Background: Cysteine desulfurases deliver sulfur from cysteine to sulfur acceptors for sulfur utilization in various biological processes.
Results: The crystal structure of CsdA-CsdE invokes a unique binding mode compared with other cysteine desulfurase and sulfur acceptor complexes.
Conclusion: Conformational flexibility is pronounced only in the region of CsdE during the sulfur transfer interaction.
Significance: This structure marks the first complex structure between CsdA/SufS-type cysteine desulfurase and the sulfur acceptor.
Keywords: Iron, Iron-Sulfur Protein, Structural Biology, Sulfur, X-ray Crystallography, CsdA, CsdE, Cysteine Desulfurase, Sulfur Acceptor, trans-Persulfuration
Abstract
In Escherichia coli, three cysteine desulfurases (IscS, SufS, and CsdA) initiate the delivery of sulfur for various biological processes such as the biogenesis of Fe-S clusters. The sulfur generated as persulfide on a cysteine residue of cysteine desulfurases is further transferred to Fe-S scaffolds (e.g. IscU) or to intermediate cysteine-containing sulfur acceptors (e.g. TusA, SufE, and CsdE) prior to its utilization. Here, we report the structures of CsdA and the CsdA-CsdE complex, which provide insight into the sulfur transfer mediated by the trans-persulfuration reaction. Analysis of the structures indicates that the conformational flexibility of the active cysteine loop in CsdE is essential for accepting the persulfide from the cysteine of CsdA. Additionally, CsdA and CsdE invoke a different binding mode than those of previously reported cysteine desulfurase (IscS) and sulfur acceptors (TusA and IscU). Moreover, the conservation of interaction-mediating residues between CsdA/SufS and CsdE/SufE further suggests that the SufS-SufE interface likely resembles that of CsdA and CsdE.
Introduction
Iron-sulfur clusters (Fe-S) as cofactors have versatile roles in a wide range of biological processes (1). Biogenesis of Fe-S involves successive transfers of iron and sulfur from sophisticated multiprotein assembly systems, and four distinct systems known as Nif (nitrogen fixation), Isc (iron-sulfur cluster formation), Suf (sulfur mobilization), and CIA (cytosolic iron-sulfur assembly) have been characterized (2–4). A pyridoxal 5′-phosphate (PLP)2-dependent cysteine desulfurase converts l-cysteine to l-alanine with sulfur released on its active cysteine as persulfide in all cases (5). Consequently, the sulfur generated by the cysteine desulfurase (donor) can either be directly transferred to Fe-S scaffolds for Fe-S assembly or to intermediate cysteine-containing sulfur acceptors via the trans-persulfuration reaction between active cysteines. The sulfur of the intermediate acceptors can be further supplied to Fe-S scaffolds as well as to the proteins of sulfur-utilizing pathways such as those involved in the synthesis of sulfur-containing cofactors or thiol modification of tRNA.
Three distinct cysteine desulfurases (IscS, SufS, and CsdA) are encoded on operons of the Isc, Suf, and Csd systems in Escherichia coli (6). Briefly, the Isc system acts as the housekeeper of Fe-S biogenesis, in which sulfurs are delivered from IscS to several sulfur acceptors. The sulfurs are employed in various biosynthetic pathways, including Fe-S assembly, thiamine/biotin synthesis, and tRNA thiolation. Among the sulfur acceptors, complex crystal structures with IscS have been reported for Zn/Fe2S2-binding scaffold IscU from E. coli (apo-form, see Ref. 7) and Archaeoglobus fulgidus (holo-form, see Ref. 8) and for E. coli TusA (7), which is an intermediate sulfur acceptor utilized during tRNA thiolation. The Suf system functions under oxidative stress or iron deprivation, which utilizes sulfurs from SufS (also known as CsdB) transferred to SufE, an intermediate sulfur acceptor. SufE further transfers sulfurs to other Fe-S proteins in the same operon (9, 10). Because SufS exhibits ∼300-fold higher activity for l-selenocysteine over l-cysteine, SufS also likely functions as a selenocysteine lyase in E. coli (11). Structures of E. coli SufS and SufE in the free forms are available (12–14).
In the most recently characterized Csd (cysteine sulfinite desulfinase) system, CsdA (also known as CSD) transfers sulfurs to CsdE (also known as YgdK) (15). However, further consequences of the CsdE sulfur are mostly unknown. A recent study suggested that CsdA/CsdE either cross-talks with the Suf system or supplies sulfur for synthesis of an unidentified thiolated compound via CsdL (16). Structure of E. coli CsdE has been determined by using NMR (17). Unlike SufS, CsdA is not specific toward l-selenocysteine.
Sequence identities between CsdA and SufS (∼43%) and between CsdE and SufE (∼35%) suggest that CsdA/CsdE and SufS/SufE are closely related (sequence identities between CsdA/SufS and IscS (less than ∼24%) or CsdE/SufE and IscU (< ∼10%) are limited). In the structures of free CsdE and SufE, the catalytic sulfur-accepting cysteines were both buried in a hydrophobic cavity (14, 17). Hence, trans-persulfuration between CsdA and CsdE (or between SufS and SufE) had been difficult to imagine without any conformational change (2, 14, 17, 18).
To gain mechanistic insight on this sulfur transfer event, we herein determined structures of the free CsdA and CsdA-CsdE complex (Table 1). Because active cysteine-containing regions in both CsdA and CsdE are well ordered in the CsdA-CsdE structure, their comparisons with free CsdA and free CsdE indicate that the conformational flexibility in the active cysteine loop of CsdE provides the basis for understanding the sulfur transfer between the two proteins.
TABLE 1.
Data collection and refinement statistics
| CsdA | CsdA-CsdE | |
|---|---|---|
| X-ray wavelength | 0.97934 ′ | 0.97934 ′ |
| Temperature | 100 K | 100 K |
| Resolution range | 50 to 1.8 ′ | 50 to 2.0 ′ |
| (highest resolution shell) | (1.86 to 1.80) | (2.07 to 2.00) |
| Space group | C2 | P21 |
| a, b, c | 168.30, 91.44, 83.89 ′ | 77.14,76.87, 89.36 ′ |
| β | 97.26° | 104.48° |
| Total reflections | 1,249,185 | 849,428 |
| Unique reflections | 116,275 | 67,595 |
| Redundancy | 3.7 (3.7) | 3.6 (3.3) |
| Completeness | 99.4% (100%) | 99.8% (99.5%) |
| Mean I/σ(I) | 31.6 (4.8) | 24.6 (5.0) |
| Rmergea | 6.9% (37.7%) | 6.9% (34.0%) |
| Wilson B-factor | 22.0 ′2 | 19.1 ′2 |
| Molecules/asymmetric units | 3 | 4 |
| Matthews coefficient, % solvent | 2.47, 50.2% | 2.17, 43.3% |
| Refinement R-factorb (Rfreec) | 0.165 (0.212) | 0.159 (0.218) |
| Residue range | CsdA | CsdA |
| 2–400 (a, a′, and a″) | a, 3–48, 55–400 | |
| a′, 3–400 | ||
| CsdE | ||
| a, 3–147 | ||
| a′, 8–60, 62–145 | ||
| No. of waters | 1107 | 685 |
| Root mean square deviation bonds | 0.028 ′ | 0.023 ′ |
| Root mean square deviation angles | 2.108° | 1.963° |
| Average B-factor (all atoms) | 24.23 ′2 | 25.11 ′2 |
| Average B-factor (main chain) | 21.33 ′2 | 23.41 ′2 |
| Average B-factor (side chain and water) | 26.76 ′2 | 26.68 ′2 |
a Rmerge = ΣhΣi|I(h,i) − 〈I(h)〉|/Σh 〈I(h)〉, where I(h,i) is the intensity of the ith measurement of reflection h, and 〈I(h)〉 is the mean value of I(h,i) for all i measurements.
