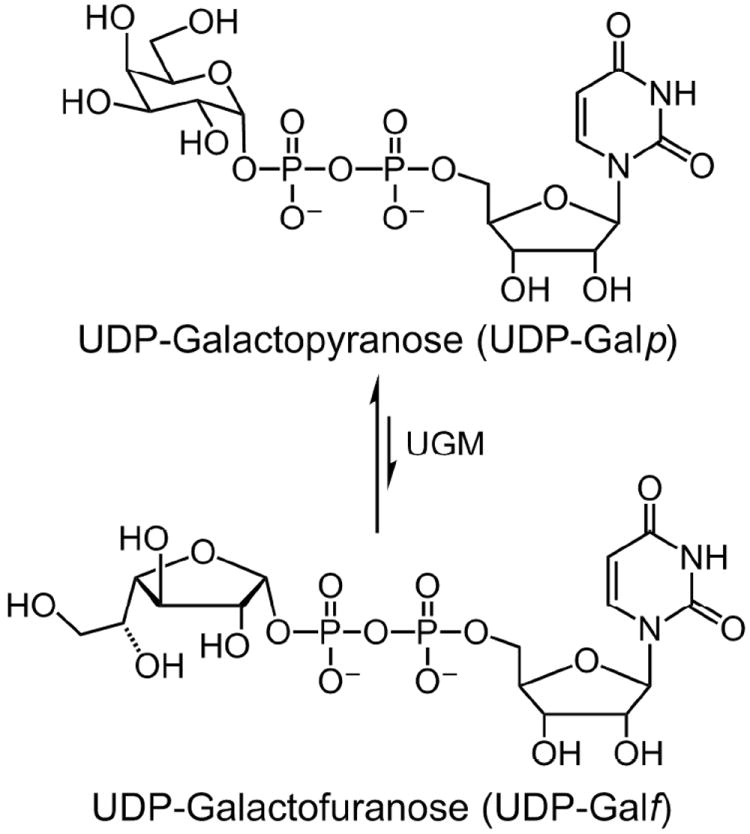
The precursor to Galf-containing glycans is UDP-Galf, which is generated by the enzyme UDP-galactopyranose mutase (UGM or Glf). UGM catalyzes the isomerization of UDP-Galp and UDP-Galf.
Evidence is emerging for the importance of UGM in the assembly of Galf-containing glycoconjugates that contribute to the viability or virulence of prokaryotic and eukaryotic pathogens. Within the mycobacterial cell wall, a polymer composed of Galf residues, termed the galactan, anchors the mycolic acids to the peptidoglycan. Genetic deletion27 or chemical inhibition28 of UGM prevents mycobacterial growth. Eukaryotic UGMs also have been investigated. For example, deletion of the gene encoding the UGM in the opportunistic fungus Aspergillus fumigatus abates virulence and decreases cell wall thickness, thereby enhancing sensitivity to antifungal agents.29 Genetic disruption of the UGM gene30,31 or a putative galactofuranosyl transferase gene (LPG1)32,33 in Leishmania major leads to attenuated infectivity of the parasite and increased susceptibility to human complement and oxidants. These reports suggest that Galf-containing glycans are essential for the viability of several human pathogens.
All organisms that incorporate galactofuranose into their glycans use the building block UDP-Galf, the product of UGM catalysis. We therefore investigated the protein encoded by the C. elegans gene glf-1 (Figure S1). Our results indicate that the glf-1 gene product (CeUGM) catalyzes the interconversion of UDP-Galp and UDP-Galf. The kinetic parameters of CeUGM are in the range of other eukaryotic homologs.34,35 In addition, we provide biochemical data that the flavin cofactor of CeUGM participates in covalent catalysis (Figure 2), a result that is consistent with previous mechanistic studies of UGM homologs from other species.36-38 These data prompted us to devise a homology model for CeUGM, and its validity is supported by the catalytic activities of CeUGM variants. Small molecule inhibitors developed against prokaryotic UGMs also block CeUGM. Thus, we report the first inhibitors of a eukaryotic UGM.28,39
Figure 2.
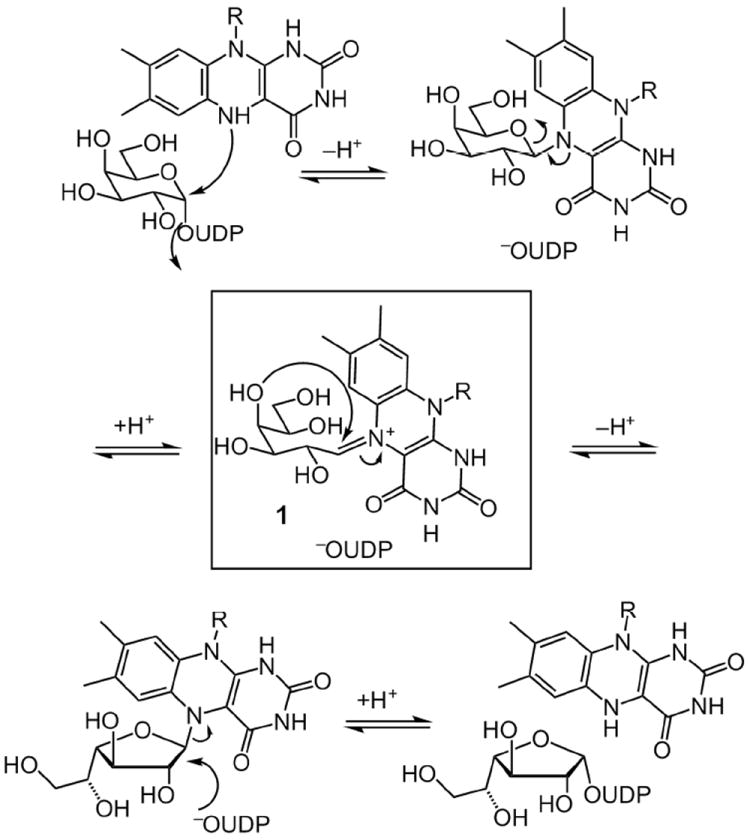
A generalized view of the proposed mechanism of UGM, depicting a covalent flavin intermediate. The arrows shown depict changes in covalent bond formation, but whether the mechanism proceeds via an SN1 or SN2 reaction is not known nor is the protonation state of the flavin cofactor. A key intermediate in this proposal is iminium ion 1.
MATERIALS AND METHODS
Cloning, Expression and Purification of CeUGM
The glf-1 ORF was amplified by PCR using PfuTurbo DNA Polymerase (Stratagene) from a pET3a:glf-1 construct19 generously provided by Professor Stephen Beverley (Washington University in St. Louis School of Medicine) using the forward primer 5’-GACCACAACGGTTTCCCTCTAGAAATAATTTTG-3’ and the reverse primer 5’-GCAGCCGGATCCGCGGCCGCTCCCCGTGGAATAGTTGG-3’. The forward and reverse primers added an XbaI and NotI restriction site, respectively. The purified PCR product and pET-24a vector were digested with XbaI and NotI restriction endonucleases (Promega). The resulting products were purified using the QIAquick PCR Purification Kit (Qiagen). Digested pET-24a vector and glf-1 insert were ligated using T4 DNA Ligase (Promega). The presence of glf-1 with a vector encoded C-terminal hexahistidine (His6) tag was confirmed by DNA sequence analysis. The pET-24a:glf-1 construct was used as template DNA for generating point mutants via site-directed mutagenesis using the QuickChange Kit (Stratagene). The oligonucleotide primer sequences used for generating each mutant can be found in Table S1 of the Supporting Information.
