Background: Poliovirus induces formation of double-membraned vesicles as the site of viral replication in infected cells.
Results: The membrane-associated protein 3AB alone produces double-membraned liposomes by invagination of purified liposomes and intracellular membranes.
Conclusion: A single poliovirus protein produces membrane remodeling similar to the formation of autophagosomes.
Significance: Similar cellular processes are critical for autophagy, multivesicular body formation, unconventional secretion, and viral pathogenesis.
Keywords: Lipoprotein, Liposomes, Membrane Bilayer, Membrane Biophysics, Membrane Reconstitution, Positive-strand RNA Viruses, Viral Protein, Viral Replication
Abstract
Infection with many positive-strand RNA viruses dramatically remodels cellular membranes, resulting in the accumulation of double-membraned vesicles that resemble cellular autophagosomes. In this study, a single protein encoded by poliovirus, 3AB, is shown to be sufficient to induce the formation of double-membraned liposomes via the invagination of single-membraned liposomes. Poliovirus 3AB is a 109-amino acid protein with a natively unstructured N-terminal domain. HeLa cells transduced with 3AB protein displayed intracellular membrane disruption; specifically, the formation of cytoplasmic invaginations. The ability of a single viral protein to produce structures of similar topology to cellular autophagosomes should facilitate the understanding of both cellular and viral mechanisms for membrane remodeling.
Introduction
Positive-strand RNA viruses of eukaryotic cells replicate their genomes on the surfaces of membranous structures of disparate origin, ultrastructure, and cellular function within infected cells. In cells infected with poliovirus (1), rhinovirus (2), West Nile virus (3), and severe acute respiratory syndrome coronavirus (4), double-membraned vesicles are formed that morphologically resemble cellular autophagosomes. During infection with hepatitis C virus, less ordered “webs” of endoplasmic reticulum-derived membranes are formed (5). Electron tomography of cells infected with Dengue virus revealed both invaginated membranes and abundant double-membraned vesicles (6). In cells infected with brome mosaic virus or flock house virus (7, 8), numerous “spherules” (invaginations of membranes that remain open to the cytoplasm) form and sequester components of the viral RNA replication complex. What these rearrangements have in common is that viral RNA replication remains, topologically, on cytoplasm-facing surfaces and that the membrane rearrangements can be induced by viral proteins. The mechanisms by which viral proteins orchestrate these rearrangements, the specific host proteins and membranes required, and the purpose of the different origins and morphologies for the different viruses are topics of intense investigation.
The membranous vesicles induced during infection with poliovirus range in diameter from 50–400 nm (1, 9). The RNA replication complexes that assemble on their cytoplasmic surfaces are composed of multiple nonstructural viral and cellular proteins (1, 10–15). The similarities of these double-membraned structures with cellular autophagosomes include their ultrastructure, with two lipid bilayers surrounding a lumen that can contain cytoplasmic organelles and other cytosolic content (1, 16, 17), and their colocalization of posttranslationally lipidated LC3 and late endosomal LAMP-1 (2, 17, 18). Differences between the poliovirus-induced vesicles and cellular autophagosomes include the smaller size of the virally induced structures and their apparent persistence as double-membraned structures. Cellular autophagosomes “mature” by the acquisition of degradative machinery via fusion with lysosomes that cause them to lose their double-membraned character as rapidly as 30 min after their formation (reviewed e.g. in Ref. 19). The subversion of components or sequelae of the autophagy pathway has also been reported for coxsackievirus (20), foot and mouth disease virus (21), Dengue virus (6, 22, 23), and hepatitis C virus (24).
The 2B, 2C, and 3A nonstructural poliovirus proteins and their stable precursors, 2BC and 3AB, possess an intrinsic membrane-binding capacity that tethers the RNA replication components to membranes. The combined expression of 2BC and 3A in transfected cells has been shown to induce double-membraned vesicles similar to those induced during poliovirus infection (17, 18). The contributions of these two proteins, and host proteins with which they associate, to the formation of double-membraned structures (25, 26) is of great interest in understanding both viral RNA replication and cellular processes, such as bona fide autophagy and multivesicular body formation, that require membrane invagination and fusion. In transfected cells, expression of 2BC alone is sufficient to induce the lipidation of autophagy-associated protein LC3 and its recruitment to membranes as well as the formation of large, single-membraned vesicles with no visible lumenal contents (12, 17, 18). Belov et al. (27) have presented evidence that, early in infection, more single-membraned than double-membraned vesicles are observed and have suggested that these early membranes become double- or multilayered during maturation of RNA replication complexes. This is also similar to cellular autophagosomes, in which there is evidence that they invaginate from single-membraned phagaphores derived from the endoplasmic reticulum (28). The 2BC protein is known to interact physically with 3A and precursors, such as 3AB (64, 65), which provides the primer (3B) for the initiation of RNA synthesis. Cells that express 3A show a distinct accumulation of dilated tubular and swollen membranes (29). However, the role of 3AB in vesicle formation or membrane ultrastructure has not been investigated, primarily because of difficulties in its exogenous expression in mammalian cells. Here, we investigate its interaction with single-membrane liposomes in solution to examine its role in the unique membrane topology achieved during viral RNA replication.
Poliovirus 3AB protein is a 109-amino acid protein (Fig. 1A) that plays multiple roles during viral infection. It contains a hydrophobic domain that is responsible for its association with the outer leaflet of lipid bilayers (25, 30), from which position it recruits the viral RNA-dependent RNA polymerase 3Dpol and the multifunctional precursor 3CDpro to membranes via the 22-amino acid carboxy-terminal 3B portion (25). Viral protein 3AB has also been reported to stimulate the RNA polymerase activity of 3Dpol (31), bind RNA nonspecifically, and, in complex with 3CD, bind specifically to both the 5′ cloverleaf and the 3′ nontranslated RNA of the poliovirus genome (32–34). Cleavage of the membrane-associated 3AB protein or its larger precursors by the viral protease 3CDpro yields 3A and 3B (also called “VPg”), the protein primer for viral RNA replication (35–37). The NMR structure of the N-terminal 59 amino acids of 3AB revealed that interaction between domains in each monomer that comprise two short α-helices connected by an α-helical hairpin forms a stable, symmetric dimer. The 14 amino acids of the N termini of both monomers are natively unstructured (38). Such domains often interact transiently with many partners.
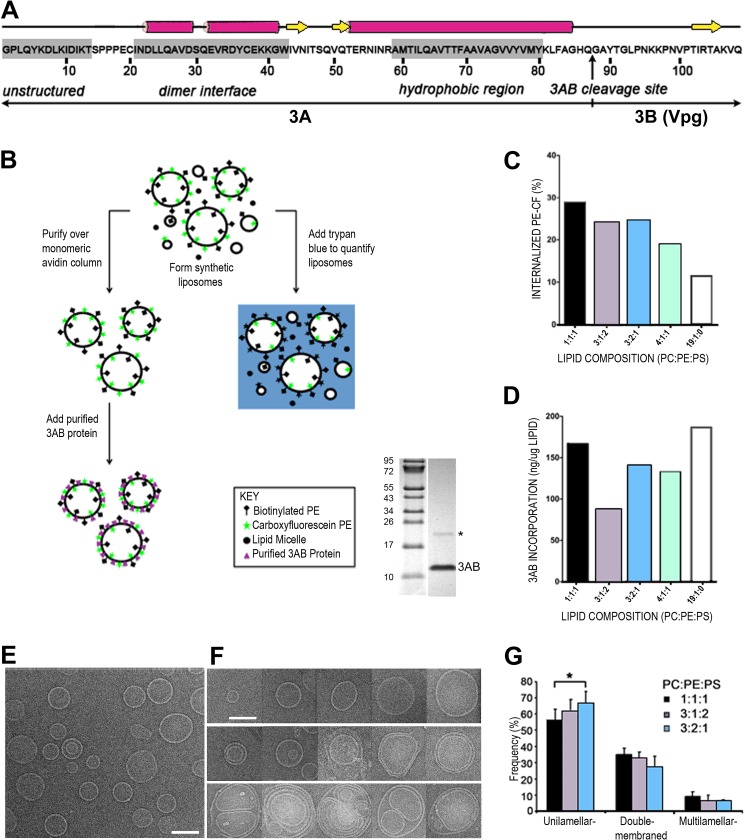
FIGURE 1.
