Background: S-Palmitoylation is an important reversible modification that involves the action of an acyl-protein thioesterase (APT).
Results: We identified an active serine hydrolase (TgASH1) specifically targeted by human APT1 inhibitors in Toxoplasma gondii.
Conclusion: TgASH1 is dispensable and cannot be solely responsible for S-depalmitoylation in Apicomplexa.
Significance: β-Lactone-based APT1 inhibitors hit multiple targets in T. gondii and severely compromise parasite survival.
Keywords: Enzyme Inhibitors, Hydrolases, Parasitology, Posttranslational Modification, Protein Palmitoylation, Acyl-protein Thioesterase, Plasmodium falciparum, Serine Hydrolase, Toxoplasma gondii, Inhibitor
Abstract
In eukaryotic organisms, cysteine palmitoylation is an important reversible modification that impacts protein targeting, folding, stability, and interactions with partners. Evidence suggests that protein palmitoylation contributes to key biological processes in Apicomplexa with the recent palmitome of the malaria parasite Plasmodium falciparum reporting over 400 substrates that are modified with palmitate by a broad range of protein S-acyl transferases. Dynamic palmitoylation cycles require the action of an acyl-protein thioesterase (APT) that cleaves palmitate from substrates and conveys reversibility to this posttranslational modification. In this work, we identified candidates for APT activity in Toxoplasma gondii. Treatment of parasites with low micromolar concentrations of β-lactone- or triazole urea-based inhibitors that target human APT1 showed varied detrimental effects at multiple steps of the parasite lytic cycle. The use of an activity-based probe in combination with these inhibitors revealed the existence of several serine hydrolases that are targeted by APT1 inhibitors. The active serine hydrolase, TgASH1, identified as the homologue closest to human APT1 and APT2, was characterized further. Biochemical analysis of TgASH1 indicated that this enzyme cleaves substrates with a specificity similar to APTs, and homology modeling points toward an APT-like enzyme. TgASH1 is dispensable for parasite survival, which indicates that the severe effects observed with the β-lactone inhibitors are caused by the inhibition of non-TgASH1 targets. Other ASH candidates for APT activity were functionally characterized, and one of them was found to be resistant to gene disruption due to the potential essential nature of the protein.
Introduction
Toxoplasma gondii and Plasmodium falciparum are protozoan parasites of the phylum Apicomplexa that have adopted an obligate intracellular lifestyle. Many species of this phylum are notorious because of their capacity to cause severe disease in humans and animals and lead to major economic burdens in developing nations. The processes implicated in establishment of infection and parasite development are tightly controlled, and reversible posttranslational modifications such as phosphorylation play a central role in cell signaling. Palmitoylation is another posttranslational modification that has received considerable attention in recent years as it notably promotes membrane association of a vast array of soluble proteins in many eukaryotic species (1–4) while also being important for function of integral membrane proteins (5, 6). Palmitoylation is an important mode of control for the accurate targeting and steady state localization of many proteins to subcellular compartments as well as for regulation of protein-protein interactions and intracellular signaling (4, 5, 7).
There is significant evidence to suggest that apicomplexan parasites capitalize on palmitoylation as a regulatory mechanism for protein function. First, 18 and 12 genes coding for putative DHHC motif-containing protein S-acyltransferases are present in T. gondii and P. falciparum genomes, respectively (8, 9), providing for the addition of palmitate to proteins in diverse subcellular compartments. Furthermore, a large number of substrates have been predicted based on bioinformatics searches and identified in a palmitome analysis of P. falciparum (9, 10). Additionally, palmitoylation of several apicomplexan proteins has been experimentally proven to be critical for correct protein localization and function (11–15). The fast reaction kinetics of palmitoylation (16) and the reversibility of this modification contribute to its effectiveness as a regulatory system for protein dynamics. For the palmitoylation cycle to act as a rapid, specific control mechanism, depalmitoylation of protein substrates must be controlled by a set of enzymes.
Protein depalmitoylation has been less extensively researched than palmitoylation; nevertheless, several enzymes with depalmitoylation activity have been identified in mammalian cells. Palmitoyl-protein thioesterases 1 and 2 (PPT1 and PPT2)4 are localized within lysosomes and critically contribute to the degradation of lipid-modified proteins (17–19). Disruption of either gene causes severe lysosomal storage disorders and cell death due to the inability to degrade fatty acid-modified material (20–23). Unlike PPT1 and PPT2, the acyl-protein thioesterases 1 and 2 (APT1 and APT2) are cytoplasmic enzymes that have been implicated in dynamic palmitoylation cycles (5, 24). As a member of the superfamily of α/β-hydrolases, specifically the serine hydrolase class of enzymes, APT1 was initially classified as a lysophospholipase (25) until the favored substrates were identified as thioacylated proteins (26). This enzyme is potentially responsible for the depalmitoylation of a number of proteins that go through a palmitoylation cycle, although only a few candidates for this activity have been identified. Confirmed targets of APT1 include intracellular messengers such as H-Ras and N-Ras (27–29), G-protein α subunits (26), calcium-activated potassium channels (6), and endothelial NOS (30). APT2 on the other hand has to date only been shown to be active on palmitoylated GAP43 and H-Ras in Chinese hamster ovary (CHO)-K1 and HeLa cells (24), indicating non-redundant roles for the two enzymes. The separated function of APT1 and APT2 is likely to be due in part to their differential expression between cell types (24, 31). To study the importance of depalmitoylation in various cell types, inhibitors were designed to block APT1 and APT2 function with the objective to specifically perturb palmitoylation dynamics and reduce the function of palmitoylated proteins in vitro and in vivo (32, 33). The dynamic palmitoylation/depalmitoylation cycle has been demonstrated for a number of substrates in mammalian cells including H- and N-Ras (16, 34) and most recently Rac1 (35).
In Apicomplexa, the anchoring of the gliding-associated protein, GAP45, in the pellicle is dependent on its myristoylation and palmitoylation and in turn is critical for parasite motility, invasion, and egress of some species from infected cells (12). These parasites also possess a family of calcium-dependent protein kinases that control important functions such as motility and invasion (36). Some of the calcium-dependent protein kinases possess consensus motifs for N-myristoylation and palmitoylation, and in the case of P. falciparum, calcium-dependent protein kinase 1 acylation is critical for the localization of the kinase (37), although its role in regulation of activity has not been investigated. In P. falciparum, calpain contributes to parasite growth and shuttles between the nucleolus and perinuclear membranes in a palmitoylation-dependent fashion (11).
In this work, we report severe effects caused by inhibitors of the hAPTs on the lytic cycle of T. gondii and P. falciparum. By using these inhibitors in combination with a serine hydrolase probe, we detected several enzymes potentially responsible for APT activity. A gene closely homologous to hAPT1 encodes one target of these inhibitors. This active serine hydrolase 1 (TgASH1) is conserved across the coccidian subgroup of the Apicomplexa and is dispensable for T. gondii survival in tissue culture. Unless protein depalmitoylation is dispensable, another enzyme must be at least in part responsible for depalmitoylation activity in T. gondii. Here we investigate the properties of TgASH1 and three distantly related putative serine hydrolases that may contribute to APT activity and compensate for the loss of TgASH1.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
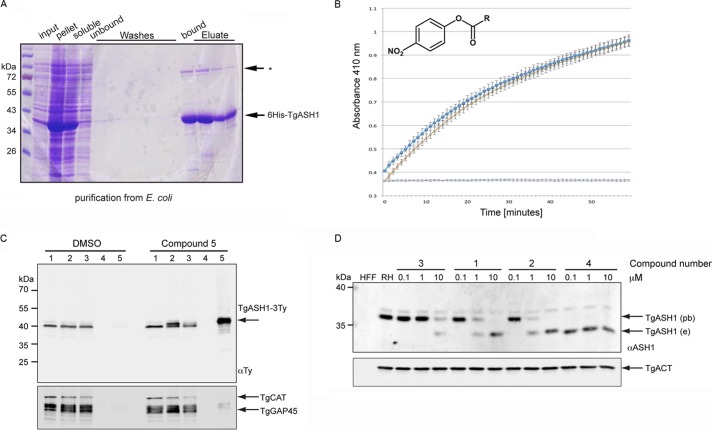
Restriction enzymes were purchased from New England Biolabs. PCR polymerases ExTaq (TaKaRa), GoTaq (Promega), and Phusion (Thermo Scientific) were used according to the manufacturers' instructions. Primers for amplification of DNA and cloning strategies are listed in supplemental Table 1. The following antibodies (Abs) were used: mouse BB2 α-Ty tag (used previously (38)), rabbit α-GAP45 (39), rabbit α-catalase (40), mouse α-TgISP1 (14), rabbit α-SAG1, and mouse α-GRA3 provided by J. F. Dubremetz. The polyclonal α-TgASH1 and α-TgARO antibodies were generated by immunization of rabbits (Eurogentec) with recombinant penta-His-tagged ASH1 that was produced in Escherichia coli and purified on nickel beads. Secondary goat α-rabbit-HRP and goat α-mouse-HRP antibodies (Molecular Probes, G21234 and G21040, respectively) were used to detect proteins by Western blot. Secondary antibodies from Molecular Probes (Alexa Fluor) were used for indirect immunofluorescence assay (IFA). Streptavidin-HRP was used at a dilution of 1:5000. T. gondii genomic DNA was prepared using the Wizard genomic DNA purification kit (Promega). The development and use of fluorophosphonate-rhodamine (FP-Rh) has been described (32, 41).
Preparation of Inhibitors
Two groups of serine hydrolase inhibitors were tested: β-lactones RM448 (1), RM449 (2), and FD242 (3) and triazole urea AA401 (4). Both classes of inhibitors covalently and irreversibly modify the catalytic serine residue in the active site of serine hydrolases. Compound 5, RM496, is a modified version of RM449.
Synthesis of Compound 4
A mixture of diphenyl(1H-1,2,3-triazol-4-yl)methanol (90 mg; 0.36 mmol), piperidine-1-carbonyl chloride (44 mg; 0.3 mmol), and 4-dimethylaminopyridine (catalyst) in 5:1 tetrahydrofuran/triethylamine was stirred for 10 h at 60 °C. The solvents were removed to yield the desired triazole urea (85 mg; 0.24 mmol; 78%) as a mixture of N2- and N1-carbamoylated regioisomers. The regioisomers were separated by silica gel chromatography (3:1 hexanes/ethyl acetate → ethyl acetate), and the major (N2-) isomer was characterized and used for subsequent biological experiments.1H NMR (400 MHz, CDCl3): δ = 7.58 (s, 1H), 7.38–7.27 (m, 10H), 3.67 (m, 4H), 1.68 (m, 6H). High resolution mass spectrometry (MS) (m/z): calculated for C21H23N4O2 [M + H]+: 363.1816; found: 363.1819.
Amino Acid Sequence Alignments and Molecular Phylogeny Analyses
Sequences used in this study have all been obtained from the EuPathDB database (42) or GenBankTM database. Multiple alignments of full-length APT1 protein sequences were computed using MUSCLE (43). Before PhyML analysis, all gap-containing sites were removed from the alignment. The final alignment used for the phylogenetic analysis is presented in supplemental Fig. 2. Phylogenetic analysis was performed by maximum likelihood (44) using PhyML (45) with approximate likelihood-ratio test (using minimum Shimodaira-Hasegawa (SH)-like and χ2-based) (46) and WAG (47) selected as the amino acid substitution model as recommended by ProtTest 3.2 (48) analysis under Akaike Information Criterion (AIC) and corrected AIC (AICc). All the maximum likelihood analysis was performed on DIVEIN (49), and phylogenetic trees were visualized with TreeDyn (50). Bootstrap values obtained from these two different phylogeny analyses are indicated on the tree presented in Fig. 3 and alignment in supplemental Fig. 2. Only values up to 95 were considered as significant, allowing formation of clusters.