The pET-24a:glf-1 construct was transformed into competent E. coli BL21(DE3) cells (Novagen). Cultures were grown in LB medium supplemented with 50 μg/L kanamycin at 37 °C until OD600 = 0.6 were reached. Cells were cooled to 20 °C and protein overexpression was induced upon the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to 0.1 mM. Cells were grown for 18 hours at 20 °C then harvested by centrifugation (5,000g). Pellets were resuspended in buffer containing 20 mM potassium phosphate (pH 7.4), 25 mM imidazole, 300 mM sodium chloride and 15% glycerol. Cells were disrupted by lysozyme, 0.1% Triton X-100, and sonication (Branson 450 sonifier). Lysates were cleared with centrifugation (22,000g) and filtration (0.45 μM). Cleared lysate was purified with immobilized nickel-ion chromatography using a HisTrap HP column (GE Healthcare) on an AKTA FPLC (Amersham Biosciences). Protein was eluted with a linear gradient of 25 to 500 mM imidazole in potassium phosphate (pH 7.4), 300 mM sodium chloride, and 15% glycerol. Typical yields were 4 mg/L. Fractions of CeUGM, >90% pure, were pooled and dialyzed against a solution of 50 mM potassium phosphate (pH 7.4), 500 mM sodium chloride, 2 mM dithiothreitol (DTT) and 15% glycerol. Protein concentration was determined by absorbance of the flavin cofactor at 450 nm (ε450 = 11,300 M-1cm-1). Protein was aliquoted, vitrified in liquid nitrogen, and stored at -80 °C.
Enzymatic Activity
The activity of CeUGM was measured using an HPLC-based assay with minor modifications.40,41 Reactions were performed in 50 mM potassium phosphate (pH 7.0) and 10 mM fresh sodium dithionite at 22 °C. The reaction was initiated with the rapid addition of chemically synthesized UDP-Galf to the enzyme solution.42 The time that each reaction was allowed to proceed was adjusted to ensure the conversion of UDP-Galf to UDP-Galp was under 10%. Reaction mixtures were quenched with the addition of an equal volume of 1:1 chloroform:methanol. The aqueous portion was removed and analyzed using a CarboPac PA-100 column (Dionex) on a Waters HPLC system. Reaction substrate (e.g. UDP-Galf) and product (e.g. UDP-Galp) were separated via isocratic elution using 200 mM ammonium acetate (pH 8.0), and the eluate was monitored at 262 nm. Initial reaction rates were calculated based on the concentration of substrate and the percentage converted as determined by integration of the HPLC chromatograph. Kinetic parameters were determined by nonlinear regression analysis using Graphpad Prism 4. Quantified error represents the standard error of the mean. The activities of CeUGM variants R187A and R336A were measured at 100 μM UDP-Galf, or ~12 times the KM value for the wild-type enzyme. The activity of the variants was decreased to a level in which the kinetic and binding constants could not be reliably determined. Data were normalized to the activity measured from the wild-type sample.
UV-Visible Spectroscopy
UV-visible absorbance spectra of wild-type, R187A, and R336A CeUGM, or free FAD were taken in a Cary 50 Bio UV-Visible Spectrophotometer (Varian) using a 1 cm cuvette. As a blank, 50 mM potassium phosphate (pH 7.4), 500 mM sodium chloride, and 15% glycerol was used. Spectra were normalized to 450 nm.
Far-UV Circular Dichroism (CD)
Wild-type, R187A, and R336A CeUGM were dialyzed into 20 mM potassium phosphate (pH 7.0) and 15% glycerol. Protein concentrations were determined using the BCA Protein Assay (Pierce). Samples were diluted to roughly 3 μM. Spectra were collected in a Model 202SF Circular Dichroism Spectrophotometer (AVIV Instruments) using a quartz cuvette with a 0.1 cm path length. Data were collected every 1 nm from 197-300 nm using seven shot averages at 22 °C. A baseline scan of a solution of 20 mM potassium phosphate (pH 7.0) and 15% glycerol was subtracted from each sample. Molar ellipticities (ME; deg cm2/decimol) were calculated using the equation ME = MRWθ/(10lC) where MRW is the mean residue weight (114.36 for CeUGM-His6), θ is the ellipticity in degrees, l is the pathlength in centimeters, and C is the concentration is g/mL. Data were plotted using Igor Pro (WaveMetrics).
Clustal W Alignment
Putative UGM proteins from three eukaryotes and three prokaryotes were selected for Clustal W analysis using MegAlign in the Lasergene 8 Suite (DNASTAR). Eukaryotic UGMs include Caenorhabditis elegans (CeUGM), H04M03.4; Leishmania major (LmUGM), AAX09638; Aspergillus fumigatus (AfUGM), CAI38754. Prokaryotic UGMs analyzed consist of Mycobacterium tuberculosis (MtUGM), NP_218326; Klebsiella pneumoniae (KpUGM), KP1_3695; Escherichia coli (EcUGM), ACD37140. Proteins were aligned using the default Clustal W Method parameters on the slow and accurate mode. Residues that were conserved in every species are highlighted in red.
Homology Model
The homology model of CeUGM was generated using the “Automated Mode” of SWISS-MODEL with default parameters.43 Full length protein was used as the query. As a target, UGM from M. tuberculosis (PDB Code: 1V0J) was selected.44 The UGM from M. tuberculosis (PDB Code: 1V0J) was chosen as a template because the 2-aminothiazole inhibitors assayed here were identified, optimized, and characterized against MtUGM. The model, residue 2-473 of CeUGM, was viewed and analyzed using PyMOL.
Intermediate Trapping
A solution of CeUGM, sodium dithionite, and UDP-Galp was allowed to equilibrate for one minute. Solid sodium cyanoborohydride was added to a concentration of 1 M. After thirty minutes, sodium chloride was added to a final concentration of 1 M to facilitate the extraction of the FAD cofactor. Protein was precipitated with heat and the soluble components were collected. The buffer was exchanged using a Sep-Pak C18 cartridge (Waters). To elute the sample, 10 mM ammonium acetate (pH 6.4) in 1:1 water:methanol was added. The eluate was concentrated to dryness using a SpeedVac SC100 (Varian) under vacuum. Samples were analyzed sequentially using a C18 column on a Shimadzu electrospray ionization HPLC-MS with the mass analyzer in both positive and negative ion mode. For mass assignment, positive ion mode was used.
