Background: BER of quadruplex structures and telomere DNA has not been well studied.
Results: Neil3 and NEIL1 remove damages from quadruplex DNA and show telomeric sequence context effects.
Conclusion: Neil3 and NEIL1 may be involved in repair of damages in quadruplex DNA.
Significance: This is the first evidence of glycosylase activity on quadruplex DNA and suggests new roles for these enzymes.
Keywords: Base Excision Repair, DNA Damage, DNA Repair, DNA Structure, Telomeres, DNA Glycosylases, NEIL1, Neil3, Quadruplex, Sequence Context
Abstract
The telomeric DNA of vertebrates consists of d(TTAGGG)n tandem repeats, which can form quadruplex DNA structures in vitro and likely in vivo. Despite the fact that the G-rich telomeric DNA is susceptible to oxidation, few biochemical studies of base excision repair in telomeric DNA and quadruplex structures have been done. Here, we show that telomeric DNA containing thymine glycol (Tg), 8-oxo-7,8-dihydroguanine (8-oxoG), guanidinohydantoin (Gh), or spiroiminodihydantoin (Sp) can form quadruplex DNA structures in vitro. We have tested the base excision activities of five mammalian DNA glycosylases (NEIL1, NEIL2, mNeil3, NTH1, and OGG1) on these lesion-containing quadruplex substrates and found that only mNeil3 had excision activity on Tg in quadruplex DNA and that the glycosylase exhibited a strong preference for Tg in the telomeric sequence context. Although Sp and Gh in quadruplex DNA were good substrates for mNeil3 and NEIL1, none of the glycosylases had activity on quadruplex DNA containing 8-oxoG. In addition, NEIL1 but not mNeil3 showed enhanced glycosylase activity on Gh in the telomeric sequence context. These data suggest that one role for Neil3 and NEIL1 is to repair DNA base damages in telomeres in vivo and that Neil3 and Neil1 may function in quadruplex-mediated cellular events, such as gene regulation via removal of damaged bases from quadruplex DNA.
Introduction
Cells are continuously exposed to endogenous and environmental insults such as reactive oxygen species, genotoxic agents, and ionizing radiation, which cause DNA damage. Reactive oxygen species are by-products of cellular respiration and play important physiological roles in healthy cells (1). Unfortunately, DNA bases are particularly susceptible to reactive oxygen species, and oxidative base damage is almost an inevitable consequence (2, 3). DNA base damages may be mutagenic and, if left unrepaired, may lead to base mispairing, blockage of DNA polymerases, and eventually result in genomic instability (2, 3). For example, thymine glycol (Tg),3 the major oxidation product of DNA thymine, is an efficient block to DNA polymerases and thus a lethal lesion in cells (4). 8-Oxo-7,8-dihydroguanine (8-oxoG), a major oxidation product of DNA guanine, and 5-hydroxyuracil (5-OHU), an oxidation product of DNA cytosine, are important premutagenic lesions both mispairing with adenine (5, 6). The further oxidation products of 8-oxoG, guanidinohydantoin (Gh), and spiroiminodihydantoin (Sp) can also mispair with adenine as well as block some DNA polymerases (7, 8).
Base excision repair is the predominant pathway for repairing oxidative DNA base damages. DNA glycosylases are the first enzymes in base excision repair that recognize damaged bases, excise the lesions, and provide substrates for later enzymes in the pathway (9). Five DNA glycosylases that are specific for oxidative DNA base damages have been identified in human cells, namely OGG1, NTH1, NEIL1, NEIL2, and NEIL3 (2, 3). Although there is much redundancy in substrate specificity, each glycosylase has a unique signature in terms of lesion type and DNA conformation. For example, the mammalian glycosylases OGG1 and NTH1 specifically remove oxidized purines and pyrimidines, respectively, from duplex DNA (10). NEIL1, NEIL2, and NEIL3 prefer oxidized pyrimidine and some purine damages (11, 12), with NEIL2 and NEIL3 preferring lesions in single-stranded DNA (13–15).
Telomeres are DNA-protein complexes at the end of chromosomes. By capping the chromosome ends, telomeres prevent degradation and undesired fusion of these ends (16). Telomeres also suppress improper activation of DNA damage response pathways (17). Both the length and integrity of this deoxyribonucleoprotein complex has to be carefully maintained. Shortening and loss of function of telomeres lead to telomere-mediated cell senescence and premature aging syndromes in humans (18, 19). Additionally, telomere lengthening due to overactive telomerase has been linked to cancer (20).
Under physiological salt conditions, human telomere repeats (TTAGGG)n can form quadruplex (G4) structures (21, 22), which are four-stranded DNA topologies mediated by guanine-rich sequences. Several quadruplex structures containing the telomere sequence have been solved in both Na+ (23) and K+ (24) solutions. The basic building blocks of G-quadruplexes are layers of guanine tetrads, which consist of four guanines that pair with each other via Hoogstein base pairs. A quadruplex consists of two or more layers of guanine tetrads, one stacking onto the other with a monovalent cation (K+ or Na+) sandwiched within the tetrads (Fig. 1D).
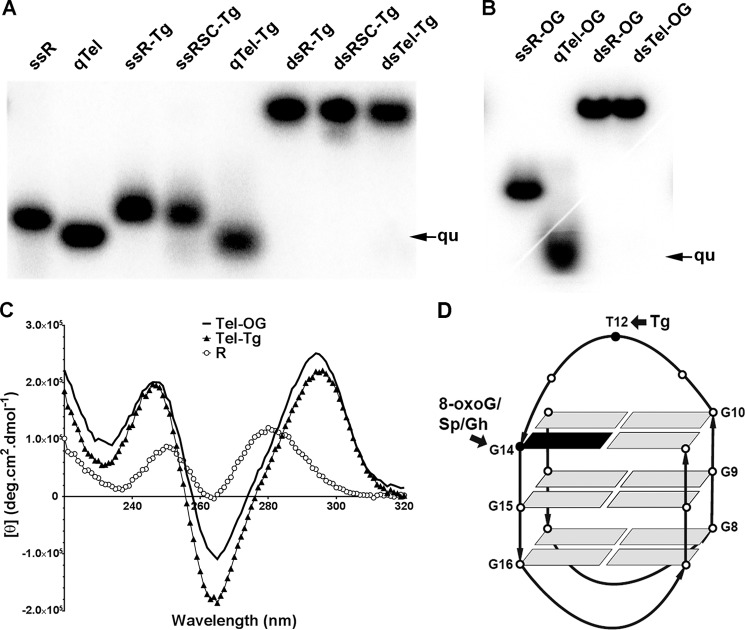
FIGURE 1.
Telomeric DNA sequences containing Tg or 8-oxoG form quadruplex DNA. All samples were prepared in 10 mm sodium phosphate (pH 7.0) and 100 mm Na+. Samples were separated using native gel electrophoresis or measured by circular dichroism as described under “Experimental Procedures.” A, Tg-containing telomeric DNA forms a quadruplex. q, quadruplex DNA; ss, single-stranded DNA; ds, double-stranded DNA. B, 8-oxoG-containing telomeric DNA forms a quadruplex. C, CD spectrum confirms formation of an antiparallel quadruplex structure. Tel-Tg and Tel-OG show two peaks at 265 and 295 nm. D, scheme of an antiparallel quadruplex structure. Arrows indicate positions of lesion substitution.