Synthesis and characterization of synthetic proteoliposomes containing purified poliovirus 3AB protein. A, the sequence of the stable precursor protein 3AB encoded by poliovirus is shown, with the vertical arrow denoting the Gln/Gly cleavage site between the 3A and 3B proteins. The unstructured N terminus (30), dimerization interface, and the membrane association domain of 3AB (38) are highlighted by gray shades. B, flow chart of liposome formation, quantitation, and recruitment of purified 3AB protein. The gel shows the purity of 3AB, and the asterisk marks the dimeric form of 3AB. C, quantitation of liposome formation from a representative experiment. Five different lipid compositions were tested: 1:1:1, 3:1:2, 3:2:1, 4:1:1, and 19:1:0 PC:PE:PS. Trypan blue was used to quench the fluorescently labeled PE in lipid micelles and in the outer leaflet of the lipid bilayer of liposomes. The remaining, topologically sequestered fluorescence was quantified to estimate the liposome formation. CF, carboxyfluorescein. D, 3AB reconstitution was quantified by measuring the amount of 3AB in the lipid bilayer after isolation of proteoliposomes by ultracentrifugation. The amount of lipid was quantified by fluorimetry, and 3AB protein was quantified by immunoblot analysis and densitometry. E, micrograph of liposomes composed of 3:2:1 PC:PE:PS. Scale bar = 100 nm. F, representative images of unilamellar (top row), double-membraned (center row), and multilamellar (bottom row) liposomes of different sizes. Liposomes were composed of 3:2:1 PC:PE:PS. G, affinity-purified liposomes of the indicated compositions were visualized by cryo-EM, and the number of single-, double-, and multimembraned liposomes was quantified. The asterisk denotes a statistically significant difference.
To determine the role of the poliovirus 3AB protein in membrane remodeling, the ultrastructure of liposomes was investigated in the absence and presence of 3AB. In this study, we show that reconstitution of purified 3AB protein onto synthetic unilamellar liposomes is sufficient to induce the formation of double-membraned liposomes via the invagination of the single-membraned liposomes. The ability of a single viral protein to induce a double-membraned topology will inform studies of other positive-strand RNA viruses that induce membrane rearrangements. Furthermore, our understanding of cellular processes has often been advanced through virus studies such as, for example, elucidation of cell entry via coated pits by studies on Semliki virus entry (39) and multivesicular body formation through investigation of HIV budding mechanisms (40, 41). We propose that cellular processes such as autophagy, multivesicular body formation, and unconventional secretion may involve mechanisms similar to that observed in 3AB-mediated liposome remodeling.
EXPERIMENTAL PROCEDURES
Preparation of Synthetic Model Membrane Vesicles
l-α-phosphatidylcholine and l-α-phosphatidylserine from porcine brain, l-α-phosphatidylethanolamine from chicken egg, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(carboxyfluorescein) (ammonium salt), and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(cap biotinyl) (sodium salt), l-α-phosphatidylethanolamine-N-(lissaminerhodamine B sulfonyl) (ammonium salt) from transphosphatidylated chicken egg were purchased from Avanti Polar Lipids (Alabaster, AL). Lipids were stored in chloroform at −20 °C under nitrogen gas, mixed at the indicated molar ratios, dried under a stream of N2 gas, washed three times with pentane, dried again with N2 gas, and dried under a vacuum for a minimum of 2 h. The lipid films were hydrated to a final concentration of 1 mg/ml in HEPES-buffered saline (50 mm HEPES (pH 7.5), 100 mm NaCl, and 5 mm MgCl2) for 1 h at 32 °C. The resulting liposomes were downsized by two cycles of freeze/thaw followed by 11 passes through a polycarbonate membrane with a pore size of 200 nm. Vesicles containing biotinylated phosphatidylethanolamine were purified by binding to a monomeric avidin-functionalized resin column, washed with HEPES-buffered saline, and then eluted with free biotin in a method similar to that developed by Peker et al. (42).
Purification of 3AB Protein
Wild-type Mahoney type 1 poliovirus 3AB was expressed from plasmid pLG-3AB (a gift from Deborah S. Wuttke, University of Colorado, Boulder, CO) in Escherichia coli BL21(DE3)pLysS according to a protocol provided by Oliver C. Richards (Salt Lake City, UT). Cells containing the expression plasmid were grown in M9 minimal medium at 37 °C to an A600 of 0.6–0.8. The cells were cooled to 20 °C, and production of the 3AB protein was induced by the addition of 0.5 mm isopropyl-β-d-thiogalactopyranoside followed by incubation for an additional 16 h at 20 °C. The following buffers were used to purify the poliovirus 3AB proteins. All buffers were kept at 4 °C and contained 1 mm EDTA, 1 mm dithiothreitol, 5% (v/v) glycerol, and the following additional ingredients. Buffer A contained 100 mm NaCl and 50 mm Tris-HCl (pH 7.5). Buffer 1A contained 0.5% (v/v) IGEPAL CA-630 (Sigma-Aldrich), 0.5 m NaCl, and 50 mm Tris-HCl (pH 7.5). Buffer D contained 0.1% IGEPAL CA-630, 20 mm NaCl, and 50 mm Tric-HCl (pH 8.0). Buffer DI contained 0.1% IGEPAL CA-630, 10 mm NaCl, and 20 mm Tris-HCl (pH 8.0). Buffer DE contained 0.1% IGEPAL CA-630, 10 mm NaCl, and 25 mm Tris-HCl (pH 8.0). Buffer DS contained 0.1% IGEPAL CA-630, 5 mm NaCl, and 25 mm MOPS (pH 7.0). Buffers S10, S100, and S200 were the same as buffer DS, except they contained 10 mm, 100 mm, and 200 mm NaCl, respectively. The following procedure was conducted at 4 °C, unless otherwise noted. The cell pellet was washed once in a solution that contained 50 mm Tris (pH 7.5) and 100 mm NaCl. The pellet was suspended and brought to a total volume of 10 ml with buffer A. Cells were lysed by passing through a French pressure cell four times at 10,000 psi. The lysate was centrifuged at 20,000 × g for 15 min. The supernatant was further centrifuged at 100,000 × g for 1 h. Then the second pellet was suspended and brought to a total volume of 10 ml in buffer A and washed for 1 h with rotation. The lysate was centrifuged at 10,000 × g for 1 h. The supernatant (S1) was saved, and the pellet was suspended in 4.5 ml of buffer 1A and incubated overnight with rotation. The lysate was centrifuged at 100,000 × g for 1 h. Again, the supernatant (S2) was saved, and the pellet was resuspended in buffer 1A and incubated for 2 h on a rotator. The last steps of centrifugation and supernatant isolation were repeated again (S3). The three supernatants were combined and dialyzed twice against 1 liter of buffer D using a dialysis membrane with a molecular weight cutoff of 3500 Da for 2 h each. The dialysate was diluted to 20 ml with buffer D, centrifuged at 20,000 × g for 30 min, diluted with 10 ml of buffer DI, and passed over a DEAE-cellulose column (Pharmacia) equilibrated with buffer DE. The flowthrough and a wash of 10 ml of buffer DE were collected and concentrated through a Vivaspin 20 3-kDa molecular weight cutoff polyethersulfone membrane filter (Bioexpress) until the retentate was 5 ml. The concentrated 3AB solution was diluted with four volumes of buffer DS and passed over a HiTrap SP FF column (Pharmacia) equilibrated with S10 buffer. Poliovirus 3AB protein was eluted with S-200 buffer and stored at −80 °C. For 3AB-proteoliposome reconstitution using the dialysis method, 3AB was purified by the same procedure, except the 0.1% IGEPAL CA-630 was substituted by 0.8% n-octyl-β-d-glucopyranoside (A.C. Scientific) in all chromatography buffers.
Reconstitution of Purified Poliovirus 3AB Protein by Controlled Dilution
Two amounts of purified 3AB protein diluted with buffer S200 to yield final concentrations of 0.38 or 1.52 mm, or S200 buffer alone, were incubated with solutions of liposomes (310 mm final phospholipid concentration in HEPES-buffered saline) at 37 °C for 15 min. Buffer S200, containing 0.1% IGEPAL CA-630, made up only 4% (v/v) of the reaction. The detergent IGEPAL CA-630 was diluted below its critical micelle concentration (0.005% v/v) to eliminate effects on vesicle integrity.
Lipid and Liposome Quantitation by Fluorescence Spectroscopy
To quantify the amount of lipid-containing structures recovered after purification over a monomeric avidin-functionalized column, the fluorescence intensity of carboxyfluorescein-PE was measured using a Gemini XS fluorometer (Molecular Devices) as follows. First, the lipid mixtures generated upon hydration at a known concentration of 1 mg/ml were serially diluted to construct a standard curve. To quantify the amount of total lipid present in the samples from one of several repetitions of these experiments, serial dilutions of the purified liposome samples were aliquoted into a 384 well plate (Corning). Upon excitation at 490 nm, the carboxyfluorescein-PE emission intensity at 515 nm was recorded. To quantify the formation of topologically closed liposomes, the dye trypan blue (Merck) diluted in HEPES-buffered saline or buffer alone was added to the preparations at 0.008% (w/v) and the carboxyfluorescein-PE emission intensity measured. The emission intensity of samples containing trypan blue was expressed as a percentage of the fluorescence of the same sample in buffer alone and referred to as “percent internalized lipid.”