Chemical Inhibition
Compounds 3-5 were synthesized as previously reported.28 For percentage inhibition and IC50 determination, solutions of CeUGM, sodium dithionite, 12 μM UDP-Galf, and compound (at various concentrations from DMSO stocks) were assessed in the aforementioned HPLC-based product formation assay for catalytic activity. Compound stocks were adjusted such that 1% dimethyl sulfoxide (DMSO) was present in each reaction. As a vehicle control, 1% DMSO was used. The IC50 values were calculated using the One Site Competition Model from Graphpad Prism 4. Quantified error represents standard error of the mean. To assess the mode of inhibition of the 2-aminothiazoles described above, reaction kinetics were determined in the presence of varying concentrations of compound 3. Data were fit using nonlinear regression analysis. The double reciprocal plot was generated and fit using Graphpad Prism 4 (GraphPad Software).
RESULTS AND DISCUSSION
Purification of CeUGM and Initial Velocity Kinetic Analysis
Based on sequence analysis and a LPS synthesis assay,19 the C. elegans genome appears to encode a UGM, glf-1. To assess directly the activity of the gene product, we produced it as a His-tagged protein in E. coli. The resulting protein gave rise to a UV-visible absorbance spectrum with maximal absorbance near 380 and 450 nm (Figure S2A), a spectral signature indicative of a flavoprotein. We then evaluated the enzymatic activity of recombinant CeUGM using an HPLC-based assay.40,41 Recombinant protein was incubated with synthetic UDP-Galf as the substrate under reducing conditions,42 because reduction of the flavin cofactor is essential for UGM catalytic activity.45 Recombinant CeUGM catalyzed the isomerization reaction (Figure S3).
Steady-state kinetic parameters were determined from the initial velocities of UDP-Galp production over a range of UDP-Galf concentrations (Figure 3). The KM and kcat values are approximately ten-fold lower for CeUGM than those reported for prokaryotic UGMs (Table 1). The catalytic efficiency (kcat/KM) of CeUGM, however, is similar to that of the UGM from L. major, the nearest homolog of CeUGM characterized to date.34 Prior protein localization studies using a CeUGM∷GFP fusion revealed that the enzyme is produced in C. elegans seam cells.25 Seam cells are involved in nematode surface glycoconjugate biosynthesis. All of the available data, therefore, suggest that the biological function of CeUGM is to generate UDP-Galf. This building block then can serve as a glycosyl donor for yet unidentified C. elegans galactofuranosyl transferases. Thus, a catalytically competent UGM is produced in a multicellular organism.
Figure 3.
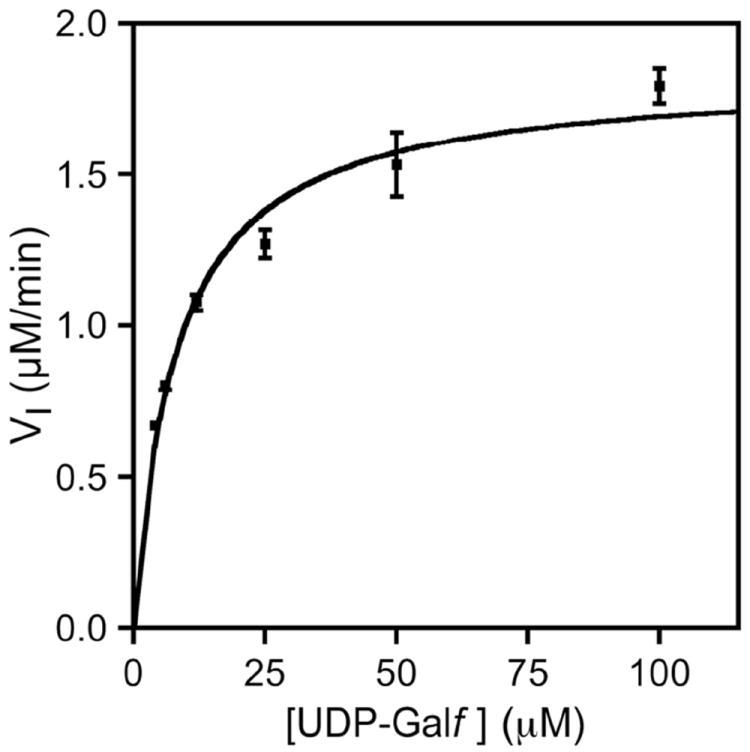
Steady-state kinetic analysis of CeUGM. Initial velocities were calculated by measuring the rate of UDP-Galp formation at increasing concentrations of UDP-Galf. Data were fit to the Michaelis-Menten equation. Error bars represent the standard deviation of measurements performed in triplicate.
Table 1.
Kinetic Parameters of UGM Homologs
| Species | Km (μM) | kcat (s-1) | kcat/Km (104 M-1s-1) |
|---|---|---|---|
| C. elegans | 8 ± 0.8 | 0.61 ± .08 | 7.6 ± 1.2 |
| L. major34 | 87 ± 11 | 5 ± 0.2 | 5.7 ± 0.6 |
| A. fumigatus35 | 110 ± 15 | 72 ± 4 | 65 ± 9 |
| K. pneumoniae57 | 43 ± 6 | 5.5 ± 0.7 | 13 ± 2 |
| E. coli41 | 27 | 22 | 81 |
All constants were determined with UDP-Galf as the substrate.
Covalent Catalysis via FAD
In 2004, our group provided direct evidence that prokaryotic UGM enzymes use their flavin cofactor to facilitate covalent catalysis (Figure 2).36 Since then, additional results have been described that are consistent with such a mechanism, including those using nuclear magnetic resonance (NMR)37, FAD analogs46,47, other spectrophotometric methods36,38, and computational approaches.48 We were interested in whether results supporting this mechanism could be obtained using CeUGM.
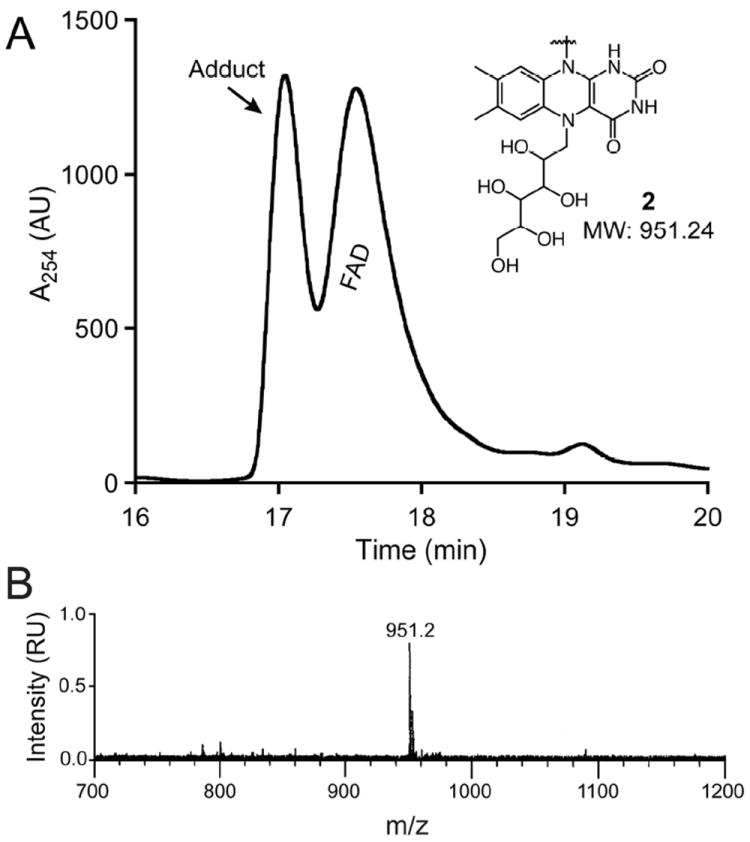
Carbohydrate ring contraction via covalent catalysis by FAD is predicted to proceed through a galactose–N5-flavin iminium ion intermediate (Figure 2). If CeUGM catalyzes isomerization by this nucleophilic mechanism, the addition of a hydride donor should trap the iminium ion to form a covalent adduct, compound 2 (Figure 4).36 Isolation and characterization of this adduct would suggest flavin engages in covalent catalysis in evolutionarily distant eukaryotic and prokaryotic proteins. To trap the iminium ion intermediate, the reaction was initiated with CeUGM, UDP-Galp and sodium dithionite. The resulting reaction mixture was then quenched with the hydride reducing agent sodium cyanoborohydride, the protein was precipitated, and the supernatant was analyzed using high-performance liquid chromatography mass spectrometry (HPLC-MS). In reactions containing sodium cyanoborohydride, a second peak of near equal abundance elutes prior to FAD (Figure 4A). Analysis of the species that gives rise to this peak using electrospray mass spectrometry indicates that the predominant ion corresponds to the covalent adduct (Figure 4B).
