Background: AB5 toxins consist of a pentameric B-subunit and a catalytic A-subunit.
Results: Crystallographic data, dissociation, and intracellular trafficking of SubAB toxin are reported.
Conclusion: SubAB architecture is similar to other AB5 toxins, whereas the B-pentamer plays an important role in assembly and intracellular trafficking.
Significance: The conserved hydrophobic ring in the B-pentamer supports the view that A- and B-subunits have evolved independently.
Keywords: Carbohydrate-binding Protein, Infectious Diseases, Intracellular Trafficking, Structural Biology, Toxins, AB5 Toxins, Cellular Trafficking, Disassembly/Assembly
Abstract
Pathogenic strains of Escherichia coli produce a number of toxins that belong to the AB5 toxin family, which comprise a catalytic A-subunit that induces cellular dysfunction and a B-pentamer that recognizes host glycans. Although the molecular actions of many of the individual subunits of AB5 toxins are well understood, how they self-associate and the effect of this association on cytotoxicity are poorly understood. Here we have solved the structure of the holo-SubAB toxin that, in contrast to other AB5 toxins whose molecular targets are located in the cytosol, cleaves the endoplasmic reticulum chaperone BiP. SubA interacts with SubB in a similar manner to other AB5 toxins via the A2 helix and a conserved disulfide bond that joins the A1 domain with the A2 helix. The structure revealed that the active site of SubA is not occluded by the B-pentamer, and the B-pentamer does not enhance or inhibit the activity of SubA. Structure-based sequence comparisons with other AB5 toxin family members, combined with extensive mutagenesis studies on SubB, show how the hydrophobic patch on top of the B-pentamer plays a dominant role in binding the A-subunit. The structure of SubAB and the accompanying functional characterization of various mutants of SubAB provide a framework for understanding the important role of the B-pentamer in the assembly and the intracellular trafficking of this AB5 toxin.
Introduction
AB5 toxins are an important family of toxins, comprising three established subfamilies: (i) cholera toxin (Ctx)6 and the Escherichia coli heat-labile enterotoxins (LT-I and LT-II); (ii) pertussis toxin (Ptx); and (iii) Shiga toxin (Stx). These AB5 toxins are important virulence factors of the bacteria that produce them: Vibrio cholerae and enterotoxigenic E. coli (Ctx and LT-I and II, respectively); Bordetella pertussis (Ptx); and Shiga toxigenic E. coli and Shigella dysenteriae (Stx) (1, 2). These human pathogens cause considerable global morbidity and mortality, accounting for 1–2 million deaths each year, particularly among children in developing countries. AB5 toxins exert their effects in a two-step process: (i) binding of the pentameric B-subunit to specific glycan receptors on the target cell surface and (ii) internalization of the AB5 toxin followed by A-subunit-mediated inhibition or corruption of essential host functions. The above AB5 toxins ultimately act on cytosolic targets, and so after internalization, they must be transported to the appropriate site and translocated across the respective organelle membrane. Although they may recognize different glycan receptors, they exploit similar trafficking pathways within the cell.
Subtilase cytotoxin (SubAB) is a recently discovered fourth AB5 toxin subfamily that, through a series of biochemical, cell-based, and structural studies, was shown to have novel activity. Namely, the catalytic A-subunit of SubAB is a protease that belongs to the peptidase_S8 (subtilisin) family (3), which was shown to be a unique bacterial protease in that it exhibited a very strict substrate preference. Namely, the only known physiological target of SubA is BiP (4), an ER-resident chaperone that is critical for ER function and homeostasis. BiP mediates folding of nascent proteins destined for secretion; it also maintains the permeability barrier of the ER membrane by sealing the lumenal end of the Sec61 translocon pore, as well as targeting terminally misfolded proteins to the Sec61 apparatus for retrotranslocation into the cytosol and degradation by the proteasome. BiP, as the ER stress-signaling master regulator (5, 6), plays a crucial role in the unfolded protein response and exhibits antiapoptotic properties through interference with caspase activation (5). Thus, SubA-mediated blockade of BiP function has inevitably fatal consequences for the cell, including ectopic accumulation of misfolded proteins in the ER lumen, a massive irresolvable ER stress response, and apoptosis.
The receptor specificity of SubB is also unique for the AB5 family as it exhibits a strong preference for glycans terminating in the sialic acid N-glycolylneuraminic acid (7). This glycan is not synthesized by humans, but is incorporated onto the cell surface as a result of dietary intake (8). The most common human sialic acid is N-acetylneuraminic acid (Neu5Ac), which is the glycan receptor for Ptx and differs from N-glycolylneuraminic acid by a single hydroxyl group.
Although the structures of the isolated A- and pentameric B-subunits have been determined (4, 7), how the SubAB holotoxin complex associates is unknown, and yet the interaction between the two subunits plays an important role in pathogenesis. All AB5 toxins, including SubAB, undergo retrograde transport via the Golgi to the ER. The A-subunits of members of the Ctx, Ptx, and Stx subfamilies have cytosolic targets and so must separate from their respective B-pentamers and be retrotranslocated into the cytosol before they can exert their action. However, as the molecular target of SubA is located in the ER lumen, it suggests that SubA may not need to dissociate from the B-subunit. To obtain structural insight into the assembly/disassembly and overall stability of the SubAB holotoxin, the crystal structure of SubAB was determined. This structure, in association with extensive mutational analysis at the SubA/SubB interface, has provided insight into the SubAB assembly and its role in pathogenesis.
EXPERIMENTAL PROCEDURES
Cloning, Site-directed Mutagenesis, Expression, and Purification of SubAB
The cloning of the wild type SubAB is described in detail elsewhere (3). In brief, the SubAB coding region was amplified by PCR and ligated into the pET23(+) expression vector. The SubAB mutants were generated by site-directed mutagenesis using the Stratagene QuikChange® mutagenesis kit. The wild type and mutant SubAB plasmids were transformed into E. coli BL21 (DE3) cells and plated on Luria-Bertani agar plates containing 50 μg/ml ampicillin. Expression of the SubAB wild type and variants with C-terminal B-subunit His6 tags was carried out in autoinduction medium (9). The cultures were grown at 37 °C for 20 h, and the cells were harvested by centrifugation for 15 min (4000 × g) at 4 °C. The wild type and mutant SubAB holotoxins were initially purified by nickel affinity chromatography, as detailed previously (3), but with the following modifications. Following loading of the clarified cell lysate, the nickel-nitrilotriacetic acid resin was washed with 50 ml of wash buffer comprising 50 mm MES, pH 6.0, containing 300 mm NaCl and 10% v/v glycerol to remove any unbound protein. The His-tagged holotoxin was eluted in 20 ml of wash buffer containing 500 mm imidazole and then loaded onto an S200 16/60 gel filtration column (GE Healthcare), which had been pre-equilibrated with 25 mm Tris-HCl, pH 7.0, containing 150 mm NaCl and 2 mm EDTA. Fractions were analyzed by SDS-PAGE, and those containing SubAB were pooled and concentrated to 7 mg/ml. The identity and purity of the final sample were assessed by MALDI-TOF mass spectrometry and SDS-PAGE before crystallization proceeded.
Crystallization and X-ray Data Collection
Rod-shaped crystals of SubAB were grown over the duration of 3 days at 20 °C by the hanging drop vapor diffusion method. Drops contained 1 μl of protein (7 mg/ml) and 1 μl of reservoir solution of 11% w/v PEG 8000 containing 0.5 m lithium sulfate and 0.1 m citrate, pH 3.1. Crystals were soaked for 10 s in cryoprotectant containing 20% v/v glycerol in mother liquor before being flash-frozen in liquid nitrogen and transported to the Advanced Photon Source (APS), Chicago, IL, for x-ray data collection. Data were collected using an Area Detector Systems Corp. Quantum 210 CCD detector on the Industrial Macromolecular Crystallography Association Collaborative Access Team (IMCA-CAT) beamline at the APS and processed using MOSFLM and SCALA, with 5% of diffraction data flagged for the calculation of Rfree. A 2.6 Å dataset was collected from a crystal belonging to space group P21 containing two holotoxin complexes in the asymmetric unit.