Quantitation of Liposome-associated 3AB
To quantify the amount of 3AB associated with liposomes, proteoliposomes were isolated by ultracentrifugation at 112,000 × g in a Beckman TLA 100.2 rotor at 4 °C for 25 min. The pellets were resuspended in 600 μl of 1× Laemmli sample buffer and subjected to SDS-PAGE through a 15% polyacrylamide gel. Following SDS-PAGE, samples were transferred to polyvinylidene difluoride membranes (Millipore). Membranes were blocked by 5% BSA (w/v) in PBS-T (a PBS solution that contained 0.1% (v/v) Tween 20 (Sigma)) for 30 min at room temperature. Membranes were then incubated with a 1:30 dilution of anti-3A monoclonal mouse antibody (29) in 5% BSA in PBS-T for 2 h at room temperature. After incubation, membranes were washed four times in PBS-T for 5 min each. Membranes were then incubated with a 1:10,000 dilution of anti-mouse alkaline phosphatase-conjugated secondary antibody (Jackson ImmunoResearch Laboratories, Inc.) in 5% BSA in PBS-T for 1 h at room temperature. After incubation, membranes were washed five times in PBS-T for 5 min each. After washing the blots were incubated with ECF reagent (Amersham Biosciences) and viewed on a Storm 860 PhosphorImager. Concentration of 3AB protein was determined by densitometry using ImageQuant software. The 3AB concentration in the liposome samples was quantified against a standard curve of 3AB concentration.
Reconstitution of Poliovirus 3AB Protein by Dialysis of Lipid Detergent-mixed Micelles
Dried lipid films were suspended in MBS (25 mm MOPS (pH 7.0), 100 mm NaCl, 1 mm EDTA, and 1 mm DTT) with 1% n-octyl-β-d-glucopyranoside to a final lipid concentration of 1 mm. The lipid solution was then mixed with an equal volume of 40 μm 3AB (purified in 0.8% n-octyl-β-d-glucopyranoside) or MBS containing 0.8% n-octyl-β-d-glucopyranoside on ice for 1 h and then dialyzed against MBS using a 3500-Da molecular weight cutoff membrane (Spectrum Laboratory) for up to 4 days. A 333-μl aliquot of the resulting proteoliposome-containing solution was mixed with 666 μl of 60% sucrose (w/v) in MBS, overlaid first with 2 ml of 20% sucrose in MBS and then with 1 ml of MBS, and centrifuged at 165,000 × g for 1 h at 4 °C. Fractions (500 μl) collected from the 0–20% interface, the 20–40% interface, and the 40% sucrose fraction were analyzed by SDS-PAGE and silver staining.
Transduction of Purified 3AB Protein into HeLa Cells
Purified 3AB was delivered into the cells using PULSin (Polyplus Transfection). The transduction mixture was incubated with HeLa cells for 4 h at 37 °C and optimized in this experiment using Alexa Fluor 488 histone H1 (Invitrogen), which has a similar molecular weight and charge (21 kDa, pI = 11.5) as 3AB (12 kDa, pI = 9.1). The recommended condition of (4 μg of protein plus 3 μl of PULSin)/100,000 cells (43) was optimal for the nuclear delivery of histone H1, and these conditions were employed for 3AB transduction.
To facilitate further chemical fixation and sectioning, 1 million HeLa cells were seeded in 10-cm tissue culture dishes in Eagle's minimal essential medium supplemented with 10% FBS (EMEM-FBS)4 and incubated at 37 °C until 70% confluency. Prior to protein delivery, cells were washed once and kept in 4.5 ml of EMEM-FBS. Purified 3AB (25 μg) was diluted into 500 μl of HEPES buffer (pH 7.2) on ice and then mixed with 25 μl of PULSin reagent. The mixture was incubated for 15 min at room temperature and then added dropwise into cultured cells to ensure a homogenized distribution of the 3AB-PULSin complex. The cells were then incubated at 37 °C for up to 2 h. The medium was then replaced by EMEM-FBS until cells were processed for fixation. As a negative control, the same amount of 3AB treated with protease K for 1 h and heat-inactivated at 80 °C for 1 h was delivered into HeLa cells by PULSin reagent in parallel.
Electron Microscopy
3AB-reconstituted liposomes were incubated at 37 °C for 20 min to facilitate membrane remodeling and then chilled on ice. For cryoelectron microscopy, 3 μl of sample was applied to a copper grid coated with continuous carbon. The grid was then blotted with filter paper (Ted Pella Inc.) for 3 s and rapidly plunge-frozen by Vitrobot (FEI, Oregon) in liquid ethane. Specimens were examined on a Philips CM12 transmission electron microscope at 120 keV at a magnification of ×28,000 and a defocus range from 1.5∼2.5 μm using a Gatan 626 cryoholder (Gatan, Inc., Warrendale, PA). Images were recorded on Kodak SO163 film, developed in full-strength Kodak D19 developer for 12 min, and scanned using Nikon Coolscan 9000 at a pixel size corresponding to 2.3 Å on the specimen.
To collect tilt data, the specimen holder was set to the eucentric height of the stage. A first exposure was taken at 0° tilt and recorded on a Tietz video and image processing system 1K X 1K CCD camera at a magnification corresponding to 6.8 Å/pixel. If vesicles were imaged in an intermediate state within the camera frame, the holder was manually tilted to 30o for a second exposure and 45o for a third exposure. The defocus for the tilted images was set at −3 to −6 μm to ensure good contrast across the whole image.
For ultrathin cell sectioning, after 2 h incubation with 3AB-PULSin complex or the negative control complex at 37 °C, cells were washed once with EMEM-FBS and fixed in 2% glutaraldehyde for 10 min at 37 °C before being scraped off the dish. The cell suspension was transferred to a glass tube and pelleted by centrifugation at 1000 × g for 5 min. After three rounds of washing in PBS, pellets were postfixed in 2% osmium tetraoxide for 2 h at 4 °C, washed three times in PBS, and then dehydrated through an ascending ethanol series of 50, 70, 95, and 100% (15 min per condition). Subsequently, the cell pellet was transferred to a glass vial in 100% ethanol, exchanged to propylene oxide, and incubated for 15 min at room temperature. The dry pellet was infiltrated with propylene/epon resin mixed at a 1:1 ratio overnight, incubated in 100% epon for 2 h at room temperature, and embedded in fresh epon. After 24 h of curing at 37 °C and then at 55 °C for another 24 h, the solid block was cut on a microtome (Bausch & Lomb Optical Co.) using a diamond knife (Microstar). Sections of ∼800-nm thickness were stained with toluidine blue to locate the specimen within the block. Ultrathin sections (100–200 nm) collected on 200-mesh copper grids and stained with 3% aqueous uranyl acetate for 15 min, rinsed in water, stained in Reynold's lead citrate for 5 min, rinsed in water, and air-dried.
RESULTS
Effect of Phospholipid Composition on Liposome Formation, Topology, and Recruitment of 3AB Protein
To determine the potential contribution of poliovirus 3AB protein to membrane rearrangement during poliovirus infection, we developed a synthetic model membrane system. Lipid composition is a major factor in membrane remodeling because it is the interplay between lipids and proteins that determines how membranes are shaped. We began by testing the effect of different lipid compositions on liposome formation and topology. The major phospholipid components of cell membranes include phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylserine (PS). Membranes containing the polar phospholipid PE are more fluid and favor the formation of nonlamellar membrane structures, thereby facilitating the fusion and fission of lipid vesicles (44, 45). PE is also an important constituent in cell membranes because it is the predominant phospholipid conjugated to LC3, marking an essential step of autophagosome formation (46). The anionic phospholipid PS is entirely localized to the inner leaflet of cellular membranes and facilitates the membrane association of proteins containing cationic motifs (47). We systematically prepared a series of mostly unilamellar (single-membraned) liposomes, ∼200 nm in diameter, composed of the following molar ratios of naturally derived phospholipids (PC:PE:PS): 1:1:1, 3:1:2, 3:2:1, 4:1:1, and 19:1:0. The last ratio was chosen because it has been used previously to prepare poliovirus 3AB-containing proteoliposomes (25).
The purity of 3AB is shown by gel electrophoresis in Fig. 1B. The faint band seen at twice the molecular weight of 3AB is expected to be the 3AB dimer. This band appears only in samples from cells expressing 3AB, has a molecular weight consistent with a 3AB dimer, copurifies with the 3AB monomer, and changes in relative abundance with alterations in denaturation conditions prior to electrophoresis. As shown schematically in Fig. 1B, each liposome mixture also contained 5 molar percent carboxyfluorescein-labeled PE to facilitate liposome quantitation and 0.2 molar percent biotinylated PE to allow liposome purification over monomeric avidin-functionalized columns.