b R-factor = Σ(|Fobs − |Fcalc|)/Σ|Fobs|.
c Rfree = R-factor for 5% of randomly selected reflections excluded from the refinement.
EXPERIMENTAL PROCEDURES
Protein Preparations
The genes of full-length E. coli CsdA (residues 1–401) and full-length CsdE (residues 1–147) were PCR-cloned into vector pET28a (Novagen) using the E. coli genomic DNA. E. coli genomic DNA was prepared with LB-cultured E. coli strain XL1-Blue (Stratagene) using a commercial kit (Qiagen). The plasmid constructs were designed with thrombin-cleavable His6 tags at the N terminus of the proteins for convenient purification. Unavoidable non-native amino acids at the N terminus after the thrombin digestion were minimized by using NdeI restriction enzyme sites at the 5′-ends of the primer. Cloned constructs were verified by DNA sequence analysis using primers of T7 promoter and terminator prior to transformation into E. coli strain BL21 (DE3) (Stratagene). Plasmid-transformed cells were grown in 1 liter of LB with kanamycin selection (25 μg/ml) at 37 °C. Recombinant protein expressions were induced with 0.5 mm isopropyl β-d-thiogalactopyranoside at A600 = ∼0.8, and the cells were further grown at 25 °C for 16 h. The cells were harvested by centrifugation (4500 × g) at 4 °C for 10 min and resuspended in ice-cold lysis buffer (20 mm Tris, pH 7.5, 500 mm NaCl, and 5 mm imidazole). Resuspended cells were homogenized by sonication, and the lysates were centrifuged (70,000 × g) at 4 °C for 30 min. Supernatants were loaded onto Ni2+-nitrilotriacetic beads and washed with wash buffer (20 mm Tris, pH 7.5, 500 mm NaCl, and 20 mm imidazole). The bound proteins were eluted with elution buffer (20 mm Tris, pH 7.5, 500 mm NaCl, and 200 mm imidazole), and the His6 tags were removed by adding bovine thrombin (Invitrogen) directly to the eluents for 16 h at 4 °C. The proteins were further purified using Superdex 200 HR26/60 sizing column on ÄKTA FPLC system (GE Healthcare) equilibrated in gel filtration buffer (50 mm Tris, pH 7.5, 150 mm NaCl, and 2 mm DTT). Proteins were concentrated by centrifugation (Amicon Millipore Centriprep). The final CsdA concentration (∼13 mg/ml) was estimated by absorption at λ = 280 nm with a molar extinction coefficient of 47,000 m−1 cm−1 calculated based upon the number of tryptophan and tyrosine residues (19). The final CsdE concentration (∼40 mg/ml) was estimated similarly with a molar extinction coefficient of 14,000 m−1 cm−1.
For the purification of the homogeneous CsdA-CsdE complex, purified CsdA was mixed with an ∼4-fold molar excess of purified CsdE. The homogeneous complex was selected over the excess CsdE by re-eluting the mixture on the same Superdex 200 sizing column. The final CsdA-CsdE concentration (∼30 mg/ml) was estimated similarly with the calculated molar extinction coefficient of 61,000 m−1 cm−1.
Crystallization and X-ray Data Collection
Conditions for growing the crystals of free CsdA and CsdA-CsdE were screened using commercial solutions (Hampton Research). Crystals were screened at 22 °C of ambient air using the hanging drop vapor diffusion in 24-well Linbro plates. Optimized CsdA crystals were obtained from reservoirs of 20–30% (w/v) PEG 8000, 0.05 m potassium di-hydrogen phosphate, and 0.1 m MES, pH 6.5 (or pH 5.5), or 20–30% (w/v) PEG 8000, 0.05 m sodium di-hydrogen phosphate, and 0.1 m MES, pH 6.5 (or 0.1 m Tris, pH 8.5). Crystals grew up to approximate dimensions of 0.5 × 0.5 × 1.5 mm in a day. Optimized CsdA-CsdE crystals were obtained from reservoirs of 22% (w/v) polyacrylic acid 5100, 0.02 m MgCl2, and 0.1 m HEPES, pH 7.5. Crystals grew up to approximate dimensions of 0.3 × 0.3 × 1.0 mm in a few weeks.
CsdA and CsdA-CsdE crystals from drops were transferred to reservoir solutions supplemented with 15% glycerol acting as a cryo-protectant and were quickly frozen in liquid nitrogen for transportation. Diffraction data were collected under a 100 K nitrogen stream at Pohang Accelerator Laboratory (Beamline 7A) on a CCD detector (ADSC Q270). Data were processed using DENZO and SCALEPACK (20). The CsdA and CsdA-CsdE crystals, respectively, belong to the space groups of C2 and P21 (Table 1).
Structure Determination and Refinement
The positions of three E. coli CsdAs in the free CsdA crystal asymmetric units were determined by molecular replacement using E. coli SufS as a search model (PDB code 1C0N) against data from all the resolution shells. PHASER (21) found two CsdAs, and by fixing these solutions MOLREP (22) further determined the third CsdA. Subsequent submission of this model (R/Rfree = 0.29/0.34) to the automated web-based model building ARP/wARP (23) server generated the partial CsdA model and the electron density map into which errors were manually changed using COOT (24). The final CsdA model contains three molecules of CsdA (CsdA residues 2–400) each with a PLP. The structure was refined with REFMAC5 (25) to a resolution of 1.8 Å with no data cutoff (Table 1).