Structure Determination, Model Building, and Refinement
Molecular replacement was carried out using PHASER (10), using the SubA and SubB structures (Protein Data Bank (PDB) codes: 2IY9 and 3DWA, respectively) as separate search models. First, two SubA molecules were located, and these were used as known solutions for a further molecular replacement search using a SubB pentamer as the next search model. Two SubB pentamers were next successfully located, thus completing the two-holotoxin molecules within the asymmetric unit. Analysis of the solution in COOT (11) confirmed that the identified holotoxins packed well, and rigid body refinement of the two-holotoxin molecules proceeded in REFMAC5 (12). Density modification (SigmaA, DM) was interspersed with Prime and Switch Phasing in RESOLVE and restrained refinement in REFMAC5, maintaining strict non-crystallographic symmetry restraints until the later stages of refinement, when these were released. Buster 2.10 (13) was then used for the latter stages of refinement. Water molecules were finally added to the model in COOT (11). Residues with no visible density were removed from the model. The final model consists of two SubAB molecules, each of ∼110 kDa, with final R and Rfree values of 22 and 25%, respectively.
Sequence Analysis
Sequence searches were carried out at the National Center for Biotechnology Information (NCBI) web site (www.ncbi.nlm.nih.gov) using the Basic Local Alignment Search Tool (BLAST) (14). The sequences were subsequently compared using Clustal Omega (15), and a structure based-sequence alignment was generated using the Indonesia program (16).
Cell Culture, Cytotoxicity Assays, and Intracellular Trafficking
All tissue culture media and reagents were obtained from Life Technologies. Vero (African green monkey kidney) cells were grown at 37 °C in DMEM supplemented with 10% (v/v) heat inactivated fetal calf serum (FCS), 50 IU/ml penicillin, and 50 μg/ml streptomycin, unless otherwise indicated. For cytotoxicity assays, cells were seeded into 96-well flat bottom trays and incubated overnight at 37 °C until confluent. Confluent monolayers were washed twice with PBS, treated with 50 μl of filter-sterilized toxin extracts, which had been serially diluted in tissue culture medium (without FCS), and incubated at 37 °C for 30 min. After incubation, 150 μl of medium supplemented with 2% (v/v) FCS was added per well. Cytotoxicity was assessed microscopically after 3 days of incubation at 37 °C. The toxin titer was defined as the reciprocal of the maximum dilution producing a cytopathic effect on at least 50% of the cells in each well (CD50/ml). For intracellular trafficking studies, Vero cells were seeded onto eight-chamber glass microscope slides (Lab-Tek, Thermo Scientific) at a density of 2 × 104 cells/chamber and incubated overnight. Cell monolayers were then treated with 3 μg/ml purified wild type SubAB or various mutant derivatives for 30 min, washed with PBS, and then fixed with 4% (w/v) paraformaldehyde in PBS for 10 min at room temperature. Slides were then washed with PBS, permeabilized with 0.1% (v/v) Triton X-100 in PBS for 5 min, washed with PBS, and then blocked with PBS, 10% (v/v) FCS, 2% (v/v) normal goat serum for 1 h at room temperature. Slides were then incubated with primary antibody (polyclonal rabbit anti-SubA diluted 1:500 or 2 μg/ml monoclonal mouse anti-His6 IgG1 (Roche Applied Science)) overnight at 4 °C, washed with PBS, incubated with secondary antibody (20 μg/ml Alexa Fluor 488 goat anti-mouse IgG and 4 μg/ml Alexa Fluor 594 goat anti-rabbit IgG (Molecular Probes)) for 45 min at room temperature, washed thoroughly with PBS, and rinsed twice in deionized water. The chamber slide gasket was then removed, and slides were air-dried; slides were then mounted in VECTASHIELD mounting medium containing DAPI (Vector Laboratories), and the coverslips were sealed with nail varnish. Fluorescence images were obtained using a Leica SP5 spectral scanning confocal microscope (Leica Microsystems) with a 63× (NA = 1.40) oil objective lens. Confocal settings used for image capture were held constant in comparison experiments. Images were processed, and colocalization was quantified using the LAS AF software (Leica Microsystems CMS GmbH). Background fluorescence was subtracted using regions of the cell devoid of staining. For ER colocalization comparisons, regions positive for SubA were selected and used as a constant. Data were processed and analyzed statistically using unpaired Student's t tests in GraphPad Prism.
In Vitro Stability of SubA Association with SubB
50 μg of purified SubAB and SubAB mutant variants was bound to nickel-Sepharose resin in 50 mm NaH2PO4 (pH 6.0), 300 mm NaCl, 10% (v/v) glycerol (buffer A). Unbound protein was removed by washing with 2 column volumes of buffer A. Bound protein was incubated for 30 min in buffer A supplemented with 30 mm 3-(decyldimethylammonio)propanesulfonate inner salt (Sigma-Aldrich). Dissociated SubA was removed by washing with 2 column volumes of buffer A. SubA and SubB proteins bound to the nickel-Sepharose were quantified by SDS-PAGE and densitometry using the Typhoon Trio (Amersham Biosciences) and IQTLTM image analysis software (GE life sciences Australia). The dissociation was calculated by dividing the quantity of SubA after incubation with detergent by the quantity of SubA before. Dissociation of each SubB mutant was normalized against the wild type SubB dissociation to calculate the relative dissociation.
RESULTS
Structural Organization of SubAB
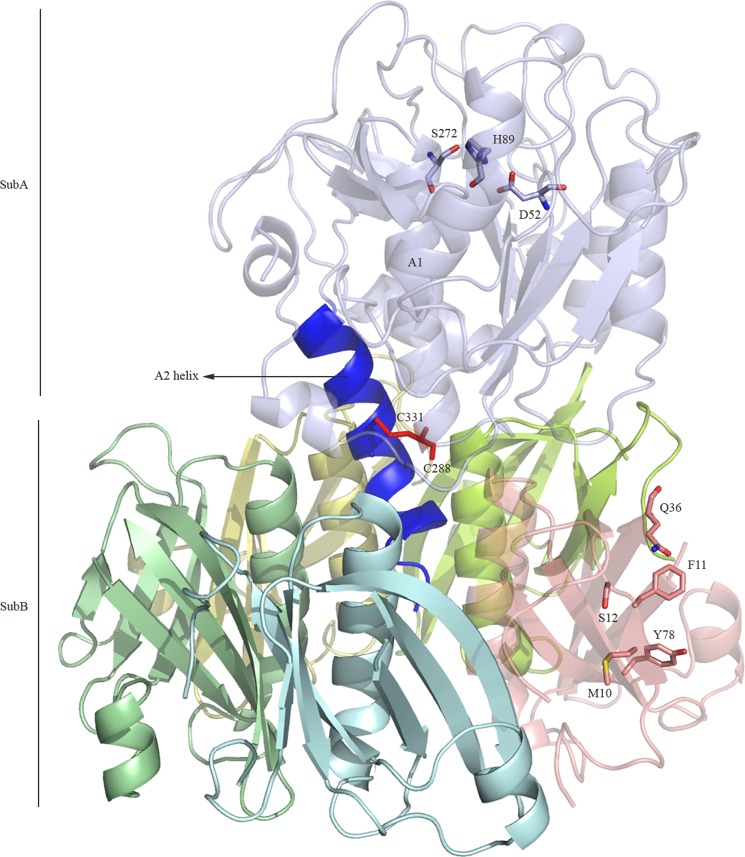
The three-dimensional structure of the SubAB holotoxin was determined to a resolution of 2.6 Å (Table 1). The crystal contains two SubAB molecules in the asymmetric unit; the two noncrystallographic symmetry-related molecules are very similar (average r.m.s.d. of 0.38 Å for 812 residues), and unless explicitly stated, only one complex will be discussed (protomers B, C, D, E, and F forming the pentameric B-subunit). The overall architecture of the SubAB holotoxin (Fig. 1) comprises an A-subunit (SubA) responsible for the catalytic activity of the toxin (4) and a B-subunit that mediates binding to the cell surface receptors (2). SubA is composed of two domains, A1 and A2, which are linked together via a disulfide bond formed by the residues Cys-288 and Cys-331 (Fig. 1). The catalytic triad (Asp-52-His-89-Ser-272) responsible for the proteolytic activity of SubA is located at the bottom of an unusual deep cleft (4) and is not occluded by the B-subunit (Fig. 1). The A2 domain consists of an α-helix of about 25 amino acid residues that penetrates into the central “pore” of the B-subunit (SubB) (Fig. 1). SubB adopts a homopentameric ring-shaped assembly as shown previously (7). This structural architecture (Fig. 1) is common to all AB5 toxin x-ray structures determined to date (2).