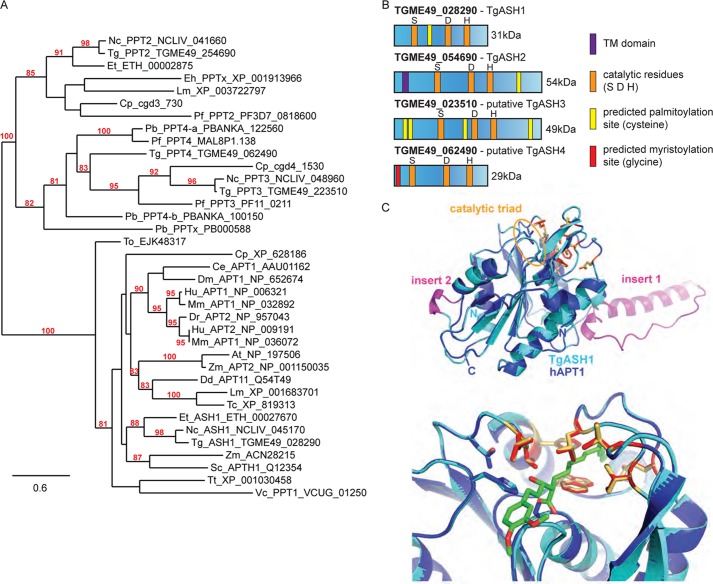
FIGURE 3.
TgASH1, the T. gondii gene most closely related to APT1, is specifically targeted by APT1 inhibitors. A, phylogenetic tree of APT1 and related proteins based on PhyML distance analysis. Only nodes supported by a bootstrap value ≥80 are indicated, and values ≥95 were considered as significant. Apicomplexan genes accession numbers are given according to the EuPathDB web site (42) and GenBank. Multiple sequence alignment used to compute the phylogenetic trees is presented in supplemental. Fig. 2A. At, Arabidopsis thaliana; Ce, Caenorhabditis elegans; Cp, Cryptosporidium parvum; Dd, Dictyostelium discoideum; Dm, Drosophila melanogaster; Dr, Danio rerio; Eh, Entamoeba histolytica; Et, Eimeria tenella; Hu, human; Lm, Leishmania major; Mm, Mus musculus; Nc, Neospora caninum; Pb, Plasmodium berghei; Pf, P. falciparum; Tc, Trypanosoma cruzi; To, Thalassiosira oceanica; Tt, Tetrahymena thermophila; Vc, Vavraia culicis; Zm, Zea mays. B, scheme representing TgASH1 and TgASH2 and the putative TgASH3 and TgASH4. The catalytic residues and predicted myristoylation and palmitoylation sites are indicated as well as the potential transmembrane domain (TM). C, homology modeling of TgASH1 using human APT1 as a template demonstrates similarity of TgASH1 to hAPT1. The two inserts (shown in magenta) most likely do not interfere with the catalytic site (upper panel). Furthermore, the putative substrate binding sites (indicated by a modeled β-lactone inhibitor molecule covalently bound to the active site serine shown in green) are conserved (lower panel; residues shown in red and yellow for hAPT1 and TgASH1, respectively).
Parasite and Cell Culture
T. gondii tachyzoites (RH hxgprt-KO or ku80-KO strains (51)) were grown in human foreskin fibroblast (HFF) monolayers in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 5% fetal calf serum (FCS), 2 mm glutamine, and 25 μg/ml gentamicin (37 °C; 5% CO2). P. falciparum strain NF54 was cultured in human erythrocytes in RPMI 1640-based medium supplemented with 0.5% AlbuMAX® I (Invitrogen) as described (52). Cultures were maintained in an atmosphere of 3% O2, 4% CO2, and 93% N2 in humidified modular chambers at 37 °C. L6 rat skeletal myoblasts cells were cultivated in RPMI 1640 medium with 10% FCS and 1.7 μm l-glutamine (37 °C; 5% CO2).
DNA Construct Cloning
cDNA for TgASH1-ASH4 was amplified from an RH hxgprt-KO cDNA pool and cloned into final vectors using primers and enzymes stated in supplemental Table 1. Genomic DNA fragments for generation of vectors to tag endogenous loci and create TgASH knock-out constructs were amplified from an RH hxgprt-KO preparation.
Parasite Transfection and Selection of Transformants
Parasite transfection and selection of transformants was performed as described previously (53). ASH1-Ty:wild type RH hxgprt-KO parasites were transfected with 80 μg of pT8ASH1Ty-HX vector (linearized with BamHI). ASH1-3Ty:ku80-KO parasites were transfected with 40 μg of pTgASH1-3Ty_HX vector (linearized with AvrII). ash1-KO:ku80-KO parasites were transfected with 50 μg of pTgASH1-KO vector (linearized with PacI). Plasmids and enzymes for generation of ASH2–4 strains can be found in supplemental Table 1. Stable transgenic parasites were selected in the presence of mycophenolic acid and xanthine and cloned under selection by limiting dilution in 96-well plates.
IFAs and Confocal Microscopy
Intracellular parasites grown in HFF monolayers seeded on coverslips were fixed with 4% paraformaldehyde (PAF) or 4% PAF, 0.005% glutaraldehyde (PAF/glutaraldehyde) in PBS depending on the antigen to be labeled. Coverslips were washed once in PBS, 0.1 m glycine before permeabilization of cells with 0.2% Triton X-100 in PBS (PBS-TX) for 20 min at room temperature. Cells were blocked with PBS-TX containing 2% BSA (PBS-TXB) for a further 20 min at room temperature before labeling with primary Abs diluted in PBS-TXB for 60 min. Coverslips were washed in PBS-TX and then stained with secondary Abs for 45 min at room temperature. Cells were washed in PBS-TX, stained for 10 min with DAPI diluted in PBS, washed in PBS, and then mounted on slides using Fluoromount G. Confocal images were taken with a Leica laser-scanning confocal microscope (TCS-NT DM/IRB and SP2) using a 100× Plan-Apochromat objective.
Plaque Assay
HFF monolayers were infected with tachyzoites and incubated for 7–10 days before being fixed with PAF and stained with Giemsa.
FP-Rhodamine Identification of Active Serine Hydrolases
Parasite or mouse brain lysate was incubated for 30 min at room temperature with 1 μm FP-Rh. The reaction was quenched by the addition of SDS loading buffer. The samples were electrophoresed on a 15 × 15-cm 12% SDS-polyacrylamide gel without preboiling. The gel was scanned using an Ettan DIGE fluorescence imager.
FP-Rh/Inhibitor Competition Assay
Parasite or mouse brain lysate was pretreated with inhibitors or DMSO as a control at various concentrations for 30 min at 37 °C. FP-Rh was added at 1 μm final concentration for a further 30 min at room temperature before the reaction was quenched by the addition of SDS loading buffer. Bands that disappeared upon treatment with the inhibitors represent targets of inhibitors.
Intracellular Growth Assay
HFF monolayers were inoculated with freshly egressed parasites and incubated for 24 h before fixation with PAF/glutaraldehyde. IFAs were completed using α-GAP45 to detect individual parasites and α-GRA3 to distinguish vacuoles. Parasites per vacuole were counted for at least 100 vacuoles per replicate. Results are the mean of three independent experiments. p < 0.05 is deemed significant.
Induced Egress Assay
Freshly egressed parasites were added to HFF monolayers and incubated for 30–32 h. Wells were washed with medium without serum before a 3 μm final concentration of calcium ionophore (A23187; Streptomyces chartreusis; Calbiochem) or an equivalent volume of DMSO was added to medium without serum for 5–10 min. Cells were fixed with PAF/glutaraldehyde, and IFA was completed using α-SAG1 to stain parasites and α-GRA3 to identify vacuoles. p < 0.05 is deemed significant.
Invasion Assay
The invasion assay was performed as described previously (54). The ratio of intracellular parasites to extracellular parasites was determined based on differential staining for SAG1 (parasite plasma membrane, extracellular) and GAP45 (inner membrane complex, intracellular).
Subcellular Fractionations
Freshly egressed tachyzoites were washed in PBS and resuspended in PBS; PBS plus 0.1, 0.5, or 1% Triton X-100; 1 m NaCl; or 0.1 m Na2CO3 (pH 11.5). Parasites were incubated for 5 min on ice, and complete lysis was ensured through sonication on ice (three times; 10 × 5 s at 30%). Pellet and soluble fractions were separated by centrifugation at 4 °C for 1 h at 14,000 rpm before detection of proteins by Western blot.
Assessment of Inhibitor Effect on Parasites and Cells
Intracellular growth and egress for T. gondii were assessed as described above except that parasites were incubated in the presence of various concentrations of inhibitors or DMSO during the intracellular growth assay (after 30 min or 12 h allowed for invasion) and for 30 min prior to inducing egress for the egress assay. To assess T. gondii invasion, a homogenous suspension of extracellular parasites was equally split into aliquots and treated for 30 min at 37 °C with either the inhibitors or DMSO. After pretreatment, parasite suspensions were diluted, added to HFF monolayers, and allowed 15 min for invasion before wells were washed. Parasites were incubated for 24 h, fixed, and stained for GAP45 and GRA3. Vacuoles per field were counted, and values represent the mean of three independent experiments.
The P. falciparum drug-sensitive NF54 strain was cultivated in a variation of the medium described previously consisting of RPMI 1640 supplemented with 0.5% AlbuMAX II, 25 mm HEPES, 25 mm NaHCO3 (pH 7.3), 0.36 mm hypoxanthine, and 100 μg/ml neomycin (52, 55). Human erythrocytes served as host cells, and cultures were maintained in an atmosphere of 3% O2, 4% CO2, and 93% N2 in humidified modular chambers at 37 °C. Compounds were dissolved in DMSO (10 mm), diluted in hypoxanthine-free culture medium, and titrated in duplicates over a 64-fold range in 96-well plates. Infected erythrocytes (1.25% final hematocrit and 0.3% final parasitemia) were added to the wells. After varying hours of incubation (8, 24, and 48 h as shown in Table 1), 0.5 μCi/well [3H] hypoxanthine was added, and plates were incubated for an additional 24 h (in the case of 8-h compound exposure, 16 h of [3H]hypoxanthine incubation time was used). Parasites were harvested onto glass fiber filters, and radioactivity was counted using a Betaplate liquid scintillation counter (Wallac, Zurich, Switzerland). Compounds were measured in duplicate, and results were recorded and expressed as a percentage of the untreated controls. Fifty percent inhibitory concentrations (IC50) were estimated by linear interpolation (56).
TABLE 1.
IC50 of β-lactone inhibitor compounds with P. falciparum NF54 strain
A cytotoxicity test was performed on L6 rat myoblast cells. All results are expressed as ng/ml. —, no data.
| Compound | Compounds added at T0 and reading taken at 24, 48, and 72 h |
Fresh compound added every 12 h; 72-ha IC50 NF54 | Compound added at T0; 72-h IC50 cytotoxicity | ||
|---|---|---|---|---|---|
| 24-h IC50 NF54 | 48-h IC50 NF54 | 72-h IC50 NF54 | |||
| 3 | 22,853 | 8,724 | 17,652 | 11,616 | 3,555 |
| 1 | 8,992 | 1,435 | 3,231 | 1,017 | 4,820 |
| 2 | 8,140 | 1,577 | 2,259 | 1,746 | 3,175 |
| Artesunate | 1.9 | 1.6 | 1.7 | 1.2 | — |
| Chloroquine | 5.1 | 4.1 | 4.3 | — | — |
| Podophyllotoxin | — | — | — | — | 6 |
a One reading taken at 72 h.