The positions of two E. coli CsdAs in the CsdA-CsdE complex crystal were determined by molecular replacement using the CsdA structure as a search model against data from all the resolution shells in PHASER. Subsequent submission of this model (R/Rfree = 0.35/0.36) to ARP/wARP generated the partial models of two CsdEs and the electron density map into which changes were manually made using COOT. The final CsdA-CsdE model contains two molecules of CsdA (residues 3–48 and 55–400 in a and 3–400 in a′) each with PLP, and two molecules of CsdE (residues 3–147 in a and 8–60 and 62–145 in a′). The structure was refined with REFMAC5 to a resolution of 2.0 Å with no data cutoff (Table 1). All structural figures were rendered with PyMOL (Schrödinger, LLC). The buried solvent-accessible surface areas per subunit were calculated using the PISA server (26).
RESULTS AND DISCUSSION
Free CsdA Structure
The structure of E. coli CsdA(2–400) was determined at 1.8-Å resolution using molecular replacement with the free E. coli SufS as a search model. Three CsdAs were found in the crystal asymmetric units. Two CsdAs form a dimer along a noncrystallographic 2-fold axis, and the third CsdA associates into an identical dimer with the crystallographic symmetry mate. Given that cysteine desulfurases are classified as members of the “aminotransferase class V of fold-type I family,” the overall α/β-fold of CsdA is very similar to SufS, IscS, and NifS (12, 13, 27, 28). The CsdA fold can be divided into a “large region” and a “small region” (12), in which the large region mediates the dimeric interface (Fig. 1A). The large region mostly consists of a seven-stranded parallel β-sheet flanked by seven α-helices and several tightly packed α-helices. The dimeric interfaces of CsdA, SufS, and IscS are mediated by similar corresponding structural elements, but the buried accessible surface areas (per subunit) between the CsdA dimer (3148 Å2) and the SufS dimer (3086 Å2) are higher than that of the IscS dimer (2320 Å2). Among the various reported structures of cysteine desulfurases, CsdA mostly resembles SufS as indicated from the high sequence identity (∼43%). Hence, the α-helices and β-strands described herein are numbered sequentially as in the E. coli SufS structure (12). Discussions of the different cysteine desulfurase folds with comparisons on the PLP-containing active sites are well documented (12, 13, 29). The only structural differences in CsdA compared with SufS are a shorter H2-H3 loop and a kink in H4 of CsdA, which are both manifested near the dimeric interface (Fig. 2).
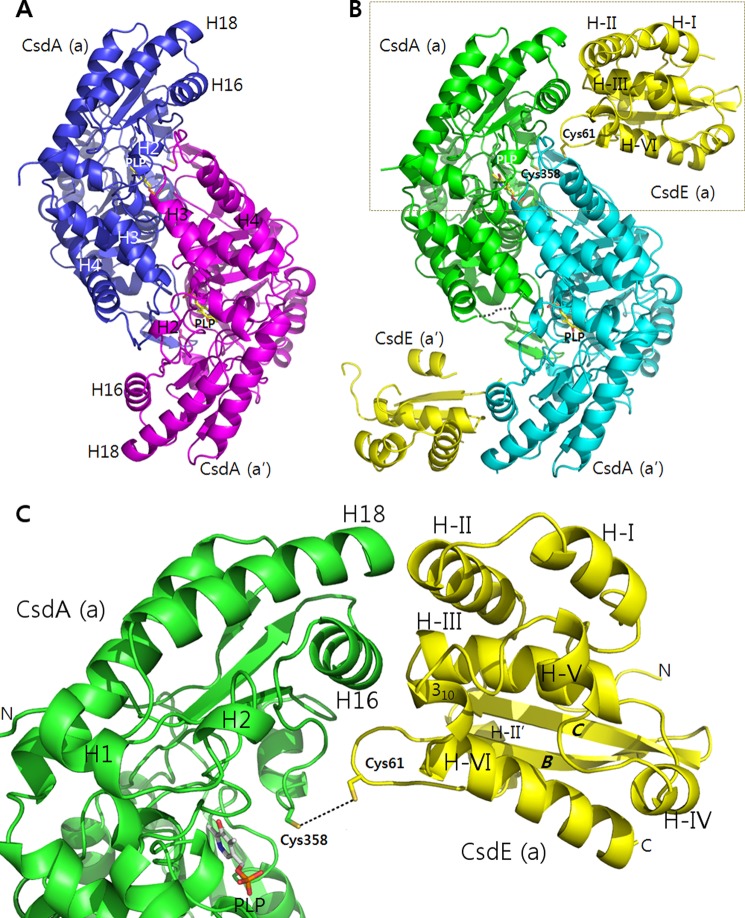
FIGURE 1.
Structures of free CsdA and CsdA-CsdE. A, dimeric CsdA is shown with designation of CsdE interaction-mediating structural elements. B, dimeric CsdA-CsdE is shown with active cysteines, indication of the missing loop in CsdA(a), and the designation of structural elements on CsdE. Regions of CsdE(a′) with higher B-factors than the average (55.9 Å2) are not included in the ribbon diagram. C, boxed region in B is magnified for clarity.
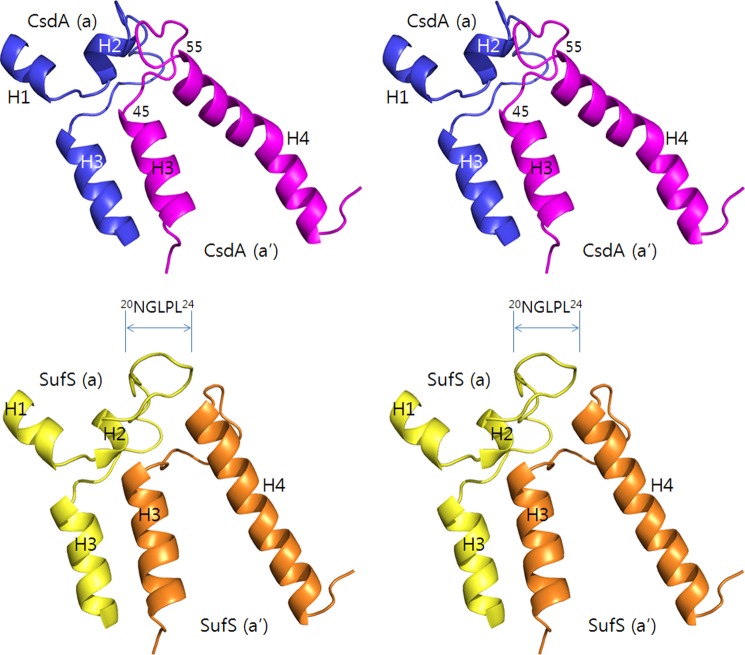
FIGURE 2.
Structural differences between CsdA and SufS. Structural differences between CsdA and SufS are pronounced at regions of H2, H3 (dimeric interface), and H4 as shown by wall-eye stereoview. These regions in the dimeric CsdA (a in blue; a′ in violet) and SufS (a in yellow; a′ in orange) are shown to visualize the differences of a shorter H2-H3 loop and an H4 helical kink in CsdA. The longer H2-H3 loop of SufS(a) (20NGLPL24) shifts the orientation of the H3-H4 loop in the interacting SufS(a′). The structure of E. coli SufS was analyzed using PDB code 1C0N.