We first investigated which phospholipid composition was most suitable for the formation of topologically closed liposomes. Using the dye trypan blue to quench the carboxyfluorescein-labeled lipid in lipid micelles and in the outer leaflet of the bilayer of liposomes, we measured the percentage of unquenched, internalized phospholipid (Fig. 1C). If all lipids hydrated to form single-membraned liposomes, then 50% of the fluorescent phospholipid would be internalized. The 1:1:1, 3:1:2, and 3:2:1 PC:PE:PS lipid mixtures displayed a substantial level of internalized fluorescent lipid, 29, 24, and 25%, respectively, indicating that ∼58, 48, and 50% of the lipids in the mixture were present in topologically closed liposomes. However, lipid mixtures composed of 4:1:1 and 19:1:0 PC:PE:PS resulted in only 19 and 12% internalized lipid, indicating that they were not optimal for liposome formation under these conditions.
Next, we examined the effect of lipid composition on the efficiency of 3AB reconstitution into liposomes. We added increasing concentrations of the purified poliovirus 3AB protein to the synthetic liposomes and quantified the point of saturation of 3AB incorporation (Fig. 1D). To reconstitute the detergent-containing 3AB protein without compromising the integrity of the liposomes, the 3AB protein was diluted below the critical micellular concentration of the IGEPAL CA-630 detergent. After 3AB-containing proteoliposomes were separated from free 3AB protein by ultracentrifugation, SDS-PAGE analysis of the pellets revealed that 3AB protein reconstituted most efficiently into liposomes composed of 1:1:1 and 19:1:0 PC:PE:PS lipid ratios, with the other ratios giving intermediate results (Fig. 1D).
We endeavored to optimize the percentage of the liposome population that was unilamellar before addition of protein to simplify the interpretation of any liposome remodeling caused by protein association. To this end, we investigated the topology of liposomes of lipid compositions 1:1:1, 3:1:2, and 3:2:1 by cryoelectron microscopy (cryo-EM). Examination of liposome topology in the resulting populations (Fig. 1, E and F) revealed that hydration of the lipid films of each mixture resulted in mostly unilamellar liposomes with a low background of double-membraned and multilamellar liposomes. We presume that the remaining lipids form small amorphous aggregates. Quantitation of these results (Fig. 1G) revealed that the 3:2:1 PC:PE:PS ratio resulted in the highest percentage of single-membraned vesicles. This ratio, which also displayed an acceptable amount of 3AB incorporation and most closely resembles the lipid composition of the mammalian endoplasmic reticulum (55% PC, 30% PE, 5% PS; Ref. 48), was chosen for the remaining experiments.
Purified Poliovirus 3AB Protein Induces the Formation of Double-membraned Liposomes
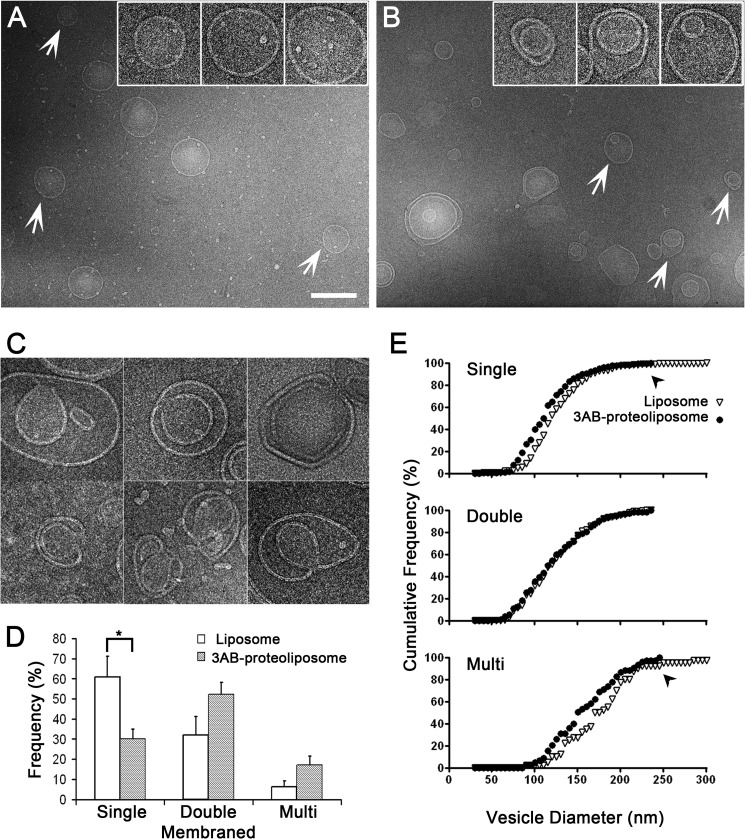
To determine the effects of the poliovirus 3AB protein on membrane structure, purified 3AB was incubated with synthetic liposomes following controlled dilution. The ultrastructure of liposomes lacking and containing poliovirus 3AB protein was examined by cryo-EM. In the absence of 3AB protein, most of the liposomes were spherical, with an average membrane through which they were extruded. Strikingly, when the 3AB protein was reconstituted into liposomes, the percentage of single-membraned liposomes was reduced from ∼60 to 30%, with a concomitant increase in the percentages of multilamellar, especially double-membraned, liposomes (Fig. 2B). The morphology of the double-membraned vesicles formed in the absence and presence of 3AB also differed. A substantial number (15%) of the 3AB-containing liposomes contained a discontinuous inner bilayer (Fig. 2C), suggesting continuity with the outer bilayer. In addition, we observed that a few vesicles formed horseshoe structures, potential intermediates in the formation of double-membraned liposomes by invagination (Fig. 2C). Such putative intermediates were absent in the liposome population lacking 3AB protein. The percentages of single-, double-, and multimembraned structures observed in liposomes lacking and containing the 3AB protein are quantified in Fig. 2D and argue that poliovirus 3AB protein can transform membranes from a single- to a double-membraned topology. This effect was not observed when the poliovirus polymerase protein 3Dpol was added to liposomes.
FIGURE 2.
Effect of 3AB protein reconstitution by controlled dilution into synthetic liposomes. A, cryo-electron micrographs of liposomes lacking 3AB protein. Arrows indicate vesicles that are enlarged ×2.5 in the inset. Scale bar = 200 nm. B, cryo-electron micrographs of reconstituted 3AB proteoliposomes. Arrows indicate vesicles that are enlarged ×2.5 in the inset. C, images of putative intermediates of double-membraned liposome formation after 3AB reconstitution. Scale bar = same as the insets in A and B. D, quantitation of the numbers of liposomes of single-, double-, and multilamellar topology in the absence and presence of 3AB protein is shown. The asterisk denotes a statistically significant difference. E, cumulative frequencies of vesicle diameters for single-, double-, and multilamellar liposomes reconstituted with buffer alone (▿) or with 3AB protein (●) are shown. Significant statistical differences between the diameters of liposomes and proteoliposomes for single-membraned vesicles (p < 0.1, Kolmogorov-Smirnov test) and multilamellar vesicles were observed.
If the double-membraned vesicles were formed by the 3AB-mediated invagination of single-membraned vesicles, 3AB incubation should result in a decrease in both the number and the size of the single-membraned vesicles. To test this hypothesis, we measured the size distribution of the single-, double-, and multimembraned liposomes in each population (Fig. 2E). A comparison of the size distribution data revealed that the sizes of single-membraned vesicles that had been incubated with 3AB protein spanned a smaller range (49–232 nm) compared with liposomes that lacked protein (32–350 nm). The statistical significance of this effect was shown using the Kolmogorov-Smirnov test (p < 0.1 as determined by a D-value of 0.17 when Dp = 0.1 = 0.16 and Dp = 0.05 = 0.21) (49). The decrease of the population of large single-membraned vesicles upon 3AB incubation was accompanied by the increase in double-membraned vesicles, arguing that 3AB protein induced the formation of the new double-membraned structures via the invagination of larger, single-membraned liposomes.