TABLE 1.
Data collection, processing, and refinement statistics
| Parameters | SubAB |
|---|---|
| Data collection and processing | |
| Space group | P21 |
| a, b, c (Å) | 59.21, 81.71, 227.79 |
| Angles (°) | 90, 100.24, 90 |
| No. of measured reflections | 252,165 |
| No. of unique reflections | 66,139 |
| Resolution rangea (Å) | 66.08–2.6 (2.74–2.6) |
| Completenessa (%) | 99.9 (99.6) |
| Mean I/σ(I)a | 11.3 (2.3) |
| Wilson B-factors (Å2) | 49 |
| Rpima(%) | 6.5 (34.2) |
| Rmergea,b (%) | 11.2 (56.5) |
| Refinement statistics | |
| Resolution rangec | 38.07–2.6 (2.67–2.6) |
| R/Rfree (%)c,d | 21.9/25 (23.2/25.7) |
| Protein atoms (residues) | 12,528 |
| Water molecules | 85 |
| B-factors (Å2) | |
| Average main chain | 59.7 |
| Average side chain | 61.8 |
| Water | 31.2 |
| Glycerol | 42.3 |
| Sulfate | 69 |
| r.m.s.d. values from ideal geometry | |
| Bonds (Å) | 0.010 |
| Angles (°) | 1.09 |
| Ramachandran plot (%) (32) | |
| Residues in favored regions | 97.42 |
| Residues in allowed regions | 2.46 |
| Outliers | 0.12 |
| MolProbity score | 1.88 (98th percentile) |
a Values shown in parentheses correspond to the high resolution shell.
b Rmerge = Σhkli ΣI | I(hkl)ii −〈 I(hkl)〉 |/ΣhklΣi I(hkl)i.
c Values in parentheses are for the outermost resolution shell.
d R and Rfree = Σ‖Fobs| − |Fcalc‖/Σ|Fobs|, where Rfree was calculated over 5% of amplitudes that were chosen at random and not used in refinement.
FIGURE 1.
Overall architecture of SubAB. The A1 component of the A-subunit (SubA) is shown in light blue, whereas the α-helix A2 is shown in blue. The disulfide tethering the A1 and A2 is shown in red. Each protomer of the pentameric B-subunit (SubB) is colored in cyan, yellow, lemon, salmon, and pale green. The figure was generated using PyMOL (29).
SubAB versus SubA and SubB
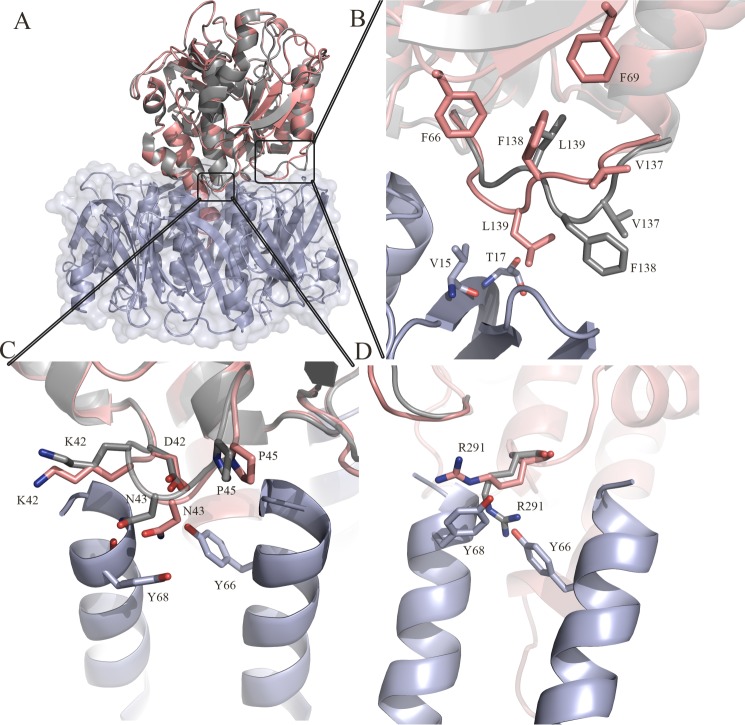
The structures of the A-subunit (SubA/PDB code: 2IY9) and B-subunit (SubB/PDB code: 3DWA), previously determined in isolation (4, 7) and in a complex form (SubAB), are very similar (Fig. 2A). Despite the close structural similarity, the loop comprising the residues Ser-135 to Pro-141 in the A-subunit undergoes structural rearrangement (r.m.s.d. >2.5 Å) upon the formation of the holotoxin (Fig. 2B). Namely, Leu-139 reorients toward the top of the B-subunit to interact with residues Val-15 and Thr-17 from the protomer C (Table 2). To compensate for the movement of this loop, Phe-138 and Val-137 from SubA flip over to form a hydrophobic pocket with Phe-69 and Phe-66. The loop Lys-42–Pro-45 from SubA is also rearranged (r.m.s.d. 1.2 Å) to optimize the hydrogen bond network of Asn-43 and Asp-41 with the SubB residues Tyr-66 (protomer C) and Tyr-68 (protomer F) (Table 2; Fig. 2C). Additionally, the side chain of Arg-291 in SubA flips by 180° to accommodate the B-subunit and in particular the two residues Tyr-68 (protomer C) and Tyr-66 (protomer E) (Fig. 2D). Superposition of SubB in its nonliganded form and in complex with SubA did not reveal any significant structural changes in SubB. In particular, the key residues (Met-10, Phe-11, Ser-12, Gln-36, and Tyr-78) involved in the binding of the glycan N-glycolylneuraminic acid and located halfway down the side of the B-subunit (Fig. 1) are not impacted by the presence of SubA. In both SubA (4) and SubAB, the disulfide bond bridging the A1 and A2 domains is conserved. Thus, limited conformational change underpins SubAB assembly.
FIGURE 2.
A, overall superposition of the isolated crystal structure of SubA and the SubAB holotoxin. The crystal structure of SubA (PDB code: 2IY9) is colored in gray, and the A-subunit in the SubAB structure is colored in salmon. The B-subunit of SubAB is shown in light blue. B, structural rearrangement of the loop (Ser-135–Pro-141). C, structural rearrangement of the loop (Asp-41–Pro-45). D, structural rearrangement of the residue Arg-291 upon formation of the holotoxin. The figures were generated using PyMOL (29).