G4-forming sequences are prevalent throughout the human genome (25, 26). Although most structural studies of quadruplexes were done in vitro, there is increasing evidence supporting the existence of quadruplex structures in cells (27, 28). Quadruplex structures have been proposed to play inhibitory roles during DNA replication, gene transcription, and mRNA translation (29). Most interestingly, quadruplex structures inhibit telomerase activity, which, together with the inhibitory role of quadruplexes on oncogenes (30), has led to research on quadruplex DNA as a target for cancer treatment (31).
Guanine has the lowest redox potential among the four bases and thus is more susceptible to oxidation. Because of their G-rich nature, G4-forming sequences are vulnerable targets for oxidation through DNA charge transport (32, 33). For example, telomeric DNA (TTAGGG)n is more sensitive to UVA radiation (34) and hydrogen peroxide (35) than non-telomeric DNA with the same nucleotide composition. Also, cellular oxidative stress induces 8-oxoG production in telomeres (36), and the production is higher than in other regions of the genome (37). DNA base damages in telomeres must be repaired to preserve telomere integrity because accumulation of base damages in telomeres may hinder telomerase activity (38) and/or disrupt the telomere-guarding shelterin complex (39).
Little biochemical work has been published on how glycosylases remove base lesions from quadruplex DNA or from the telomeric sequence context. Here, we tested the glycosylase activity of five mammalian oxidative DNA glycosylases, OGG1, NTH1, NEIL1, NEIL 2, and mNeil3, on quadruplex DNA containing base damages. We also investigated the impact of the telomeric sequence context on the activity of these glycosylases. Our results suggest that Neil3 and Neil1 may function in repair of damages harbored in quadruplex DNA and may play a role in base excision repair in telomeres.
EXPERIMENTAL PROCEDURES
Substrates
Sequences of oligodeoxyribonucleotides used in this study and a brief description of each are listed in Table 1. The telomeric DNA sequence (Tel) used was d(AGGGTTAGGGTTAGGGTTAGGG). A random sequence (R) with the same nucleotide composition as the telomeric sequence was used as a control. The random sequence containing the telomeric sequence context (RSC) has the same backbone sequence as R, except that there is a “telomeric sequence patch” surrounding the damage. Oligodeoxyribonucleotides with Tg, 8-oxoG, or 5-OHU as well as all of the non-lesion-containing oligodeoxyribonucleotides were purchased from Midland Certified Reagent Co. (Midland, TX). Gh- and Sp-containing substrates were synthesized as described previously (40). The Tel-Sp oligodeoxyribonucleotides used here were a mixture of Sp1 and Sp2 due to the inability to resolve these diastereomers under the purification method utilized.
TABLE 1.
Oligodeoxynucleotide sequences used in this study
| Name | Sequence | Description |
|---|---|---|
| Tel | AGGGTTAGGGTTAGGGTTAGGG | Telomeric sequence without lesion |
| Tel-Tg | AGGGTTAGGGT (Tg)AGGGTTAGGG | Telomeric sequence with Tg |
| Tel-OG | AGGGTTAGGGTTA (8OG)GGTTAGGG | 8-oxoG at 5′ of GGG tetrad |
| R | GGTGTGAGTGTGAGTGTGAGAG | Random sequence without lesion |
| R-Tg | GGTGTGAGTG (Tg)GAGTGTGAGAG | Random sequence with Tg |
| RSC-Tg | GGTGTGAGTGGT (Tg)AGGTGAGAG | Random sequence with telomeric sequence flanking Tg |
| Tel-Sp | TGTTAGGGTTAGGGTTA (Sp)GGTTAGGGCCAT | Telomeric sequence with Sp |
| Tel-Gh | TGTTAGGGTTAGGGTTA (Gh)GGTTAGGGCCAT | Telomeric sequence with Gh |
| Tel-5-OHU | TGTCAATCCCTAA (5-OHU)CCTAACCCTAACCCTGAGTCT | Telomeric sequence with 5-OHU |
| R-5-OHU | TGTCAATAGCAAG (5-OHU)GGAGAAGTCAATCGTGAGTCT | Random sequence with 5-OHU |
All of the oligodeoxyribonucleotides were gel purified and quantified by NanoDrop spectrophotometry. The damage-containing oligodeoxyribonucleotides were 32P-labeled and ethanol precipitated using our standard protocol as described previously (41). The telomeric quadruplex DNA substrates were prepared as described previously (21, 38) with minor modifications. Briefly, oligodeoxyribonucleotides were prepared in 10 mm sodium phosphate (pH 7.0) plus 100 mm NaCl in Milli-Q water. Potassium phosphate buffer (pH 7.0) and KCl of the same concentrations were used when preparing K+-containing quadruplexes. The mixture was boiled for 5 min and gradually cooled down to room temperature in a water bath for 2 h. The same procedure was used for preparing the other oligodeoxyribonucleotide substrates. In some experiments, a 1.2-fold excess of the complementary strand was added to ensure that all the lesion-containing strands were annealed into duplexes.
Enzymes
Details of the cloning, expression, and purification of the glycosylase domain of the Mus musculus Neil3 were described previously (42). Unless it was specifically pointed out, the mNeil3 enzyme used in this study was the glycosylase domain of M. musculus Neil3 (M. musculus Neil3Δ324). NEIL1, NEIL2, NTH1, and OGG1 enzymes were from our laboratory stocks and were purified as described previously (43, 44). Protein concentrations were determined by the BCA protein assay kit (Thermo Scientific Pierce). The percentage of active DNA glycosylase was determined by Schiff base assay or by the molecular accessibility method (45), and all protein concentrations used in this study were corrected to active enzyme fraction.
Native Gel Electrophoresis and CD Spectra
DNA substrates were prepared as described above. Samples were loaded with 5% glycerol on to a 16% acrylamide (29:1) gel with 0.5× TBE and 100 mm NaCl, and the gel was run at 60 V overnight at 4 °C. The native gel was then dried and exposed to a PhosphorImager screen for detection.
CD spectra were recorded by a JASCO-810 spectropolarimeter, using 1-mm path length cuvette in a volume of 300 μl at 37 °C. The DNA oligodeoxyribonucleotides (20 μm) were prepared by the same procedure as described under the “Substrates” section above. For each sample, four scans were taken at wavelengths from 190 to 350 nm. An average value was then calculated from the four scans and corrected for the spectrum of the buffer control. The spectrum was then zero-corrected at 330 nm and finally normalized to the concentration of the oligodeoxyribonucleotides.
Glycosylase Activity Assay
Enzymes and substrates were incubated at 37 °C in various glycosylase assay buffers as described previously (13). To measure the glycosylase activity only, reactions were terminated by adding NaOH to a final concentration of 0.33 n, and samples were immediately put on ice. The quenched reactions were then heated at 95 °C for 4 min. An equal volume of formamide with dyes (bromphenol blue and xylene cyanol) was added to the reactions before loading on to a 12% urea gel for separation. The gel was dried and exposed to a phosphoimaging screen. Finally, bands from the screen were scanned by Molecular Imager FX (Bio-Rad) and quantified by Quantity One software (Bio-Rad).
Single Turnover Kinetics
For single-turnover kinetics analysis of mNeil3, the concentration of substrates was set at 10 nm, and the concentration of enzyme was 4 μm or 6 μm, under which conditions the reaction rate reached the maximum (data not shown). The glycosylase activity assay was performed as described above. The intensity of bands was quantified by Quantity One (Bio-Rad) and analyzed by Prism software (version 5). The catalytic rate (kobs) was determined by fitting the data points to either a one or a two phase association model, depending on which was preferred.