Topographical Analysis of 3AB Proteoliposomes
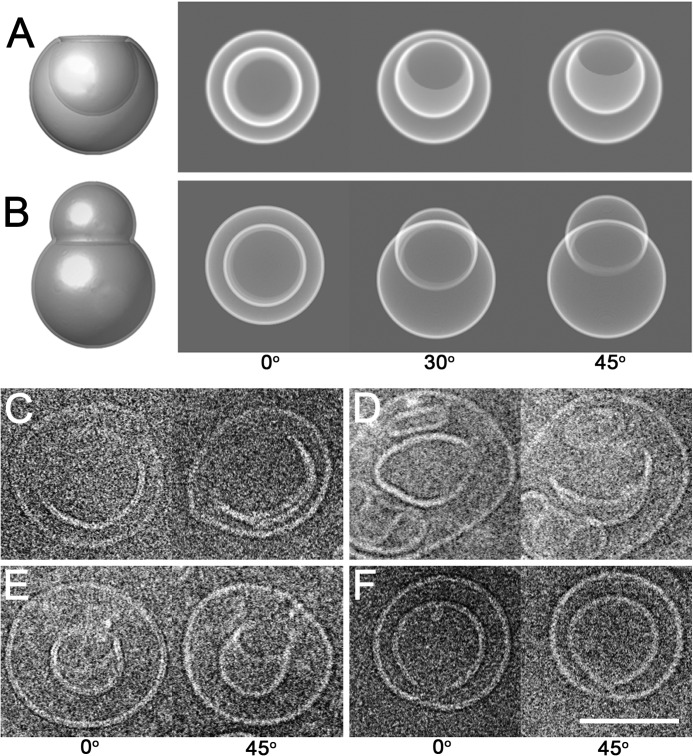
An apparently double-membraned vesicle could be generated either from the invagination of a single-membraned liposome or from engulfment of a smaller single-membraned liposome by a larger one, as shown schematically in Fig. 3. To discriminate between these two scenarios and to further investigate the mechanism of double membrane formation induced by the 3AB protein, we examined the detailed ultrastructure of double-membraned proteoliposomes, focusing on those that contained irregularly shaped internal bilayers that might represent structural intermediates. Tilt pairs of cryo-EM images were collected, with the vitrified 3AB proteoliposomes tilted from 0o to 30o and then to 45o. Depending on whether a liposome contains an invaginated or a protruding deformation, it will give rise to different projections in such a tilt series. Some projections will be identical for the two structures, as shown in the left panels of Fig. 3, A and B. However, further images in a tilt series will discriminate between the two structures. For an invaginated deformation, the inner membrane of a double-membraned vesicle will always remain contained within the outer membrane (Fig. 3A, right panel). In fact, the inner membrane will appear to transform into a crescent but still remain bounded by the outer membrane (Fig. 3A). On the other hand, a single-membraned vesicle that is in the process of engulfing another vesicle will produce a tilt series of images in which the “inner membrane” can shift outside of the outer membrane boundary (Fig. 3B, right panel). The 0o and 45o tilt images of four representative 3AB proteoliposomes (Fig. 3, C–E) show that the inner membranes of all four structures remained confined by the outer membranes. Furthermore, three of the four tilted images reveal a transformation of the inner membrane into a crescent shape, arguing that these structures contained invaginations. Such structures are potential intermediates of double-membrane formation orchestrated by poliovirus 3AB protein.
FIGURE 3.
Tilt series of putative intermediates of double-membraned vesicles induced by poliovirus 3AB. A, schematic side view of an invaginating vesicle (left panel). An electron beam coming from above the virtual vesicle would generate the two-dimensional projection shown in the 0o image in the right panel. Rotation of the model invaginated structure by 30o and 45o around a horizontal tilt axis is shown. B, schematic side view of a liposome that is engulfing a second liposome (left panel). The modeled vesicle was rotated 0o, 30o, and 45o around a horizontal axis, and the corresponding two-dimensional projections are shown in the right panel. C–F, four proteoliposomes that contained 3AB were rotated 0o and 45o around a tilt axis perpendicular to the electron beam and imaged by cryo-electron microscopy. Scale bar = 100 nm.
Liposome Formation in the Presence of Viral Protein 3AB
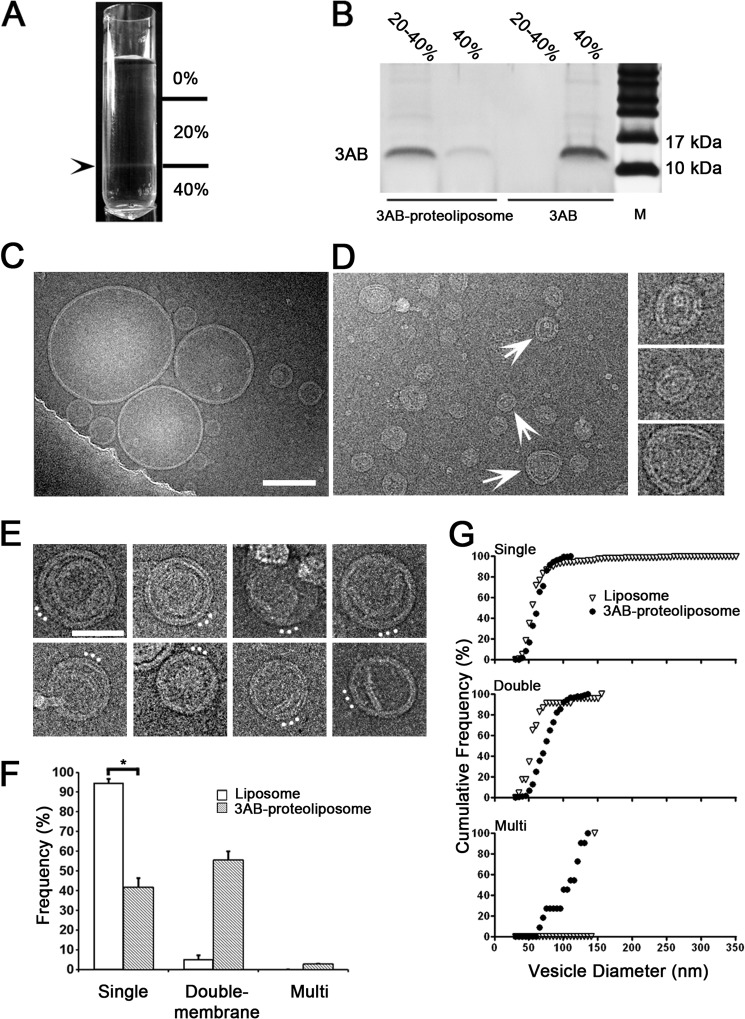
Although 3AB induction of double-membraned liposomes from preformed liposomes was quite evident, the amount of 3AB protein present, approximately one protein for every 100 lipid molecules (Fig. 1D) was limited by the dilution method used. To increase the ratio of 3AB protein to lipid and, thus, perhaps, to increase the efficiency of double-membraned vesicle formation, we reconstituted 3AB proteoliposomes by directly mixing detergent-suspended 3AB with mixed micelles of lipid and detergent at a protein-to-lipid ratio of 1:25, followed by removal of the β-octylglucoside detergent by dialysis. Sucrose gradient fractionation and SDS-PAGE analysis of the resulting protein-lipid mixtures (Fig. 4, A and B) showed that although 3AB in the absence of lipid remained in the 40% sucrose fraction, a majority of the protein comigrated with lipids following reconstitution, localizing primarily at the 20–40% sucrose interface. Given that 3AB associated efficiently with lipids (Fig. 4B), it was likely that, if liposomes were present, they would contain 3AB. Electron microscopy confirmed that liposomes were observed in the absence (Fig. 4C) or presence (D and E) of 3AB protein. Most liposomes generated upon dialysis in the absence of protein were single-membraned and ranged from 21–352 nm in diameter (Fig. 4C). A likely explanation for the larger sizes of unilamellar membranes obtained is that, in contrast to Fig. 2, liposome formation by the dialysis method began with lipid micelles instead of the stacked lipid films formed during lipid hydration and extrusion. Liposomes formed in the presence of 3AB were smaller and displayed a more restricted size range, from 18–130 nm in diameter (Fig. 4D). The relative amounts of double-membraned liposomes were greatly increased in the presence of 3AB, as were the amounts of more complex, multilamellar structures (Fig. 4E). The changes in morphology upon liposome formation in the presence of 3AB are quantified in Fig. 4F, and the changes in vesicle diameter of the unilamellar, double-membraned and multilamellar liposomes are shown in G. Liposome formation in the presence of 3AB reduced the size and frequency of unilamellar samples and increased the size and frequency of more complex structures. Unlike the experiments shown in Fig. 2, in which 3AB protein was added to preformed liposomes, these changes argue that poliovirus 3AB protein induces the formation of double-membraned vesicles de novo, at least in part by stabilizing increased membrane curvature.
FIGURE 4.
