Background: The C-type lectin macrophage galactose-type lectin (MGL) specifically dampens effector T cell function.
Results: Activation through the ERK and calcineurin pathways leads to dramatic changes in T cell surface glycosylation.
Conclusion: MGL-binding glycans are expressed only on recently activated T cells.
Significance: This study provides mechanistic insight into the signaling pathways that regulate T cell surface glycosylation.
Keywords: ERK, Glycosylation, Lectin, Signaling, T Cell, C-type Lectin
Abstract
The C-type lectin macrophage galactose-type lectin (MGL) exerts an immunosuppressive role reflected by its interaction with terminal GalNAc moieties, such as the Tn antigen, on CD45 of effector T cells, thereby down-regulating T cell receptor signaling, cytokine responses, and induction of T cell death. Here, we provide evidence for the pathways that control the specific expression of GalNAc moieties on human CD4+ T cells. GalNAc epitopes were readily detectable on the cell surface after T cell activation and required de novo protein synthesis. Expression of GalNAc-containing MGL ligands was completely dependent on PKC and did not involve NF-κB. Instead, activation of the downstream ERK MAPK pathway led to decreased mRNA levels and activity of the core 1 β3GalT enzyme and its chaperone Cosmc, favoring the expression of Tn antigen. In conclusion, expression of GalNAc moieties mirrors the T cell activation status, and thus only highly stimulated T cells are prone to the suppressive action of MGL.
Introduction
Protein glycosylation is one of the most common forms of post-translational modifications. About 1–3% of the mammalian genome is dedicated to the glycosylation machinery, defined by the set of enzymes, chaperones, regulatory molecules, and co-factors involved in the glycan biosynthesis (1). It is estimated that over 90% of all proteins on the cell surface carry glycosidic residues. These surface glycans create the cell glycocalyx that acts as a shield to protect cells from injury, but it also has a structural role in maintaining tissue integrity. In addition, surface glycans can be recognized by carbohydrate-specific lectin receptors, thus mediating cell-cell adhesion or communication.
The importance of glycosylation is emphasized in several syndromes in which genetic mutations occur in key molecules of the glycosylation pathway. These congenital disorders of glycosylation can lead to malformations, mental retardation, and even embryonic death (2). Glycosylation defects can also instigate the onset of systemic autoimmune diseases (3, 4). In chronic inflammatory conditions or during cancer progression, glycosylation is altered (5), significantly contributing to disease progression and pathology. Clearly, glycosylation is a tightly regulated process whereby cellular development, differentiation, and activation have an enormous impact on the glycosylation of cells.
New insights indicate that subtle glycosylation differences appear in T cells during development, peripheral activation, and aging (6). These variations regulate not only the threshold for T cell receptor activation but also apoptosis susceptibility through the interaction with lectins (7, 8). For instance, CD4+ T helper subsets display distinct glycosylation patterns, resulting in specific galectin-1-mediated apoptosis of T helper 1 and T helper 17 cells. In contrast, α2–6 sialylation of T helper 2 cells protects them from galectin-1-induced cell death (9, 10). T cell function is likewise regulated via N-glycan branching through metabolic supply of the hexosamine pathway (11). We have demonstrated that the C-type lectin macrophage galactose-type lectin (MGL)4 specifically interacts with a terminal N-acetylgalactosamine (GalNAc) epitope on CD45 present only on human effector T cells (12). Binding of MGL to CD45 reduced CD45 phosphatase activity and inhibited several signaling pathways downstream of the T cell receptor, resulting in decreased secretion of pro-inflammatory cytokines, lowered T cell proliferation, and the induction of T cell death (12).
Although the qualitative nature of the T cell glycan changes following activation are well characterized, little is known about the cellular pathways that direct the specific exposure of GalNAc epitopes on activated human T cells. We establish here that expression of GalNAc moieties and subsequent MGL binding is controlled via the Src-PKC-ERK axis, whereas NF-κB is not involved. Activation of the ERK MAPK pathway led to a down-regulation of core 1 β3GalT and Cosmc mRNA levels, thus favoring the expression of MGL ligands on the surface of activated T cells.
EXPERIMENTAL PROCEDURES
Cells
Human CD4+ and CD8+ T cells were isolated from buffy coats (Sanquin, Amsterdam, The Netherlands) of healthy volunteers (after informed consent) through Ficoll gradient centrifugation, followed by negative selection of the CD4+ or CD8+ T cells using MACS isolation (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer's instructions. Human immature monocyte-derived dendritic cells (DCs) were cultured for 4–7 days in RPMI 1640 medium (Invitrogen) containing 10% fetal calf serum from monocytes obtained after informed consent from the buffy coats of healthy donors (Sanquin) in the presence of IL-4 and GM-CSF (500 units/ml and 800 units/ml respectively, BIOSOURCE). Jurkat cells were maintained in RPMI 1640 medium containing 10% fetal calf serum.