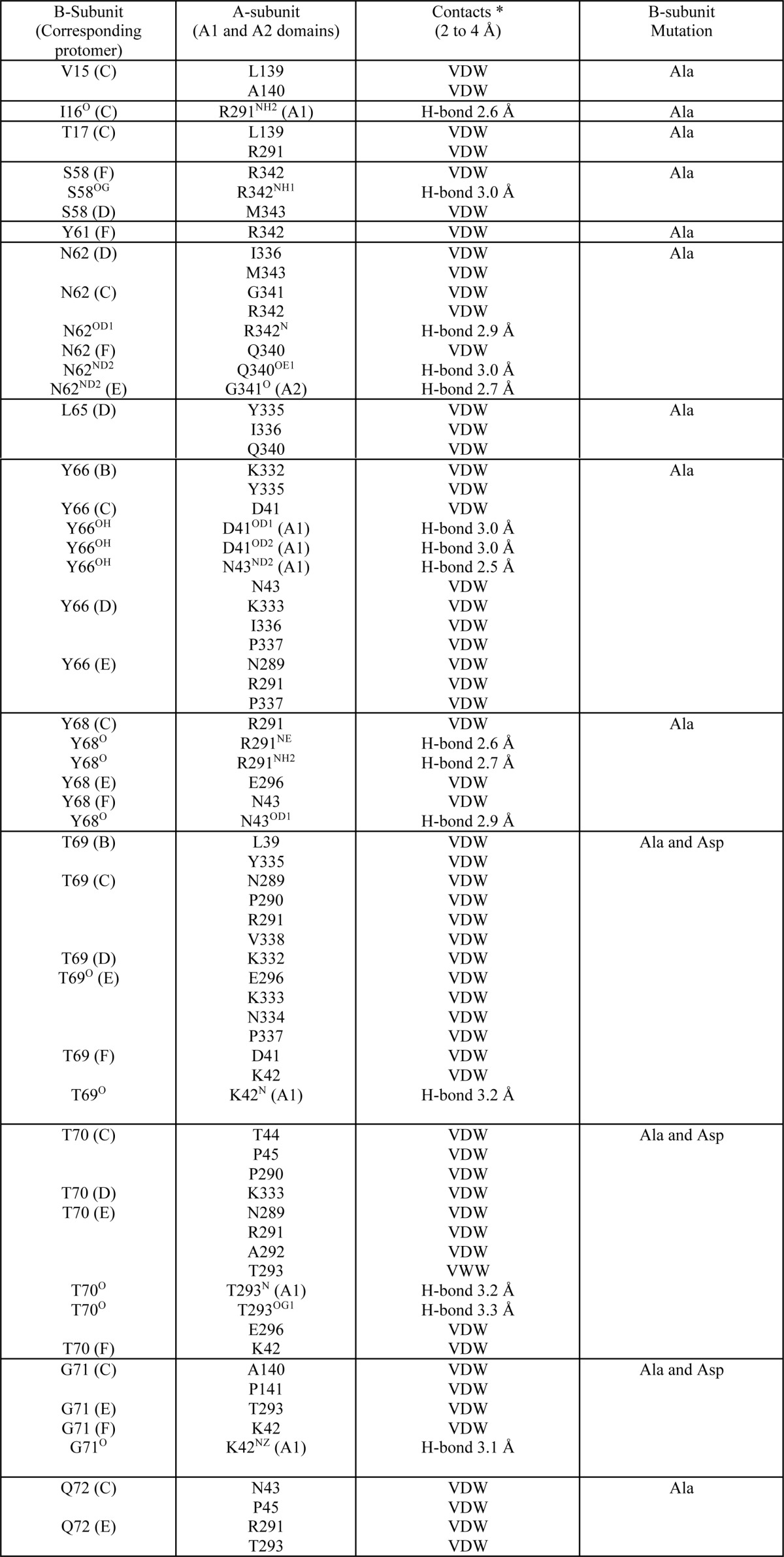
TABLE 2.
Table of contacts for interface of A- and B-subunits
*, Contacts were determined with the CCP4 program CONTACT (33) and at a cutoff of 4.0 ′. van der Waals interactions (VDW) are defined as non-H-bond contact distances of 4 ′ or less; H-bond interactions are defined as contact distances of 3.5 ′ or less.
SubA/SubB Interface
The catalytic A-subunit of SubAB interacts closely with the pentameric B-subunit (SubB) to form the holotoxin. A total of 13 SubB residues make direct contacts with SubA (Table 2). The total buried surface area (BSA) at the SubA/SubB interface is ∼3800 Å2. Although the five SubB protomers contact SubA, the nature and occurrence of those interactions vary from one protomer to the other, reflecting the asymmetry of the SubA/SubB interactions. Namely, the protomer C interacts most extensively with SubA and contributes 14.5% to the overall BSA at the SubA/SubB interface (Table 3). In particular, the hydrophobic residues Val-15, Ile-16, and Gly-71 and the polar residues Thr-69, Thr-70, Asn-62, Gln-72, and Tyr-66 have more than 50% of their individual surface buried when complexed with SubA. SubA contributes 15.3% to the total BSA at this interface, and the contributors are predominantly the hydrophobic residues (Pro-45, Leu-139, Ala-140, Pro-141, Pro-290, and Val-338). The SubB Tyr-68 (protomer C) packs well against the side chain Arg-291 (SubA), whereas Gly-71 forms hydrophobic interactions with Pro-141. The contacts also include a number of hydrogen bonds as described in Fig. 3 and Table 2. Protomers E and F contribute to a lesser extent to the SubA/SubB interface with 11.2 and 9.6% BSA, respectively. However, similar to protomer C, the residues Asn-62, Thr-69, Thr-70, and Gly-71 (protomers E and F) contribute predominantly to the SubA/SubB interface. The corresponding SubA interface (21% BSA) comprises the hydrophobic residues Pro-337 and Gly-341 from the A2 helix and Ala-292 (A1 domain) that contribute 3% to the BSA of the SubA/SubB interface, whereas the polar residues Arg-339, Gln-340, and Arg-342 (A2 helix) contribute 6.8% BSA. In protomer E, there are only three hydrogen bonds with SubA mediated by residues Thr-70, Thr-293, and Asn-62 (Table 2; Fig. 3), although in protomer F, there are there also three main chain hydrogen bonds mediated by Tyr-68, Thr-69, and Gly-71. The last two protomers B and D contribute 6.2 and 7.4% to the overall BSA of the SubA/SubB interface, respectively.
TABLE 3.
Buried surface area (Å2) of the A- and B-subunits at the A- and B-subunit interface
| Family 1 SubAB | Family 2 |
Family 3 Ptx (PDB code: 1PRT) | Family 4 |
||||
|---|---|---|---|---|---|---|---|
| LT-I (PDB code: 1LTI) | LT-IIb (PDB code: 1TII) | Ctx (PDB code: 1S5E) | Stx (PDB code: 1R4P) | Stx2 (PDB code: 1DM0) | |||
| Protomer Ba | 237 (247)b | 395 (389) | 235 (237) | 261 (240) | 260 (283) | 217 (224) | 375 (358) |
| Protomer C | 548 (586) | 619 (656) | 376 (367) | 445 (437) | 371 (385) | 331 (338) | 496 (486) |
| Protomer D | 284 (316) | 318 (350) | 393 (448) | 481 (506) | 422 (461) | 206 (226) | 566 (571) |
| Protomer E | 427 (399) | 295 (297) | 320 (340) | 473 (497) | 655 (618) | 287 (295) | 537 (590) |
| Protomer F | 365 (402) | 624 (609) | 183 (196) | 302 (327) | 664 (627) | 277 (278) | 475 (457) |
| Total | 1861 [48.5%]c (1950) | 2251 [49.4%] (2301) | 1507 [48.7%] (1588) | 1962 [49.4%] (2007) | 2372 [50%] (2374) | 1318 [49.2%] (1361) | 2449 [49.8%] (2462) |
a The protomer chain name refers only to SubB.
b Values in parentheses are for the corresponding A-subunit buried surface area at the holotoxin interface. The buried surface area was calculated using the Protein Interfaces, Surfaces and Assemblies Service (PISA) at European Bioinformatics Institute (16).
c Relative BSA contribution (%) of the B-subunits to the SubA/SubB interface.
FIGURE 3.
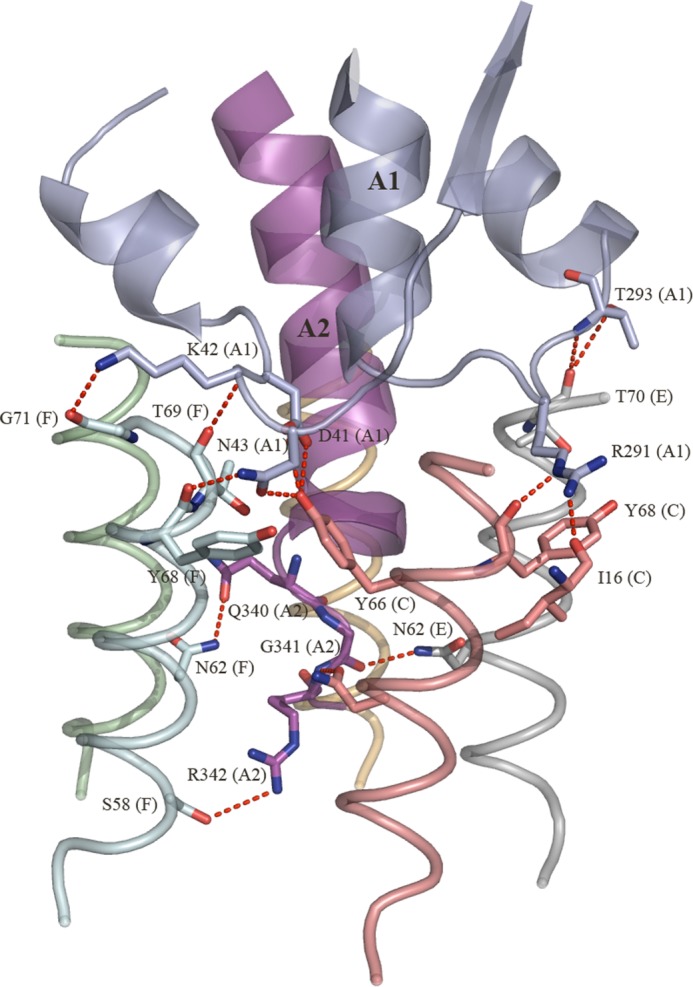
Hydrogen bond network between the SubA and SubB residues. The hydrogen bonds are shown as red dashed lines.