RESULTS
The Human Telomeric Sequence with a Tg or 8-oxoG Lesion Forms an Antiparallel Quadruplex Structure in the Presence of Na+
We first asked the question: do damage-containing oligodeoxyribonucleotides form quadruplex structures? We addressed this question using both native gel electrophoresis and circular dichroism. Because of its more compact structure, intramolecular quadruplex DNA runs faster in a non-denaturing gel than single-stranded DNA with the same nucleotide composition. In our experiments, we used the random sequence (R) as a control for the single-stranded DNA structure. As expected, the telomere sequence (Tel) had a higher mobility than R (Fig. 1A), and this compact structure in Na+ solution was likely to be an intramolecular quadruplex structure as demonstrated previously (46). Similar to Tel, telomeric DNA containing a Tg lesion (Tel-Tg, Fig. 1A) also ran faster on a native gel than the random sequence containing Tg (R-Tg and RSC-Tg, Fig. 1A). This pattern indicates that Tel-Tg is able to form a compact, quadruplex-like structure in Na+ solution. We did the same experiment using the Tel-OG oligodeoxyribonucleotide, which has an 8-oxoG substitution at the 5′ of the third GGG triad, and a very similar running pattern was obtained (Fig. 1B). This is consistent with the observation of Szalai et al. (32) who showed that substitution of the 5′-G of the GGG triad with 8-oxoG did not disrupt the formation of the intramolecular quadruplex structure although a significant drop in thermal melting temperature has been reported (47).
Considering that the formation of a quadruplex structure is critical for our later experiments, we confirmed quadruplex formation by examining the structural details of our substrates using circular dichroism. The CD spectrum of Tel-Tg displayed a positive band at 295 nm and a negative band at 265 nm (Fig. 1C), a characteristic CD pattern of an antiparallel quadruplex structure in NaCl solution (46). The spectrum of Tel-OG was very similar to that of Tel and Tel-Tg, indicating that Tel-OG also formed an antiparallel quadruplex structure (Fig. 1C), similar to reported spectra (47). As a control, the random sequence R did not show the two-banded spectroscopic pattern (Fig. 1C). Therefore, the CD spectra confirmed that Tel-Tg and Tel-OG fold into antiparallel quadruplex structures. These data agree with those of others that telomeric DNA forms an antiparallel structure in Na+ solution and that substitution of a single base does not hinder its formation (38, 46, 47). Based on the antiparallel quadruplex structures solved in Na+ (23), we conclude that the Tg of qTel-Tg is located in the diagonal TTA loop and the 8-oxoG of qTel-OG is in an outer G-tetrad (Fig. 1D).
mNeil3 Glycosylase Efficiently Excises a Tg Lesion from Quadruplex DNA
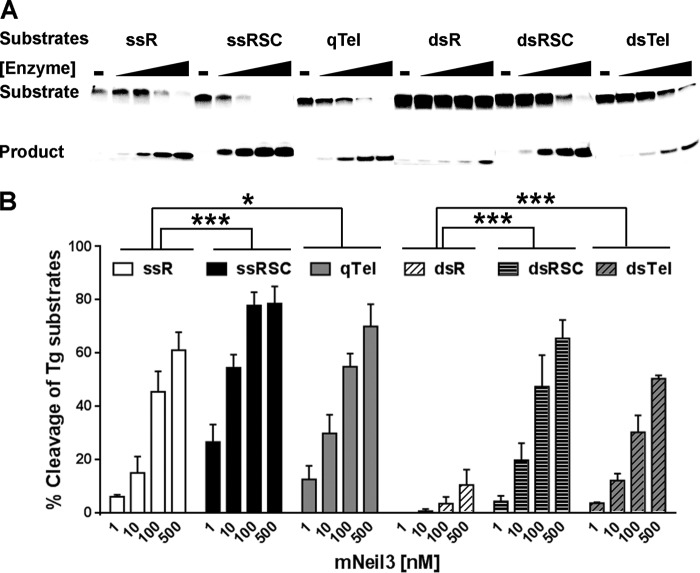
Despite the fact that the G-rich quadruplex-forming sequences are susceptible to oxidation, there have been no biochemical studies to look at repair of base damages in quadruplex DNA structures. To examine this question, we used the five mammalian DNA glycosylases that recognize oxidative damages, NEIL1, NEIL2, mNeil3, NTH1, and OGG1, and measured their activity on Tg-containing quadruplex DNA (Fig. 2A and data not shown). In these experiments, NaOH was added to the reactions after 15 min, and samples were heated to measure glycosylase activity only. Of the five glycosylases, only mNeil3 showed significant activity on the Tg-containing quadruplex (qTel-Tg). NEIL1 and NEIL2 showed very weak glycosylase activity on the qTel-Tg substrate; there was only a trace amount of product after incubating qTel-Tg with a 10-fold excess of NEIL1 or NEIL2 for 15 min. NTH1 had no glycosylase activity on qTel-Tg (Fig. 2A), although Tg itself is a good substrate for NTH1. The activity of mNeil3 was not a result of the quadruplex structure being disrupted as the quadruplex structure of undamaged Tel was still intact after incubation with the mNeil3 enzyme. As can be seen in Fig. 2C, the qTel substrate still ran as a unique band ahead of the single-stranded control (ssR) after incubation with mNeil3. To compare the activities of the glycosylases on qTel-Tg, the percentage of cleaved substrate by each enzyme was calculated (Fig. 2B). Clearly, mNeil3 was the most active glycosylase for the Tg-containing quadruplex.
FIGURE 2.
mNeil3 has significant glycosylase activity on quadruplex DNA containing a Tg lesion. A, glycosylase activity of NEIL1, NEIL2, mNeil3, and NTH1 on the qTel-Tg substrate. qTel-Tg (10 nm) was incubated with buffer only or with increasing concentrations of each enzyme (1, 10, and 100 nm) for 15 min. Reactions were terminated by NaOH and heating to measure glycosylase activity only. B, quantification of substrate (qTel-Tg) cleavage. Mean and S.D. calculated from three replicates are shown. C, native gel electrophoresis showing the intact quadruplex structure of qTel (without a lesion) after incubation with mNeil3.
mNeil3, but Not NEIL1, Prefers Thymine Glycol in the Telomeric Sequence Context
Next, we asked how the quadruplex substrate, qTel-Tg, compared with single-stranded or duplex DNA as a substrate for mNeil3. Single-stranded substrate (ssR-Tg) and two double-stranded substrates (dsR-Tg and dsTel-Tg) were used as controls. ssR-Tg, qTel-Tg, dsR-Tg, or dsTel-Tg were incubated with mNeil3 at four different substrate-to-enzyme ratios (10:1, 1:1, 1:10, 1:50) for 15 min, and the glycosylase assay was carried out (Fig. 3, A and B). Surprisingly, mNeil3 showed more robust cleavage of the Tg lesion in quadruplex DNA than in single-stranded DNA (qTel versus ssR).
FIGURE 3.
mNeil3 prefers the Tg lesion in the telomeric sequence context. A, six Tg-containing substrates (10 nm) were incubated with buffer only or with increasing concentrations of mNeil3 (1, 10, 100, 500 nm) for 15 min. Reactions were terminated by NaOH and heating to reveal glycosylase activity only. B, quantification of substrate cleavage by mNeil3. Mean and S.D. from three replicates are shown. Fisher's combined probability test was performed between telomeric and non-telomeric substrate pairs to reveal the effect of sequence context. The asterisks indicate the combined p value of the four t tests performed for the four individual concentrations. *, p < 0.05; ***, p < 0.0001.