T Cell Stimulation Assays
T cells were cultured in 24-well plates in RPMI 1640 medium supplemented with 10% fetal bovine serum (Lonza, Basel, Switzerland) at a density of 1 × 106 cells/ml. T cells were stimulated for the indicated times using PMA (10 ng/ml) and ionomycin (500 ng/ml both from Sigma) or using αCD3/CD28-coated beads (Invitrogen, bead/cell ratio of 1:1). T cell activation was monitored by staining for the activation markers CD25 and CD69 (data not shown). In some experiments, T cells were pretreated for 30 min with cell signaling inhibitors at the indicated final concentrations prior to the activation with PMA/ionomycin or αCD3/CD28 beads. All inhibitors were routinely titrated to avoid cell death, checked for functionality by assessing their ability to block IL-2 production by the T cells (data not shown), and kept present during the whole stimulation procedure. The following inhibitor concentrations were used: PP2 (Src kinases, 12.5 μm), GO6983 (PKC, 10 μm), SR11302 (AP-1, 10 μm), FK506 (calcineurin, 10 nm, all from Tocris Biosciences, Bristol, UK), U0126 (MEK/ERK, 25 μm), SB203580 (P38 MAPK, 12.5 μm), SP600125 (JNK, 25 μm) and Bay11-7082 (IκB-α, 1 μm, all from Invivogen, San Diego). The AP-1 blocking peptide and scrambled control were described before (13) and used at 25 μm. Peptides were synthesized on Protein Technologies, Inc. Symphony peptide synthesizers (Tucson, AZ) at the 50-μmol scale using a 5-fold excess of Fmoc-amino acids (400 mm) relative to the resin (Rinkamide-HMPB-ChemMatrix resin, Sigma). O-(7-azabenzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate was purchased from Iris Biotech (Marktredwitz, Germany), and all solvents and Fmoc amino acids were from Biosolve (Valkenswaard, The Netherlands). The side chain-protecting groups for the amino acids were as follows: Trt for cysteine, histidine, asparagine, and glutamine; tBu for aspartic acid, glutamic acid, tyrosine, serine, and threonine; pentamethyldihydrobenzofuran-5-sulfonyl for arginine; and tBoc for lysine and tryptophan. Coupling was performed using a 1:1:2 mixture of amino acid/O-(7-azabenzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate/N,N-diisopropylethylamine in dimethylformamide. Deprotection was performed using 20% piperidine/dimethylformamide. Cleavage reactions were carried out in a 82.5:5:5:5:2.5 mixture of TFA/water/anisole/phenol/triisopropylsilane for 2 h.
DC-CD4+ T Cell Co-culture Using HD7 T Cells
HD7 (a kind gift from Dr. Lanzavecchia) is a human CD4+ T cell clone that recognizes an epitope derived from murine IgG1 in the context of HLA-DR0101/DQw1 (14). Immature DCs (20,000 cells/well) from a typed donor were preincubated for 1 h with antibody and subsequently co-cultured with 80,000 HD7 cells/well. The AZN-D1 antibody (IgG1 isotype, directed against DC-expressed DC-SIGN) was used as the model antigen at concentrations indicated in the figures. After 48 h, supernatants were harvested and analyzed for IFNγ secretion by ELISA CytoSetsTM ELISA kits (BIOSOURCE), according to the manufacturer's protocol. In addition, the HD7 cells were assessed for their cell surface glycosylation pattern by MGL-Fc staining.
Flow Cytometry, Lectin Profiling, and MGL-Fc Staining
The following biotinylated lectins were used: Helix pomatia agglutinin (HPA, α-GalNAc/Tn antigen, Sigma); soybean agglutinin (SBA, α/β-GalNAc); concanavalin A (mannose structures and di-antennary N-glycans); peanut agglutinin (PNA, Galβ1–3GalNAc); Sambucus nigra agglutinin (SNA, α2–6-sialic acid), and Maackia amurensis agglutinin II (MAA II, α2–3-sialic acid, all from Vector Laboratories, Burlingame, CA). MGL-Fc consisting of the extracellular domains of MGL fused to the human IgG1-Fc tail was generated as described previously (15). T cells were stained for 30 min at 37 °C with 10 μg/ml MGL-Fc or 5 μg/ml of the biotinylated lectins in Hanks' buffered saline solution containing 0.5% BSA. Lectins were counterstained using Alexa Fluor 488-labeled streptavidin (Molecular Probes). After washing, bound MGL-Fc was counterstained with FITC-conjugated goat anti-human Fc (Jackson ImmunoResearch, Suffolk, UK). Cells were analyzed by flow cytometry (FACSCalibur, BD Biosciences). Dead cells were excluded by 7-AAD (Molecular Probes) staining.
For surface expression analysis, cells were incubated for 30 min at 4 °C with the following primary antibodies: anti-CD45 (MEM-28), anti-CD43 (MEM-59, provided by Dr. V. Horesji, Academy of Sciences of the Czech Republic, Prague, Czech Republic), and with a Tn-specific antibody ((16), kindly provided by Dr. R. Cummings, Emory University School of Medicine, Atlanta, GA). After washing, antibodies were counterstained for 30 min at 4 °C with the secondary antibodies F(ab′)2 goat anti-mouse Alexa 488 (Molecular Probes) or F(ab′)2 goat anti-mouse IgM-FITC (Jackson ImmunoResearch). Cells were analyzed by flow cytometry (FACS Calibur, BD Biosciences). Dead cells were excluded by 7-AAD staining.