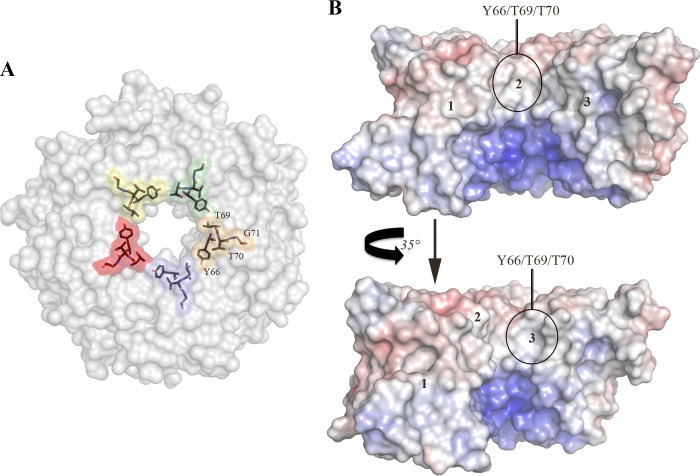
Accordingly, in comparison with protomers E and C, respectively, the contribution of these promoters to the SubAB interface is more moderate. This is also reflected by a more limited number of contacts (no H-bonds) between the two SubB protomers and SubA. Although it is now evident that the five SubB protomers contribute differently (BSA, H-bonds, and van der Waals interactions) to the SubA/SubB interface, the residues Thr-69 and Thr-70 are the only two residues in which 50% or more of their surface is buried upon the formation of the holotoxin in all five protomers. Interestingly, Thr-69 is also the only residue that contacts SubA (<4 Å) in the five SubB protomers (Table 2). The residues Thr-69, Thr-70, and Gly-71 are positioned at the top of the individual α-helix (OB fold) of the SubB protomers (Fig. 4A). The calculated molecular electrostatic surface potential shows that Tyr-66 and Thr-69/Thr-70 have their aromatic and methyl side chains (neutral surface), respectively, exposed to the surface of the central pore (Fig. 4B). Positively charged surface residues dominate the lower portion of the pore. The five-fold symmetry of this exposed surface forms a hydrophobic ring that could play a role in the overall stability and assembly/disassembly of the holotoxin.
FIGURE 4.
A, The molecular surfaces of residues Tyr-66, Thr-69, Thr-70, and Gly-71 are colored by protomer, and the positions of those residues are also shown as sticks. B, calculated molecular electrostatic surface potential of three SubB protomers shown in two orientations. The Tyr-66, Thr-69, Thr-70, and Gly-71 surface locations are indicated. Negative surfaces are in red (−10 kT), neutral surfaces are in white (0 kiloteslas), and positive surfaces are in blue (10 kiloteslas). The surface electrostatic potential map was calculated using the nonlinear mode of the Adaptive Poisson Boltzmann Solver (APBS) (30, 31). The figures were generated using PyMOL (29).
A- to B-subunit Interface of AB5 Toxins
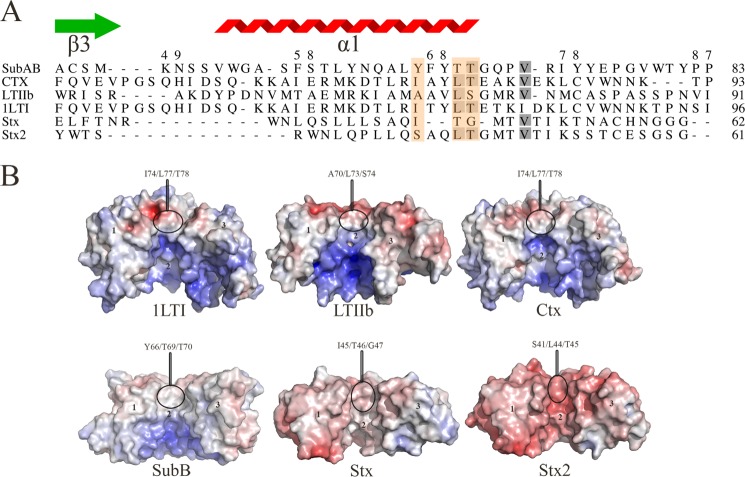
Despite sharing a common structural architecture, the B-subunits of the various AB5 subfamilies exhibit a very low amino acid sequence identity (2). However, they do have conserved structural features such as the OB fold and hydrophobic residues dominating the upper portion of the central pore of the B-pentamer (Ctx, LT-I, and LT-IIb) (Fig. 5, A and B) (17). The conservation of these hydrophobic residues is proposed to be important for the assembly of the A-subunit with the B pentamer and, in particular, in the cholera AB5 toxin (Ctx) (18). It is analogous in the SubAB x-ray structure, which contains the residues at positions Tyr-66, Thr-69, Thr-70, and Gly-71 (Fig. 5A). In Ctx, the residues Ile-74/Leu-77/Thr-78 (Fig. 5, A and B) form this hydrophobic surface and exhibit a BSA of 740 Å2 upon complex formation (38% of the B-subunit BSA). In the closely related E. coli heat-labile enterotoxins (LT-I and LT-IIb), the corresponding residues Ile-74/Leu-77/Thr-78 and Ala-70/Leu-73/Ser-74 contribute 31% (700 Å2) and 45% (680 Å2) of the BSA of the B-subunit at the interface, respectively. LT-IIb contains the polar residue serine (Ser-74) in the motif, but its hydroxyl moiety points inwards exposing mainly the carbon Cβ. Shiga toxigenic E. coli and S. dysenteriae AB5 toxins (Stx/Stx1 and Stx2) do exhibit a slightly more negatively charged exposed surface. In SubB, residues Tyr-66/Thr-69/Thr-70 contribute (920 Å2 BSA at the SubA/SubB interface). The extent of the exposed hydrophobic surface in SubB is more pronounced than any other AB5 toxin mainly due to the aromatic platform of residue Tyr-66 (Figs. 4B and 5B). Finally, although the A-subunit overall fold of the bacterial AB5 toxins varies from one family to the other (2), all A-subunits contribute similarly (∼48–50% BSA) to the overall A- and B-subunits interface (Table 3). Accordingly, a common core of residues underpins AB5 toxin assembly.
FIGURE 5.
A, structure-based sequence alignment of the B-subunit α1-helix of members of the four AB5 toxin families. The residues that form the hydrophobic patch are boxed in orange. The gray shadowing indicates the sequence identity level between the six sequences. The SubB secondary structural elements are shown. The figure was generated using the Indonesia program (16). B, calculated molecular electrostatic surface potential of members of the four AB5 toxin families (SubAB, LT-I, LT-IIb, Ctx, Stx, and Stx2). For clarity, the surface of three protomers for each AB5 toxins (1 to 3) is shown. The molecular electrostatic surface potential was calculated as described in the legend for Fig. 4. The figure was generated using PyMOL (29).