We also observed that dsTel-Tg was a better substrate than dsR-Tg (Fig. 3), suggesting that sequence context might be playing a role. To examine the effect of telomeric sequence context on mNeil3 activity, we designed another random sequence RSC-Tg, which was similar to R-Tg except for the three bases on each side of the Tg lesion. In the RSC-Tg sequence, the Tg was in the same surrounding sequence context as that of telomeric DNA Tel-Tg (GGT-Tg-AGG) (Table 1). If the telomeric sequence context makes dsTel-Tg a better substrate compared with dsR-Tg, then RSC-Tg, which mimics the sequence context of Tel-Tg, should be a comparable substrate with Tel-Tg. This turned out to be the case. Similar to dsTel-Tg, the dsRSC-Tg was cleaved much more rapidly by mNeil3 than dsR-Tg (Fig. 3A). For example, the reaction was close to completion for dsRSC-Tg when it was incubated with a 10-fold excess of mNeil3 for 15 min, whereas the dsR-Tg substrate was barely cleaved. Similarly, ssRSC-Tg was a much better substrate than ssR-Tg (Fig. 3A). When the experiments were quantified, it can be clearly seen that the telomeric sequence context makes the duplex DNA containing Tg (dsRSC-Tg and dsTel-Tg) remarkably better substrates for mNeil3 (Fig. 3B). Similar results were obtained for single-stranded substrates (ssRSC-Tg versus ssR-Tg, Fig. 3B). Statistical analysis using Fisher's combined probability test shows that there are significant differences in the following comparisons: ssR versus qTel, ssR versus ssRSC, dsR versus dsTel, and dsR versus dsRSC, and the results are summarized in Table S1. Although the difference between ssR and qTel was significant, the difference between the means was much smaller than that of other comparisons, as can be seen in Fig. 3B by visual inspection and by the statistical results in supplemental Table S1.
We then measured the catalytic rates of mNeil3 on the six substrates (Table 2). The rates (kobs) were measured under single turnover conditions, where the enzyme concentration was much higher than the substrate concentration in order to measure the chemistry step only. Surprisingly, we found that ssRSC-Tg had a catalytic rate (kfast) of 6.66 min−1, which was >25-fold faster than ssR-Tg (0.25 min−1). Similar to ssRSC-Tg, the telomeric quadruplex substrate qTel-Tg had a kfast of 4.95 min−1, which was close to ssRSC-Tg and nearly 20 times greater than that of ssR-Tg. More strikingly, when we compared the duplex substrate with or without the telomeric sequence context, we found the kobs of dsRSC-Tg (7.49 min−1) to be >80 times greater than that of dsR-Tg (0.09 min−1). In fact, the kfast of dsRSC-Tg (7.49 min−1) was in the same range as its corresponding single-stranded DNA ssRSC-Tg (6.66 min−1). It is important to note that the only difference between the two single-stranded substrates was the sequence context surrounding the Tg lesion. The RSC-Tg has a telomeric sequence context surrounding Tg (GGT-Tg-AGG), whereas the R-Tg does not (GTG-Tg-GAG) (Table 1). The other duplex substrate dsTel-Tg (1.36 min−1) was also a much better substrate than dsR-Tg (0.09 min−1) (Table 2).
TABLE 2.
Single turnover kinetics of mNeil3 on lesion-containing substrates with or without the telomeric sequence context
Rates are calculated from at least three independent experiments. NA, not applicable.
| Substrates | kfast | kslow | Fast phase |
|---|---|---|---|
| min−1 | min−1 | % | |
| ssR-Tg | 0.25 ± 0.01 | NA, one phase | NA, one phase |
| ssRSC-Tg | 6.7 ± 1.0 | 0.50 ± 0.06 | 50 ± 3 |
| qTel-Tg | 5.0 ± 0.7 | 0.62 ± 0.05 | 39 ± 4 |
| dsR-Tg | 0.09 ± 0.01 | NA, one phase | NA, one phase |
| dsRSC-Tg | 7.5 ± 1.4 | 0.19 ± 0.01 | 24 ± 2 |
| dsTel-Tg | 1.4 ± 0.2 | 0.15 ± 0.02 | 44 ± 5 |
| dsR-5-OHU | 0.58 ± 0.20 | 0.04 ± 0.02 | 58 ± 9 |
| dsTel-5-OHU | 1.7 ± 0.8 | 0.10 ± 0.05 | 53 ± 12 |
The kobs values were calculated by Prism (version 5) using its “one-phase association” or “two-phase association” model, depending on which one fit the data better. Most of the reactions with the Tg substrates turned out to be double-exponentials, giving a kfast and a kslow when fitted to a two-phase association model. The double exponential curves were most likely due to the presence of two isomers of Tg, which may be processed differently by different glycosylases. As observed by us (48) and others (49, 50), both bacterial and mammalian Nth and Nei glycosylases exhibit strong stereospecificity toward the two isomers of thymine glycol. In contrast, the ssR-Tg and dsR-Tg were best fit as a single exponential (Table 2). It is likely that the activity of mNeil3 was so slow on one of the two Tg isomers in these two substrates that we were not able to detect the second phase (kslow) during the time frame of our assay.
We then tested whether mNeil3 preferentially removed 5-OHU in the telomeric sequence context. The 5-OHU is a common oxidative damage of cytosine, which can occur on the opposite strand of (TTAGGG)n. We found that mNeil3 also preferred 5-OHU in duplex DNA containing the telomeric sequence, although the difference in kobs was less striking with 5-OHU in the telomeric sequence context being only about twice that in the non-telomeric sequence (Table 2, dsTel-5OHU versus dsR-5OHU).
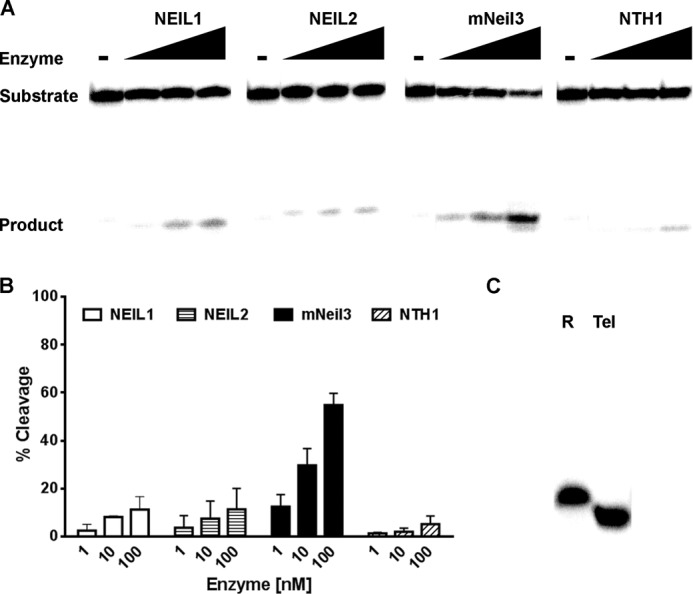
We also asked whether the effect of telomere sequence was unique to mNeil3 or whether it was shared by other glycosylases. Accordingly, similar experiments were done with NEIL1, NTH1, and OGG1 glycosylases. Fig. 4A clearly shows that the activity of NEIL1 on Tg was not affected by the presence of the telomere sequence (also see supplemental Table S1 for statistical results). OGG1 was also not sensitive to the sequence surrounding 8-oxoG in duplex DNA (Fig. 4B), which is consistent with a previous telomeric sequence context study of OGG1 (51). Moreover, NTH1 glycosylase preferred the random sequence over the telomere sequence (Fig. 4C). Thus, we concluded that the strong preference toward thymine glycol in the telomeric sequence context was unique to Neil3.