Glycosidase Assays
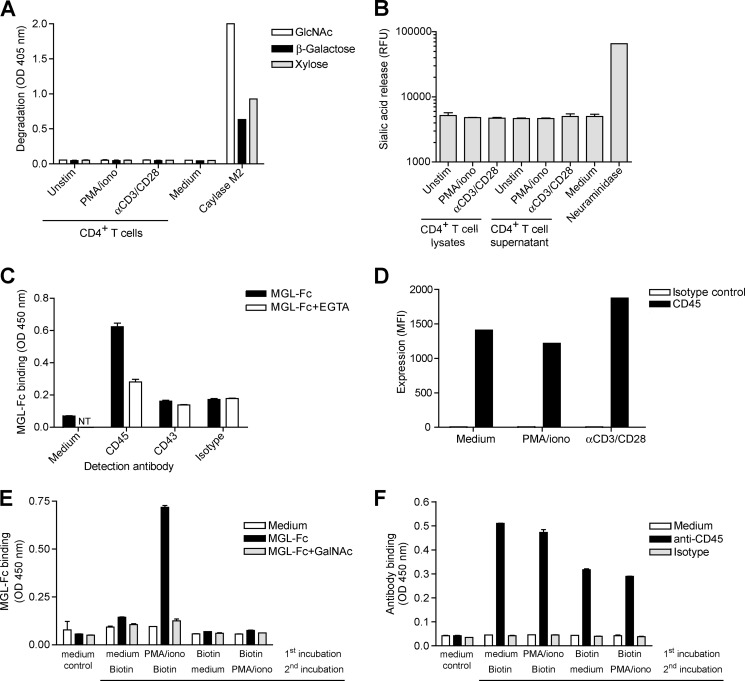
Glycosyl hydrolase activity was tested as described (17). In brief, supernatants of activated T cells were harvested 24 h after stimulation. Cell extracts were prepared by resuspending cells in TBS (1:8 v/v) containing protease inhibitors (Roche Applied Science) and 0.5% Triton X-100 (Sigma) followed by five consecutive sonication steps of 3 s. The extracts were incubated on ice for 20 min and subsequently centrifuged at 1000 × g for 5 min at 4 °C. Protein concentrations were determined by BCA protein assay (Pierce). T cell supernatants were tested for their ability to cleave p-nitrophenyl (pNp)-coupled carbohydrates (Sigma) as a substrate. Reaction mixtures (60 μl final volume) consisted of 6 μl of 1 m NaAc, pH 4.5, 8 μl of a 5 mm pNp/sugar substrate, and 20-μl supernatants or 2 μl of Caylase M2 as a positive control (5 mg/ml, kindly provided by Dr. A. F. J. Ram, originally from Societe Cayla, Toulouse, France). Samples were incubated for 6.5 h at 37 °C. The reaction was stopped by the addition of 240 μl of 0.25 m NaOH. Free(d) pNp was determined by optical density at 405 nm.
T cell supernatants and cellular extracts were tested for their ability to cleave 4-methylumbelliferone (MU)-coupled sialic acid (Sigma) as a substrate. Reaction mixtures consisted of 19 μl of 50 mm MES/NaOH, pH 6.8, 5 μl of a 1 mm 4-MU/sialic acid substrate, 5 μl 20 mm MnCl2, 1 μl of 0.2% Triton X-100, and 20-μl supernatants or cell extracts. 0.01 U Vibrio cholera Neuraminidase (Roche Applied Science) was used as a positive control and free 4-MU as a standard. Samples were incubated for 30 min at 37 °C. The reaction was stopped by the addition of 100 μl of 1 m glycine-NaOH, pH 10.0. Free(d) 4-MU was determined by fluorescence measurement at excitation 360 nm and emission 460 nm.
Core 1 β3GalT/Cosmc Activity Assay
To analyze the activity of the core 1 β3GalT-Cosmc complex, we sorted the MGL-binding and MGL-nonbinding cells using a MoFlo XDP cell sorter (Beckman Coulter, Indianapolis, IN) and performed an enzymatic activity assay on these subsets of cells as described previously (18). Activities were normalized to the protein content of the samples, which was determined using the micro BCA protein assay kit (Pierce). The Cosmc-deficient cell line Jurkat was included as a negative control.