Effects of the Selected SubB Mutations on Assembly and Cytotoxicity
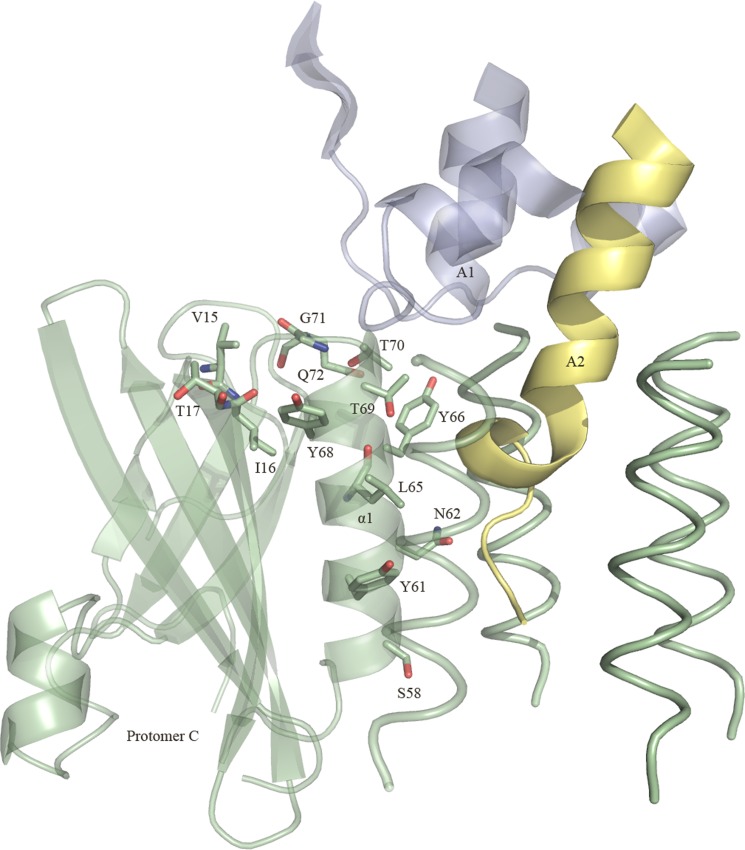
We have shown that the SubAB toxin presents all the hallmarks that define an AB5 toxin such as a disulfide bond bridging the A1 and A2 domains and an A2 helix that penetrates into the central pore of a pentameric B-subunit that contains a conserved hydrophobic patch at the top of the pentamer. In addition to the residues forming the hydrophobic ring (Tyr-66, Thr-69, Thr-70, and Gly-71), the SubAB crystal structure enabled us to identify nine other SubB residues (Val-15, Ile-16, Thr-17, Ser-58, Tyr-61, Asn-62, Leu-65, Tyr-68, and Gln-72) that contact the A-subunit (SubA) (Table 2) (Figs. 3 and 6). The conserved hydrophobic patch observed in the B-subunits is proposed to aid in assembly with the A-subunit, and the strength of the interaction can determine the level of cytotoxicity by individual AB5 toxins (Rodighiero et al. (28), Tinker et al. (18)). To determine whether the hydrophobic patch or any of the other contact residues in SubB has a role in the assembly or cytotoxicity of SubAB, we mutated these contact residues to alanine. In addition, residues of the hydrophobic patch Tyr-66, Thr-69, Thr-70, and Gly-71 were mutated to aspartate (Figs. 3 and 6). The Asp substitution was specifically chosen as the charged residue will effectively disrupt the hydrophobic patch. All SubB mutants were expressed and could assemble with SubA with the exception of Y68A and G71D; expression of holotoxin variants carrying these mutations was not detected, suggesting a role for Tyr-68 and Gly-71 in the formation of the SubAB holotoxin. Interestingly, residue Tyr-68OH is hydrogen-bonded to the neighboring protomer residue Gln-72NE2 in all SubB protomers; an Ala substitution could indeed prevent the assembly of the pentameric B-subunit and subsequently of SubAB.
FIGURE 6.
Positions of the 13 mutated SubB residues. The locations of the residues belonging to protomer C are shown. Segments of the A1 domain and the A2 helix (SubA) are shown and colored in blue and yellow, respectively. For clarity, only the α-helix of the other four protomers is shown (in green). The figure was generated using PyMOL (29).
The selected mutants were evaluated for their cytotoxic effect on Vero cells at different time points (Fig. 7A). Trafficking and cytotoxicity defects were apparent at all time points. However, as shown by our previous studies, the 30-min time point is most informative for assessing the efficiency/kinetics of trafficking; therefore, all results are reported for the 30-min time point (19).
FIGURE 7.
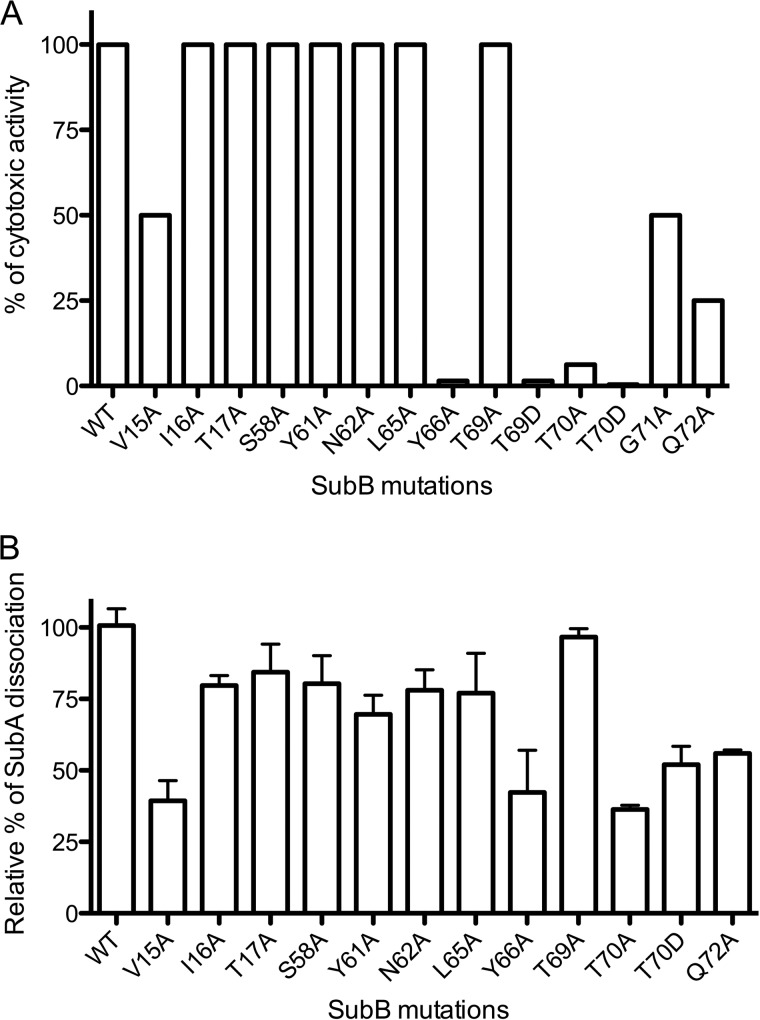
Effect of SubB mutations on cytotoxic activity of SubAB. A, SubAB cytotoxicity was assayed on Vero cell monolayers as described under “Experimental Procedures.” Data are expressed as a percentage of wild type activity. Data are representative of two independent experiments. B, relative dissociation of SubA bound to SubB and SubB mutants after incubation in detergent (30 mm 3-(decyldimethylammonio)propanesulfonate inner salt). SubA quantities were analyzed by SDS-PAGE and densitometry. SubA quantities for each SubB mutant were normalized to the wild type SubA dissociation. Error bars indicate mean ± S.E. of triplicate samples.
Out of the 16 mutations, seven (I16A, T17A, S58A, Y61A, N62A, L65A, and T69A) retained the same cytotoxic activity for Vero cells as the wild type SubAB holotoxin (Fig. 7A). SubA contacts the SubB residues Ile-16, Thr-17, Tyr-61, and Leu-65 via van der Waals interactions and/or via a single main chain carbonyl hydrogen bond (Ile-16). These residues interact with SubA through only one protomer. Residues Ser-58 and Asn-62 exhibit more extensive interactions with SubA (Table 2 and Fig. 3). The V15A, G71A, and Q72A substitutions had relatively modest impact on SubAB cytotoxicity with 50, 50, and 75% inhibition, respectively.
In contrast, the Y66A, T69D, T70A, and T70D mutations almost completely abolished SubAB cytotoxicity for Vero cells, thereby indicating a crucial role for these residues in maintaining the stability of the SubAB complex. As described previously, Thr-69 is the only residue that contacts SubA in all five protomers. Although the T69A mutation had no impact on cytotoxicity, the substitution to an aspartate had a profound effect (98.5% inhibition). The insertion of a charged residue (Asp) could indeed disrupt the hydrophobic ring formed by Tyr-66, Thr-69, and Thr-70. The T70D could interfere with the neighboring residue Tyr-66, forcing this residue to move away, and consequently would impact indirectly on the overall stability of the SubB.