FIGURE 4.

NEIL1, OGG1, and NTH1 do not prefer lesions in telomere sequence context. A, four Tg-containing substrates (10 nm) were incubated with buffer only or with increasing concentrations of NEIL1 (1, 10, 100 nm) for 15 min. Reactions were terminated by NaOH and heating to reveal glycosylase activity only. The same statistical analysis was performed as described in Fig. 3B. There is no significant difference between ssR and qTel or between dsR and dsTel. See supplemental Table S1 for a more detailed analysis. The same experimental conditions and analysis as described in A were performed for OGG1 on 8-oxoG-containing substrates (B) and NTH1 on Tg-containing substrates (C).
mNeil3 and NEIL1 Remove Sp and Gh from Quadruplex DNA
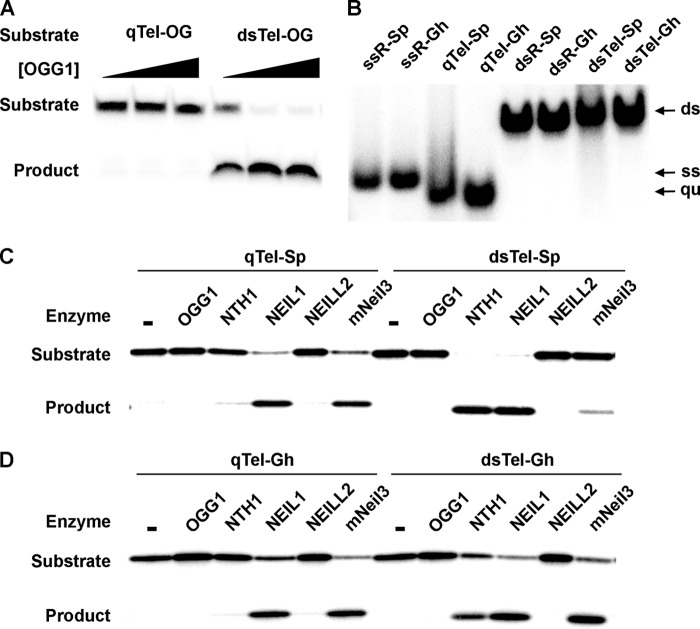
Considering that guanine has the lowest redox potential of the four bases and is thus the most susceptible base to oxidation, we tested the activity of the glycosylases on quadruplex DNA containing oxidized guanine lesions. We started with an 8-oxoG substitution in one of the two outer G-tetrads. Because the 5′-G of the GGG triad is the major target for oxidation (52, 53), we substituted a 5′-G with 8-oxoG (Tel-OG, Table 1). The oligodeoxyribonucleotide folded into an antiparallel quadruplex structure as discussed above (Fig. 1, B and C). Glycosylase assays were performed using qTel-OG substrate and the five mammalian glycosylases (OGG1, NEIl1, NEIL2, mNeil3, NTH1). None of the five enzymes showed glycosylase activity on the 8-oxoG-containing quadruplex substrate (qTel-OG) (Fig. 5A and data not shown), despite the fact that 8-oxoG is the major substrate for OGG1.
FIGURE 5.
mNeil3 and NEIL1 recognize Sp and Gh in telomeric quadruplex DNA. A, OGG1 does not remove 8-oxoG from quadruplex DNA. qTel-OG and dsTel-OG (10 nm) were incubated with buffer only or increasing concentrations of OGG1 (1, 10, 100 nm) for 15 min. B, native gel electrophoresis shows that Tel-Sp and Tel-Gh sequences form quadruplexes. C, mNeil3 and NEIL1 remove Sp from quadruplex DNA. qTel-Sp or dsTel-Sp substrate (10 nm) was incubated with each enzyme (10 nm) at 37 °C for 15 min. D, mNeil3 and NEIL1 remove Gh from quadruplex DNA.
Sp and Gh are further oxidation products of 8-oxoG. Our recent data show that Sp and Gh are two major oxidation products formed after treating telomeric G-quadruplex DNA with several oxidant systems in vitro (54). Moreover, Sp and Gh are the two best substrates for mNeil3 (13) and NEIL1 (40); NEIL2 (55) and Nth (56) also excise the two hydantoin lesions. We thus tested the activity of the glycosylases on Sp-containing and Gh-containing quadruplex substrates (qTel-Sp and qTel-Gh). Folding of the Tel-Sp and Tel-Gh to quadruplexes was confirmed by native gel electrophoresis (Fig. 5B). Glycosylase activity assays with the five enzymes were then performed using qTel-Sp and qTel-Gh as substrates. Both mNeil3 and NEIL1 very efficiently removed Sp (Fig. 5C) and Gh (Fig. 5D) from quadruplex DNA. However, we did not see any activity of NEIL2, NTH1, or OGG1 on either qTel-Sp (Fig. 5C) or qTel-Gh (Fig. 5D).
NEIL1, but Not mNeil3, Prefers Gh in the Telomere Sequence Context
To determine whether Gh is a preferred substrate in the telomere sequence, the glycosylase activity of mNeil3 and NEIL1 was performed on ssR-Gh, qTel-Gh, dsR-Gh, and dsTel-Gh. When comparing dsR-Gh with dsTel-Gh (Fig. 6A), mNeil3 clearly does not prefer Gh in the telomere sequence. In addition, when measured under single turnover conditions, the kobs of dsR-Gh and dsTel-Gh were 5.01 ± 0.30 min−1 and 0.26 ± 0.01 min−1, respectively. mNeil3 also does not prefer the Sp lesion in the duplex telomere sequence (data not shown). However, NEIL1 showed slightly enhanced activity on dsTel-Gh (Fig. 6B), as evidenced by comparing dsR and dsTel at 10 nm of NEIL1. The results of our statistical analysis are summarized in supplemental Table S1, C and D.
FIGURE 6.
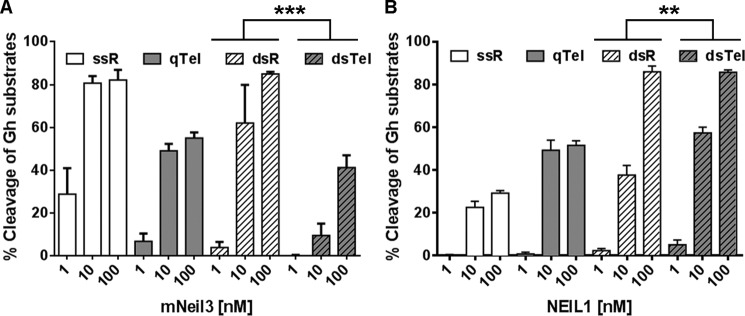
mNeil3 does not prefer Gh in the telomeric sequence context. A, quantification of substrate cleavage by mNeil3. Gh-containing substrates (10 nm) were incubated with increasing concentrations of mNeil3 (1, 10, 100 nm) for 5 min. Reactions were terminated by NaOH and heating to reveal glycosylase activity only. The same experiments were done using NEIL1, and results are shown in B. Mean and S.D. from four replicates are shown. Fisher's combined probability test was performed between dsR and dsTel to reveal the effect of sequence context. The asterisks indicate the combined p values of the four t tests performed for the four individual concentrations. **, p < 0.005; ***, p < 0.0001. See supplemental Table S1 for a more detailed analysis.