Figure 4.
HPLC-MS analysis of the products of trapping the CeUGM-catalyzed reaction of UDP-Galp with sodium cyanoborohydride. (A) A reverse phase HPLC chromatograph obtained from analysis of the soluble reaction products monitored at 254 nm. Inset shows the structure and mass of the predicted galactose–N5-FAD adduct, 2, which is the product of reduction of iminium ion 1 (Figure 2). (B) Mass spectral analysis of product eluting as adduct in the chromatograph above. The predicted mass of unmodified FADox is 785.16 Da.
The proposal that UGM uses its flavin cofactor in covalent catalysis was initially controversial, as it invoked a new catalytic role for flavin.36,49 Our data from CeUGM provides additional support that UGM-catalyzed ring isomerization proceeds via covalent catalysis. A related adduct was recently reported to be trapped from the Trypanosoma cruzi UGM, although the mass and structure correspond to a C4a hydroxylated species.38 Position C4a of the flavin isoalloxazine is the site of molecular oxygen addition during flavin reoxidation, a step not consistent with the proposed non-redox UGM mechanism and likely a byproduct of the isolation procedure. The galactose–N5-flavin covalent adduct we isolated suggests that CeUGM uses a nucleophilic flavin cofactor to catalyze isomerization of UDP-Galp and UDP-Galf (Figure 2). We and others50 posit that the nucleophilic character of the UGM flavin can be targeted by small molecules for potent and selective inhibition of UDP-Galf biosynthesis.
Proposed Structure and Active Site of CeUGM
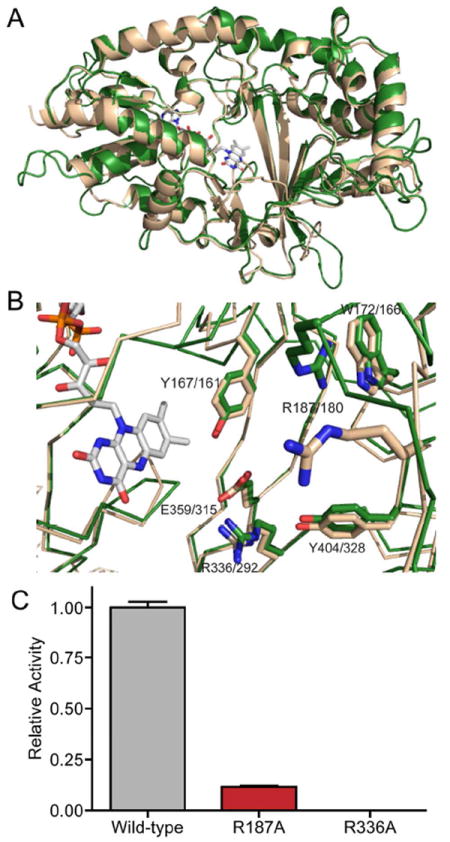
Structural data have been invaluable for understanding substrate binding and the catalytic mechanism of prokaryotic UGM proteins.37,45,51-53 Recently, two independent groups applied protein x-ray crystallography to determine the structure and investigate a potential biological reducing agent of the A. fumigatus UGM.54-56 These studies revealed that despite differences in the prokaryotic and A. fumigatus genes, the overall structure and folds of prokaryotic and eukaryotic UGMs are similar.54,55 Our ability to trap a catalytic intermediate (compound 2) further suggested that the active site of CeUGM is similar to that of its prokaryotic homologs. To this end, we generated a homology model43 of CeUGM using the UGM from M. tuberculosis (MtUGM) (PDB code: 1V0J)44 as a template. The model predicts that the overall architecture of CeUGM and MtUGM is similar. It also indicates that the locations of many residues involved in substrate binding are structurally conserved (Figure 5A&B). The primary sequence alignment and the homology model collectively suggest that arginines 187 and 336 from CeUGM correspond to the two arginine residues essential for substrate binding by prokaryotic proteins.57 To test the accuracy of the homology model, two CeUGM variants were generated in which either Arg187 or Arg336 was substituted with alanine (Table S2). The proper folding and flavin binding of the variants was assessed by circular dichroism and UV-visible absorbance spectroscopy, respectively. The resulting spectra were nearly identical to those from wild-type enzyme (Figure S2).
Figure 5.
Proposed structure and active site of CeUGM. (A) CeUGM homology model (green) generated using SWISS-MODEL superimposed with the structure of M. tuberculosis UGM (PDB Code: 1V0J; wheat). (B) A comparison of residues in the active site. Select conserved residues predicted (CeUGM) or known (M. tuberculosis UGM) involved in substrate binding are highlighted, with C. elegans residue numbers denoted first. (C) Relative activity of wild-type, the R187A variant, and the R336A variant CeUGM at a UDP-Galf concentration of approximately 12-fold above the Km of the wild-type enzyme. Error bars represent the standard deviation of triplicate measurements. Relative activity is derived from normalizing to wild-type enzyme.
The homology model led us to predict that both arginine variants would exhibit significantly diminished catalytic activity. Indeed, under standard conditions, replacement of either arginine residue drastically hampers catalysis. To quantify the activity of these enzyme variants, a high substrate concentration (approximately 12-fold above the Km for the wild-type enzyme) and extended periods of incubation were employed. Because the catalytic rate of wild-type CeUGM was not linear during the extended incubations, we measured bulk catalytic activity by integration of the substrate and product HPLC trace peaks. The total fraction of UDP-Galf converted to UDP-Galp by the R187A variant was roughly one tenth of that catalyzed by the wild-type enzyme (Figure 5C). This diminished, but detectable activity at a high substrate concentration indicates that our model can identify active site residues. The observation that the R187A variant shows activity when substrate concentration is high is consistent with a role for arginine 187 in substrate binding. In contrast, no activity was detected from the R336A variant even under these permissive conditions, suggesting a more critical role in enzyme catalysis. Our data are consistent with results from the A. fumigatus UGM;54,55 the collective experiments suggest a role for these arginine resides in coordinating the negatively charged pyrophosphoryl group of the substrate. Cumulatively, the kinetic and spectrophotometric data we present here supports our model of CeUGM. Given the similarities of the enzyme active sites of our homology model and crystallized prokaryotic enzymes, we explored whether previously identified inhibitors of the prokaryotic homologs might also inhibit CeUGM.