To verify that the observed cytotoxicity effects were due to a weaker association between SubA and SubB and not to some transport defect, we measured the strength of the interaction by taking advantage of the fact that the interaction is detergent-sensitive. We have previously shown that SubA can be completely dissociated from SubB in the presence of high concentrations of the detergent 3-(decyldimethylammonio)propanesulfonate inner salt (4). The stability of the association of SubA with various SubB mutants was tested in the presence of 30 mm 3-(decyldimethylammonio)propanesulfonate inner salt (Fig. 7B), and the level of SubA dissociation was measured by SDS-PAGE and densitometry. Overall, the results were in agreement with the cytotoxicity data. The mutations with the greatest impact on cytotoxicity (Y66A, T70A, and T70D) also had the strongest dissociation effect between SubA and SubB, whereas mutations in other SubB residues (I16A, T17A, S58A, Y61A, N62A, L65A, and T69A) had minimal effects on SubA/SubB dissociation relative to the wild type (Fig. 7B). It should be noted that mutants T69D and G71A could not purified in sufficient amounts to perform these experiments.
Intracellular Trafficking of the Mutants with Reduced Cytotoxicity
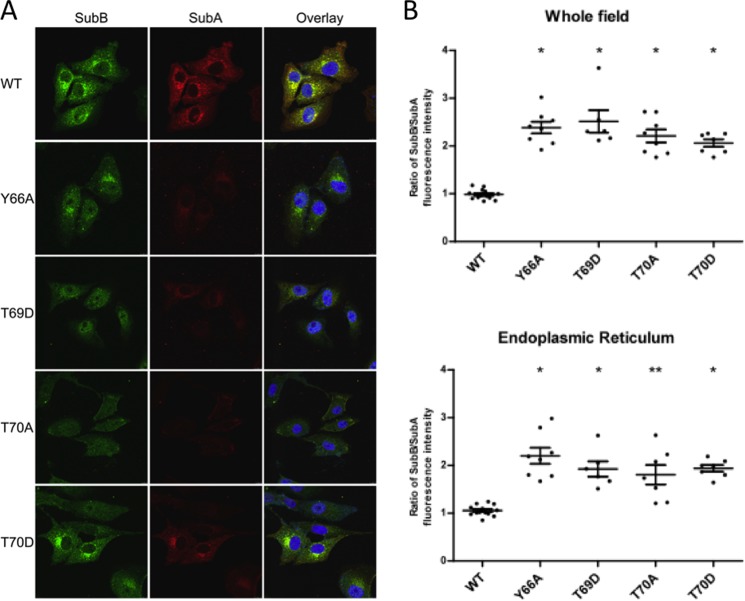
For SubAB to be cytotoxic, SubA needs to be delivered to the ER to cleave BiP, and SubB is largely responsible for both binding of the holotoxin to the cell surface and for triggering this retrograde intracellular trafficking (19). Thus, a possible explanation for why SubB mutations Y66A, T69D, T69A, and T70D severely reduced the cytotoxicity of SubAB is that the mutations cause a defect in intracellular trafficking. Therefore, Vero cells were treated with the various mutant toxins or wild type SubAB for 30 min, the earliest time point at which significant trafficking to the ER compartment is observed for wild type SubAB-treated cells (19). Cells were then examined by confocal fluorescence microscopy after differential staining with SubA- and SubB-specific antibodies (Fig. 8A). All mutants exhibited defects in terms of the amount of SubA that reached the ER compartment relative to wild type toxin. Moreover, quantitation of the SubB/SubA fluorescence ratio, either in whole microscopic fields or specifically in the ER compartment, revealed a significant increase for all four mutants tested relative to that for the wild type SubAB (p < 0.0001) (Fig. 8B). Thus, although the mutant holotoxins were sufficiently stable in vitro to enable co-purification from recombinant E. coli, significant instability of the holotoxin complexes is evident during intracellular trafficking. Moreover, although the B-subunits of most of the mutants were trafficked to a perinuclear location consistent with the ER, albeit less efficiently than the WT and with less associated A-subunit, the SubB staining pattern seen for T70A showed no concentration of label in the vicinity of the ER, consistent with a major defect in trafficking.
FIGURE 8.
Intracellular trafficking of SubAB mutants. Vero cells were treated with 3 μg ml−1 purified wild type SubAB or the indicated mutant derivatives for 30 min, and then fixed, permeabilized, and differentially stained for SubA (red) or SubB (green) using immunofluorescence, as described under “Experimental Procedures.” A, separate and overlaid confocal images are shown for SubA and SubB; in the overlay, nuclei are stained with DAPI (blue). B, SubB/SubA fluorescence intensity ratios were calculated either on whole fields or on the ER compartment. Data are the mean ± S.D. ratios for at least six fields/cells (*, p < 0.0001; **, p = 0.0001; unpaired Student's t test (relative to wild type ratio)).
DISCUSSION
The crystal structure of SubAB revealed the conserved structural features of AB5 toxins in which the A-subunit is divided into two domains with the A1 domain containing the catalytic domain and the A2 helix interacting with the central pore of the B-pentamer. Linking the two domains is a disulfide bond, which is conserved in all AB5 toxins (2). This disulfide bond serves an important role in AB5 toxin biology. Namely, all AB5 toxins studied to date, with the exception of SubAB, target molecules located in the cytosol, and therefore, their A-subunits have to be separated from the B-pentamer, unfolded, and then retrotranslocated from the endoplasmic reticulum lumen into the cytosol, where they cause cellular disruption (20). During the trafficking of AB5 toxins, the A-subunits are cleaved by proteases at residues located between the disulfide bond, providing a means to free A-subunit from B-pentamer by the action of a protein disulfide isomerase in the ER (21, 22). However, although SubA is not required to dissociate from SubB to cleave its molecular target BiP, the disulfide bond in SubA could have a structural role in assembly of the holotoxin as there is no evidence for proteolytic cleavage in the residues between the disulfide bond in SubAB. Studies with E. coli heat-labile enterotoxin showed that if the cysteines were mutated to serine, there was a severe decrease in the amount of assembled toxin, and the assembled mutant toxin also had reduced cytotoxicity (23). The reduced toxicity was attributed to a decrease in stability of the holotoxin, and hence reduced delivery of the A-subunit to the ER and inefficient dissociation of A-subunit from the B-pentamer (23).
The N-terminal end of the SubA A2 helix (residues Lys-333, Lys-334, Tyr-335, Ile-336, Pro-337, and Val-338) formed predominantly hydrophobic interactions with a ring composed of the hydrophobic or neutral residues (Leu-65, Tyr-66, Thr-69, and Thr-70) located at the top of the SubB pentamer (Figs. 4, A and B, and 5A). This interaction has been observed in the cholera AB5 holotoxin and has been proposed to aid in assembly of AB5 toxins by overcoming non-pentaradiate symmetry of the A2 domain. Indeed, an elegant structural study showed that mutations of the cholera toxin B-subunit in the conserved hydrophobic residues Ile-74, Leu-77, and Thr-78 resulted in B-subunits that could not form B-pentamers or assemble with the A-subunit (18). However, in the present study, mutating the corresponding residues in SubB did not affect the assembly of B-pentamer or the holotoxin, suggesting that these residues are not critical for assembly of SubAB toxin. Interestingly, the predominance of hydrophobic interactions between the A-subunit and the B-pentamer has enabled the formation of hybrid toxins. For example, cholera toxin A-subunit has been assembled with the B-subunit of LT-I both in vitro and in vivo. However, not all combinations of A-subunits and B-subunits can assemble into functional toxins, suggesting that surface complementation between the A2 helix and the B-pentamer pore is also important (24–27).
The only exception to this conserved hydrophobic ring is the presence of the hydrophilic Ser-41 in the Stx2 toxin. However, Stx2 still has a dominant but smaller overall hydrophobic patch. The conserved hydrophobic ring in the B-pentamer also supports the view that A- and B-subunits have evolved independently from common ancestral A- and B-subunits. This enables the B-pentamers to recruit different A-subunits with distinct catalytic activity as in the SubAB holotoxin case.