MGL Ligand ELISA
NUNC Maxisorb plates were coated with goat anti-human-Fc antibody (4 μg/ml for 1 h at 37 °C, Jackson ImmunoResearch), followed by a 1% BSA blocking step (30 min at 37 °C) and MGL-Fc (1 μg/ml for 1 h at 37 °C). MGL-Fc coated plates were incubated overnight at 4 °C with cell lysates (30·106 cells/ml in lysis buffer (10 mm triethanolamine, pH 8.2, 150 mm NaCl, 1 mm MgCl2, 1 mm CaCl2, 1% (v/v) Triton X-100) containing EDTA-free protease inhibitors (Roche Applied Science)). After extensive washing with TSM (20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 2 mm MgCl2, 1 mm CaCl2), anti-CD45 (MEM-28) or anti-CD43 (MEM-59) was added at a concentration of 1 μg/ml for 2 h at RT. Binding was detected using a peroxidase-labeled goat anti-mouse antibody (Jackson ImmunoResearch).
To specifically test newly synthesized proteins, T cells were biotinylated for 30 min at 4 °C using 0.5 mg/ml sulfo-NHS-biotin (Pierce) in PBS prior to or after stimulation with medium or PMA/ionomycin. Cells were lysed at a concentration of 30·106 cells/ml in lysis buffer. Streptavidin-coated plates (Pierce) were incubated overnight at 4 °C with a 1:1 dilution of T cell lysates in PBS. After extensive washing with TSM, wells were subsequently probed for 2 h at RT with MGL-Fc (0.5 μg/ml) or anti-CD45 antibodies (MEM28, 1 μg/ml). After extensive washing with TSM, 0.05% Tween, binding was determined by peroxidase-labeled goat anti-human Fc or goat anti-mouse Fc (both from Jackson ImmunoResearch).
mRNA Isolation and cDNA Synthesis
mRNA isolation and cDNA synthesis were performed using the mRNA Capture (Roche Applied Science) and Reverse Transcription System kit (Promega, Madison, WI) according to the manufacturer's instructions. Briefly, cells were washed twice with ice-cold PBS and resuspended in 100 μl of lysis buffer. Lysates were incubated with biotin-labeled oligo(dT)20 for 5 min at 37 °C, transferred to streptavidin-coated tubes, and incubated for another 5 min at 37 °C. After washing with 200 μl of washing buffer, 30 μl of the reverse transcription mix (5 mm MgCl2, 1× reverse transcription buffer, 1 mm dNTPs, 0.4 units of recombinant RNasin ribonuclease inhibitor, 0.4 units of avian myeloblastosis virus reverse transcriptase, 0.5 μg of random hexamers in nuclease-free water) was added and incubated for 10 min at room temperature followed by 90 min at 42 °C and a denaturing step at 99 °C for 5 min. cDNA samples were stored at −20 °C.
Quantitative Real Time PCR
Primers were designed using Primer Express 2.0 (Applied Biosystems, Invitrogen) and synthesized by Invitrogen. PCRs were performed with FAST SYBR Green method in an ABI 7900HT sequence detection system (Applied Biosystems). The reaction mixture consisted of 4 μl of FAST SYBR Green Master Mix (Applied Biosystems), 0.2 μl of the primer solution containing 5 nmol/μl of both primers, 1.8 μl of H2O, and 2 μl of a 1:2 dilution of cDNA solution. PCRs were run for 2 min at 50 °C, followed by 10 min at 95 °C and 40 cycles of 15 s at 95 °C and 1 min 60 °C. The Ct value was defined as the number of PCR cycles where the fluorescence signal exceeds the threshold value, which is fixed above 10 times the standard deviation of the fluorescence during the first 15 cycles and typically corresponds to 0.2 relative fluorescence units. GAPDH served as an endogenous reference gene (19). Because of the low expression of glycosylation-related genes, the results are shown as 100 times the relative abundance. The primers used to measure mRNA levels of Cosmc and core 1 β3GalT have been described before (20).
Immunohistochemistry
Cryosections of human multiple sclerosis tissue (7 μm, kind gift of Dr. J. van Horssen, VU University Medical Center, Amsterdam, The Netherlands) were fixed with 100% acetone and stained with CD3 (clone T3b, 10 μg/ml) and HPA (5 μg/ml) for 1 h at 37 °C. Sections were counterstained with Alexa 594-labeled goat anti-mouse and Alexa 488-labeled streptavidin (both from Molecular Probes). Nuclei were visualized using Hoechst (Molecular Probes).
Statistical Analysis
Significant differences were evaluated using Graphpad Prism software. Student's t test and one-way analysis of variance with Dunn's multiple comparisons were used for statistical analysis. Significance was accepted at the p < 0.05 level.