The small region in CsdA consists of a four-stranded anti-parallel β-sheet flanked by three α-helices. The enzymatically essential PLP was observed between the large and small regions (Fig. 1A). The Schiff base linkage of PLP to CsdA Lys-222 was observed near the active Cys-358. The active Cys-358 was well ordered with the same conformation in all three CsdAs of the asymmetric units (Fig. 3). Interestingly, glycerol used as a cryo-protectant was observed in the active site (Fig. 3). This same region in E. coli SufS (12) and Thermotoga maritima NifS-like protein (29) has been reported to be occupied by acetate and sulfate, respectively, which were both from the crystallization conditions. This region is also where the substrates cysteine (29), selenocysteine (13), and cysteine persulfide (13, 28) are located in SufS and related cysteine desulfurase structures. In our structure, neither DTT modification nor polysulfide modification of active Cys-358 reported in other studies (9, 30, 31) was seen.
FIGURE 3.
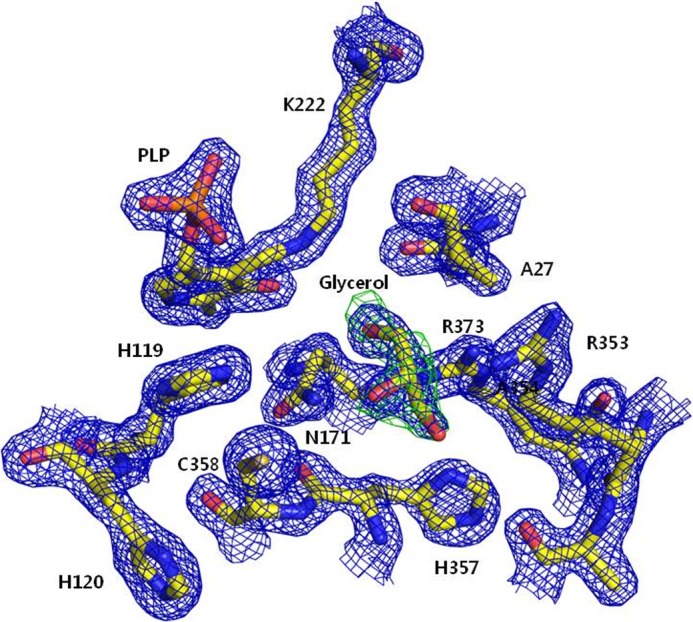
Electron densities near the active Cys-358 of CsdA. The view of Cys-358 active site in one representative CsdA subunit is shown in ball-and-stick representation with electron density maps. 2Fo − Fc density in blue is contoured at 1.5σ, and Fo − Fc density of the glycerol OMIT map is contoured at 4σ. PLP and glycerol are observed in all the three CsdA subunits.
Structural comparisons between CsdA and SufS have failed to explain how CsdA and SufS are able to discriminate sulfur and selenium, respectively. The residues composing the active sites in CsdA and SufS are more or less conserved. Because the ionic radius differences of sulfide and selenide are only ∼1 Å, the molecular determinants for S/Se selectivity must rely on the subtle differences in the micro-environment around the substrate (cysteine or selenocysteine). The structure of a rat selenocysteine lyase suggested that the slight difference in the substrate-binding modes, which was mediated by the active cysteine, confers the S/Se selectivity (32). A more recent study using a structure-guided bioinformatics approach reported that the single aspartate near the PLP-binding site of human selenocysteine lyase was responsible for the enzymatic specificity (33). Interestingly, these aspartates that are conserved among selenocysteine lyases are histidines in both CsdA and SufS. In any case, the molecular determinants of S/Se selectivity yet unrecognized between CsdA and SufS merit further investigation.
Structural Analysis of the CsdA-CsdE
The structure of E. coli CsdA-CsdE was determined at 2.0-Å resolution using molecular replacement with free E. coli CsdA as a search model, followed by automated model building, which further located the two CsdEs. CsdA-CsdE consists of a CsdA dimer (each subunit denoted as “a” and “a′”) and two CsdEs (a and a′), which independently interact with the cognate CsdA (Fig. 1, B and C). Unlike the continuous electron density observed throughout the free CsdA structure, part of the H3-H4 loop (CsdA residues 49–54) of one CsdA(a) was not observed. However, all regions were ordered in the other CsdA(a′). The active Cys-358 was ordered in both subunits. For CsdE, continuous electron density was also observed for only one CsdE(a). The sulfur-accepting active Cys-61-containing loop was clearly discernible in CsdE(a) with only the side chain of Cys-61 disordered. In the other CsdE(a′), the entire Cys-61 was disordered. Judging from the overall temperature B-factors, CsdE(a) was more ordered than CsdE(a′) (30.0 versus 55.9 Å2). Several regions of CsdE(a′), which include helices II′, IV, VI, loop of β-sheets B–C, and loop of helices V–VI have higher B-factors than the 55.9-Å2 average.
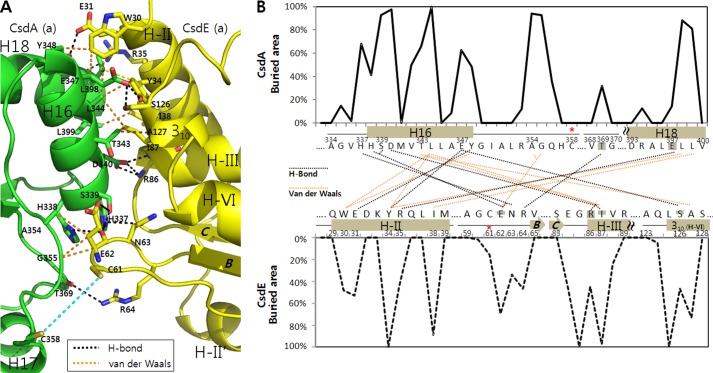
Analysis of the CsdA and CsdE interface indicates that the small region (described above) of CsdA mediates the contact with the cognate CsdE (Fig. 1, B and C). In particular, three CsdA elements (H16, Cys-358–loop, and H18) are dedicated for interactions with CsdE (H-II, Cys-61–loop, H-III and a partial 310-helix of H-VI) (detailed interaction analyzed in Table 2 and Fig. 4). Briefly, the N-terminal residues of CsdA H16 (337HHS339) interact with the CsdE Cys-61–loop (62EN63), whereas the following residues of H16 interact with multiple regions of CsdE (N-terminal half of H-II, N-terminal end of H-III, and 310-helix of H-VI). The CsdA Cys-358–loop (354AG355) interacts solely with Glu-62 of the CsdE Cys-61–loop. The C-terminal end of CsdA H18 (397ELL399) interacts with the C-terminal half of CsdE H-II.