Rat skeletal myoblasts (L6 cells; 100 μl of 2 × 104 cells/ml) were added to each well (RPMI 1640 medium with 10% FCS and 1.7 μm l-glutamine) of a 96-well microtiter plate. Cells were allowed to attach overnight and incubated at 37 °C under a 5% CO2 atmosphere for 24 h. Then serial drug dilutions were prepared; and plates were incubated as described for 70 h. Afterward, 10 μl of AlamarBlue (12.5 mg of resazurin dissolved in 100 ml of distilled water) was added to each well, and incubation continued for a further 1–3 h. Then the plates were analyzed with a Spectramax Gemini XS microplate fluorometer (Molecular Devices, Sunnyvale, CA) using an excitation wavelength of 530 nm and an emission wavelength of 590 nm. Data were analyzed using the software Softmax Pro (Molecular Devices). A decrease of fluorescence (=inhibition) was expressed as the percentage of the fluorescence of control cultures and plotted against the drug concentrations. From the sigmoidal inhibition curves, the IC50 values were calculated.
Inhibitor Target Pulldown
T. gondii parasites were treated with 25 μm compound 5 or an equivalent volume of DMSO for 20 min at room temperature. Parasites were washed with PBS to remove excess probe and then resuspended at a concentration of ∼1 mg of total parasite protein in 1 ml of PBS containing 0.2% Triton X-100. The suspension was sonicated on ice to ensure complete lysis of the cells. The click chemistry reaction for coupling a biotin moiety to the inhibitor-target complex has been described previously (57) and was adapted for T. gondii as follows. After incubation with compound 5, proteins were precipitated for 1 h at −20 °C in ice-cold acetone before washing three times in methanol with sonication (3 × 10 s at 30%) to resuspend. Proteins were resuspended in 1 ml of PBS, 0.2% SDS using sonication, and the protein solution was incubated with streptavidin beads for 1 h at room temperature. Beads were washed extensively using PBS, 0.2% SDS; 6 m urea; and PBS. SDS loading buffer was then added to the beads before boiling. Eluate was electrophoresed on a 12% SDS-polyacrylamide gel.
Enzymatic Analysis
TgASH1 was purified from E. coli BL-21 strain in lysis buffer containing 1% Triton X-100. Elution of protein from nickel-nitrilotriacetic acid beads was performed using 250 mm imidazole before overnight dialysis. Recombinant TgASH1 was investigated for esterase activity in a biochemical assay where p-nitrophenol palmitate (R = C15H31) was used as substrate. The conditions for monitoring the reaction progress by measuring the increase of absorbance (λabs = 410 nm) were optimized using 100 nm enzyme and 1 mm substrate. The assay was developed in a 96-well format on a Tecan plate reader and performed in buffer containing 10 mm phosphate buffer (pH 7.54), 150 mm NaCl, and 0.5% CHAPS. The formation of p-nitrophenolate (PNP) was recorded (λabs = 410 nm; bandwidth = 9 nm) over 60 min at 60-s intervals at 37 °C. During the incubation and in between measurements, the reaction mixture was shaken for 5 s with 60-s intervals. 100 nm recombinant human APT1 was purified as described (57) and measured on the same plate under identical conditions. Both experiments were repeated in triplicate.
RESULTS
Description of Inhibitors
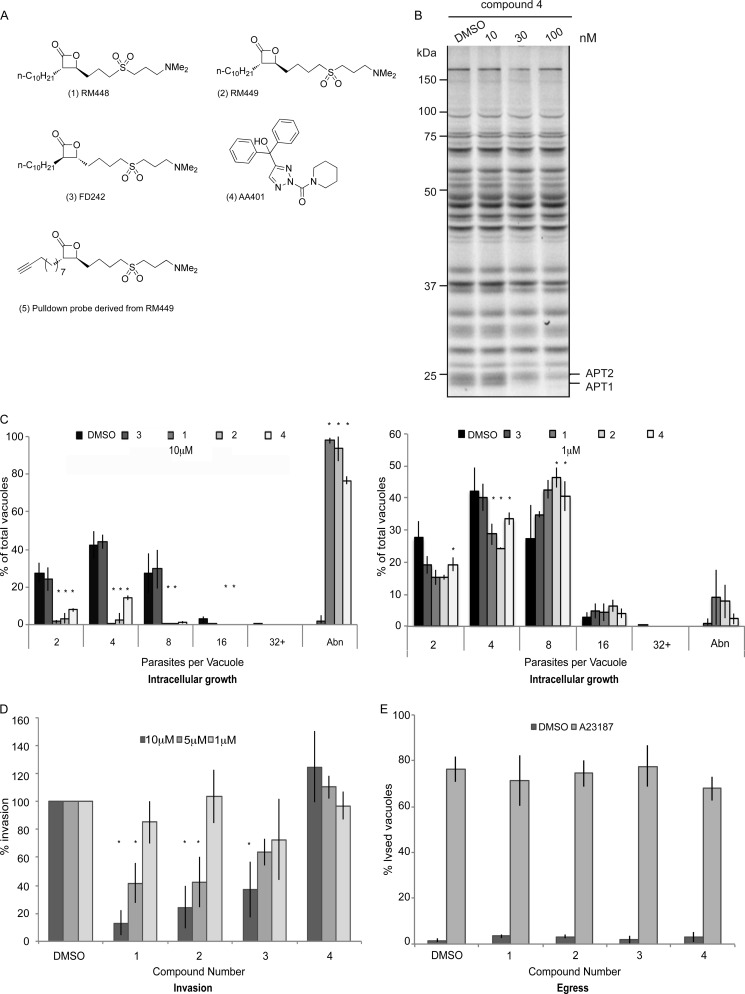
Two structurally distinct classes of serine hydrolase inhibitors were exploited to investigate the importance of a dynamic palmitoylation cycle in T. gondii and P. falciparum (Fig. 1A): β-lactones RM448 (1), RM449 (2), and FD242 (3) and triazole urea AA401 (4). RM496 (5) is a modified version of compound 2 that is click chemistry-competent.
FIGURE 1.
Human APT1 inhibitors disrupt the lytic cycle of apicomplexan parasites. A, structures of compounds used in this study. Compounds 1–3 and 5 are β-lactones, whereas compound 4 is a triazole urea. Compound 3 is the enantiomer of compound 2 and has reduced hAPT1 binding efficiency in biochemical assays where recombinant hAPT1 is the substrate (47, 48). All compounds target the active site serine of hAPT1 to inhibit the enzyme. Inhibition by the β-lactones is temporary as the inhibitor hydrolyzes the enzyme, whereas inhibition by the triazole urea is more stable. B, gel of activity-based pulldown using FP-Rh ABPP probe (59) shows 4 inhibition of APT activity after 30 min of treatment. C, left, intracellular growth of T. gondii treated with 10 μm hAPT1 inhibitors over 24 h showed a severe defect. Parasites were allowed to invade host cells and then were treated for the duration of the experiment. Right, intracellular growth of T. gondii treated with 1 μm hAPT1 inhibitors over 24 h showed little growth defect. Parasites were allowed to invade host cells and were then treated for the duration of the experiment. Samples were then fixed and stained with α-GAP45 to allow quantification of the number of parasites per vacuole. Those with morphological defects that could not be counted were classified as abnormal (Abn). D, extracellular parasites treated with inhibitor compounds or with DMSO were tested for invasion efficiency following 30 min of treatment with inhibitors. Infected cultures were washed and incubated for 24 h before fixation and counting of the number of vacuoles per field. Treatment with 10 and 5 μm β-lactone inhibitors led to a significant reduction in invasion. In contrast, compound 4 caused a slight increase in invasion at higher concentration, although this was not statistically significant. E, induced egress of tachyzoites from host cells by treatment with the calcium ionophore A23187 was not affected by pretreatment for 30 min with 10 μm inhibitors compared with DMSO. Lysed versus intact vacuoles were quantified. The results presented correspond to three independent biological experiments, and values are represented as mean ± S.D. (error bars).
β-Lactones 1-3 inactivate hAPTs by reversible covalent modification of the serine residue in the enzyme active site. The enzyme is acylated by the inhibitor through nucleophilic opening of the β-lactone electrophile followed by regeneration of the active enzyme by hydrolysis of the acylated enzyme-inhibitor complex. Thus, β-lactones 1-3 are selective small molecule inhibitors that behave as slowly converted, apparently competitive substrates and thus temporarily inhibit the APT enzyme activity. Compound 1 has been shown to inhibit hAPT1/2 selectively compared with other intracellular phospholipases such as phospholipases A1, A2, Cβ, and D. For the β-lactones, compounds 1 and 2 are highly potent inhibitors of hAPTs, whereas compound 3 is the enantiomer of compound 2, which shows an order of magnitude lower potency in a biochemical assay using recombinant hAPT1 (57, 58).
The specificity of compound 4 for mammalian APT1 and APT2 was assessed using a small molecule probe, FP-Rh, developed for identification of active serine hydrolases (41, 59). The APT inhibitors covalently bind to the active site serine of their targets and therefore block the catalytic pocket, precluding binding of the FP-Rh probe, which leads to disappearance of the associated band when samples are analyzed by SDS-PAGE. Compound 4 was tested in an FP-Rh/inhibitor competition assay using mouse brain lysate as the source of serine hydrolases and shown to selectively block APT1 and APT2 at 30 nm (Fig. 1B) as reported previously for other similar compounds (32).
Both classes of compounds (β-lactones and triazole urea) covalently modify the catalytic active site serine residue to irreversibly deactivate the target enzymes. We reasoned that if dynamic palmitoylation cycles were essential for the parasites then treatment with the inhibitors would affect parasite growth and/or propagation. The effects of the inhibitors at various stages of the lytic cycle were measured using standard assays for parasite fitness and are described below.
APT1 Inhibitors Block the Lytic Cycle of T. gondii
The growth of T. gondii tachyzoites treated with 10 μm compounds 1, 2, and 4 was severely impaired compared with DMSO-treated controls and parasites treated with compound 3 (Fig. 1C, left panel). Greater than 75% of parasites treated with 10 μm compounds 1, 2, and 4 for 24 h exhibited a severe morphological defect that hampered T. gondii replication; most were only able to divide once (data not shown). At 1 μm, only 10% of parasites had an abnormal morphology upon treatment with compounds 1 and 2, and this dropped further to 1% upon treatment with 1 μm compound 4 (Fig. 1C, right panel).
The capacity to invade host cells was greatly reduced in parasites treated with compounds 1, 2, and 3. Parasites pretreated for 30 min with 10 μm compounds 1 and 2 showed an 80% reduction in invasion efficiency compared with DMSO-treated controls, whereas parasites treated with compound 4 appeared to invade slightly more efficiently than the control, although the difference was not statistically significant (Fig. 1D). Parasites invaded normally when pretreated with all compounds at a concentration of 1 μm. The fact that compound 3 affects invasion, although it binds APT1 much less efficiently, indicates that the targets of this inhibitor are critical for the invasion process, and even low level inhibition has fatal consequences for the parasites. No impairment in egress from infected host cells was detectable when parasites were treated for 30 min with any of the compounds at 10 μm before stimulation of egress with calcium ionophore (A23187) when compared with DMSO-pretreated parasites (Fig. 1E).