RESULTS
MGL Ligands Are Up-regulated on Recently Activated CD4+ T Cells
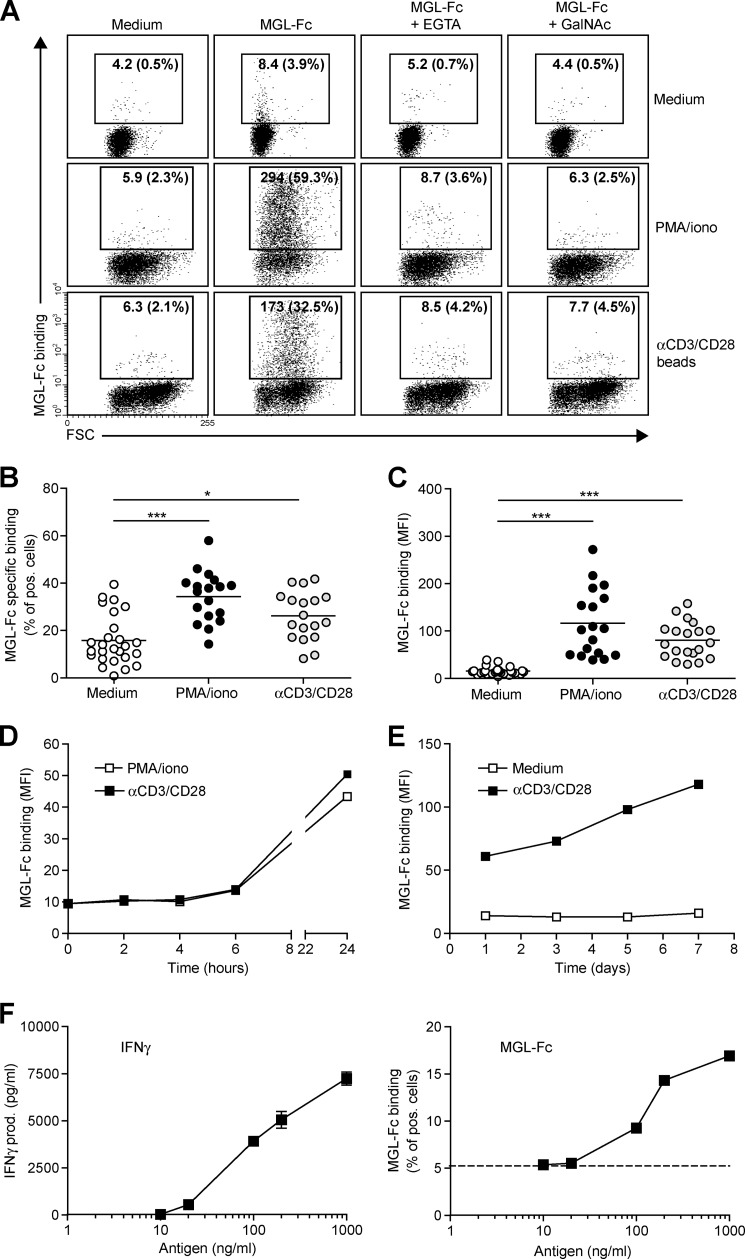
We have previously demonstrated a preferential Tn antigen-dependent interaction of the C-type lectin MGL with human effector T cells that results in T cell apoptosis (12). To gain more insight in the mechanisms that control expression of terminal GalNAc epitopes, human CD4+ T cells were exposed to the T cell stimulatory agents, PMA/ionomycin or αCD3/CD28-coated beads, of which the latter closely mimics the interaction with antigen-presenting cells. Overnight stimulation resulted in a robust increase in expression of MGL ligands on the cell surface of CD4+ T cells, as visualized by MGL-Fc staining (Fig. 1A). MGL binding could be blocked by the addition of the Ca2+-chelator EGTA or an excess amount of free ligand (GalNAc monosaccharides) confirming the specificity of the interaction. Polyclonal T cell activation augmented both the number of CD4+ T cells carrying MGL ligands as well as the amount of ligand on a per cell basis (Fig. 1, B and C, respectively). MGL binding glycans appeared on the T cell surface 24 h after activation and steadily increased for the next days after αCD3/CD28 triggering (Fig. 1, D and E). Additionally, we wanted to confirm also that a direct antigen-specific interaction between antigen-presenting cells and T cells could up-regulate the expression of MGL-binding epitopes. Therefore, human CD4+ T cells (clone HD7) that recognize a peptide derived from mouse IgG1 in the context of MHC class II (14) were co-cultured with DCs primed with mouse IgG1 antibodies as the model antigen. Antigen-specific T cell activation, as measured by IFNγ release (Fig. 1F, left panel), completely coincided with enhanced MGL binding (Fig. 1F, right panel), indicating that antigen experienced T cell up-regulated MGL-binding glycan structures within 24 h after stimulation.
FIGURE 1.
MGL ligands are up-regulated on recently activated CD4+ T cells. CD4+ T cells were left untreated or stimulated with PMA/ionomycin or αCD3/CD28 beads. A, after overnight incubation, MGL-Fc binding in the presence or absence of EGTA or free GalNAc was measured by flow cytometry. Mean fluorescence and the % of positive cells are indicated in the plots. Data are from one representative donor out of 20. FSC, forward scatter. B and C, both the % of cells expressing MGL binding epitopes as well as the magnitude of expression increases after overnight CD4+ T cell stimulation using PMA/ionomycin or αCD3/CD28 beads. At least 18 donors are plotted for each condition. *, p < 0.05; ***, p < 0.001. D and E, MGL ligands appear after 24 h of stimulation but subsequently remain stable on the cell surface for days. CD4+ T cells were stimulated for indicated time points with medium, PMA/ionomycin, or αCD3/CD28 beads. At each time point, MGL-Fc binding was measured by flow cytometry. Experiments were repeated three times with different donors, yielding similar results. Data are from one representative donor. F, antigen-specific stimulation of memory T cells increases expression of MGL ligands. HD7 T cells were stimulated with HLA-matched DCs that had been pulsed with the appropriate antigen. After 48 h, IFNγ secretion, as a measure for T cell activation, was determined by ELISA (left panel) and MGL ligands were assessed by MGL-Fc binding (right panel). Dashed line indicates the amount of MGL-Fc-positive cells when no antigen was added. One independent experiment out of two is shown.