DISCUSSION
Despite the susceptibility of the guanine-rich quadruplex-forming sequence to oxidation and the biological importance of quadruplex DNA, no study has addressed the question of how damaged bases in quadruplex DNA are repaired. In this study, we show that telomeric DNA containing Tg, 8-oxoG, Sp, or Gh can form quadruplex structures in the presence of physiological salts in vitro. We further demonstrate that the mouse ortholog of human NEIL3 can efficiently remove Tg, Sp, and Gh lesions from quadruplex DNA; NEIL1 is also able to efficiently excise Sp and Gh lesions from quadruplex DNA. Interestingly, the mNeil3 glycosylase shows a marked preference toward thymine glycol (but not Sp or Gh) in telomeric DNA sequences, whereas NEIL1 appears to have a complementary role, with a preference for Gh in the telomeric sequence.
The structures of quadruplexes have been intensely studied for the past 20 years, but the impact of base damages on the stability of quadruplexes and how these base damages are repaired in quadruplexes remain to be investigated. The fact that Tg, 8-oxoG, Sp, and Gh can be folded into quadruplex DNA structures implies that an oxidative base lesion may stably exist in quadruplex DNA in vivo.
The excision activities of mNeil3 and NEIL1 on lesion-containing quadruplexes led us to speculate on their biological function in cells. Quadruplex DNA has been proposed to exist during important biological functions such as DNA replication (29), during which time the two strands of the duplex DNA molecule become separated. Interestingly, both NEIL3 (58) and NEIL1 (59) have been linked to DNA replication. A further speculation is that genes might be regulated by glycosylases via modulation of quadruplex structures at promoter regions. Quadruplex-forming sequences are prevalent in the human genome and are enriched in promoters (25, 26). These guanine-rich sequences will be inevitably oxidized to create damage-containing quadruplex DNA (60). Glycosylases may function in gene regulation by removing DNA base damages from the quadruplex and creating transient strand breaks. This action may result in collapse of the quadruplex structures in promoters, which would relieve the inhibitory effect of the quadruplex on transcription. A similar mechanism has been proposed by Gillespie et al. (61) who found that the base excision repair enzymes OGG1 and APE1 were recruited to G4 sequences in promoters of those genes exhibiting altered expression after hypoxia treatment. The recruitment of OGG1 and APE1 to the promoter regions coincides with increased 8-oxoG production and DNA strand breaks in these G4 sites (60). Interestingly, we did not find any glycosylase activity of OGG1 on 8-oxoG-containing quadruplex DNA (Fig. 5A).
Another interesting observation is the strong sequence context effect governing the activity of mNeil3 on thymine glycol. The kobs (Table 2) implies that a faster chemistry step is significantly contributing to the preference for the telomere sequence because the kobs only reflects the catalytic step but not binding. According to the transition state theory (62), a lower activation barrier (Gibbs free energy of activation, ΔG‡) due to the difference in the surrounding DNA sequence may explain the increased rates. Moreover, although the data shown in Fig. 3B do not support a formal estimation of binding affinities and catalytic rates for the Tg-containing substrates, they suggest qualitative conclusions. The general impression is that mNeil3 binding to all of these substrates is weak, a conclusion consistent with numerous EMSA experiments where we have been unable to show Neil3 to bind to Tg-containing DNA. However, the increasing extent of cleavage as enzyme: substrate increases from 1:1 to 10:1 and 50:1 implies that more binding occurs with higher protein concentrations, and therefore the differences in the extent of cleavage on the same substrate do reflect differences in binding. This raises the possibility that differences between observations on different substrates also reflect differences in binding. However, inspection of the non-quadruplex substrates in Fig. 3B also shows that both DNA strandedness and context play a role. First, the extents of cleavage are greater for each single-stranded representative by comparison with its double-stranded mate. This is consistent with the possibility that availability of Tg for insertion into the active site pocket is greater in the single-stranded substrates due to the absence of the opposite strand and is in keeping with our prior biochemical studies of Neil3 showing its preference for lesions in single-stranded substrates (13) and with its structure (63) showing why. Similarly, there is a strong effect of context, that is, the extents of cleavage are greater for each RSC context by comparison with its R mate. This is consistent with both biochemical and crystallographic studies from our laboratory with DNA polymerases that have shown that the methyl and hydroxyl groups present on the 5′ position of thymine glycol severely alter stacking of the 5′ base and in the case of guanine, a hydrogen bond is formed with Tg. Moreover, there is an additional interaction with a 3′-purine via a water molecule (64). Thus, in the case of the random sequence we used, 5′-G Tg G-3′, the Tg would be less able to be extruded into the Neil3 substrate binding pocket than in the telomere sequence context 5′-T Tg G-3′. So even though the enzyme is bound to the DNA, poorer extrusion of Tg into the active site pocket would explain the slower chemistry step (Table 2) observed with the random sequence compared with the telomere sequence. The preference of mNeil3 for the telomeric sequence appears to be restricted to the oxidized pyrimidines, especially thymine glycol, as there is no sequence context effect with Sp and Gh.
The question remains as to why NEIL1 does not exhibit a preference for Tg in the telomere sequence. It turns out that NEIL1, similar to most members of the Fpg/Nei family, contains three void-filling residues that help extrude the lesion from the DNA into the active site pocket, whereas two of them are missing in mNeil3 (63). Although NEIL1 has no preference for Tg in the telomere sequence context, it did show a slight preference for Gh (Fig. 6B). This complementary telomere sequence context effect of mNeil3 and NEIL1 suggests that they may play special roles in telomere damage repair.
Base damages have been shown to be deleterious to telomeres. For example, 8-oxoG attenuates the binding of telomeric proteins such as TRF1 and TRF2, loss of which results in telomere dysfunction and fusions (39). In addition, Tg, Sp, and Gh are efficient blocks to some DNA polymerases (4, 7, 8). A mechanism for oxidative stress-induced telomere shortening has been proposed, which argues that blocking of DNA polymerases by unrepaired bases or nucleotides is a major contributor to this shortening (57).
In summary, the data presented here show that Neil3 and NEIL1 may play important roles in telomere maintenance by preventing accumulation of base damages in telomeres, thus protecting their integrity. However, we were unable to colocalize NEIL3 to telomere markers (TRF1, TRF2, and POT1) in HeLa or HEK293 cells or demonstrate any physical interaction between NEIL3 and TRF1 or TRF2 by co-immunoprecipitation. Because Neil3 appears to function in a cell cycle-dependent and tissue-specific manner (58), we suspect it may do the same for repair in telomeres. Future work is needed to address these questions.
Supplementary Material
Acknowledgments
We thank Dr. Jeffery Bond for help in enzyme kinetics analysis and Dr. Martin Case for help with the CD experiments. We also thank Drs. Susan Robey-Bond, Scott Kathe, and Jany Chan for helpful discussions and April Averill and Lauren Harvey for purifying the enzymes used in this study.
This work was supported by National Institutes of Health Grants P01 CA 098993 (to S. S. W.) and CA 090689 (to C. J. B.) from the NCI.

This article contains supplemental Table S1.