To investigate the effective dose of the β-lactone inhibitors against P. falciparum, an in vitro test was performed and indicated that IC50 values of compounds 1 and 2 in P. falciparum were essentially equivalent to their IC50 values in L6 rat myoblast cells, indicating that they have high levels of toxicity (Table 1). Interestingly, the compounds also seem to exert only a static effect on parasite growth, which is consistent with their temporary inhibitory activity as shown previously (58).
Identification of T. gondii Serine Hydrolases as Targets of Serine Hydrolase Inhibitors
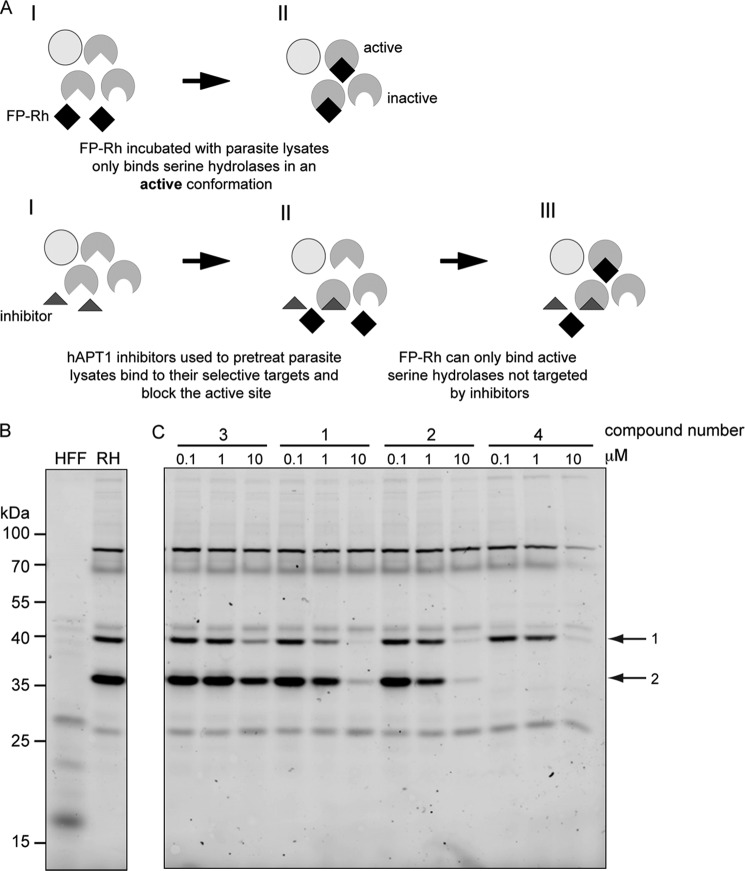
We used the small molecule probe FP-Rh to visualize the repertoire of active serine hydrolases in T. gondii (Fig. 2A, top panel). Over 20 bands of varying intensities were detected (Fig. 2B); however, three prominent bands were apparent likely due to a higher level of expression and/or an increased affinity for the probe. In a second step, the APT inhibitors were used in combination with the probe to identify their serine hydrolase targets in parasite lysates (Fig. 2A, bottom panel). Two active serine hydrolases were clearly visualized as targets in the presence of 10 μm compounds 1, 2, and 4 (arrows; labeled 1 and 2). Moreover, compound 4 depleted the activity of one of these completely at 0.1 μm (Fig. 2C). Treatment with compound 3 (the enantiomer of compound 2, which has a lower affinity for recombinant hAPT1) caused a slight decrease in the levels of the same bands as the other compounds; however, none were greatly reduced, indicating that this compound has a significantly lower affinity for these enzymes. We cannot exclude that other enzymes, below the detection limit of this experiment, might be affected upon treatment with the inhibitors.
FIGURE 2.
Identification of active serine hydrolases and targets of hAPT1 inhibitors. A, scheme depicting the labeling of cell lysates with FP-Rh, a serine hydrolase probe (upper panel). FP-Rh was incubated with lysates for 30 min, and then active enzymes were visualized by SDS-PAGE and fluorescence imaging of the gel. Pretreatment of the lysate with the inhibitor compounds for 30 min before labeling with FP-Rh leads to competition for binding to the enzyme (lower panel). B, incubation of wild type RH T. gondii parasite lysate with FP-Rh labels ∼20 bands at various intensities and sizes, all of which represent active serine hydrolases. C, lysates pretreated with the inhibitor compounds and then labeled with FP-Rh present a different profile compared with untreated lysates, indicating that FP-Rh probe binding to active serine hydrolases is blocked by the interaction between the inhibitor compound and a target protein. The two main bands sensitive to inhibitors are marked with arrows and labeled 1 and 2. Compound numbers corresponding to those in Fig. 1A are indicated above the concentrations used. RH, wild type parasite line.
TgASH1 Is Encoded by a Close Homologue of hAPT1
As a means to determine the identity of the inhibitor targets, a modified version of the FP probe containing biotin in place of rhodamine was used to pull down probe targets on streptavidin beads. Proteins contained in the bands that correspond to inhibitor targets identified in the FP-Rh/inhibitor competition experiments above were then analyzed by MS. Three proteins that contain α/β-hydrolase domains were identified (Table 2 and supplemental Fig. 1). Furthermore, a search of the T. gondii genome database for α/β-hydrolase domain-containing proteins revealed 20 genes that could act as serine hydrolases and potentially exhibit an APT activity (Table 3).
TABLE 2.
Inhibitor compound targets identified by mass spectrometry that contain an annotated α/β-hydrolase (Hyd) domain
ER, endoplasmic reticulum.
| Size | Domains | SDH | Conservation across Apicomplexa | Information from orthologous genes | |
|---|---|---|---|---|---|
| kDa | |||||
| TGME49_028290 | 34 | α/β-Hyd, phospholipase/carboxylesterase | Yes | Coccidia | Acyl-protein thioesterase 1/2 |
| TGME49_054690 | 54 | α/β-Hyd, phospholipase/carboxylesterase | Yes | Neospora, Eimeria, P. falciparum, Cryptosporidium, Theileria | BEM46-like protein, ER localization, putative role in signal transduction and cell polarity |
| TGME49_018540 | 45 | α/β-Hyd | Yes | Coccidia | S15 X-Pro dipeptidyl-peptidase |
TABLE 3.
Repertoire of genes coding for α/β-hydrolase domain-containing proteins in T. gondii that could potentially function as APTs
T. gondii ME49 has 57 significant domains in 54 proteins. 20 of these are lipases/esterases/thioesterases that could potentially function as APTs. Bold text indicates genes investigated in this study.
| Sequence ID | E value | Region | Family | Annotation |
|---|---|---|---|---|
| gb|TGME49_003300 | 4.81e−06 | 1183–1278 | Hypothetical protein VC1974 | Hypothetical |
| gb|TGME49_018540 | 4.26e−35 | 32–213, 273–346 | Fungal lipases | Putative S15 peptidase |
| gb|TGME49_023510 | 5.63e−28 | 194–364, 407–439 | Haloalkane dehalogenase | Hypothetical (TgASH3) |
| gb|TGME49_026390 | 3.7e−13 | 370–430, 509–558 | Haloperoxidase | Hypothetical |
| 0.0186 | 615–635, 669–744 | Carboxylesterase/thioesterase 1 | ||
| 0.0231 | 505–531, 620–680 | Hypothetical protein TT1662 | ||
| gb|TGME49_026470 | 2.09e−07 | 1562–1632 | Thioesterase domain of polypeptide, polyketide, and fatty-acid synthases | Hypothetical |
| 3.39e−05 | 2583–2677 | Carbon-carbon bond hydrolase | ||
| gb|TGME49_028290 | 1.35e−40 | 23–162, 202–282 | Carboxylesterase/thioesterase 1 | Phospholipase/carboxylesterase (TgASH1) |
| gb|TGME49_034350 | 6e−19 | 148–220, 325–372 | Bacterial lipase | Hypothetical |
| 1.96e−15 | 325–362, 607–674 | Proline iminopeptidase-like | ||
| gb|TGME49_038200 | 3.64e−28 | 504–661 | Carboxylesterase | α/β-Hydrolase domain-containing protein |
| gb|TGME49_040830 | 4.6e−28 | 10–153, 225–335 | Pancreatic lipase, N-terminal domain | α/β fold family protein |
| gb|TGME49_049300 | 1.05e−07 | 6–229 | Carbon-carbon bond hydrolase | Hypothetical |
| gb|TGME49_050360 | 3.6e−35 | 272–434, 488–536, 666–702 | Carboxylesterase | α/β-Hydrolase domain-containing protein |
| gb|TGME49_054010 | 2.41e−13 | 273–401, 583–708 | Carboxylesterase | Serine carboxypeptidase |
| gb|TGME49_054690 | 7.58e−46 | 40–324 | Carboxylesterase | Phospholipase (TgASH2) |
| gb|TGME49_054710 | 2.27e−16 | 1099–1284 | Bacterial lipase | Serine esterase DUF676 |
| gb|TGME49_056840 | 2.14e−15 | 378–449, 482–657 | Carbon-carbon bond hydrolase | Hypothetical |
| gb|TGME49_062490 | 1.23e−34 | 14–237 | Haloperoxidase | α/β-Hydrolase putative (TgASH4) |
| gb|TGME49_071800 | 3.59e−11 | 187–314 | Bacterial lipase | Serine esterase DUF676 |
| gb|TGME49_077820 | 6.25e−10 | 1087–1129, 1247–1369 | Carboxylesterase/thioesterase 1 | Hypothetical |
| gb|TGME49_089880 | 4.8e−06 | 1006–1114 | Bacterial lipase | Hypothetical |
| gb|TGME49_104500 | 1.4e−06 | 54–206 | Carbon-carbon bond hydrolase | Hypothetical |
Two candidates identified in the MS experiment (TGME49_028290 and TGME49_054690) and two further top candidates from the list in Table 3 (TGME49_023510 and TGME49_062490) were subjected to phylogenetic analysis along with their putative homologues in Apicomplexa, human APT1 and APT2 and their homologues in diverse eukaryotic species (Fig. 3, A and B, and supplemental Fig. 2A).
TGME49_028290, which we have called ASH1 (GenBank accession number KC473455), identified by MS is the only one of the four genes that clusters with hAPT1 in the phylogenetic tree (Fig. 3A) and exhibits 33% identity and 46% similarity at the amino acid level (supplemental Fig. 2B). ASH1 is found only in the coccidian subgroup of the Apicomplexan phylum and hence is absent in the malaria parasite P. falciparum. However, putative APT homologues are well represented in other lower eukaryotic species, being present in kinetoplastids, amoeba, and ciliates (supplemental Fig. 2).
The amino acid sequence alignment of ASH1 and hAPT1 (supplemental Fig. 2B) highlights a highly charged region of low complexity that sits between the catalytic serine, aspartate, and histidine residues of ASH1. This insertion is also present in the ASH1 sequence of different T. gondii strains as well as in other Coccidia (ToxoDB). Homology modeling of ASH1 using hAPT1 (Protein Data Bank code 1fj2) as a template suggests that this large insertion can exhibit different conformations or even be disordered (one model with a helix/loop conformation (“insert 1”) is shown in Fig. 3C, top). Similar regions in proteins with a known crystal structure tend to be flexible or disordered. However, according to the alignments/models, the insert is far away from the active site and leaves the catalytic residues undisturbed. Furthermore, the residues forming the putative substrate binding site of hAPT1 are completely conserved (Fig. 3C, bottom), thus strongly emphasizing the similarity to hAPT1 and suggesting a similar substrate specificity.