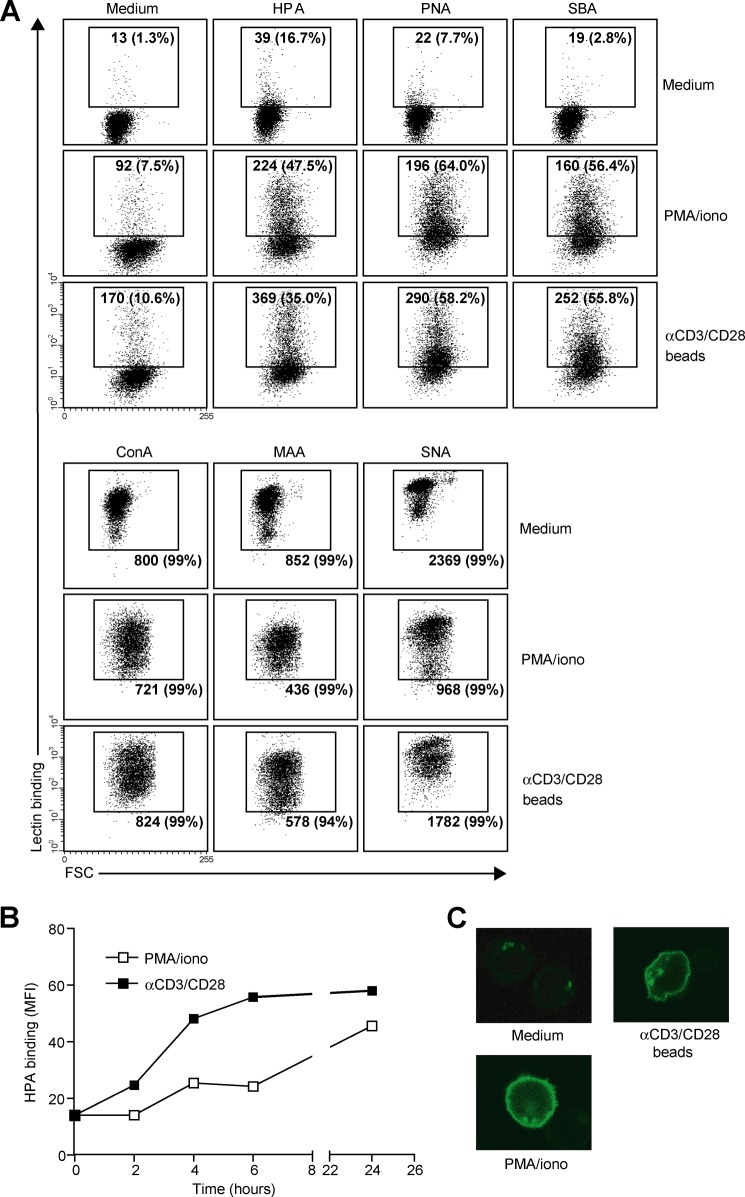
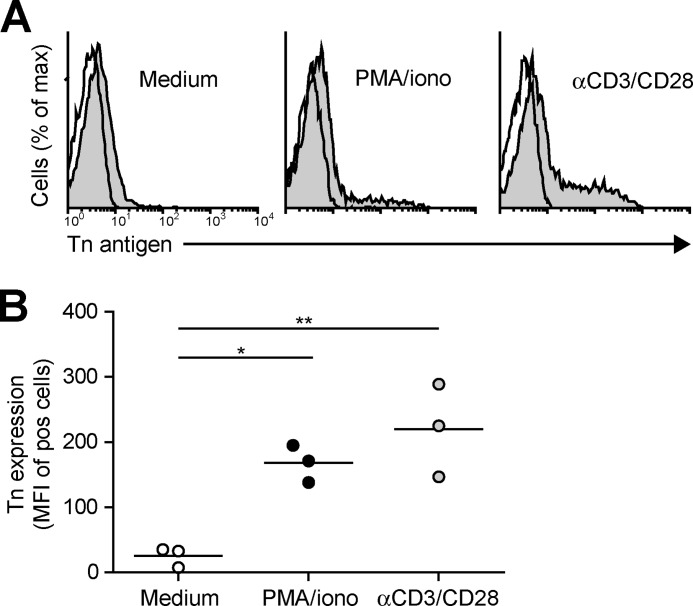
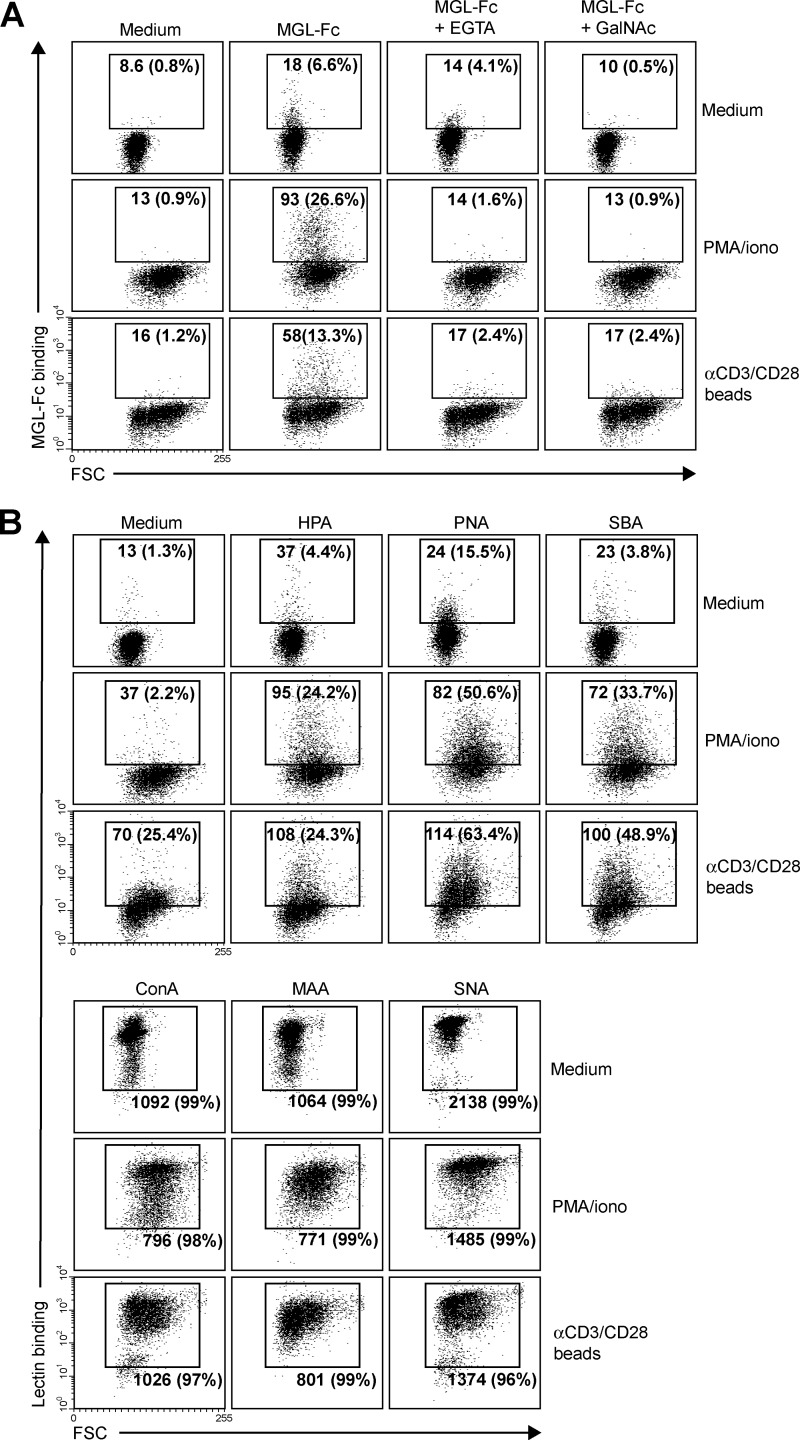
We next assessed whether the enhanced MGL binding is accompanied by concomitant changes in cell surface glycosylation and especially by the up-regulation of the MGL-binding glycan epitope Tn antigen (αGalNAc-Ser/Thr). Glycan changes on cells can be visualized through the use of plant/invertebrate lectins, which have well defined carbohydrate specificities (21). CD4+ T cell activation was characterized by a time-dependent increase in binding of HPA (Tn antigen), (SBA (terminal α- and β-GalNAc, including Tn antigen), and PNA (Galβ1–3GalNAc) (Fig. 2, A and B). Whereas on resting T cells HPA staining displayed predominantly an intracellular Golgi-like pattern, a clear cell surface localization could be detected on activated T cells (Fig. 2C). This expression pattern was confirmed by the increased staining with a Tn-specific antibody after CD4+ T cell activation (Fig. 3). T cell stimulation also resulted in a loss of both α2–3- and α2–6-sialylation, as depicted by the reduced staining with the lectins MAA II and SNA, respectively (Fig. 2A). No major changes were observed on concanavalin A binding (high mannose and di-antennary glycans). Similar changes in cell surface glycosylation and induction of MGL ligands was observed on activated CD8+ T cells (Fig. 4, A and B). Glycan changes were visible not only on in vitro triggered T cells but we could also detect GalNAc epitopes on activated T cells in vivo within an active lesion of a multiple sclerosis patient (data not shown). In summary, human T cell activation is complemented by an altered cell surface glycosylation profile, displaying enhanced Tn antigen expression and an up-regulation of MGL-binding ligands.