Effect of 3AB protein on reconstitution of synthetic liposomes by direct mixing and dialysis. A, following dialysis, 3AB-containing proteoliposomes were fractionated by ultracentrifugation through discontinuous sucrose gradients. The arrowhead denotes the position of liposomes in the gradient at the 20–40% sucrose interface. B, fractions obtained from the indicated sucrose gradient position were analyzed by SDS-PAGE and silver staining to detect poliovirus 3AB. M designates molecular weight markers. C, cryo-electron micrograph of liposomes lacking 3AB protein displaying single-membraned liposomes of various sizes. Scale bar = 100 nm for C and D. D, cryo-electron micrograph of reconstituted 3AB-containing proteoliposomes. Selected double-membraned vesicles from this image (arrows) are enlarged 2-fold in the right panel. E, additional images of double-membraned vesicles and more complex structures after reconstitution in the presence of 3AB. White dots mark the presumed directionality of the membrane invagination. Scale bar = 50 nm. F, frequencies of different membrane topologies formed in the absence (white bar) and presence (gray bar) of 3AB protein. The asterisk denotes a statistically significant difference. G, cumulative frequency plot of vesicle diameters for liposomes alone (▿) or proteoliposomes containing purified 3AB protein (●) as visualized by cryo-electron microscopy. Data show significant statistical differences for single-, double-, and multimembraned vesicles (p < 0.1, Kolmogorov-Smirnov test).
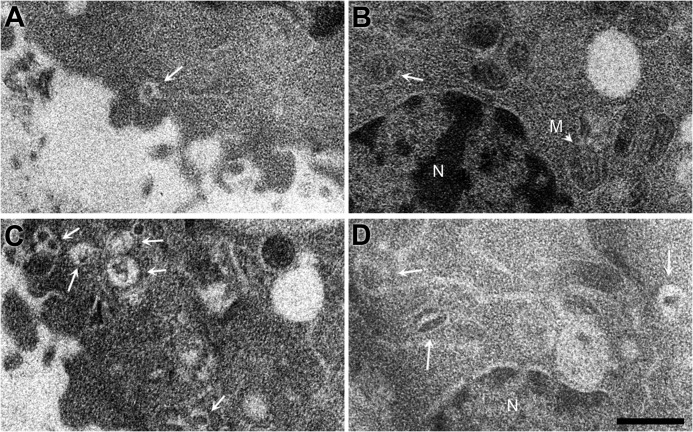
The Effect of 3AB Transduction on Intracellular Membranes
To investigate the effect of 3AB protein in mammalian cells, we transduced purified 3AB protein directly rather than the more toxic procedure of expressing 3AB from plasmids. Direct transduction was accomplished using a protein transduction reagent (PULSin; Polyplus Transfection) we have used previously to introduce poliovirus 3D polymerase directly into cells (50). As a control mock transduction, heat-inactivated 3AB was transduced into HeLa cells using PULSin. To investigate the effect of 3AB on intracellular membranes, ultrathin sectioning was performed on fixed HeLa cells that had been incubated for 2 h at 37 °C in the presence or absence of 3AB protein or inactivated 3AB protein. In mock-transduced cells, vesicular structures near the plasma membrane (Fig. 5A) or nucleus (B) that contained cytoplasm inclusions were occasionally observed. Upon 3AB transduction, however (Fig. 5, C and D), the frequency of such cytoplasmic invaginations increased significantly. This effect was quantitated by an increase in the number of observed cytoplasmic inclusions from 1.2 (± 1.3) invaginations/field in mock-transduced cells to 3.1 (± 2.3) invaginations/field in 3AB-transduced cells, with significance confirmed by a p value of 0.008.
FIGURE 5.
Effect of transducing purified 3AB protein into human cells in tissue culture. Shown are cytoplasmic (A) and perinuclear regions (B) of HeLa cells transduced with inactivated 3AB protein and cytoplasmic (C) and perinuclear regions (D) of HeLa cells transduced with purified 3AB protein. The presence of 3AB caused the numbers of observed cytoplasmic inclusions to increase significantly (p = 0.008) from 1.2 (± 1.3) to 3.1 (± 2.3) invaginations/field. White arrows mark cytoplasmic inclusions. N, nucleus; M, representative mitochondrion. Scale bars = 500 nm.
DISCUSSION
In this study, we show that purified poliovirus 3AB protein reconstituted into synthetic lipid bilayers is sufficient to induce the formation of double-membraned vesicles independent of any additional viral or host proteins. Upon incubation of purified 3AB protein with single-membraned liposomes, ultrastructural analysis revealed a decrease in both the average vesicle size and the number of single-membraned liposomes that was concomitant with the increase in double-membraned vesicles. We conclude that the 3AB protein induces the formation of double-membraned structures via the invagination of larger single-membraned liposomes. Topological analysis revealed that putative structural intermediates resembled invaginating vesicles, supporting this hypothesis. This process resembles the invagination of small vesicles into liposomes demonstrated for the ESCRT I and II complexes, which comprise seven different proteins (66) and whose three-dimensional structure reveals a curved complex (51). Subsequent vesicle scission requires an additional multisubunit complex, ESCRT III. This work reports for the first time that a single protein induces the formation of membrane invaginations or double-membraned vesicles.
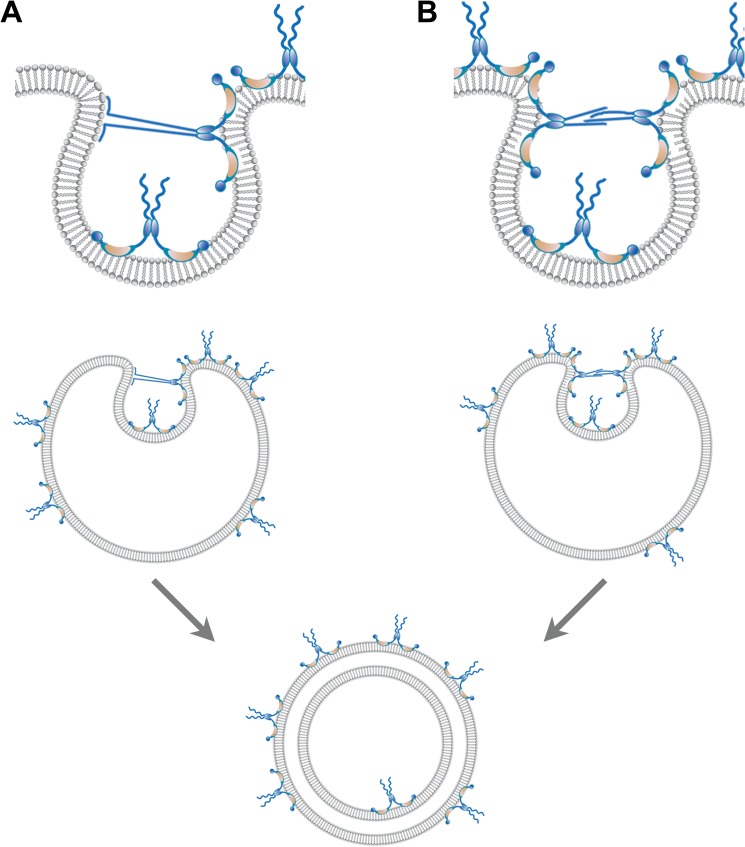
Fig. 6 details two hypotheses for how a small protein such as 3AB could induce the formation of a double-membraned vesicle. The 3AB protein is known to insert into the outer leaflet of the bilayer in a wedge-like fashion (25). It has been shown that partial membrane insertion of proteins displaces lipids in only the outer leaflet of the bilayer, thereby generating a localized positive membrane curvature (53–55). Such a localized deformation of the membrane can produce lateral crowding in the outer leaflet that will change the curvature of membranes to compensate for the resulting asymmetry of surface density (reviewed in Ref. 54). The resulting lipid crowding in the outer leaflet can be relieved by invagination (Fig. 6). Invagination relies on the neck of the invaginating membrane being transiently stabilized by interactions. Such interactions could be between the previously unstructured amino-terminal sequences of 3AB and either phospholipid headgroups (Fig. 6A) or other 3AB molecules (B). Lipids would then flow into the growing, invaginating membrane, whereas protein would be excluded because of the high surface pressure on the neck of the invaginating membrane. In line with either mechanism, it has been well documented that unstructured protein domains can become ordered upon either phospholipid or protein binding (reviewed in Refs. 53, 56, and 57). Thus, we suggest that the initially unstructured amino-terminal domain of 3AB plays a role in stabilizing intermediates in membrane invagination. Supporting the hypothesis that the amino-terminal sequences of 3AB protein play a direct role in vesicle formation, a mutant poliovirus that contained an insertion of a serine residue between Thr-14 and Ser-15 of the 3A sequence was shown to induce double-membraned vesicles with larger diameters than the wild type (58). In the proposed models, stabilization of membrane apposition in close proximity can, furthermore, reduce the thermodynamic barrier for spontaneous membrane fusion, facilitating the closure of these double-membraned structures.
FIGURE 6.