Characterization of TgASH1
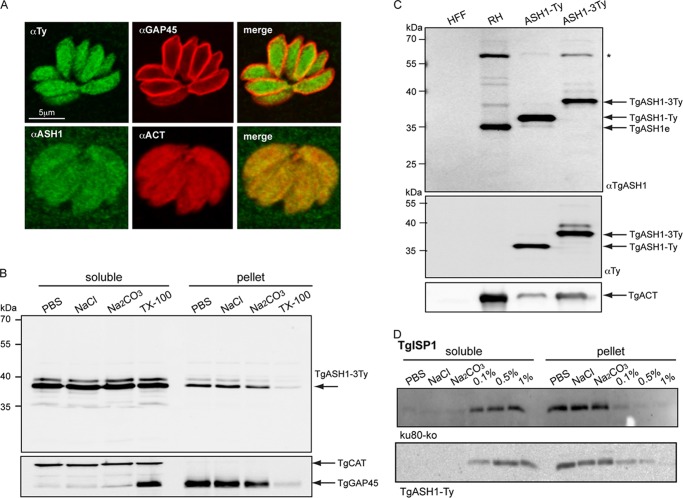
Based on epitope tagging of ASH1 at the endogenous locus in T. gondii (knock-in strain ASH1-3Ty) as well as expression of a second epitope-tagged copy (strain ASH1-Ty), TgASH1 was found to be a cytosolic enzyme as reported for hAPT1 (Fig. 4A). Subcellular fractionation experiments indicated that TgASH1 was readily soluble in PBS (Fig. 4B). Rabbit polyclonal antibodies were raised against recombinant TgASH1 purified from E. coli to confirm the size and localization of the endogenous protein by Western blot and IFA, respectively (Fig. 4, A and C). The knock-in strain expressed ASH1-3Ty at a level comparable with that of the untagged endogenous gene but at a substantially lower level than the second copy ASH1-Ty controlled by the tubulin promoter (Fig. 4C). Overexpression of TgASH1 did not lead to altered membrane association of palmitoylated proteins such as TgISP1 (Fig. 4D).
FIGURE 4.
APT1 homologue present in coccidian species only. A, indirect immunofluorescence microscopy of T. gondii tachyzoites. The upper panel shows endogenous epitope Ty-tagged TgASH1 in green (TgASH1-3Ty) and parasite pellicle shown with α-TgGAP45 in red. The lower panel shows endogenous TgASH1 detected by specific α-ASH1 antibodies in green and co-localized with cytoplasmic α-actin in red. B, subcellular fractionation of parasites indicates that TgASH1 is readily soluble in PBS, in high salt (1 m NaCl), and at high pH (0.1 m Na2CO3, pH 11.5). Complete solubilization occurs in the presence of 1% Triton X-100. C, Western blot analysis of an epitope-tagged second copy of TgASH1 (TgASH1-Ty) and epitope-tagged endogenous TgASH1 (knock-in TgASH1-3Ty) strains indicate a higher level of expression compared with endogenous TgASH1 (TgASH1e) in wild type strain (RH). The size shifts observed are due to the addition of one or three Ty tags. * indicates an unspecific band identified by the α-ASH1 Ab. D, subcellular fractionation of ku80-KO and TgASH1-Ty parasites in different salt and detergent conditions indicates that overexpression of TgASH1 does not impact the solubility of the palmitoylated protein TgISP1.
TgASH1 Exhibits an Esterase Activity
To determine whether TgASH1 exhibits an esterase activity, the enzymatic activity and substrate specificity were examined in vitro using recombinant protein produced and purified from E. coli (Fig. 5A). The isolated protein was assayed for esterase activity using p-nitrophenyl palmitate as substrate and detecting the released PNP absorbance at 410 nm (Fig. 5B). The results indicated that TgASH1 is as active as hAPT1, confirming the prediction of the homology modeling and suggesting that TgASH1 is a bona fide acyl-protein thioesterase.
FIGURE 5.
TgASH1 has thioesterase activity and interacts specifically with APT1 inhibitors. A, purification of recombinant His6-tagged full-length TgASH1 from E. coli. Lysis was performed in 1% Triton X-100 with sonication. * indicates an unspecific band identified by the α-ASH1 Ab. B, TgASH1 hydrolyzes PNP-palmitate with activity comparable with that of human APT1. The detection is based on the absorbance measurement associated with release of PNP (measured at 410 nm). Dark blue squares represent TgASH1, light blue crosses with an orange line represent hAPT1. The light blue line is the control without protein. C, pulldown of inhibitor targets with compound 5, a clickable compound modified to accept biotin, indicates a specific interaction between the inhibitor compound and TgASH1 as shown by Western blot. TgASH1-3Ty parasites were incubated with compound 5 to allow it to bind to its protein targets. Following precipitation of proteins and performing the click reaction to add the biotin to the compound 5-target complex, inhibitor targets were enriched on streptavidin beads and analyzed by Western blot. TgGAP45 and catalase (TgCAT) were used as negative controls, and a second sample that was treated with DMSO only instead of compound 5 was run. Numbers 1–5 represent the different fractions: 1, input; 2, precipitated proteins; 3, postclick reaction sample; 4, wash; 5, eluate. D, FP-Rh binds specifically to TgASH1, increasing its size and causing it to migrate more slowly on SDS-PAGE. Western blot analysis demonstrates the shift of endogenous TgASH1 (TgASH1e) bound to the probe (TgASH1pb). In the presence of an increasing concentration of inhibitors that interfere with binding to the probe, migration reverts back to the size of unbound TgASH1-3Ty. Actin (TgACT) was used as loading control. RH, wild type parasite line.
To confirm that TgASH1 is a target of the inhibitors, enrichment experiments were performed using “clickable” inhibitors that allow attachment of biotin and a subsequent biotin-streptavidin pulldown to purify the inhibitor targets (57). TgASH1 was enriched in the pulldown eluate only in the presence of compound 5 but not DMSO, whereas unrelated and abundant proteins such as GAP45 and catalase were not detectable in this fraction (Fig. 5C, lane 5). Furthermore, specific competition for binding to TgASH1 was evident. Western blots performed on parasite lysate treated with different concentrations of inhibitors before incubation with FP-Rh showed a transition from probe-bound (low inhibitor concentration) to probe-unbound TgASH1 (high inhibitor concentration) (Fig. 5D). Taken together, these results indicate that an active APT1 homologue exists in T. gondii, and it is a specific target of both the β-lactone and triazole urea hAPT1 inhibitors.
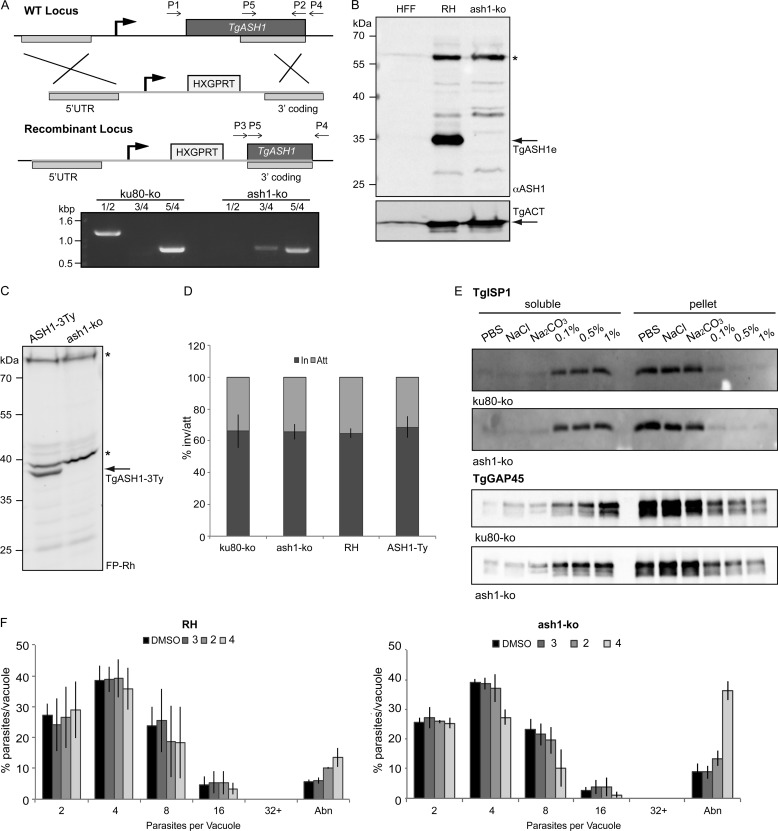
TgASH1 Gene Is Dispensable for Tachyzoite Survival
Functional analysis of TgASH1 was performed by double homologous recombination to replace the endogenous genomic locus with a drug resistance gene (Fig. 6A). The loss of TgASH1 gene was assessed by genomic PCR (Fig. 6A), and the absence of the protein was confirmed by Western blot, which validated the specificity of the rabbit polyclonal antibodies produced in this study (Fig. 6B). Additionally, loss of protein in the ash1-KO strain coincided with a loss of detectable activity with the FP-Rh probe (Fig. 6C). The phenotypic consequence of the absence of TgASH1 was first examined by the size of the plaque of lysis caused by the propagation of a single parasite over 7 days (plaque assay), which recapitulates all the steps of the lytic cycle. The ash1-KO mutant showed no alteration in plaque formation compared with wild type parasites (data not shown). Moreover, a more detailed investigation revealed no defect in induced egress in the presence of the calcium ionophore A23187, intracellular growth (data not shown), or invasion capacity as determined using the attachment/invasion assay that determines the ratio of intracellular to extracellular parasites (Fig. 6D).
FIGURE 6.
TgASH1 is dispensable for T. gondii tachyzoite survival. A, scheme of TgASH1 knock-out strategy with primers used for analysis of genomic locus indicated. Deletion of TgASH1 by homologous recombination was confirmed by genomic PCR using primers described in A and supplemental Table 1. Primer sets are as follows: 1/2, for full coding sequence (present in WT only); 3/4, for 3′ integration of resistance cassette; 5/4, as a control present in both strains. B, Western blot analysis of ash1-KO strain using α-ASH1 antibodies. * indicates an unspecific band identified by the α-ASH1 Ab. C, parasite lysates treated with FP-Rh probe indicate loss of serine hydrolase activity in ash1-KO strain compared with ASH1-3Ty strain. * indicates active serine hydrolases detected with the FP-Rh probe that are unaffected in the ash1-KO strain. D, the red/green attachment (Att)-invasion (In) assay performed using both wild type (RH) and ku80-KO control parasites and ash1-KO and ASH1-Ty-overexpressing parasites shows that all strains are equivalent. E, subcellular fractionation of parasites lysates in various conditions indicates no difference in solubility of palmitoylated proteins TgISP1 (upper panel) and TgGAP45 (lower panel) in the ash1-KO strain compared with the ku80-KO control. F, comparison of intracellular growth between wild type (RH) (left panel) and ash1-KO (right panel) after 12 h of treatment in the presence of 1 μm inhibitors shows a slight increase in sensitivity to compound 4 in the ash1-KO strain. The results presented correspond to three independent biological experiments, and values are represented as mean ± S.D. (error bars). Abn, abnormal.