- Tg
- thymine glycol
- Tel
- telomere sequence
- R
- random sequence
- RSC
- random sequence with telomeric sequence context
- G4
- G-quadruplex
- 8-oxoG
- 8-oxo-7,8-dihydroguanine
- OGG1
- 8-oxoguanine DNA glycosylase
- TRF1
- telomere repeat-binding factor 1
- NEIL1
- human endonuclease VIII-like glycosylase 1
- NEIL2
- human endonuclease VIII-like glycosylase 2
- ss
- single-stranded
- ds
- double-stranded
- mNeil3
- mouse endonuclease VIII-like glycosylase 3
- NTH1
- human endonuclease III glycosylase 1
- Sp
- spiroiminodihydantoin
- Gh
- guanidinohydantoin
- 5-OHU
- 5-hydroxyuracil.
REFERENCES
- 1. Sena L. A., Chandel N. S. (2012) Physiological roles of mitochondrial reactive oxygen species. Mol. Cell 48, 158–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wallace S. S. (2002) Biological consequences of free radical-damaged DNA bases. Free Radic. Biol. Med. 33, 1–14 [DOI] [PubMed] [Google Scholar]
- 3. Duclos S., Doublié S., Wallace S. S. (2012) Consequences and repair of oxidative DNA damage in The Cellular Response to the Genotoxic Insult: The Question of Threshold for Genotoxic Carcinogens (Greim H., Albertini R. J., eds), pp. 15–159, The Royal Society of Chemistry, Cambridge, United Kingdom [Google Scholar]
- 4. Ide H., Kow Y. W., Wallace S. S. (1985) Thymine glycols and urea residues in M13 DNA constitute replicative blocks in vitro. Nucleic Acids Res. 13, 8035–8052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cheng K. C., Cahill D. S., Kasai H., Nishimura S., Loeb L. A. (1992) 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G—-T and A—-C substitutions. J. Biol. Chem. 267, 166–172 [PubMed] [Google Scholar]
- 6. Kreutzer D. A., Essigmann J. M. (1998) Oxidized, deaminated cytosines are a source of C → T transitions in vivo. Proc. Natl. Acad. Sci. U.S.A. 95, 3578–3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Duarte V., Muller J. G., Burrows C. J. (1999) Insertion of dGMP and dAMP during in vitro DNA synthesis opposite an oxidized form of 7,8-dihydro-8-oxoguanine. Nucleic Acids Res. 27, 496–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Henderson P. T., Delaney J. C., Muller J. G., Neeley W. L., Tannenbaum S. R., Burrows C. J., Essigmann J. M. (2003) The hydantoin lesions formed from oxidation of 7,8-dihydro-8-oxoguanine are potent sources of replication errors in vivo. Biochemistry 42, 9257–9262 [DOI] [PubMed] [Google Scholar]
- 9. Hegde M. L., Hazra T. K., Mitra S. (2008) Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 18, 27–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McCullough A. K., Dodson M. L., Lloyd R. S. (1999) Initiation of base excision repair: glycosylase mechanisms and structures. Annu. Rev. Biochem. 68, 255–285 [DOI] [PubMed] [Google Scholar]
- 11. Prakash A., Doublié S., Wallace S. S. (2012) The Fpg/Nei family of DNA glycosylases: substrates, structures, and search for damage. Prog. Mol. Biol. Transl. Sci. 110, 71–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu M., Doublié S., Wallace S. S. (2013) Neil3, the final frontier for the DNA glycosylases that recognize oxidative damage. Mutat. Res. 743, 4–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu M., Bandaru V., Bond J. P., Jaruga P., Zhao X., Christov P. P., Burrows C. J., Rizzo C. J., Dizdaroglu M., Wallace S. S. (2010) The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 107, 4925–4930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wallace S. S., Bandaru V., Kathe S. D., Bond J. P. (2003) The enigma of endonuclease VIII. DNA Repair 2, 441–453 [DOI] [PubMed] [Google Scholar]
- 15. Hazra T. K., Kow Y. W., Hatahet Z., Imhoff B., Boldogh I., Mokkapati S. K., Mitra S., Izumi T. (2002) Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J. Biol. Chem. 277, 30417–30420 [DOI] [PubMed] [Google Scholar]
- 16. Hackett J. A., Feldser D. M., Greider C. W. (2001) Telomere dysfunction increases mutation rate and genomic instability. Cell 106, 275–286 [DOI] [PubMed] [Google Scholar]
- 17. de Lange T. (2009) How telomeres solve the end-protection problem. Science 326, 948–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Armanios M. (2009) Syndromes of telomere shortening. Annu. Rev. Genomics Hum. Genet. 10, 45–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baird D. M., Rowson J., Wynford-Thomas D., Kipling D. (2003) Extensive allelic variation and ultrashort telomeres in senescent human cells. Nat. Genet. 33, 203–207 [DOI] [PubMed] [Google Scholar]
- 20. Artandi S. E., Alson S., Tietze M. K., Sharpless N. E., Ye S., Greenberg R. A., Castrillon D. H., Horner J. W., Weiler S. R., Carrasco R. D., DePinho R. A. (2002) Constitutive telomerase expression promotes mammary carcinomas in aging mice. Proc. Natl. Acad. Sci. U.S.A. 99, 8191–8196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Williamson J. R., Raghuraman M. K., Cech T. R. (1989) Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell 59, 871–880 [DOI] [PubMed] [Google Scholar]
- 22. Smith F. W., Feigon J. (1992) Quadruplex structure of Oxytricha telomeric DNA oligonucleotides. Nature 356, 164–168 [DOI] [PubMed] [Google Scholar]
- 23. Wang Y., Patel D. J. (1993) Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure 1, 263–282 [DOI] [PubMed] [Google Scholar]
- 24. Parkinson G. N., Lee M. P., Neidle S. (2002) Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 417, 876–880 [DOI] [PubMed] [Google Scholar]
- 25. Todd A. K., Johnston M., Neidle S. (2005) Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 33, 2901–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huppert J. L., Balasubramanian S. (2005) Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 33, 2908–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang Q., Xiang J., Yang S., Zhou Q., Li Q., Tang Y., Xu G. (2009) Verification of specific G-quadruplex structure by using a novel cyanine dye supramolecular assembly: I. recognizing mixed G-quadruplex in human telomeres. Chem. Commun. 9, 1103–1105 [DOI] [PubMed] [Google Scholar]
- 28. Biffi G., Tannahill D., McCafferty J., Balasubramanian S. (2013) Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 5, 182–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lipps H. J., Rhodes D. (2009) G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 19, 414–422 [DOI] [PubMed] [Google Scholar]
- 30. Siddiqui-Jain A., Grand C. L., Bearss D. J., Hurley L. H. (2002) Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 99, 11593–11598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Neidle S., Parkinson G. (2002) Telomere maintenance as a target for anticancer drug discovery. Nat. Rev. Drug Discov. 1, 383–393 [DOI] [PubMed] [Google Scholar]
- 32. Merino E. J., Boal A. K., Barton J. K. (2008) Biological contexts for DNA charge transport chemistry. Curr. Opin. Chem. Biol. 12, 229–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Genereux J. C., Boal A. K., Barton J. K. (2010) DNA-mediated charge transport in redox sensing and signaling. J. Am. Chem. Soc. 132, 891–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oikawa S., Tada-Oikawa S., Kawanishi S. (2001) Site-specific DNA damage at the GGG sequence by UVA involves acceleration of telomere shortening. Biochemistry 40, 4763–4768 [DOI] [PubMed] [Google Scholar]