TABLE 2.
Residue-by-residue interactions between CsdA and CsdE
H means hydrogen bond, and VDW means van der Waals interaction.
| CsdA | CsdE | Distance | ||
|---|---|---|---|---|
| Å | ||||
| His-337 | NE2 | Glu-62 | O | 3.5 (H) |
| Asn-63 | OD1 | 3.7 (H) | ||
| His-338 | ND1 | Glu-62 | O | 3.5 (H) |
| Ser-339 | OG | Glu-62 | O | 2.3 (H) |
| OG | OE2 | 3.7 (H) | ||
| Asp-340 | OD1 | Ile-87 | N | 2.6 (H) |
| OD2 | Arg-86 | N | 2.6 (H) | |
| Thr-343 | CG2 | Ile-87 | CD1 | 4.2 (VDW) |
| Ala-127 | CB | 3.9 (VDW) | ||
| Leu-344 | CD2 | Trp-30 | CZ3 | 4.5 (VDW) |
| Tyr-34 | CD2 | 3.7 (VDW) | ||
| Ile-87 | CD1 | 3.9 (VDW) | ||
| Glu-347 | OE2 | Tyr-34 | OH | 2.5 (H) |
| Ser-126 | N | 2.9 (H) | ||
| OG | 2.7 (H) | |||
| CB | Trp-30 | CH2 | 3.6 (VDW) | |
| Tyr-348 | CE1 | Trp-30 | CH2 | 3.3 (VDW) |
| OH | Glu-31 | OE2 | 2.6 (H) | |
| Ala-354 | CB | Glu-62 | CB | 4.1 (VDW) |
| Gly-355 | CA | Glu-62 | CA | 3.4 (VDW) |
| CB | 3.6 (VDW) | |||
| Thr-369 | O | Arg-64 | NH2 | 3.8 (H) |
| Glu-397 | O | Arg-35 | NH1 | 3.0 (H) |
| Leu-398 | CD2 | Tyr-34 | CB | 4.3 (VDW) |
| Leu-399 | CD2 | Ile-38 | CD1 | 3.6 (VDW) |
FIGURE 4.
Interfacial residues of CsdA and CsdE. A, residues of CsdA and CsdE that mediate the interaction surface are indicated with bond types. Hydrogen bonds (black dotted lines) and van der Waal interactions (orange dotted lines), which are within the documented distances, are indicated as shown in Table 2. The distance between the two active cysteines of CsdA and CsdE is shown (in cyan) for clear perspective in reference to the previous figure. B, residue-by-residue buried area percentages for the CsdA and CsdE residues were calculated using the web-based PISA server. In the interaction diagram, residue pairs that undergo either the hydrogen bonding (black) or the van der Waals interaction (orange) are shown with dotted lines. Interactions mediated by the main chains (solely or partly) are noted by shading (complete or partial, respectively) on the participating residues.
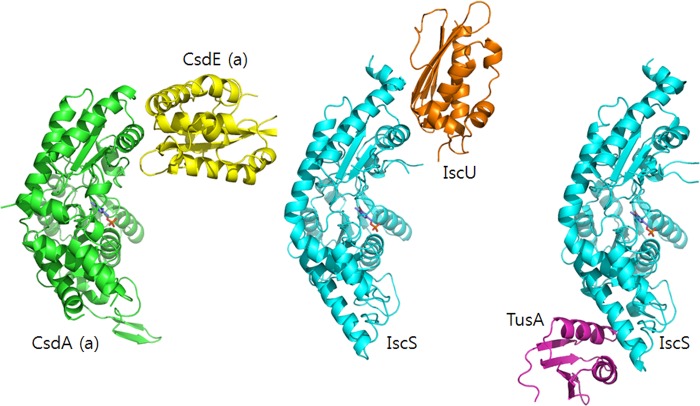
The buried accessible surface area between CsdA and CsdE (731 Å2) was similar to IscS and its interacting IscU (767 Å2) or TusA (702 Å2). However, interfacial modes are different between the interaction pairs (Fig. 5). In IscS-TusA, completely different structural elements of IscS in comparison with CsdA are utilized for the TusA interaction. In IscS-IscU, similar regions of IscS corresponding to that of CsdA are utilized for the IscU interaction. This similarity may originate from the fact that unlike the fold of TusA, the α/β-sandwich folds of CsdE (14, 17) and IscU (34) are alike (35). Nevertheless, the active cysteines of CsdE and IscU are contained on different structural elements. Hence, the mode of CsdE surface projecting to CsdA is distinct from that of IscU and IscS (Fig. 5). The different interaction modes seen in IscS-TusA and IscS-IscU indicate the versatility of IscS in using multiple surfaces for the protein interactions. Hence, CsdA may also invoke other regions than the CsdE interaction surface for other unknown interactions.
FIGURE 5.
Different interfaces of cysteine desulfurases and the sulfur acceptors. Protein interaction modes of CsdA-CsdE, IscS-IscU, and IscS-TusA are shown on one monomeric subunit of CsdA (and IscS) for clarity. The E. coli IscS-IscU and IscS-TusA are from PDB accession codes, 3LVL and 3LVK, respectively.
Analysis of the CsdA upon CsdE Interaction
Analysis of free CsdA and CsdA in CsdA-CsdE indicated that no significant change was observed in CsdA. Interestingly, the active CsdA Cys-358-containing loop in both cases had identical conformation with the Cys-358 side chain completely solvent-exposed (Fig. 4B). It is interesting to observe that the active cysteine of CsdA needs no conformational oscillation for sulfur delivery to the acceptor. In the structure of E. coli IscS, the corresponding active cysteine (Cys-328) was disordered (27); however, the accessibility of the nearby ordered residue (Ala-327) suggests that the cysteine is also solvent-exposed. Unfortunately, this cysteine-containing loop of E. coli IscS in complex with IscU in the apo-form was also found disordered (7). However, the active IscS cysteine (Cys-321) in the Fe2S2-containing A. fulgidus IscS-IscU holo-structure was ordered and became ∼55% buried upon IscU interaction (8). Additionally, Marinoni et al. (8) report that the Cys-321–loop of A. fulgidus IscS undergoes significant structural movement of ∼14 Å toward IscU when compared with the ordered active cysteine of the Synechocystis SufS structure (8, 28). Hence, the oscillation of the IscS cysteine loop seems essential for the sulfur delivery in the case of Fe-S generation within IscU. In E. coli IscS-TusA, the IscS cysteine loop underwent no significant change upon TusA interaction as in CsdA. However, the active cysteine was ∼33% buried upon TusA interaction (7).