Given the presumed activity of ASH1 as an acyl-protein thioesterase, we have examined whether the ash1-KO induces changes in the membrane association of proteins known to be palmitoylated such as TgISP1 and TgGAP45. No difference in the solubility of TgISP1 or TgGAP45 was observed between wild type and ash1-KO strains (Fig. 6E).
Increased Sensitivity of ash1-KO Strain to APT Inhibitors
Because ASH1 was identified here as a major target of the APT1 inhibitor compounds, interacting specifically with both the β-lactone and triazole urea classes, we examined whether loss of ASH1 caused any change in sensitivity to these inhibitors. We reasoned either that the ash1-KO parasites would become resistant to the inhibitors as their main target is absent or that without their main target the inhibitors will be free to interact with other targets.
The ash1-KO parasites appeared to have a slightly increased sensitivity to the inhibitors after prolonged treatment. At 10 μm, ash1-KO parasites and ASH1-Ty-overexpressing parasites were comparably sensitive to the compounds, whereas at 1 μm, the triazole urea inhibitor had a greater toxicity compared with the control and the β-lactone compounds (Fig. 6F).
Characterization of Alternative Candidates for APT Activity in T. gondii
The dispensable nature of ASH1 in T. gondii and its absence in malaria parasites (Plasmodium spp.) suggests the existence of an alternative enzyme responsible for protein depalmitoylation in Apicomplexa. To gain further information, the three genes most closely related to TgASH1 and present across Apicomplexa were characterized. TGME49_054690 (TgASH2; GenBank accession number KF114274) was identified in the MS of the inhibitor targets (supplemental Fig. 1 and Table 2), whereas TGME49_023510 (putative TgASH3; GenBank accession number KF114275) and TGME49_062490 (putative TgASH4; GenBank accession number KF114276) were annotated as containing α/β-hydrolase domains and clear sequence similarity to TgASH1 (supplemental Fig. 2A and Table 3).
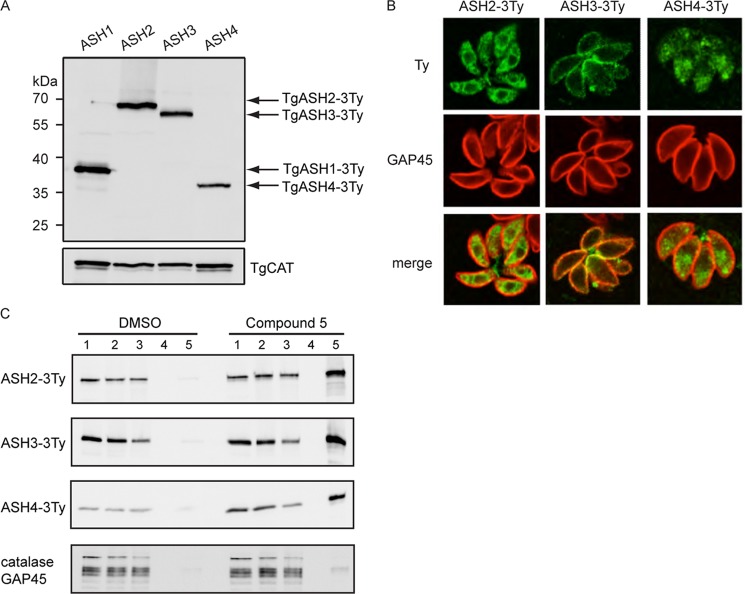
Transgenic parasites were generated to tag the proteins at the endogenous locus, and clones were analyzed by Western blot. The three genes were expressed at levels comparable with that of TgASH1 in the tachyzoite stage (Fig. 7A), and IFAs revealed distinct subcellular localizations (Fig. 7B). TgASH2 is targeted to the endoplasmic reticulum, TgASH3 is more concentrated toward the pellicle, and TgASH4 is predominantly localized at the apical pole.
FIGURE 7.
Characterization of TgASH2, TgASH3, and TgASH4. A, Western blot analysis of transgenic parasites expressing carboxyl-terminally 3Ty-tagged TgASH1–4. B, subcellular localization of 3Ty-tagged TgASH2, TgASH3, and TgASH4 by indirect immunofluorescence microscopy using α-Ty antibodies (green). TgASH2-3Ty localizes to the endoplasmic reticulum, TgASH3-3Ty localizes to the periphery of the parasite, and TgASH4-3Ty localizes to the apical pole. Parasites were stained with α-GAP45 (red). E, pulldown experiments to confirm interaction between the β-lactone inhibitors and TgASH2–4 using compound 5. Numbers 1–5 represent the different fractions: 1, input; 2, precipitated proteins; 3, postclick reaction sample; 4, wash; 5, eluate. TgCAT, catalase.
To determine whether these alternative APT candidates interact with the inhibitors, we performed pulldown experiments as done for TgASH1. The clickable inhibitor, compound 5, specifically enriched all three candidates in the pulldown eluates, whereas GAP45 and catalase were absent (Fig. 7C).
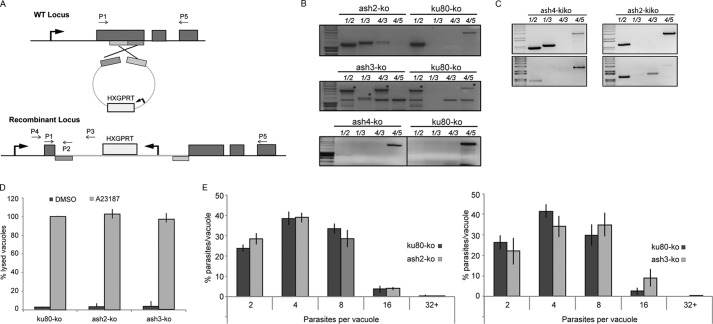
TgASH2 and TgASH3 Are Dispensable, Whereas TgASH4 Is Refractory to Genetic Disruption
To determine whether these candidates were essential for parasite survival, we performed gene disruption by single homologous recombination (Fig. 8A). Genomic PCR analysis confirmed replacement of the endogenous locus for both TgASH2 and TgASH3 (Fig. 8B). Plaque assays were performed with knock-out strains, and no growth defect was detected over 10 days (data not shown). Furthermore, neither stimulated egress (Fig. 8D) nor intracellular growth (Fig. 8E) of these strains was negatively impacted, although the ash3-KO strain appeared to grow slightly faster than the control over the 24-h period (Fig. 8E, right panel). Several attempts were made to disrupt TGME49_062490 (Fig. 8, B and C); however, all attempts failed. To investigate whether this was a technical issue or a true phenotype, we completed PCR analysis of fresh transfectants 2 days after transfection with the aim of detecting integrated KO plasmid and again after 1 week with the aim of demonstrating that all parasites with the integrated ash4-KO plasmid were lost, whereas those with the ash2-KO plasmid remained viable (Fig. 8C). We show that the amount of integrated plasmid at this initial time point was below the level of detection (Fig. 8C). Later samples revealed integration for the ash2-KO plasmid but not for ash4-KO (Fig. 8C), indicating that TgASH4 is refractory to genetic deletion. Furthermore, IFA analysis on ash4-KO-transfected parasites showed no expression of the Ty tag used to replace the ash4 locus, indicating that negative PCRs are accurate and that there is no ash4-KO integration (data not shown). Importantly, recombination leading to the insertion of a tag at the carboxyl terminus was readily obtained for TgASH4 (Fig. 7B), indicating that the locus is accessible to genetic manipulation. Overall, these data strongly suggest that the refractoriness of TgASH4 to gene deletion is due to its importance for T. gondii survival.
FIGURE 8.
TgASH2 and TgASH3 are dispensable, whereas TgASH4 is essential for parasite survival. A, scheme explaining the gene truncation strategy used to disrupt TgASH2–4. The strategy used allows a single homologous recombination in the middle of the gene that leads to truncation. Hypoxanthine-xanthine-guanine phosphoribosyltransferase (HXGPRT) serves as a selectable marker. B and C, disruption of TgASH2 and TgASH3, but not TgASH4, was confirmed by genomic PCR analysis using primers indicated in A and supplemental Table 1. C, top panels represent PCRs completed on DNA extracted from parasites 2 days after transfection. Bottom panels represent PCRs completed on DNA extracted from parasites 1 week after transfection. D, calcium ionophore A23187 induced egress of ash2-KO and ash3-KO from infected host cells was comparable with that of the ku80-KO control strain. E, intracellular growth of ash2-KO was comparable with that of the ku80-KO control strain (left panel). ash3-KO has a slightly faster growth rate than ku80-KO (statistically significant at four and 16 parasites per vacuole only) in a 24-h period (right panel). The results presented in panels D and E represent the mean of three independent biological experiments ± S.D. (error bars).
DISCUSSION
S-Palmitoylation is a versatile and powerful control system for protein dynamics because of the tight membrane association it confers to substrates and the fast kinetics of the palmitoylation/depalmitoylation reactions (16). In this work, we exploited β-lactone- and triazole urea-based human APT1 enzyme inhibitors to explore the relevance of a protein palmitoylation cycle in Apicomplexa and search for their targets to identify an acyl-protein thioesterase.
The first class of inhibitors, the β-lactones, has severe effects on the invasion capacity and development of the parasite but does not disrupt induced egress. Using the fluorescent probe FP-Rh, which binds covalently to its substrates, in combination with the inhibitors, we determined that the inhibitors have multiple targets in T. gondii, and only at 10 μm concentrations did they manage to deplete significantly the activity of the target proteins. The second class, the triazole urea compound 4, also disrupted growth at 10 μm, but invasion was slightly increased at this concentration. Inhibitor 4 was more efficient and specific than the β-lactones, managing to fully deplete the activity of TgASH1 at less than 0.1 μm. At this concentration, neither growth nor invasion was disrupted. This suggests that the other targets of the β-lactone inhibitors or their combination is in fact responsible for the deleterious phenotype observed. By targeting multiple enzymes at once, any compensation by functional orthologues would be blocked. Interestingly, the inhibitors disrupted invasion but not egress, providing further evidence to support the notion that signaling mechanisms controlling these two essential steps in the parasite lytic cycle are not identical (60).
MS analysis of proteins identified as inhibitor targets revealed several α/β-hydrolase domain-containing proteins as candidates for APT activity (Table 2). TGME49_054690 is a putative α/β-hydrolase that is conserved across the Apicomplexa phylum, and potential homologues are present in numerous other eukaryotic species such as yeast, fungi, amoeba, and humans. A number of putative orthologues are described as being similar to the bud emergence (BEM) 46 gene. BEM46 has been described as an endoplasmic reticulum-localized protein with a role in signal transduction and cell polarity (61). TGME49_018540 is less well conserved, and the function remains unknown. Putative orthologues exist in algae, fungi, and some bacteria, although their identity is low. In this work, TGME49_028290 was identified as TgASH1, a prominent serine hydrolase targeted by all APT inhibitors. TgASH1 is present only in the coccidian subgroup of the Apicomplexa, although orthologues can be found in other protists.
As the best candidate for APT activity based on similarity to known APTs, TgASH1 was investigated functionally. Overexpression of TgASH1 did not stimulate a change in parasite growth nor did it cause changes in solubility of known palmitoylated proteins. Disruption of the TgASH1 gene also failed to cause a detectable phenotype either in growth or egress. Additionally, there was no detectable difference in the solubility of TgISP1 or TgGAP45 upon disruption of TgASH1. However, the membrane association of TgGAP45 is not only mediated through palmitoylation but also through association with integral membrane proteins, so the interpretation of these results should be taken with caution. Other palmitoylated proteins would have to be assessed before we could claim that TgASH1 has no effect on palmitoylated proteins in general.