Two hypotheses for the formation of double-membraned vesicles induced by poliovirus 3AB. The 3AB integral membrane protein inserts into the outer leaflet of the bilayer in a wedge-like fashion, with the N- and C-terminal domains both protruding from the cytoplasmic surface (25, 30). To promote membrane invagination, we propose that the 3AB protein dimers displace lipids in the outer leaflet, generating a localized deformation of the bilayer. This localized deformation leads to lateral crowding and an imbalance in the leaflets of the bilayer, stabilizing the curvature required for invagination. As more 3AB molecules associate with the vesicle surface, lateral crowding and stress increase until the protein-sparse region collapses inward. The neck of the invaginating structure is stabilized by interactions between the unstructured domains of 3AB either with (A) the negatively charged phospholipid head groups (A) or the unstructured domains of 3AB dimers (B) on opposite sides of the neck. It is likely that this close opposition, together with the membrane disruption, promotes fusion. Note that most 3AB molecules remain on the outside of the vesicle, as expected for the participation of 3AB in viral RNA replication. Random coil, unstructured N-terminal domain; blue oval, dimerization domain; orange crescent, hydrophobic domain; blue circle, C-terminal 3B domain.
Although we show here that poliovirus protein 3AB is sufficient to induce the formation of double-membraned liposomes, previous studies suggest joint participation with the 2BC protein in mammalian cells (17, 18). Isolated 2BC expression in tissue culture cells has been shown to lead to the accumulation of single-membraned vesicles, which are likely intermediates in poliovirus-induced double membrane formation (12, 27). It is plausible that, in infected and transfected cells, the 2BC protein generates single-membraned vesicles and recruits 3A, 3AB, or a larger precursor for membrane insertion and rearrangement.
A principal requirement of both autophagosome and poliovirus-induced double membrane formation is membrane deformation to induce membrane curvature. To date, experimental evidence has suggested several possible candidates that may drive the curvature of preautophagosome isolation membranes: lipidated LC3 protein, which can, in isolation, induce membrane fusion (61, 62); the Atg16L complex required for isolation membrane elongation; and the BAR domain-containing protein BIF-1 (63) (reviewed in Ref. 59). Perhaps the strongest candidate is the Atg17-Atg31-Atg29 complex, whose recently solved structure displays a curved double crescent that preferentially binds membranes with a distinct curvature (60). This complex is tethered to membranes by Atg1, whose mammalian homolog ULK1 is predicted to contain a long, intrinsically disordered domain (52). Given the ability of 3AB to transform single-membraned liposomes into double membranes via invagination, it is plausible that, in infected cells, this protein works in concert with or in place of these candidate autophagy proteins to induce membrane curvature and facilitate scission to complete a double-membraned structure. Therefore, understanding the mechanism of poliovirus 3AB-induced double-membraned vesicle formation may also yield mechanistic insights into the membrane rearrangements and regulation of membrane curvature required for autophagosome formation.
Acknowledgments
We thank Dr. Donald Small (Boston University) for critical reading and suggestions on statistical analysis, Dr. Peter Sarnow (Stanford University) for comments on the manuscript, and Drs. Jean-François Ménetret and Christopher Akey (Boston University) for advice on cryo-EM.
This work was supported, in whole or in part, by the National Institutes of Health Director's Pioneer Award (to K. K.) and National Institutes of Health Grant GM102474 (to E. B.). This work was also supported by the American Cancer Society (to J. B. P.).
- EMEM-FBS
- Eagle's minimal essential medium with 10% FBS
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PS
- phosphatidylserine
- cryo-EM
- cryoelectron microscopy.
REFERENCES
- 1. Schlegel A., Giddings T. H., Jr., Ladinsky M. S., Kirkegaard K. (1996) Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 70, 6576–6588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jackson W. T., Giddings T. H., Jr., Taylor M. P., Mulinyawe S., Rabinovitch M., Kopito R. R., Kirkegaard K. (2005) Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 3, e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Uchil P. D., Satchidanandam V. (2003) Architecture of the flaviviral replication complex. Protease, nuclease, and detergents reveal encasement within double-layered membrane compartments. J. Biol. Chem. 278, 24388–24398 [DOI] [PubMed] [Google Scholar]
- 4. Gosert R., Kanjanahaluethai A., Egger D., Bienz K., Baker S. C. (2002) RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76, 3697–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gosert R., Egger D., Lohmann V., Bartenschlager R., Blum H. E., Bienz K., Moradpour D. (2003) Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77, 5487–5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Welsch S., Miller S., Romero-Brey I., Merz A., Bleck C. K., Walther P., Fuller S. D., Antony C., Krijnse-Locker J., Bartenschlager R. (2009) Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5, 365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lanman J., Crum J., Deerinck T. J., Gaietta G. M., Schneemann A., Sosinsky G. E., Ellisman M. H., Johnson J. E. (2008) Visualizing flock house virus infection in Drosophila cells with correlated fluorescence and electron microscopy. J. Struct. Biol. 161, 439–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schwartz M., Chen J., Janda M., Sullivan M., den Boon J., Ahlquist P. (2002) A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell. 9, 505–514 [DOI] [PubMed] [Google Scholar]
- 9. Bienz K., Egger D., Troxler M., Pasamontes L. (1990) Structural organization of poliovirus RNA replication is mediated by viral proteins of the P2 genomic region. J. Virol. 64, 1156–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bienz K., Egger D., Pasamontes L. (1987) Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology 160, 220–226 [DOI] [PubMed] [Google Scholar]
- 11. Bienz K., Egger D., Pfister T., Troxler M. (1992) Structural and functional characterization of the poliovirus replication complex. J. Virol. 66, 2740–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cho M. W., Teterina N., Egger D., Bienz K., Ehrenfeld E. (1994) Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology 202, 129–145 [DOI] [PubMed] [Google Scholar]
- 13. Egger D., Pasamontes L., Bolten R., Boyko V., Bienz K. (1996) Reversible dissociation of the poliovirus replication complex. Functions and interactions of its components in viral RNA synthesis. J. Virol. 70, 8675–8683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Belov G. A., Feng Q., Nikovics K., Jackson C. L., Ehrenfeld E. (2008) A critical role of a cellular membrane traffic protein in poliovirus RNA replication. PLoS Pathog. 4, e1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hsu N.-Y., Ilnytska O., Belov G., Santiana M., Chen Y.-H., Takvorian P. M., Pau C., van der Schaar H., Kaushik-Basu N., Balla T., Cameron C. E., Ehrenfeld E., van Kuppeveld F. J., Altan-Bonnet N. (2010) Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 141, 799–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dales S. (1965) Replication of animal viruses as studied by electron microscopy. Am. J. Med. 38, 699–715 [DOI] [PubMed] [Google Scholar]
- 17. Suhy D. A., Giddings T. H., Jr., Kirkegaard K. (2000) Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins. An autophagy-like origin for virus-induced vesicles. J. Virol. 74, 8953–8965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Taylor M. P., Kirkegaard K. (2007) Modification of cellular autophagy protein LC3 by poliovirus. J. Virol. 81, 12543–12553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Haas A. (2007) The phagosome. Compartment with a license to kill. Traffic 8, 311–330 [DOI] [PubMed] [Google Scholar]
- 20. Wong J., Zhang J., Si X., Gao G., Mao I., McManus B. M., Luo H. (2008) Autophagosome supports coxsackievirus B3 replication in host cells. J. Virol. 82, 9143–9153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. O'Donnell V., Pacheco J. M., LaRocco M., Burrage T., Jackson W., Rodriguez L. L., Borca M. V., Baxt B. (2011) Foot-and-mouth disease virus utilizes an autophagic pathway during viral replication. Virology 410, 142–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Heaton N. S., Randall G. (2010) Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe 8, 422–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mateo R., Nagamine C. M., Spagnolo J., Méndez E., Rahe M., Gale M., Jr., Yuan J., Kirkegaard K. (2013) Inhibition of cellular autophagy deranges dengue virion maturation. J. Virol. 87, 1312–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ferraris P., Blanchard E., Roingeard P. (2010) Ultrastructural and biochemical analyses of hepatitis C virus-associated host cell membranes. J. Gen. Virol. 91, 2230–2237 [DOI] [PubMed] [Google Scholar]