As described above, part of the H3-H4 loop in one CsdA(a) of the CsdA-CsdE was disordered. This loop, which was ordered in the free CsdA, is mostly important for the dimeric CsdA interaction. No significant chemical interactions between CsdA(a) and the CsdE(a′) were seen; however, the helices and loops of CsdA H3 and H4 were spatially close to CsdE (Fig. 1). Further analysis on the structural differences of CsdA in the free form and in CsdA-CsdE suggests that the disorder of the H3-H4 loop in CsdA(a) transformed H2 of the interacting CsdA(a′) into a short 310-helix (Fig. 6). Hence, the propagated change in H2 of the CsdA(a′) with its proximity to CsdA H16, which is at the center of CsdE interaction, may result in the disordering of regions of CsdE(a′) in the crystal (Fig. 1B). It remains to be seen whether this H3-H4 loop has any influence in controlling the turnover of the enzyme by changing the affinity between CsdA and CsdE.
FIGURE 6.
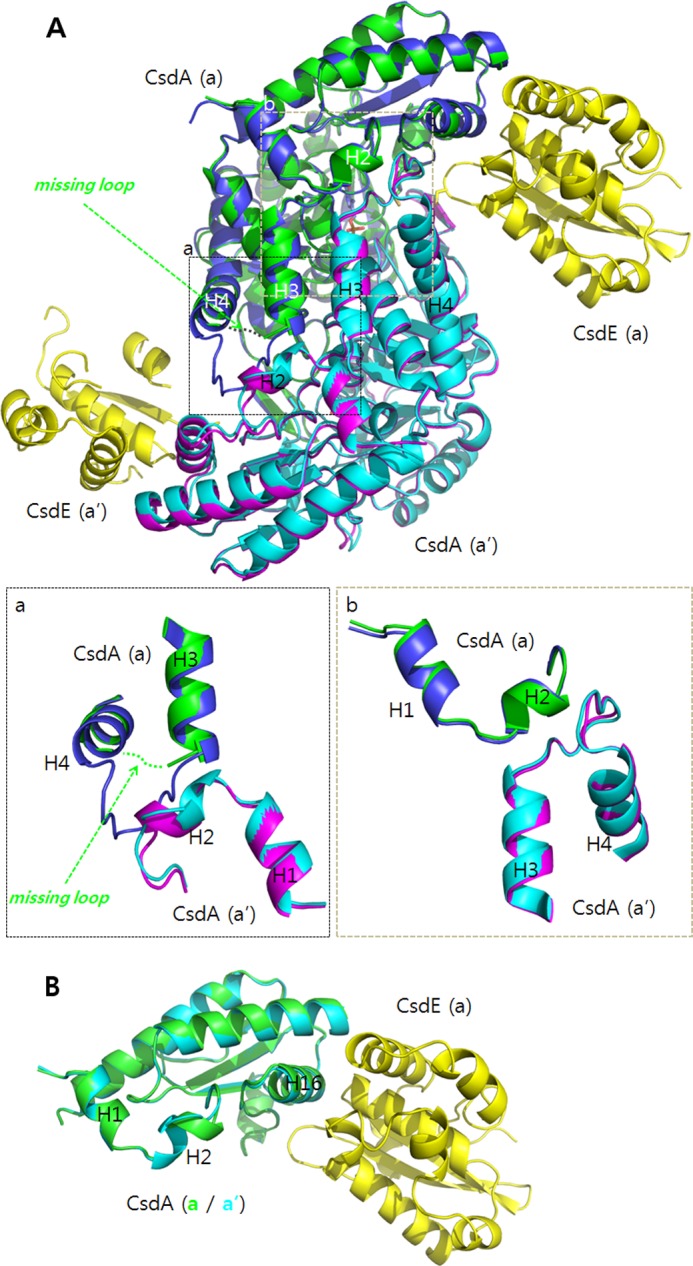
Structural differences between the free CsdA and the CsdA from CsdA-CsdE. A, two dimeric subunits of free CsdA (a in blue; a′ in violet) and CsdA from CsdA-CsdE (a in green; a′ in cyan) are structurally aligned. Both CsdEs are shown in yellow. Significant differences are seen near the dimeric interface. For one CsdA(a), part of the H3-H4 loop (CsdA residues 49–54) is disordered. The corresponding residues are ordered for CsdA(a′). The ordering in H3-H4 loop of CsdA(a′) changes the helical content of H2 in the interacting CsdA(a) (magnified in the inset, a and b). B, changes in H2 are clearly seen when the two subunits of CsdA from the CsdA-CsdE are superimposed for comparison.
Analysis of the CsdE upon CsdA Interaction
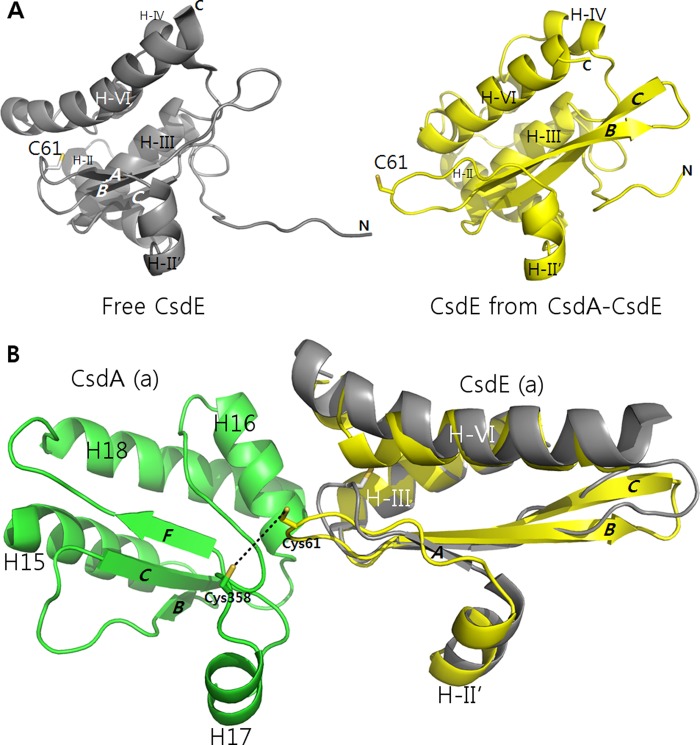
The fold of free CsdE, which was uncovered in a solution structure, is a compact two-layered α/β-sandwich with a central three-stranded anti-parallel β-sheet and seven α-helices (24). In contrast to CsdA, the structural changes taking place in CsdE upon the CsdA interaction were more pronounced (Fig. 7A). In the free CsdE, the sulfur-accepting Cys-61 is located at the low accessible hydrophobic cavity of a loop that connects the two anti-parallel β-strands (β-strands A and B). This internally positioned cysteine along with the loop shifts significantly upon CsdA interaction moving ∼11 Å in distance (Fig. 7B). This conformational oscillation of CsdE Cys-61 is unique when compared with TusA and IscU interactions to IscS. Unlike in the case of CsdE, the active cysteine of free TusA is not buried, and no major conformational change in the structural elements occurs in TusA upon IscS interaction (7). For an Fe-S sulfur acceptor IscU, sulfur from IscS is delivered to complete the Fe-S within IscU. Hence, as described previously, only the movement of the cysteine loop of IscS was pronounced (8). Thus, the conformational flexibility of CsdE newly observed in CsdA-CsdE provides the framework for understanding sulfur transfer in yet another cysteine desulfurase and sulfur acceptor.