Because TgASH1 emerged as the major target of the inhibitors, we examined whether parasites lacking TgASH1 would exhibit an altered sensitivity to the compounds. We found that the effect of the inhibitors on invasion is equivalent in the wild type and in the ash1-KO strains. However, ash1-KO parasites treated with compound 4 at 1 μm exhibited irregular growth, whereas the DMSO- and β-lactone-treated parasites were normal. Compound 4 is the only inhibitor effective at this low concentration, suggesting that in the absence of TgASH1 this inhibitor is free to interact with other targets.
Studies of the impact of disrupting the APTs with inhibitors, RNAi, or overexpression have been confined to the analysis of a few individual substrates. Functional analysis of APT1 through gene disruption was initially performed in Saccharomyces cerevisiae (62). This study demonstrated that 1) APT1 has a preference for acyl proteins as substrates, 2) no growth defect is apparent in the absence of the gene, and 3) APT1 apparently plays no role in lipid metabolism. In enzymatic assays carried out using yeast extracts from wild type or apt1-KO strains and purified substrates, thioesterase activity against Gα subunits, but not palmitoylated Ras proteins, could be attributed to APT1. These data indicate that an alternative enzyme is likely responsible for the depalmitoylation of Ras proteins and that APTs exhibit substrate specificity, which might be relevant for the APT activity of TgASH1. In oncogenic H-RasG12V-transformed fibroblasts, treatment with inhibitors that interfere with APT1 and APT2 dynamics causes a general mislocalization of H-Ras and redistribution throughout the cellular membranes, thereby reducing oncogenic Ras signaling and activity (33). These experiments have been performed using inhibitors similar to those used here. It is possible that the observed effect on H-Ras redistribution is due to the lack of inhibitor specificity because these results have not yet been confirmed by the disruption of the APT1 or APT2 genes. A recent study identified rat APT1 as a target of a microRNA that binds in the 3′-UTR and reduces APT1 function in neurons (63). In this context, APT1 activity on Gα subunits is associated to the overall synaptic plasticity in neuronal cells. Finally, APT1 and its orthologue LYPLAL1 have been reported to control potassium channel dynamics based on an overexpression strategy (6).
In this study, neither the deletion nor the overexpression of TgASH1 produced any significant phenotypic change in contrast to the severe effects observed in the presence of 10 μm hAPT1 inhibitors. Instead, the knock-out of TgASH1 recapitulated the effect of treatment with 0.1 μm compound 4, which specifically inhibited TgASH1, confirming that the β-lactone inhibitors at all concentrations and compound 4 at higher concentrations target other vital enzymes in Apicomplexa.
Enzymatic assays were performed in vitro to determine the substrate preferences of TgASH1. In these assays, TgASH1 was able to cleave PNP-palmitate with the same efficiency as human APT1, suggesting that TgASH1 is a bona fide acyl-protein thioesterase, confirming the prediction of the homology modeling.
We hypothesize that other targets of the inhibitors, which are possibly divergent from hAPT1, could still function as APT-like thioesterases and compensate for loss of TgASH1. Other enzymes with the capacity for depalmitoylation have been identified in mammalian cells: APT2, PPT1, and PPT2. In T. gondii, a search for an APT2 homologue recovered TgASH1. The mammalian lysosomal PPT1 enzyme has a putative homologue in T. gondii (TGME49_097880). This protein is expected to be a dense granule protein based on similarity to a Sarcocystis muris dense granule antigen (64, 65). It shares only 12% identity and 40% similarity with the mammalian PPT1 and has no predicted serine hydrolase activity based on its sequence. Finally, BLAST searching PPT2 in EuPathDB (66) recovered a Cryptosporidium protein with predicted esterase activity. When used to search for homologues in T. gondii and P. falciparum, proteins with putative serine esterase/α/β-hydrolase activity that were also identified in the search for α/β-hydrolases in the genome (Table 3) were recovered.
Of the 54 T. gondii proteins that contain α/β-hydrolase domains, 20 belong to families that are potentially relevant for APT activity using hidden Markov models (67) (Table 3). TgASH2 and TgASH3 further characterized here were shown to be dispensable, highlighting the possible issue of redundancy that renders gene function analysis problematic. In contrast, TgASH4, a gene present in all Apicomplexa, was refractory to gene deletion and thus is likely essential for parasite survival.
To summarize, we identified an APT1 homologue in T. gondii that was dispensable for parasite survival and appears to have a substrate specificity comparable with that of hAPT1. Inhibitors designed against hAPT1 targeted multiple parasite enzymes and were detrimental to parasite survival. The absence of a TgASH1 homologue in P. falciparum provides further evidence that these inhibitors must have other targets possibly shared between the two species. Among several candidates examined thus far, TgASH4 emerges as a primary target that deserves further biochemical and functional investigation.
Supplementary Material
Acknowledgments
We thank Dr. Karine Frenal and Leonardo Capponi for assistance during the early stage of this project. C. H. and M. R. thank Prof. Herbert Waldmann, Max Planck Institute for Molecular Physiology, Dortmund, Germany for unconditional support.

This article contains supplemental Figs. 1 and 2 and Table 1.
- PPT
- palmitoyl-protein thioesterase
- APT
- acyl-protein thioesterase
- ASH
- active serine hydrolase
- hAPT
- human APT
- FP-Rh
- fluorophosphonate-rhodamine
- HFF
- human foreskin fibroblast
- IFA
- immunofluorescence assay
- PAF
- paraformaldehyde
- PNP
- p-nitrophenolate
- BEM
- bud emergence
- Tg
- T. gondii.
REFERENCES
- 1. Charollais J., Van Der Goot F. G. (2009) Palmitoylation of membrane proteins. Mol. Membr. Biol. 26, 55–66 [DOI] [PubMed] [Google Scholar]
- 2. Resh M. D. (2006) Trafficking and signaling by fatty-acylated and prenylated proteins. Nat. Chem. Biol. 2, 584–590 [DOI] [PubMed] [Google Scholar]
- 3. Zeidman R., Jackson C. S., Magee A. I. (2009) Protein acyl thioesterases. Mol. Membr. Biol. 26, 32–41 [DOI] [PubMed] [Google Scholar]
- 4. Linder M. E., Deschenes R. J. (2007) Palmitoylation: policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 8, 74–84 [DOI] [PubMed] [Google Scholar]
- 5. Resh M. D. (2006) Palmitoylation of ligands, receptors, and intracellular signaling molecules. Sci. STKE 2006, re14. [DOI] [PubMed] [Google Scholar]
- 6. Tian L., McClafferty H., Knaus H. G., Ruth P., Shipston M. J. (2012) Distinct acyl protein transferases and thioesterases control surface expression of calcium-activated potassium channels. J. Biol. Chem. 287, 14718–14725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baekkeskov S., Kanaani J. (2009) Palmitoylation cycles and regulation of protein function. Mol. Membr Biol. 26, 42–54 [DOI] [PubMed] [Google Scholar]
- 8. Frénal K., Tay C. L., Mueller C., Bushell E. S., Jia Y., Graindorge A., Billker O., Rayner J. C., Soldati-Favre D. (2013) Global analysis of apicomplexan protein S-acyl transferases reveals an enzyme essential for invasion. Traffic 14, 895–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jones M. L., Tay C. L., Rayner J. C. (2012) Getting stuck in: protein palmitoylation in Plasmodium. Trends Parasitol. 28, 496–503 [DOI] [PubMed] [Google Scholar]
- 10. Jones M. L., Collins M. O., Goulding D., Choudhary J. S., Rayner J. C. (2012) Analysis of protein palmitoylation reveals a pervasive role in Plasmodium development and pathogenesis. Cell Host Microbe 12, 246–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Russo I., Oksman A., Goldberg D. E. (2009) Fatty acid acylation regulates trafficking of the unusual Plasmodium falciparum calpain to the nucleolus. Mol. Microbiol. 72, 229–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Frénal K., Polonais V., Marq J. B., Stratmann R., Limenitakis J., Soldati-Favre D. (2010) Functional dissection of the apicomplexan glideosome molecular architecture. Cell Host Microbe. 8, 343–357 [DOI] [PubMed] [Google Scholar]
- 13. Rees-Channer R. R., Martin S. R., Green J. L., Bowyer P. W., Grainger M., Molloy J. E., Holder A. A. (2006) Dual acylation of the 45 kDa gliding-associated protein (GAP45) in Plasmodium falciparum merozoites. Mol. Biochem. Parasitol. 149, 113–116 [DOI] [PubMed] [Google Scholar]
- 14. Beck J. R., Rodriguez-Fernandez I. A., Cruz de Leon J., Huynh M. H., Carruthers V. B., Morrissette N. S., Bradley P. J. (2010) A novel family of Toxoplasma IMC proteins displays a hierarchical organization and functions in coordinating parasite division. PLoS Pathog. 6, e1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cabrera A., Herrmann S., Warszta D., Santos J. M., John Peter A. T., Kono M., Debrouver S., Jacobs T., Spielmann T., Ungermann C., Soldati-Favre D., Gilberger T. W. (2012) Dissection of minimal sequence requirements for rhoptry membrane targeting in the malaria parasite. Traffic 13, 1335–1350 [DOI] [PubMed] [Google Scholar]
- 16. Rocks O., Gerauer M., Vartak N., Koch S., Huang Z. P., Pechlivanis M., Kuhlmann J., Brunsveld L., Chandra A., Ellinger B., Waldmann H., Bastiaens P. I. (2010) The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell 141, 458–471 [DOI] [PubMed] [Google Scholar]
- 17. Camp L. A., Hofmann S. L. (1993) Purification and properties of a palmitoyl-protein thioesterase that cleaves palmitate from H-Ras. J. Biol. Chem. 268, 22566–22574 [PubMed] [Google Scholar]
- 18. Camp L. A., Verkruyse L. A., Afendis S. J., Slaughter C. A., Hofmann S. L. (1994) Molecular cloning and expression of palmitoyl-protein thioesterase. J. Biol. Chem. 269, 23212–23219 [PubMed] [Google Scholar]
- 19. Soyombo A. A., Hofmann S. L. (1997) Molecular cloning and expression of palmitoyl-protein thioesterase 2 (PPT2), a homolog of lysosomal palmitoyl-protein thioesterase with a distinct substrate specificity. J. Biol. Chem. 272, 27456–27463 [DOI] [PubMed] [Google Scholar]
- 20. Gupta P., Soyombo A. A., Atashband A., Wisniewski K. E., Shelton J. M., Richardson J. A., Hammer R. E., Hofmann S. L. (2001) Disruption of PPT1 or PPT2 causes neuronal ceroid lipofuscinosis in knockout mice. Proc. Natl. Acad. Sci. U.S.A. 98, 13566–13571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vesa J., Hellsten E., Verkruyse L. A., Camp L. A., Rapola J., Santavuori P., Hofmann S. L., Peltonen L. (1995) Mutations in the palmitoyl protein thioesterase gene causing infantile neuronal ceroid lipofuscinosis. Nature 376, 584–587 [DOI] [PubMed] [Google Scholar]