- 25. Fujita K., Krishnakumar S. S., Franco D., Paul A. V., London E., Wimmer E. (2007) Membrane topography of the hydrophobic anchor sequence of poliovirus 3A and 3AB proteins and the functional effect of 3A/3AB membrane association upon RNA replication. Biochemistry 46, 5185–5199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Teterina N. L., Pinto Y., Weaver J. D., Jensen K. S., Ehrenfeld E. (2011) Analysis of poliovirus protein 3A interactions with viral and cellular proteins in infected cells. J. Virol. 85, 4284–4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Belov G. A., Nair V., Hansen B. T., Hoyt F. H., Fischer E. R., Ehrenfeld E. (2012) Complex dynamic development of poliovirus membranous replication complexes. J. Virol. 86, 302–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hayashi-Nishino M., Fujita N., Noda T., Yamaguchi A., Yoshimori T., Yamamoto A. (2009) A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11, 1433–1437 [DOI] [PubMed] [Google Scholar]
- 29. Doedens J. R., Giddings T. H., Jr., Kirkegaard K. (1997) Inhibition of endoplasmic reticulum-to-Golgi traffic by poliovirus protein 3A. Genetic and ultrastructural analysis. J. Virol. 71, 9054–9064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Towner J. S., Ho T. V., Semler B. L. (1996) Determinants of membrane association for poliovirus protein 3AB. J. Biol. Chem. 271, 26810–26818 [DOI] [PubMed] [Google Scholar]
- 31. Plotch S. J., Palant O. (1995) Poliovirus protein 3AB forms a complex with and stimulates the activity of the viral RNA polymerase, 3Dpol. J. Virol. 69, 7169–7179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Harris K. S., Xiang W., Alexander L., Lane W. S., Paul A. V., Wimmer E. (1994) Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J. Biol. Chem. 269, 27004–27014 [PubMed] [Google Scholar]
- 33. Paul A. V., Cao X., Harris K. S., Lama J., Wimmer E. (1994) Studies with poliovirus polymerase 3Dpol. Stimulation of poly(U) synthesis in vitro by purified poliovirus protein 3AB. J. Biol. Chem. 269, 29173–29181 [PubMed] [Google Scholar]
- 34. Xiang W., Cuconati A., Paul A. V., Cao X., Wimmer E. (1995) Molecular dissection of the multifunctional poliovirus RNA-binding protein 3AB. RNA 1, 892–904 [PMC free article] [PubMed] [Google Scholar]
- 35. Lama J., Paul A. V., Harris K. S., Wimmer E. (1994) Properties of purified recombinant poliovirus protein 3aB as substrate for viral proteinases and as co-factor for RNA polymerase 3Dpol. J. Biol. Chem. 269, 66–70 [PubMed] [Google Scholar]
- 36. Molla A., Harris K. S., Paul A. V., Shin S. H., Mugavero J., Wimmer E. (1994) Stimulation of poliovirus proteinase 3Cpro-related proteolysis by the genome-linked protein VPg and its precursor 3AB. J. Biol. Chem. 269, 27015–27020 [PubMed] [Google Scholar]
- 37. Paul A. V., Mugavero J., Molla A., Wimmer E. (1998) Internal ribosomal entry site scanning of the poliovirus polyprotein: implications for proteolytic processing. Virology 250, 241–253 [DOI] [PubMed] [Google Scholar]
- 38. Strauss D. M., Glustrom L. W., Wuttke D. S. (2003) Towards an understanding of the poliovirus replication complex. The solution structure of the soluble domain of the poliovirus 3A protein. J. Mol. Biol. 330, 225–234 [DOI] [PubMed] [Google Scholar]
- 39. Dales S. (1973) Early events in cell-animal virus interactions. Bacteriol. Rev. 37, 103–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. VerPlank L., Bouamr F., LaGrassa T. J., Agresta B., Kikonyogo A., Leis J., Carter C. A. (2001) Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. U.S.A. 98, 7724–7729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ehrlich L. S., Medina G. N., Carter C. A. (2011) ESCRT machinery potentiates HIV-1 utilization of the PI(4,5)P(2)-PLC-IP3R-Ca2+ signaling cascade. J. Mol. Biol. 413, 347–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Peker B., Wuu J. J., Swartz J. R. (2004) Affinity purification of lipid vesicles. Biotechnol. Prog. 20, 262–268 [DOI] [PubMed] [Google Scholar]
- 43. Weill C. O., Biri S., Adib A., Erbacher P. (2008) A practical approach for intracellular protein delivery. Cytotechnology 56, 41–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Seddon J. M. (1990) Structure of the inverted hexagonal (HII) phase, and non-lamellar phase transitions of lipids. Biochim. Biophys. Acta 1031, 1–69 [DOI] [PubMed] [Google Scholar]
- 45. Jan Wilschut D. H., Hoekstra D. (1991) Membrane Fusion. pp. 89–126, CRC Press, New York [Google Scholar]
- 46. Sou Y. S., Tanida I., Komatsu M., Ueno T., Kominami E. (2006) Phosphatidylserine in addition to phosphatidylethanolamine is an in vitro target of the mammalian Atg8 modifiers, LC3, GABARAP, and GATE-16. J. Biol. Chem. 281, 3017–3024 [DOI] [PubMed] [Google Scholar]
- 47. McLaughlin S., Murray D. (2005) Plasma membrane phosphoinositide organization by protein electrostatics. Nature 438, 605–611 [DOI] [PubMed] [Google Scholar]
- 48. van Meer G., Voelker D. R., Feigenson G. W. (2008) Membrane lipids. Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pasieka T. J., Woolson R. F., Grose C. (2003) Viral induced fusion and syncytium formation. Measurement by the Kolmogorov-Smirnov statistical test. J. Virol. Methods 111, 157–161 [DOI] [PubMed] [Google Scholar]
- 50. Spagnolo J. F., Rossignol E., Bullitt E., Kirkegaard K. (2010) Enzymatic and nonenzymatic functions of viral RNA-dependent RNA polymerases within oligomeric arrays. RNA 16, 382–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Boura E., Rżycki B., Chung H. S., Herrick D. Z., Canagarajah B., Cafiso D. S., Eaton W. A., Hummer G., Hurley J. H. (2012) Solution structure of the ESCRT-I and -II supercomplex. Implications for membrane budding and scission. Structure 20, 874–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Khan S. H., Kumar R. (2012) Role of an intrinsically disordered conformation in AMPK-mediated phosphorylation of ULK1 and regulation of autophagy. Mol. Biosyst. 8, 91–96 [DOI] [PubMed] [Google Scholar]
- 53. McMahon H. T., Gallop J. L. (2005) Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 438, 590–596 [DOI] [PubMed] [Google Scholar]
- 54. Zimmerberg J., Kozlov M. M. (2006) How proteins produce cellular membrane curvature. Nat. Rev. Mol. Cell Biol. 7, 9–19 [DOI] [PubMed] [Google Scholar]
- 55. Prinz W. A., Hinshaw J. E. (2009) Membrane-bending proteins. Crit. Rev. Biochem. Mol. Biol. 44, 278–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dafforn T. R., Smith C. J. (2004) Natively unfolded domains in endocytosis: hooks, lines and linkers. EMBO Rep. 5, 1046–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Thaa B., Levental I., Herrmann A., Veit M. (2011) Intrinsic membrane association of the cytoplasmic tail of influenza virus M2 protein and lateral membrane sorting regulated by cholesterol binding and palmitoylation. Biochem. J. 437, 389–397 [DOI] [PubMed] [Google Scholar]
- 58. Dodd D. A., Giddings T. H., Jr., Kirkegaard K. (2001) Poliovirus 3A protein limits interleukin-6 (IL-6), IL-8, and β interferon secretion during viral infection. J. Virol. 75, 8158–8165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Longatti A., Tooze S. A. (2009) Vesicular trafficking and autophagosome formation. Cell Death Differ. 16, 956–965 [DOI] [PubMed] [Google Scholar]
- 60. Ragusa M. J., Stanley R. E., Hurley J. H. (2012) Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell 151, 1501–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nakatogawa H., Ichimura Y., Ohsumi Y. (2007) Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130, 165–178 [DOI] [PubMed] [Google Scholar]
- 62. Weidberg H., Shpilka T., Shvets E., Abada A., Shimron F., Elazar Z. (2011) LC3 and GATE-16 N termini mediate membrane fusion processes required for autophagosome biogenesis. Dev. Cell 20, 444–454 [DOI] [PubMed] [Google Scholar]
- 63. Takahashi Y., Meyerkord C. L., Wang H. G. (2009) Bif-1/endophilin B1. A candidate for crescent driving force in autophagy. Cell Death Differ. 16, 947–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Xiang W., Cuconati A., Hope D., Kirkegaard K., Wimmer E. (1998) Complete protein linkage map of poliovirus P3 proteins: interaction of polymerase 3Dpol with VPg and with genetic variants of 3AB. J. Virol. 72, 6732–6741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Teterina N. L., Levenson E., Rinaudo M. S., Egger D., Bienz K., Gorbalenya A. E., Ehrenfeld E. (2006) Evidence for functional protein interactions required for poliovirus RNA replication. J. Virol. 80, 5327–5337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wollert T., Wunder C., Lippincott-Schwartz J., Hurley J. H. (2009) Membrane scission by the ESCRT-III complex. Nature 458, 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]