FIGURE 7.
Changes in the structure of CsdE upon CsdA interaction. A, free CsdE and CsdE from CsdA-CsdE are shown at an angle to illustrate the structural changes particularly on the CsdE Cys-61–loop. B, free CsdE is superimposed onto CsdE from CsdA-CsdE to emphasize movement of the CsdE Cys-61–loop. Free E. coli CsdE is from PDB accession code 1NI7.
Although the exact orientation of the CsdE Cys-61 side chain in CsdA-CsdE was not discernible, the closest distance between the two active cysteines of CsdA and CsdE was estimated to be ∼6 Å. Given that the transition state of the trans-persulfuration reaction contains three sulfur atoms and that the disulfide bond length is 2.1–2.3 Å, the two captured cysteines of CsdA and CsdE in the structure must be of the intermediate stage during the trans-persulfuration reaction. Of note, the thiol groups of active cysteines in IscS-TusA are positioned in closer proximity with a distance of ∼4.5 Å (7).
Notably, further changes occurred in CsdE with the CsdA interaction, including β-sheet A transformation into an extended form, and elongation of the β-sheets B and C (Fig. 7). Additional changes were also observed in CsdE H-VI whose N-terminal end was modified into a short 310-helix. Furthermore, the 10 flexible N-terminal residues of CsdE, which were observed with multiple main chain conformations in the solution structure, became ordered in the crystal CsdA-CsdE structure. However, whether this is due to the CsdA interaction or from a crystallization artifact is inconclusive.
Prediction of the SufS-SufE Interaction
As noted earlier, the overall structure of CsdA is very similar to SufS except for some regions mediating the dimerization interface. Additionally, as indicated from the ∼35% sequence identity between CsdE and SufE, the folds of the two proteins are identical with a root mean square deviation of ∼2.3 Å for aligning ∼137 Cα atoms. Similar to Cys-61 of the free CsdE, the sulfur-accepting cysteine of SufE was also located in a deep cleft (14). The conservation pattern of the interaction-mediating residues in CsdA/SufS (H16, Cys-358–loop, and H18) and in CsdE/SufE (H-II, H-III, and H-VI) suggests that the interface of SufS-SufE likely resembles that of CsdA-CsdE (Fig. 8). Sequence alignment of CsdE/SufE near the sulfur-accepting cysteines also indicates that conformational oscillation, which is observed for the CsdE Cys-61–loop, is likely to occur on the corresponding SufE region. Thus, the structural analysis of CsdA-CsdE further supports the notion that CsdA/CsdE and SufS/SufE work similarly and that the proteins may have been derived from a common ancestor. Thus, CsdE and SufE likely share some of the protein interactomes as demonstrated by others (36).
FIGURE 8.

Sequence alignments of CsdA/SufS and CsdE/SufE at the CsdA-CsdE interface. Sequences of E. coli CsdA/SufS and CsdE/SufE are aligned at the interfacial regions that mediate the CsdA-CsdE interaction. Interaction types are indicated with different colored boxes (hydrogen bonding in blue; van der Waals interactions in red). Residues mediating interactions by main chains (solely or partly) are indicated with shading (complete or partial, respectively) in the boxes.
This work was supported by Basic Science Research Program Grant 2013R1A1A2005276 through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (MEST). Experiments performed at the Pohang Light Source were supported in part by MEST and the Pohang University of Science and Technology.
The atomic coordinates and structure factors (codes 4LW2 and 4LW4) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- PLP
- pyridoxal 5′-phosphate
- PDB
- Protein Data Bank.
REFERENCES
- 1. Johnson D. C., Dean D. R., Smith A. D., Johnson M. K. (2005) Structure, function and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74, 247–281 [DOI] [PubMed] [Google Scholar]
- 2. Fontecave M., Ollagnier-de-Choudens S. (2008) Iron-sulfur cluster biosynthesis in bacteria, mechanisms of cluster assembly and transfer. Arch. Biochem. Biophys. 474, 226–237 [DOI] [PubMed] [Google Scholar]
- 3. Py B., Barras F. (2010) Building Fe-S proteins: bacterial strategies. Nat. Rev. Microbiol. 8, 436–446 [DOI] [PubMed] [Google Scholar]
- 4. Lill R., Mühlenhoff U. (2005) Iron-sulfur-protein biogenesis in eukaryotes. Trends Biochem. Sci. 30, 133–141 [DOI] [PubMed] [Google Scholar]
- 5. Zheng L., White R. H., Cash V. L., Jack R. F., Dean D. R. (1993) Cysteine desulfurase activity indicates a role for NIFS in metallocluster biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 90, 2754–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mihara H., Kurihara T., Yoshimura T., Soda K., Esaki N. (1997) Cysteine sulfinate desulfinase, a NIFS-like protein of Escherichia coli with selenocysteine lyase and cysteine desulfurase activities. Gene cloning, purification, and characterization of a novel pyridoxal enzyme. J. Biol. Chem. 272, 22417–22424 [DOI] [PubMed] [Google Scholar]
- 7. Shi R., Proteau A., Villarroya M., Moukadiri I., Zhang L., Trempe J. F., Matte A., Armengod M. E., Cygler M. (2010) Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions. PLoS Biol. 8, e1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marinoni E. N., de Oliveira J. S., Nicolet Y., Raulfs E. C., Amara P., Dean D. R., Fontecilla-Camps J. C. (2012) (IscS-IscU)2 complex structures provide insights into Fe2S2 biogenesis and transfer. Angew. Chem. Int. Ed. Engl. 51, 5439–5442 [DOI] [PubMed] [Google Scholar]
- 9. Sendra M., Ollagnier de Choudens S., Lascoux D., Sanakis Y., Fontecave M. (2007) The SUF iron-sulfur cluster biosynthetic machinery: sulfur transfer from the SUFS-SUFE complex to SUFA. FEBS Lett. 581, 1362–1368 [DOI] [PubMed] [Google Scholar]
- 10. Layer G., Gaddam S. A., Ayala-Castro C. N., Ollagnier-de Choudens S., Lascoux D., Fontecave M., Outten F. W. (2007) SufE transfers sulfur from SufS to SufB for iron-sulfur cluster assembly. J. Biol. Chem. 282, 13342–13350 [DOI] [PubMed] [Google Scholar]