- 22. Gupta P., Soyombo A. A., Shelton J. M., Wilkofsky I. G., Wisniewski K. E., Richardson J. A., Hofmann S. L. (2003) Disruption of PPT2 in mice causes an unusual lysosomal storage disorder with neurovisceral features. Proc. Natl. Acad. Sci. U.S.A. 100, 12325–12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tardy C., Sabourdy F., Garcia V., Jalanko A., Therville N., Levade T., Andrieu-Abadie N. (2009) Palmitoyl protein thioesterase 1 modulates tumor necrosis factor α-induced apoptosis. Biochim. Biophys. Acta 1793, 1250–1258 [DOI] [PubMed] [Google Scholar]
- 24. Tomatis V. M., Trenchi A., Gomez G. A., Daniotti J. L. (2010) Acyl-protein thioesterase 2 catalyzes the deacylation of peripheral membrane-associated GAP-43. PLoS One 5, e15045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sugimoto H., Hayashi H., Yamashita S. (1996) Purification, cDNA cloning, and regulation of lysophospholipase from rat liver. J. Biol. Chem. 271, 7705–7711 [DOI] [PubMed] [Google Scholar]
- 26. Duncan J. A., Gilman A. G. (1998) A cytoplasmic acyl-protein thioesterase that removes palmitate from G protein α subunits and p21RAS. J. Biol. Chem. 273, 15830–15837 [DOI] [PubMed] [Google Scholar]
- 27. Ahearn I. M., Tsai F. D., Court H., Zhou M., Jennings B. C., Ahmed M., Fehrenbacher N., Linder M. E., Philips M. R. (2011) FKBP12 binds to acylated H-ras and promotes depalmitoylation. Mol. Cell 41, 173–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rocks O., Peyker A., Kahms M., Verveer P. J., Koerner C., Lumbierres M., Kuhlmann J., Waldmann H., Wittinghofer A., Bastiaens P. I. (2005) An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science 307, 1746–1752 [DOI] [PubMed] [Google Scholar]
- 29. Dekker F. J., Hedberg C. (2011) Small molecule inhibition of protein depalmitoylation as a new approach towards downregulation of oncogenic Ras signalling. Bioorg. Med. Chem. 19, 1376–1380 [DOI] [PubMed] [Google Scholar]
- 30. Yeh D. C., Duncan J. A., Yamashita S., Michel T. (1999) Depalmitoylation of endothelial nitric-oxide synthase by acyl-protein thioesterase 1 is potentiated by Ca2+-calmodulin. J. Biol. Chem. 274, 33148–33154 [DOI] [PubMed] [Google Scholar]
- 31. D'Alessandro A., Righetti P. G., Zolla L. (2010) The red blood cell proteome and interactome: an update. J. Proteome Res. 9, 144–163 [DOI] [PubMed] [Google Scholar]
- 32. Adibekian A., Martin B. R., Wang C., Hsu K. L., Bachovchin D. A., Niessen S., Hoover H., Cravatt B. F. (2011) Click-generated triazole ureas as ultrapotent in vivo-active serine hydrolase inhibitors. Nat. Chem. Biol. 7, 469–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dekker F. J., Rocks O., Vartak N., Menninger S., Hedberg C., Balamurugan R., Wetzel S., Renner S., Gerauer M., Schölermann B., Rusch M., Kramer J. W., Rauh D., Coates G. W., Brunsveld L., Bastiaens P. I., Waldmann H. (2010) Small-molecule inhibition of APT1 affects Ras localization and signaling. Nat. Chem. Biol. 6, 449–456 [DOI] [PubMed] [Google Scholar]
- 34. Baker T. L., Zheng H., Walker J., Coloff J. L., Buss J. E. (2003) Distinct rates of palmitate turnover on membrane-bound cellular and oncogenic H-ras. J. Biol. Chem. 278, 19292–19300 [DOI] [PubMed] [Google Scholar]
- 35. Navarro-Lérida I., Sánchez-Perales S., Calvo M., Rentero C., Zheng Y., Enrich C., Del Pozo M. A. (2012) A palmitoylation switch mechanism regulates Rac1 function and membrane organization. EMBO J. 31, 534–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Billker O., Lourido S., Sibley L. D. (2009) Calcium-dependent signaling and kinases in apicomplexan parasites. Cell Host Microbe. 5, 612–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Möskes C., Burghaus P. A., Wernli B., Sauder U., Dürrenberger M., Kappes B. (2004) Export of Plasmodium falciparum calcium-dependent protein kinase 1 to the parasitophorous vacuole is dependent on three N-terminal membrane anchor motifs. Mol. Microbiol. 54, 676–691 [DOI] [PubMed] [Google Scholar]
- 38. Brecht S., Carruthers V. B., Ferguson D. J., Giddings O. K., Wang G., Jakle U., Harper J. M., Sibley L. D., Soldati D. (2001) The Toxoplasma micronemal protein MIC4 is an adhesin composed of six conserved apple domains. J. Biol. Chem. 276, 4119–4127 [DOI] [PubMed] [Google Scholar]
- 39. Plattner F., Yarovinsky F., Romero S., Didry D., Carlier M. F., Sher A., Soldati-Favre D. (2008) Toxoplasma profilin is essential for host cell invasion and TLR11-dependent induction of an interleukin-12 response. Cell Host Microbe 3, 77–87 [DOI] [PubMed] [Google Scholar]
- 40. Ding M., Clayton C., Soldati D. (2000) Toxoplasma gondii catalase: are there peroxisomes in toxoplasma? J. Cell Sci. 113, 2409–2419 [DOI] [PubMed] [Google Scholar]
- 41. Liu Y., Patricelli M. P., Cravatt B. F. (1999) Activity-based protein profiling: the serine hydrolases. Proc. Natl. Acad. Sci. U.S.A. 96, 14694–14699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Aurrecoechea C., Heiges M., Wang H., Wang Z., Fischer S., Rhodes P., Miller J., Kraemer E., Stoeckert C. J., Jr., Roos D. S., Kissinger J. C. (2007) ApiDB: integrated resources for the apicomplexan bioinformatics resource center. Nucleic Acids Res. 35, D427–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Edgar R. C. (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guindon S., Gascuel O. (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 [DOI] [PubMed] [Google Scholar]
- 45. Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 [DOI] [PubMed] [Google Scholar]
- 46. Anisimova M., Gascuel O. (2006) Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55, 539–552 [DOI] [PubMed] [Google Scholar]
- 47. Whelan S., Goldman N. (2001) A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18, 691–699 [DOI] [PubMed] [Google Scholar]
- 48. Darriba D., Taboada G. L., Doallo R., Posada D. (2011) ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Deng W., Maust B. S., Nickle D. C., Learn G. H., Liu Y., Heath L., Kosakovsky Pond S. L., Mullins J. I. (2010) DIVEIN: a web server to analyze phylogenies, sequence divergence, diversity, and informative sites. BioTechniques 48, 405–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chevenet F., Brun C., Bañuls A. L., Jacq B., Christen R. (2006) TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7, 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Huynh M. H., Carruthers V. B. (2009) Tagging of endogenous genes in a Toxoplasma gondii strain lacking Ku80. Eukaryot. Cell 8, 530–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Trager W., Jensen J. B. (1976) Human malaria parasites in continuous culture. Science 193, 673–675 [DOI] [PubMed] [Google Scholar]
- 53. Santos J. M., Ferguson D. J., Blackman M. J., Soldati-Favre D. (2011) Intramembrane cleavage of AMA1 triggers Toxoplasma to switch from an invasive to a replicative mode. Science 331, 473–477 [DOI] [PubMed] [Google Scholar]
- 54. Huynh M. H., Rabenau K. E., Harper J. M., Beatty W. L., Sibley L. D., Carruthers V. B. (2003) Rapid invasion of host cells by Toxoplasma requires secretion of the MIC2-M2AP adhesive protein complex. EMBO J. 22, 2082–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dorn A., Stoffel R., Matile H., Bubendorf A., Ridley R. G. (1995) Malarial haemozoin/β-haematin supports haem polymerization in the absence of protein. Nature 374, 269–271 [DOI] [PubMed] [Google Scholar]
- 56. Huber W., Koella J. C. (1993) A comparison of three methods of estimating EC50 in studies of drug resistance of malaria parasites. Acta Trop. 55, 257–261 [DOI] [PubMed] [Google Scholar]
- 57. Rusch M., Zimmermann T. J., Bürger M., Dekker F. J., Görmer K., Triola G., Brockmeyer A., Janning P., Böttcher T., Sieber S. A., Vetter I. R., Hedberg C., Waldmann H. (2011) Identification of acyl protein thioesterases 1 and 2 as the cellular targets of the Ras-signaling modulators palmostatin B and M. Angew. Chem. Int. Ed. Engl. 50, 9838–9842 [DOI] [PubMed] [Google Scholar]
- 58. Hedberg C., Dekker F. J., Rusch M., Renner S., Wetzel S., Vartak N., Gerding-Reimers C., Bon R. S., Bastiaens P. I., Waldmann H. (2011) Development of highly potent inhibitors of the Ras-targeting human acyl protein thioesterases based on substrate similarity design. Angew. Chem. Int. Ed. Engl. 50, 9832–9837 [DOI] [PubMed] [Google Scholar]
- 59. Patricelli M. P., Giang D. K., Stamp L. M., Burbaum J. J. (2001) Direct visualization of serine hydrolase activities in complex proteomes using fluorescent active site-directed probes. Proteomics 1, 1067–1071 [DOI] [PubMed] [Google Scholar]
- 60. Lourido S., Tang K., Sibley L. D. (2012) Distinct signalling pathways control Toxoplasma egress and host-cell invasion. EMBO J. 31, 4524–4534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mercker M., Kollath-Leiss K., Allgaier S., Weiland N., Kempken F. (2009) The BEM46-like protein appears to be essential for hyphal development upon ascospore germination in Neurospora crassa and is targeted to the endoplasmic reticulum. Curr. Genet. 55, 151–161 [DOI] [PubMed] [Google Scholar]
- 62. Duncan J. A., Gilman A. G. (2002) Characterization of Saccharomyces cerevisiae acyl-protein thioesterase 1, the enzyme responsible for G protein α subunit deacylation in vivo. J. Biol. Chem. 277, 31740–31752 [DOI] [PubMed] [Google Scholar]
- 63. Siegel G., Obernosterer G., Fiore R., Oehmen M., Bicker S., Christensen M., Khudayberdiev S., Leuschner P. F., Busch C. J., Kane C., Hübel K., Dekker F., Hedberg C., Rengarajan B., Drepper C., Waldmann H., Kauppinen S., Greenberg M. E., Draguhn A., Rehmsmeier M., Martinez J., Schratt G. M. (2009) A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat. Cell Biol. 11, 705–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Freyer B., Eschenbacher K. H., Mehlhorn H., Rueger W. (1998) Isolation and characterization of cDNA clones encoding a 32-kDa dense-granule antigen of Sarcocystis muris (Apicomplexa). Parasitol Res. 84, 583–589 [DOI] [PubMed] [Google Scholar]
- 65. Freyer B., Hansner T., Mehlhorn H., Rüger W. (1999) Characterization of a genomic region encoding the 32-kDa dense granule antigen of Sarcocystis muris (Apicomplexa). Parasitol. Res. 85, 923–927 [DOI] [PubMed] [Google Scholar]
- 66. Aurrecoechea C., Heiges M., Wang H., Wang Z., Fischer S., Rhodes P., Miller J., Kraemer E., Stoeckert C. J., Jr., Roos D. S., Kissinger J. C. (2007) ApiDB: integrated resources for the apicomplexan bioinformatics resource center. Nucleic Acids Res. 35, D427–D430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gough J., Karplus K., Hughey R., Chothia C. (2001) Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 313, 903–919 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.