Chemical Inhibition of CeUGM
If the prokaryotic inhibitors are shape selective, small molecules previously shown to inhibit prokaryotic UGMs28,39 would also block CeUGM. Still, differences in active site sequences might make this possibility unlikely. We tested two known inhibitors of prokaryotic UGMs, compounds 3 and 4, with CeUGM (Figure 6A). For comparison, a compound that is inactive against prokaryotic enzymes, compound 5, was also assessed. Using the HPLC assay that monitors the production of UDP-Galp, compounds 3 and 4 were shown to be potent inhibitors of CeUGM. The IC50 values of compounds 3 and 4 were 3.3 μM and 1.8 μM respectively (Figure 6B, Figure S4, and Table 2). When tested at 10 μM, compound 5 had only a modest effect on CeUGM activity. The mode of inhibition was determined by monitoring the kinetics of UDP-Galp formation by CeUGM in the presence of varying concentrations of 3. Analysis of the double reciprocal linear regression plots indicates that the active 2-aminothiazole inhibitors compete with UDP-Galf for the active site (Figure S5).
Figure 6.
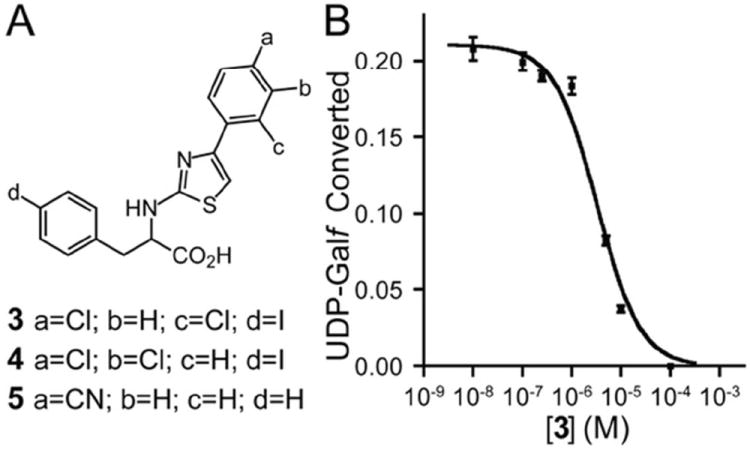
Small molecule inhibition of CeUGM. (A) Structures of 2-aminothiazole inhibitors used in this study. (B) Inhibition of UDP-Galf isomerization by CeUGM with increasing concentrations of compound 3. No UDP-Galp formation could be detected at 100 μM 3. Error bars represent the standard deviation of triplicate measurements.
Table 2.
Inhibition and Binding Constants of 2-Aminothiazoles for CeUGMa
| % Inhibition at 10 μM | IC50 (μM) | Ki (μM) | |
|---|---|---|---|
| 3 | 83 ± 6 | 3.3 ± 1.1 | 1.3 ± 0.4 |
| 4 | 95 ± 1 | 1.8 ± 1.2 | 0.7 ± 0.5 |
| 5 | 16 ± 3 | n/d | n/d |
n/d represents not determined. Ki values were calculated using the Cheng-Prusoff equation.68
The identification of small molecule inhibitors of CeUGM indicates that inhibitors of eukaryotic UGMs can be found. Of the small panel of potential 2-aminothiazole based compounds we tested, the most effective inhibitor, compound 4, had an IC50 of 1.8 μM. Compounds such as these could be used to illuminate the effects of perturbing nematode glycans. Indeed, C. elegans serves as a useful model to investigate helminth biology58-60 — specifically the biosynthesis of glycoconjugates, their structure, and their physiological roles.15,61-63 Glycomic analysis supports this approach as glycoconjugates in C. elegans and parasitic species are similar.26,64,65 Additionally, a putative UGM was recently identified in a protein mass spectrometry study of the parasitic nematode Brugia malayi, the causative agent of human lymphatic filariasis.66 UGM inhibitors could be used in these species and others to illuminate the roles of Galf-containing glycoconjugates in nematodes.
Eukaryotic UGM inhibitors may have other useful roles. It has been previously demonstrated that Galf contributes to virulence29 of the human fungal pathogen A. fumigatus, and remarkably, Galf constitutes ~5% of the dry weight of the fungus.67 The temporal control afforded with chemical genetics would allow researchers to probe the role of Galf during specific stages of infection or host colonization. We therefore anticipate that these compounds will serve as chemical probes and chemotherapeutic leads in other eukaryotes that also utilize Galf.
Conclusions
In summary, the glf-1 gene from C. elegans encodes an enzyme that catalyzes the isomerization of UDP-Galp and UDP-Galf. Our ability to trap a galactose–N5-flavin adduct implicates a nucleophilic flavin in the catalytic mechanism. The structural homology model we generated suggests that CeUGM possesses an active site similar to those described for other UGM proteins, a conclusion supported by results from site-directed mutagenesis. The high level of structural similarity between the predicted active sites from the CeUGM homology model and crystallized prokaryotic UGMs prompted us to test previously described inhibitors of prokaryotic proteins as inhibitors of CeUGM.28 Intriguingly, 2-aminothiazole based compounds are competitive inhibitors of CeUGM, indicating eukaryotic UGMs are amenable to small molecule inhibition. Though our data and others19,25 suggest UDP-Galf biosynthesis by nematodes, identification of Galf in a nematode glycan and the galactofuranosyltransferases responsible for addition of Galf remains critical for fully understanding and capitalizing on nematode glycoconjugate biosynthesis.
Supplementary Material
Acknowledgments
The authors thank Drs. Matthew R. Levengood, Rebecca A. Splain and Adam H. Courtney for advice and feedback on the manuscript. We would also like to acknowledge Dr. Jonathon Hudon for assistance with the collection of LCMS data.
Funding Sources
This research was supported by National Institutes of Health grants R01-AI063596 to L.L.K. D.A.W was supported by a National Science Foundation Graduate Research Fellowship and by the NIH-supported Chemistry–Biology Interface Training Program (T32 GM008505) at the University of Wisconsin–Madison. J.F.M. was supported by a NSF Graduate Research Fellowship and the NIH-supported Molecular Biosciences Training Grant (T32 GM007215). Circular dichroism spectra were obtained at the UW-Madison Biophysics Instrumentation Facility, which is supported by UW-Madison, NSF Grant BIR-9512577, and NIH Grant S10 RR13790. LCMS data was collected in the UW-Madison Chemistry Instrumentation Center Mass Spectrometry Facility on a Shimadzu LCMS-2010A that was purchased with funds from a Keck grant, a Shimadzu grant, and the Chemistry Department.