- 11. Mihara H., Maeda M., Fujii T., Kurihara T., Hata Y., Esaki N. (1999) A nifS-like gene, csdB, encodes an Escherichia coli counterpart of mammalian selenocysteine lyase. Gene cloning, purification, characterization, and preliminary x-ray crystallographic studies. J. Biol. Chem. 274, 14768–14772 [DOI] [PubMed] [Google Scholar]
- 12. Fujii T., Maeda M., Mihara H., Kurihara T., Esaki N., Hata Y. (2000) Structure of an NifS homologue: x-ray structure analysis of CsdB, an Escherichia coli counterpart of mammalian selenocysteine lyase. Biochemistry 39, 1263–1273 [DOI] [PubMed] [Google Scholar]
- 13. Lima C. D. (2002) Analysis of the E. coli NifS CsdB protein at 2.0 Å reveals the structural basis for perselenide and persulfide intermediate formation. J. Mol. Biol. 315, 1199–1208 [DOI] [PubMed] [Google Scholar]
- 14. Goldsmith-Fischman S., Kuzin A., Edstrom W. C., Benach J., Shastry R., Xiao R., Acton T. B., Honig B., Montelione G. T., Hunt J. F. (2004) The SufE sulfur acceptor protein contains a conserved core structure that mediates interdomain interactions in a variety of redox protein complexes. J. Mol. Biol. 344, 549–565 [DOI] [PubMed] [Google Scholar]
- 15. Loiseau L., Ollagnier-de Choudens S., Lascoux D., Forest E., Fontecave M., Barras F. (2005) Analysis of the heteromeric CsdA-CsdE cysteine desulfurase, assisting Fe-S cluster biogenesis in Escherichia coli. J. Biol. Chem. 280, 26760–26769 [DOI] [PubMed] [Google Scholar]
- 16. Trotter V., Vinella D., Loiseau L., Ollagnier de Choudens S., Fontecave M., Barras F. (2009) The CsdA cysteine desulphurase promotes Fe/S biogenesis by recruiting Suf components and participates to a new sulphur transfer pathway by recruiting CsdL (ex-YgdL), a ubiquitin-modifying-like protein. Mol. Microbiol. 74, 1527–1542 [DOI] [PubMed] [Google Scholar]
- 17. Liu G., Li Z., Chiang Y., Acton T., Montelione G. T., Murray D., Szyperski T. (2005) High-quality homology models derived from NMR and x-ray structures of E. coli proteins YgdK and SufE suggest that all members of the YgdK/Suf E protein family are enhancers of cysteine desulfurases. Protein Sci. 14, 1597–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Adinolfi S., Rizzo F., Masino L., Nair M., Martin S. R., Pastore A., Temussi P. A. (2004) Bacterial IscU is a well folded and functional single domain protein. Eur. J. Biochem. 271, 2093–2100 [DOI] [PubMed] [Google Scholar]
- 19. Gill S. C., von Hippel P. H. (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182, 319–326 [DOI] [PubMed] [Google Scholar]
- 20. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 21. McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., Read R. J. (2005) Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 61, 458–464 [DOI] [PubMed] [Google Scholar]
- 22. Vagin A., Teplyakov A. (1997) MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 23. Perrakis A., Morris R., Lamzin V. S. (1999) Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6, 458–463 [DOI] [PubMed] [Google Scholar]
- 24. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 25. Vagin A. A., Steiner R. S., Lebedev A. A., Potterton L., McNicholas S., Long F., Murshudov G. N. (2004) REFMAC5 dictionary: organisation of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr. 60, 2284–2295 [DOI] [PubMed] [Google Scholar]
- 26. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 27. Cupp-Vickery J. R., Urbina H., Vickery L. E. (2003) Crystal structure of IscS, a cysteine desulfurase from Escherichia coli. J. Mol. Biol. 330, 1049–1059 [DOI] [PubMed] [Google Scholar]
- 28. Clausen T., Kaiser J. T., Steegborn C., Huber R., Kessler D. (2000) Crystal structure of the cystine C-S lyase from synechocystis: stabilization of cysteine persulfide for FES cluster biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 97, 3856–3861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kaiser J. T., Clausen T., Bourenkow G. P., Bartunik H. D., Steinbacher S., Huber R. (2000) Crystal structure of a NifS-like protein from Thermotoga maritima: implications for iron sulphur cluster assembly. J. Mol. Biol. 297, 451–464 [DOI] [PubMed] [Google Scholar]
- 30. Smith A. D., Agar J. N., Johnson K. A., Frazzon J., Amster I. J., Dean D. R., Johnson M. K. (2001) Sulfur transfer from IscS to IscU: the first step in iron-sulfur cluster biosynthesis. J. Am. Chem. Soc. 123, 11103–11104 [DOI] [PubMed] [Google Scholar]
- 31. Ollagnier-de-Choudens S., Lascoux D., Loiseau L., Barras F., Forest E., Fontecave M. (2003) Mechanistic studies of the SufS-SufE cysteine desulfurase: evidence for sulfur transfer from SufS to SufE. FEBS Lett. 555, 263–267 [DOI] [PubMed] [Google Scholar]
- 32. Omi R., Kurokawa S., Mihara H., Hayashi H., Goto M., Miyahara I., Kurihara T., Hirotsu K., Esaki N. (2010) Reaction mechanism and molecular basis for selenium/sulfur discrimination of selenocysteine lyase. J. Biol. Chem. 285, 12133–12139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Collins R., Johansson A. L., Karlberg T., Markova N., van den Berg S., Olesen K., Hammarström M., Flores A., Schüler H., Schiavone L. H., Brzezinski P., Arnér E. S., Högbom M. (2012) Biochemical discrimination between selenium and sulfur 1: a single residue provides selenium specificity to human selenocysteine lyase. PLoS One 7, e30581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shimomura Y., Wada K., Fukuyama K., Takahashi Y. (2008) The asymmetric trimeric architecture of [2Fe-2S] IscU: implications for its scaffolding during iron-sulfur cluster biosynthesis. J. Mol. Biol. 383, 133–143 [DOI] [PubMed] [Google Scholar]
- 35. Katoh E., Hatta T., Shindo H., Ishii Y., Yamada H., Mizuno T., Yamazaki T. (2000) High precision NMR structure of YhhP, a novel Escherichia coli protein implicated in cell division. J. Mol. Biol. 304, 219–229 [DOI] [PubMed] [Google Scholar]
- 36. Bolstad H. M., Botelho D. J., Wood M. J. (2010) Proteomic analysis of protein-protein interactions within the cysteine sulfinate desulfinase Fe-S cluster biogenesis system. J. Proteome Res. 9, 5358–5369 [DOI] [PMC free article] [PubMed] [Google Scholar]