- 35. Oikawa S., Kawanishi S. (1999) Site-specific DNA damage at GGG sequence by oxidative stress may accelerate telomere shortening. FEBS Lett. 453, 365–368 [DOI] [PubMed] [Google Scholar]
- 36. Wang Z., Rhee D. B., Lu J., Bohr C. T., Zhou F., Vallabhaneni H., de Souza-Pinto N. C., Liu Y. (2010) Characterization of oxidative guanine damage and repair in mammalian telomeres. PLoS Genet. 6, e1000951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. O'Callaghan N., Baack N., Sharif R., Fenech M. (2011) A qPCR-based assay to quantify oxidized guanine and other FPG-sensitive base lesions within telomeric DNA. BioTechniques 51, 403–411 [DOI] [PubMed] [Google Scholar]
- 38. Szalai V. A., Singer M. J., Thorp H. H. (2002) Site-specific probing of oxidative reactivity and telomerase function using 7,8-dihydro-8-oxoguanine in telomeric DNA. J. Am. Chem. Soc. 124, 1625–1631 [DOI] [PubMed] [Google Scholar]
- 39. Opresko P. L., Fan J., Danzy S., Wilson D. M., 3rd, Bohr V. A. (2005) Oxidative damage in telomeric DNA disrupts recognition by TRF1 and TRF2. Nucleic Acids Res. 33, 1230–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Krishnamurthy N., Zhao X., Burrows C. J., David S. S. (2008) Superior removal of hydantoin lesions relative to other oxidized bases by the human DNA glycosylase hNEIL1. Biochemistry 47, 7137–7146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Robey-Bond S. M., Barrantes-Reynolds R., Bond J. P., Wallace S. S., Bandaru V. (2008) Clostridium acetobutylicum 8-oxoguanine DNA glycosylase (Ogg) differs from eukaryotic Oggs with respect to opposite base discrimination. Biochemistry 47, 7626–7636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu M., Bandaru V., Holmes A., Averill A. M., Cannan W., Wallace S. S. (2012) Expression and purification of active mouse and human NEIL3 proteins. Protein Expr. Purif. 84, 130–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bandaru V., Blaisdell J. O., Wallace S. S. (2006) Oxidative DNA glycosylases: recipes from cloning to characterization. Methods Enzymol. 408, 15–33 [DOI] [PubMed] [Google Scholar]
- 44. Bandaru V., Sunkara S., Wallace S. S., Bond J. P. (2002) A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair 1, 517–529 [DOI] [PubMed] [Google Scholar]
- 45. Blaisdell J. O., Wallace S. S. (2007) Rapid determination of the active fraction of DNA repair glycosylases: a novel fluorescence assay for trapped intermediates. Nucleic Acids Res. 35, 1601–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li W., Wu P., Ohmichi T., Sugimoto N. (2002) Characterization and thermodynamic properties of quadruplex/duplex competition. FEBS Lett. 526, 77–81 [DOI] [PubMed] [Google Scholar]
- 47. Vorlícková M., Tomasko M., Sagi A. J., Bednarova K., Sagi J. (2012) 8-oxoguanine in a quadruplex of the human telomere DNA sequence. FEBS J. 279, 29–39 [DOI] [PubMed] [Google Scholar]
- 48. Guo Y., Bandaru V., Jaruga P., Zhao X., Burrows C. J., Iwai S., Dizdaroglu M., Bond J. P., Wallace S. S. (2010) The oxidative DNA glycosylases of Mycobacterium tuberculosis exhibit different substrate preferences from their Escherichia coli counterparts. DNA Repair 9, 177–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Katafuchi A., Nakano T., Masaoka A., Terato H., Iwai S., Hanaoka F., Ide H. (2004) Differential specificity of human and Escherichia coli endonuclease III and VIII homologues for oxidative base lesions. J. Biol. Chem. 279, 14464–14471 [DOI] [PubMed] [Google Scholar]
- 50. Miller H., Fernandes A. S., Zaika E., McTigue M. M., Torres M. C., Wente M., Iden C. R., Grollman A. P. (2004) Stereoselective excision of thymine glycol from oxidatively damaged DNA. Nucleic Acids Res. 32, 338–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rhee D. B., Ghosh A., Lu J., Bohr V. A., Liu Y. (2011) Factors that influence telomeric oxidative base damage and repair by DNA glycosylase OGG1. DNA Repair 10, 34–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Saito I., Takayama M., Sugiyama H., Nakatani K. (1995) Photoinduced DNA cleavage via electron-transfer - demonstration that guanine residues located 5′ to guanine are the most electron-donating sites. J. Am. Chem. Soc. 117, 6406–6407 [Google Scholar]
- 53. Sugiyama H., Saito I. (1996) Theoretical studies of GC-specific photocleavage of DNA via electron transfer: Significant lowering of ionization potential and 5′-localization of HOMO of stacked GG bases in B-form DNA. J. Am. Chem. Soc. 118, 7063–7068 [Google Scholar]
- 54. Fleming A. M., Burrows C. J. (2013) G-Quadruplex Folds of the Human Telomere Sequence Alter the Site Reactivity and Reaction Pathway of Guanine Oxidation Compared to Duplex DNA. Chem. Res. Toxicol. 26, 593–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hailer M. K., Slade P. G., Martin B. D., Rosenquist T. A., Sugden K. D. (2005) Recognition of the oxidized lesions spiroiminodihydantoin and guanidinohydantoin in DNA by the mammalian base excision repair glycosylases NEIL1 and NEIL2. DNA Repair 4, 41–50 [DOI] [PubMed] [Google Scholar]
- 56. Hazra T. K., Muller J. G., Manuel R. C., Burrows C. J., Lloyd R. S., Mitra S. (2001) Repair of hydantoins, one electron oxidation product of 8-oxoguanine, by DNA glycosylases of Escherichia coli. Nucleic Acids Res. 29, 1967–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. von Zglinicki T. (2002) Oxidative stress shortens telomeres. Trends Biochem. Sci. 27, 339–344 [DOI] [PubMed] [Google Scholar]
- 58. Neurauter C. G., Luna L., Bjørås M. (2012) Release from quiescence stimulates the expression of human NEIL3 under the control of the Ras dependent ERK-MAP kinase pathway. DNA Repair 11, 401–409 [DOI] [PubMed] [Google Scholar]
- 59. Dou H., Theriot C. A., Das A., Hegde M. L., Matsumoto Y., Boldogh I., Hazra T. K., Bhakat K. K., Mitra S. (2008) Interaction of the human DNA glycosylase NEIL1 with proliferating cell nuclear antigen. The potential for replication-associated repair of oxidized bases in mammalian genomes. J. Biol. Chem. 283, 3130–3140 [DOI] [PubMed] [Google Scholar]
- 60. Clark D. W., Phang T., Edwards M. G., Geraci M. W., Gillespie M. N. (2012) Promoter G-quadruplex sequences are targets for base oxidation and strand cleavage during hypoxia-induced transcription. Free Radic. Biol. Med. 53, 51–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gillespie M. N., Pastukh V. M., Ruchko M. V. (2010) Controlled DNA “damage” and repair in hypoxic signaling. Respir. Physiol. Neurobiol. 174, 244–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Berg J. M., Tymoczko J. L., Stryer L., Stryer L. (2007) Biochemistry, 6th Ed., W.H. Freeman, New York [Google Scholar]
- 63. Liu M., Imamura K., Averill A. M., Wallace S. S., Doublié S. (2013) Structural characterization of a mouse ortholog of human NEIL3 with a marked preference for single-stranded DNA. Structure 21, 247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Aller P., Rould M. A., Hogg M., Wallace S. S., Doublié S. (2007) A structural rationale for stalling of a replicative DNA polymerase at the most common oxidative thymine lesion, thymine glycol. Proc. Natl. Acad. Sci. U.S.A. 104, 814–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.