ABBREVIATIONS
- CD
circular dichroism
- DMSO
dimethyl sulfoxide
- DTT
dithiothreitol
- FAD
flavin adenine dinucleotide
- FMN
flavin mononucleotide
- FPLC
fast protein liquid chromatography
- Galf
galactofuranose
- Galp
galactopyranose
- His6
hexahistidine
- HPLC
high performance liquid chromatography
- IC50
50% inhibitory concentration
- LB
Luria-Bertani
- LPS
lipopolysaccharide
- mP
millipolarization
- MS
mass spectrometry
- OD600
optical density at 600 nm
- PC
phosphorylcholine
- PCR
polymerase chain reaction
- PDB
Protein Data Bank
- UDP
uridine 5’-diphosphate
- UDP-Galf
UDP-d-galactofuranose
- UDP-Galp
UDP-d-galactopyranose
- UGM
uridine 5’-diphosphate galactopyranose mutase
- UV
ultraviolet
Footnotes
Author Contributions
DAW, JFM, and LLK designed the experiments and interpreted the results. DAW and JFM performed the experiments. EMH synthesized and characterized the inhibitors. DAW and LLK wrote the manuscript with suggestions from all authors
Supporting Information. Supporting material and methods; Clustal W sequence alignment of UGM proteins from eukaryotic and prokaryotic species (Figure S1); spectrophotometric analysis of wild-type and CeUGM variants (Figure S2); CeUGM catalyzed isomerization of UDP-Galf and UDP-Galp (Figure S3); inhibition of CeUGM by compound 4 (Figure S4); mechanism of inhibition on CeUGM by compound 3 (Figure S5); oligonucleotide primer sequences used to generate CeUGM point mutants (Table S1); conserved active site residues between CeUGM and MtUGM (Table S2). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Sasser J. Root-knot nematodes - A global menace to crop production. Plant Dis. 1980;64:36–41. [Google Scholar]
- 2.Kaplan R. Drug resistance in nematodes of veterinary importance: a status report. Trends Parasitol. 2004;20:477–481. doi: 10.1016/j.pt.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Hotez PJ, Kamath A. Neglected Tropical Diseases in Sub-Saharan Africa: Review of Their Prevalence, Distribution, and Disease Burden. PLoS Neglected Tropical Diseases. 2009;3:e412. doi: 10.1371/journal.pntd.0000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jasmer DP, Goverse A, Smant G. Parasitic nematode interactions with mammals and plants. Annu Rev Phytopathol. 2003;41:245–270. doi: 10.1146/annurev.phyto.41.052102.104023. [DOI] [PubMed] [Google Scholar]
- 5.Hotez PJ, Molyneux DH, Fenwick A, Kumaresan J, Sachs SE, Sachs JD, Savioli L. Control of neglected tropical diseases. N Engl J Med. 2007;357:1018–1027. doi: 10.1056/NEJMra064142. [DOI] [PubMed] [Google Scholar]
- 6.Maizels RM, Hewitson JP, Grainger JR. Helminth immunoregulation: The role of parasite secreted proteins in modulating host immunity. Mol Biochem Parasit. 2009;167:1–11. doi: 10.1016/j.molbiopara.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harnett W, Harnett MM. Molecular basis of worm-induced immunomodulation. Parasite Immunol. 2006;28:535–543. doi: 10.1111/j.1365-3024.2006.00893.x. [DOI] [PubMed] [Google Scholar]
- 8.Riddle DL. C elegans II. Cold Spring Harbor Laboratory Press; Plainview, N.Y.: 1997. [PubMed] [Google Scholar]
- 9.van Die I, Cummings RD. Glycan gimmickry by parasitic helminths: a strategy for modulating the host immune response? Glycobiology. 2010;20:2–12. doi: 10.1093/glycob/cwp140. [DOI] [PubMed] [Google Scholar]
- 10.Harn DA, McDonald J, Atochina O, Da’dara AA. Modulation of host immune responses by helminth glycans. Immunol Rev. 2009;230:247–257. doi: 10.1111/j.1600-065X.2009.00799.x. [DOI] [PubMed] [Google Scholar]
- 11.Davies KG, Curtis RH. Cuticle surface coat of plant-parasitic nematodes. Annu Rev Phytopathol. 2011;49:135–156. doi: 10.1146/annurev-phyto-121310-111406. [DOI] [PubMed] [Google Scholar]
- 12.Mendonca-Previato L, Todeschini AR, Heise N, Agrellos OA, Dias WB, Previato JO. Chemical structure of major glycoconjugates from parasites. Curr Org Chem. 2008;12:926–939. [Google Scholar]
- 13.van Die I, Cummings RD. Glycans modulate immune responses in helminth infections and allergy. Chem Immunol Allergy. 2006;90:91–112. doi: 10.1159/000088883. [DOI] [PubMed] [Google Scholar]
- 14.Hewitson JP, Grainger JR, Maizels RM. Helminth immunoregulation: the role of parasite secreted proteins in modulating host immunity. Mol Biochem Parasit. 2009;167:1–11. doi: 10.1016/j.molbiopara.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tawill S, Le Goff L, Ali F, Blaxter M, Allen JE. Both free-living and parasitic nematodes induce a characteristic Th2 response that is dependent on the presence of intact glycans. Infect Immun. 2004;72:398–407. doi: 10.1128/IAI.72.1.398-407.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Stijn CM, Meyer S, van den Broek M, Bruijns SC, van Kooyk Y, Geyer R, van Die I. Schistosoma mansoni worm glycolipids induce an inflammatory phenotype in human dendritic cells by cooperation of TLR4 and DC-SIGN. Mol Immunol. 2010;47:1544–1552. doi: 10.1016/j.molimm.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 17.Osei-Atweneboano MY, Eng JKL, Boakye DA, Gyapong JO, Prichard RK. Prevalence and intensity of Onchocerca volvulus infection and efficacy of ivermectin in endemic communities in Ghana: a two-phase epidemiological study. Lancet. 2007;369:2021–2029. doi: 10.1016/S0140-6736(07)60942-8. [DOI] [PubMed] [Google Scholar]
- 18.Kaminsky R, Ducray P, Jung M, Clover R, Rufener L, Bouvier J, Weber SS, Wenger A, Wieland-Berghausen S, Goebel T, Gauvry N, Pautrat F, Skripsky T, Froelich O, Komoin-Oka C, Westlund B, Sluder A, Mäser P. A new class of anthelmintics effective against drug-resistant nematodes. Nature. 2008;452:176–180. doi: 10.1038/nature06722. [DOI] [PubMed] [Google Scholar]
- 19.Beverley SM, Owens KL, Showalter M, Griffith CL, Doering TL, Jones VC, McNeil MR. Eukaryotic UDP-galactopyranose mutase (GLF gene) in microbial and metazoal pathogens. Eukaryotic Cell. 2005;4:1147–1154. doi: 10.1128/EC.4.6.1147-1154.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bakker H, Kleczka B, Gerardy-Schahn R, Routier FH. Identification and partial characterization of two eukaryotic UDP-galactopyranose mutases. Biol Chem. 2005;386:657–661. doi: 10.1515/BC.2005.076. [DOI] [PubMed] [Google Scholar]