Out of the 16 SubAB mutations, only two did not lead to the formation of the holotoxin, suggesting their importance in the assembly of SubAB or the SubB pentamer architectures. Mutations of the conserved hydrophobic residues in SubB resulted in no defect in assembly of either the B-pentamer or the holotoxin, but the mutations affected the level of cytotoxicity. In particular, mutation of Thr-69 or Thr-70, which are part of the conserved hydrophobic ring of residues (Fig. 4, A and B), almost completely abolished cytotoxicity. These mutations resulted in reduced levels of SubA, and a higher ratio of SubB/SubA, reaching the ER, suggesting that the mutations affected the stability and or intracellular trafficking of the SubAB complex. The stability of AB5 complexes has previously been shown to be important for cytotoxicity (18). Cholera toxin was shown to be more toxic to polarized human epithelial (Thr-84) cells than the closely related E. coli heat-labile enterotoxin (LT-I) (28). The basis for this difference in toxicity was shown to be an increase in stability of cholera toxin due to the A2 helix, which stabilizes the holotoxin during uptake and intracellular transport (28).
In summary, we have shown that the novel AB5 toxin SubAB is assembled in a similar manner as other described AB5 toxin complexes. The individual A- and B-subunits interact via the helical A2 domain located at the C-terminal end of SubA. Despite not having to be exported from the ER to cause cellular disruption, SubAB toxin displayed conserved features with AB5 toxins such as the disulfide bond linking the A1 and A2 domains and a hydrophobic ring of residues in the B-pentamer. Mutation of these residues did not affect assembly of the holotoxin, but cytotoxicity of the toxin was impacted due to decreased stability of the interaction between SubA and SubB. Future studies will aim to exploit the conserved hydrophobic ring in the B-pentamers for the development of novel therapeutics against these toxins.
Acknowledgments
We thank the Advanced Photon Source staff for assistance in data collection at the Argonne National Laboratory and Manisha Dias for technical assistance in the production of the SubB variants.
This work was supported by Program Grant 565526 and Project Grant 1002792 from the National Health and Medical Research Council of Australia (NHMRC) and Discovery Grants DP1095420 and DP120103178 from the Australian Research Council.
The atomic coordinates and structure factors (code 4BWG) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- Ctx
- cholera toxin
- Ptx
- pertussis toxin
- Stx
- Shiga toxin
- LT
- labile enterotoxin
- SubAB
- subtilase cytotoxin
- ER
- endoplasmic reticulum
- r.m.s.d.
- root mean square deviation
- BSA
- buried surface area.
REFERENCES
- 1. Fan E., Merritt E. A., Verlinde C. L., Hol W. G. (2000) AB5 toxins: structures and inhibitor design. Curr. Opin. Struct. Biol. 10, 680–686 [DOI] [PubMed] [Google Scholar]
- 2. Beddoe T., Paton A. W., Le Nours J., Rossjohn J., Paton J. C. (2010) Structure, biological functions, and applications of the AB5 toxins. Trends Biochem. Sci. 35, 411–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Paton A. W., Srimanote P., Talbot U. M., Wang H., Paton J. C. (2004) A new family of potent AB5 cytotoxins produced by Shiga toxigenic Escherichia coli. J. Exp. Med. 200, 35–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Paton A. W., Beddoe T., Thorpe C. M., Whisstock J. C., Wilce M. C. J., Rossjohn J., Talbot U. M., Paton J. C. (2006) AB5 subtilase cytotoxin inactivates the endoplasmic reticulum chaperone BiP. Nature 443, 548–552 [DOI] [PubMed] [Google Scholar]
- 5. Rao R. V., Ellerby H. M., Bredesen D. E. (2004) Coupling endoplasmic reticulum stress to the cell death program. Cell Death Differ. 11, 372–380 [DOI] [PubMed] [Google Scholar]
- 6. Hendershot L. M. (2004) The ER function BiP is a master regulator of ER function. Mt. Sinai. J. Med. 71, 289–297 [PubMed] [Google Scholar]
- 7. Byres E., Paton A. W., Paton J. C., Löfling J. C., Smith D. F., Wilce M. C. J., Talbot U. M., Chong D. C., Yu H., Huang S., Chen X., Varki N. M., Varki A., Rossjohn J., Beddoe T. (2008) Incorporation of a non-human glycan mediates human susceptibility to a bacterial toxin. Nature 456, 648–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tangvoranuntakul P., Gagneux P., Diaz S., Bardor M., Varki N., Varki A., Muchmore E. (2003) Human uptake and incorporation of an immunogenic nonhuman dietary sialic acid. Proc. Natl. Acad. Sci. U.S.A. 100, 12045–12050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Studier F. W. (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 10. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 12. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 13. Bricogne G, B. E., Brandl M, Flensburg C., Keller P, Paciorek W, Roversi P, Sharff A, Smart O. S., Vonrhein C., Womack T. O. (2011) autoBUSTER, Version 1.6.0, Global Phasing Ltd., Cambridge, United Kingdom [Google Scholar]
- 14. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Madsen D., Johansson P., Johansson N., Arent S., Harris M. R., Kleywegt G. J. (2005) Indonesia: An Integrated Workbench for Sequence and Structure Alignment and Analysis, Uppsala University, Uppsala, Sweden [Google Scholar]
- 17. van den Akker F., Sarfaty S., Twiddy E. M., Connell T. D., Holmes R. K., Hol W. G. (1996) Crystal structure of a new heat-labile enterotoxin, LT-IIb. Structure 4, 665–678 [DOI] [PubMed] [Google Scholar]
- 18. Tinker J. K., Erbe J. L., Hol W. G. J., Holmes R. K. (2003) Cholera holotoxin assembly requires a hydrophobic domain at the A-B5 interface: mutational analysis and development of an in vitro assembly system. Infect. Immun. 71, 4093–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chong D. C., Paton J. C., Thorpe C. M., Paton A. W. (2008) Clathrin-dependent trafficking of subtilase cytotoxin, a novel AB5 toxin that targets the endoplasmic reticulum chaperone BiP. Cell. Microbiol. 10, 795–806 [DOI] [PubMed] [Google Scholar]
- 20. Lencer W. I., Saslowsky D. (2005) Raft trafficking of AB5 subunit bacterial toxins. Biochim. Biophys. Acta 1746, 314–321 [DOI] [PubMed] [Google Scholar]
- 21. Orlandi P. A. (1997) Protein-disulfide isomerase-mediated reduction of the A subunit of cholera toxin in a human intestinal cell line. J. Biol. Chem. 272, 4591–4599 [PubMed] [Google Scholar]
- 22. Tsai B., Rodighiero C., Lencer W. I., Rapoport T. A. (2001) Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell 104, 937–948 [DOI] [PubMed] [Google Scholar]
- 23. Okamoto K., Nomura T., Fujii Y., Yamanaka H. (1998) Contribution of the disulfide bond of the A subunit to the action of Escherichia coli heat-labile enterotoxin. J. Bacteriol. 180, 1368–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Connell T. D., Holmes R. K. (1992) Characterization of hybrid toxins produced in Escherichia coli by assembly of A and B polypeptides from type I and type II heat-labile enterotoxins. Infect. Immun. 60, 1653–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ito H., Yutsudo T., Hirayama T., Takeda Y. (1988) Isolation and some properties of A and B subunits of Vero toxin 2 and in vitro formation of hybrid toxins between subunits of Vero toxin 1 and Vero toxin 2 from Escherichia coli O157:H7. Microb. Pathog. 5, 189–195 [DOI] [PubMed] [Google Scholar]
- 26. Weinstein D. L., Jackson M. P., Perera L. P., Holmes R. K., O'Brien A. D. (1989) In vivo formation of hybrid toxins comprising Shiga toxin and the Shiga-like toxins and role of the B subunit in localization and cytotoxic activity. Infect. Immun. 57, 3743–3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Takeda Y., Honda T., Taga S., Miwatani T. (1981) In vitro formation of hybrid toxins between subunits of Escherichia coli heat-labile enterotoxin and those of cholera enterotoxin. Infect. Immun. 34, 341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rodighiero C., Aman A. T., Kenny M. J., Moss J., Lencer W. I., Hirst T. R. (1999) Structural basis for the differential toxicity of cholera toxin and Escherichia coli heat-labile enterotoxin: construction of hybrid toxins identifies the A2-domain as the determinant of differential toxicity. J. Biol. Chem. 274, 3962–3969 [DOI] [PubMed] [Google Scholar]
- 29. DeLano W. L. (2006) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 30. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Holst M., Baker N., Wang F. (2000) Adaptive multilevel finite element solution of the Poisson-Boltzmann equation I: Algorithms and examples. J. Comput. Chem. 21, 1319–1342 [Google Scholar]
- 32. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]