- 21.Weston A, Stern RJ, Lee RE, Nassau PM, Monsey D, Martin SL, Scherman MS, Besra GS, Duncan K, McNeil MR. Biosynthetic origin of mycobacterial cell wall galactofuranosyl residues. Tuber Lung Dis. 1997;78:123–131. doi: 10.1016/s0962-8479(98)80005-1. [DOI] [PubMed] [Google Scholar]
- 22.Pedersen LL, Turco SJ. Galactofuranose metabolism: a potential target for antimicrobial chemotherapy. Cell Mol Life Sci. 2003;60:259–266. doi: 10.1007/s000180300021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richards MR, Lowary TL. Chemistry and biology of galactofuranose-containing polysaccharides. ChemBioChem. 2009;10:1920–1938. doi: 10.1002/cbic.200900208. [DOI] [PubMed] [Google Scholar]
- 24.Tefsen B, Ram AF, van Die I, Routier FH. Galactofuranose in eukaryotes: aspects of biosynthesis and functional impact. Glycobiology. 2012;22:456–469. doi: 10.1093/glycob/cwr144. [DOI] [PubMed] [Google Scholar]
- 25.Novelli JF, Chaudhary K, Canovas J, Benner JS, Madinger CL, Kelly P, Hodgkin J, Carlow CKS. Characterization of the Caenorhabditis elegans UDP-galactopyranose mutase homolog glf-1 reveals an essential role for galactofuranose metabolism in nematode surface coat synthesis. Dev Biol. 2009;335:340–355. doi: 10.1016/j.ydbio.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 26.Paschinger K, Gutternigg M, Rendic D, Wilson IB. The N-glycosylation pattern of Caenorhabditis elegans. Carbohydr Res. 2008;343:2041–2049. doi: 10.1016/j.carres.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 27.Pan F, Jackson M, Ma Y, McNeil M. Cell wall core galactofuran synthesis is essential for growth of mycobacteria. J Bacteriol. 2001;183:3991–3998. doi: 10.1128/JB.183.13.3991-3998.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dykhuizen EC, May JF, Tongpenyai A, Kiessling LL. Inhibitors of UDP-galactopyranose mutase thwart mycobacterial growth. J Am Chem Soc. 2008;130:6706–6707. doi: 10.1021/ja8018687. [DOI] [PubMed] [Google Scholar]
- 29.Schmalhorst PS, Krappmann S, Vervecken W, Rohde M, Müller M, Braus GH, Contreras R, Braun A, Bakker H, Routier FH. Contribution of galactofuranose to the virulence of the opportunistic pathogen Aspergillus fumigatus. Eukaryotic Cell. 2008;7:1268–1277. doi: 10.1128/EC.00109-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kleczka B, Lamerz AC, van Zandbergen G, Wenzel A, Gerardy-Schahn R, Wiese M, Routier FH. Targeted gene deletion of Leishmania major UDP-galactopyranose mutase leads to attenuated virulence. J Biol Chem. 2007;282:10498–10505. doi: 10.1074/jbc.M700023200. [DOI] [PubMed] [Google Scholar]
- 31.Madeira da Silva L, Owens KL, Murta SM, Beverley SM. Regulated expression of the Leishmania major surface virulence factor lipophosphoglycan using conditionally destabilized fusion proteins. Proc Natl Acad Sci U S A. 2009;106:7583–7588. doi: 10.1073/pnas.0901698106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spath G, Epstein L, Leader B, Singer S, Avila H, Turco S, Beverley S. Lipophosphoglycan is a virulence factor distinct from related glycoconjugates in the protozoan parasite leishmania major. Proc Natl Acad Sci U S A. 2000;97:9258–9263. doi: 10.1073/pnas.160257897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spath G, Garraway L, Turco S, Beverley S. The role(s) of lipophosphoglycan (LPG) in the establishment of Leishmania major infections in mammalian hosts. Proc Natl Acad Sci U S A. 2003;100:9536–9541. doi: 10.1073/pnas.1530604100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oppenheimer M, Valenciano AL, Sobrado P. Isolation and characterization of functional Leishmania major virulence factor UDP-galactopyranose mutase. Biochem Biophys Res Commun. 2011;407:552–556. doi: 10.1016/j.bbrc.2011.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oppenheimer M, Poulin MB, Lowary TL, Helm RF, Sobrado P. Characterization of recombinant UDP-galactopyranose mutase from Aspergillus fumigatus. Arch Biochem Biophys. 2010;502:31–38. doi: 10.1016/j.abb.2010.06.035. [DOI] [PubMed] [Google Scholar]
- 36.Soltero-Higgin M, Carlson EE, Gruber TD, Kiessling LL. A unique catalytic mechanism for UDP-galactopyranose mutase. Nat Struct Mol Biol. 2004;11:539–543. doi: 10.1038/nsmb772. [DOI] [PubMed] [Google Scholar]
- 37.Gruber TD, Westler WM, Kiessling LL, Forest KT. X-ray crystallography reveals a reduced substrate complex of UDP-galactopyranose mutase poised for covalent catalysis by flavin. Biochemistry. 2009;48:9171–9173. doi: 10.1021/bi901437v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oppenheimer M, Valenciano AL, Kizjakina K, Qi J, Sobrado P. Chemical Mechanism of UDP-Galactopyranose Mutase from Trypanosoma cruzi: A Potential Drug Target against Chagas’ Disease. PLoS One. 2012;7:e32918. doi: 10.1371/journal.pone.0032918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soltero-Higgin M, Carlson EE, Phillips JH, Kiessling LL. Identification of inhibitors for UDP-galactopyranose mutase. J Am Chem Soc. 2004;126:10532–10533. doi: 10.1021/ja048017v. [DOI] [PubMed] [Google Scholar]
- 40.Lee R, Monsey D, Weston A, Duncan K, Rithner C, McNeil M. Enzymatic synthesis of UDP-galactofuranose and an assay for UDP-galactopyranose mutase based on high-performance liquid chromatography. Anal Biochem. 1996;242:1–7. doi: 10.1006/abio.1996.0419. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Q, Liu H. Studies of UDP-galactopyranose mutase from Escherichia coli: An unusual role of reduced FAD in its catalysis. J Am Chem Soc. 2000;122:9065–9070. [Google Scholar]
- 42.Marlow AL, Kiessling LL. Improved chemical synthesis of UDP-galactofuranose. Org Lett. 2001;3:2517–2519. doi: 10.1021/ol016170d. [DOI] [PubMed] [Google Scholar]
- 43.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 44.Beis K, Srikannathasan V, Liu H, Fullerton SWB, Bamford VA, Sanders DAR, Whitfield C, McNeil MR, Naismith JH. Crystal structures of Mycobacteria tuberculosis and Klebsiella pneumoniae UDP-galactopyranose mutase in the oxidised state and Klebsiella pneumoniae UDP-galactopyranose mutase in the (active) reduced state. J Mol Biol. 2005;348:971–982. doi: 10.1016/j.jmb.2005.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanders DA, Staines AG, McMahon SA, McNeil MR, Whitfield C, Naismith JH. UDP-galactopyranose mutase has a novel structure and mechanism. Nat Struct Mol Biol. 2001;8:858–863. doi: 10.1038/nsb1001-858. [DOI] [PubMed] [Google Scholar]
- 46.Huang Z, Zhang Q, Liu H. Reconstitution of UDP-galactopyranose mutase with 1-deaza-FAD and 5-deaza-FAD: analysis and mechanistic implications. Bioorg Chem. 2003;31:494–502. doi: 10.1016/j.bioorg.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 47.Sun HG, Ruszczycky MW, Chang WC, Thibodeaux CJ, Liu HW. Nucleophilic participation of reduced flavin coenzyme in mechanism of UDP-galactopyranose mutase. J Biol Chem. 2012;287:4602–4608. doi: 10.1074/jbc.M111.312538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang WJ, Gauld JW. Tautomerization in the UDP-Galactopyranose Mutase Mechanism: A DFT-Cluster and QM/MM Investigation. J Phys Chem B. 2012;116:14040–14050. doi: 10.1021/jp310952c. [DOI] [PubMed] [Google Scholar]
- 49.Ghisla S, Massey V. Mechanisms of flavoprotein-catalyzed reactions. Eur J Biochem. 1989;181:1–17. doi: 10.1111/j.1432-1033.1989.tb14688.x. [DOI] [PubMed] [Google Scholar]
- 50.Ansiaux C, N’go I, Vincent SP. Reversible and Efficient Inhibition of UDP-Galactopyranose Mutase by Electrophilic, Constrained and Unsaturated UDP-Galactitol Analogues. Chem --Eur J. 2012;18:14860–14866. doi: 10.1002/chem.201202302. [DOI] [PubMed] [Google Scholar]
- 51.Partha SK, van Straaten KE, Sanders DAR. Structural basis of substrate binding to UDP-galactopyranose mutase: crystal structures in the reduced and oxidized state complexed with UDP-galactopyranose and UDP. J Mol Biol. 2009;394:864–877. doi: 10.1016/j.jmb.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 52.Yuan Y, Wen X, Sanders DA, Pinto BM. Exploring the mechanism of binding of UDP-galactopyranose to UDP-galactopyranose mutase by STD-NMR spectroscopy and molecular modeling. Biochemistry. 2005;44:14080–14089. doi: 10.1021/bi0513406. [DOI] [PubMed] [Google Scholar]
- 53.Gruber TD, Borrok MJ, Westler WM, Forest KT, Kiessling LL. Ligand binding and substrate discrimination by UDP-galactopyranose mutase. J Mol Biol. 2009;391:327–340. doi: 10.1016/j.jmb.2009.05.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Straaten KE, Routier FH, Sanders DA. Structural insight into the unique substrate binding mechanism and flavin redox state of UDP-galactopyranose mutase from Aspergillus fumigatus. J Biol Chem. 2012;287:10780–10790. doi: 10.1074/jbc.M111.322974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dhatwalia R, Singh H, Oppenheimer M, Karr DB, Nix JC, Sobrado P, Tanner JJ. Crystal Structures and Small-angle X-ray Scattering Analysis of UDP-galactopyranose Mutase from the Pathogenic Fungus Aspergillus fumigatus. J Biol Chem. 2012;287:9041–9051. doi: 10.1074/jbc.M111.327536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dhatwalia R, Singh H, Solano LM, Oppenheimer M, Robinson RM, Ellerbrock JF, Sobrado P, Tanner JJ. Identification of the NAD(P)H Binding Site of Eukaryotic UDP-Galactopyranose Mutase. J Am Chem Soc. 2012;134:18132–18138. doi: 10.1021/ja308188z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chad JM, Sarathy KP, Gruber TD, Addala E, Kiessling LL, Sanders DAR. Site-directed mutagenesis of UDP-Galactopyranose mutase reveals a critical role for the active-site, conserved arginine residues. Biochemistry. 2007;46:6723–6732. doi: 10.1021/bi7002795. [DOI] [PubMed] [Google Scholar]
- 58.Burglin T, Lobos E, Blaxter M. Caenorhabditis elegans as a model for parasitic nematodes. Int J Parasitol. 1998;28:395–411. doi: 10.1016/s0020-7519(97)00208-7. [DOI] [PubMed] [Google Scholar]
- 59.Politz SM, Philipp M. Caenorhabditis elegans as a model for parasitic nematodes: a focus on the cuticle. Parasitol Today. 1992;8:6–12. doi: 10.1016/0169-4758(92)90302-i. [DOI] [PubMed] [Google Scholar]
- 60.Jones AK, Buckingham SD, Sattelle DB. Chemistry-to-gene screens in Caenorhabditis elegans. Nat Rev Drug Discovery. 2005;4:321–330. doi: 10.1038/nrd1692. [DOI] [PubMed] [Google Scholar]
- 61.Gerdt S, Lochnit G, Dennis RD, Geyer R. Isolation and structural analysis of three neutral glycosphingolipids from a mixed population of Caenorhabditis elegans (Nematoda:Rhabditida) Glycobiology. 1997;7:265–275. doi: 10.1093/glycob/7.2.265. [DOI] [PubMed] [Google Scholar]
- 62.Griffitts JS, Whitacre JL, Stevens DE, Aroian RV. Bt toxin resistance from loss of a putative carbohydrate-modifying enzyme. Science. 2001;293:860–864. doi: 10.1126/science.1062441. [DOI] [PubMed] [Google Scholar]
- 63.Griffitts JS, Haslam SM, Yang TL, Garczynski SF, Mulloy B, Morris H, Cremer PS, Dell A, Adang MJ, Aroian RV. Glycolipids as receptors for Bacillus thuringiensis crystal toxin. Science. 2005;307:922–925. doi: 10.1126/science.1104444. [DOI] [PubMed] [Google Scholar]
- 64.Cipollo JF, Costello CE, Hirschberg CB. The fine structure of Caenorhabditis elegans N-glycans. J Biol Chem. 2002;277:49143–49157. doi: 10.1074/jbc.M208020200. [DOI] [PubMed] [Google Scholar]
- 65.Cipollo JF, Awad AM, Costello CE, Hirschberg CB. N-Glycans of Caenorhabditis elegans are specific to developmental stages. J Biol Chem. 2005;280:26063–26072. doi: 10.1074/jbc.M503828200. [DOI] [PubMed] [Google Scholar]
- 66.Bennuru S, Meng ZJ, Ribeiro JMC, Semnani RT, Ghedin E, Chan K, Lucas DA, Veenstra TD, Nutman TB. Stage-specific proteomic expression patterns of the human filarial parasite Brugia malayi and its endosymbiont Wolbachia. Proc Natl Acad Sci U S A. 2011;108:9649–9654. doi: 10.1073/pnas.1011481108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Latge JP. Galactofuranose containing molecules in Aspergillus fumigatus. Med Mycol. 2009;47:S104–S109. doi: 10.1080/13693780802258832. [DOI] [PubMed] [Google Scholar]
- 68.Cheng Y, Prusoff W. Relationship between the inhibition constant (KI) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