FIGURE 2.
MGL ligand up-regulation is accompanied by changes in cell surface glycosylation and exposure of GalNAc-containing epitopes. CD4+ T cells were left untreated or stimulated with PMA/ionomycin or αCD3/CD28 beads. A, T cell activation is characterized by increased exposure of Thomsen-Friedenreich (TF) and Tn antigen and a decrease in overall sialylation. After overnight stimulation, binding of a well characterized panel of plant/invertebrate lectins was assessed by flow cytometry. Glycan specificities of the lectins used are as follows: HPA, α-GalNAc/Tn, PNA, Galβ1–3GalNAc, SBA, α/β-GalNAc, concanavalin A, mannose structures and di-antennary N-glycans, MAA II, α2–3-sialic acid, and SNA, α2–6-sialic acid. Mean fluorescence and the % of positive cells are indicated in the plots. Experiments were repeated six times with different donors, yielding similar results. Data are from one representative donor. FSC, forward scatter. B, Tn antigens appear 4 h after T cell stimulation with PMA/ionomycin or αCD3/CD28 beads. At each time point HPA binding was measured by flow cytometry. One independent donor out of three is shown. C, Tn antigens are localized to the surface of activated T cells. HPA staining of umstimulated as well as PMA/ionomycin or αCD3/CD28 beads activated T cells was evaluated by HPA staining and confocal microscopy.
FIGURE 3.
CD4+ T cell activation is associated with an increase in Tn expression. A, after overnight T cell activation, Tn expression was measured by flow cytometry using a Tn-specific antibody. Representative flow cytometry plots for one out of three donors are shown. Open histograms indicate the isotype control staining. Tn expression is represented by the gray histograms. B, combined mean fluorescence intensities (MFI) as measured by flow cytometry are plotted for three independent donors. *, p < 0.05; **, p < 0.01.
FIGURE 4.
MGL ligands are up-regulated on recently activated CD8+ T cells. CD8+ T cells were left untreated or stimulated with PMA/ionomycin or αCD3/CD28 beads. After overnight incubation, binding of MGL-Fc (A) and a panel of plant/invertebrate lectins (B) was determined by flow cytometry. Glycan specificities of the lectins used are as follows: HPA, α-GalNAc/Tn, PNA, Galβ1–3GalNAc, SBA, α/β-GalNAc, concanavalin A, mannose structures and di-antennary N-glycans, MAA II, α2–3-sialic acid and SNA, and α2–6-sialic acid. Mean fluorescence and the % of positive cells are indicated in the plots. Experiments were repeated three times with different donors, yielding similar results. One representative donor is shown. FSC, forward scatter.
MGL Ligands Appear on Newly Synthesized Proteins and Are Not the Consequence of Glycosidase Activity
Strikingly, the appearance of Tn antigen preceded the expression of MGL ligands (compare Figs. 1D and 2B), suggesting that MGL ligands may be carried by newly synthesized glycoproteins that carry de novo glycan epitopes. However, the altered T cell surface glycosylation could also be the result of exogenous glycosidase trimming. During the biosynthesis of O-glycans, Tn antigens were elongated with either GlcNAc, β-galactose, or α2–6-sialic acid. Using an assay based on the release of pNp or MU groups upon cleavage from a glycoconjugate, we assessed the presence of exoglycosidase activity toward GlcNAc; β-galactose, sialic acid, or xylose as a negative control was present in the supernatants of overnight stimulated CD4+ T cells. However, we could not measure any degradation of pNp-GlcNAc, β-galactose, or xylose, although the positive control Caylase M2, isolated from H. jecorina, showed glycosidase activity toward these substrates (Fig. 5A). Although we detected some sialidase activity in medium containing fetal bovine serum, no additional sialidase activity was measured in T cell supernatants or lysates (Fig. 5B). Therefore, we postulate that the exposure of MGL ligands must be derived from newly synthesized proteins.
FIGURE 5.
MGL ligands appear on newly synthesized proteins and are not due to exogenous cleavage of glycans by glycosidases. A and B, no β-galactosidase or N-acetylglucosaminidase (A) or sialidase (B) activity could be detected in culture supernatants and/or cellular lysates of unstimulated (unstim), PMA/ionomycin (iono), or αCD3/CD28-activated T cells using the glycosidase assays. Background levels were subtracted, and mean + S.D. of three independent T cells donors measured in duplicate is shown. C, MGL binds CD45 on activated human CD4+ T cells. Capture of ligands from CD4+ T cells on MGL-Fc coated plates was detected by specific antibodies to CD43, CD45, or an isotype control in a MGL-ligand ELISA. MGL-Fc binding was blocked by the addition of 10 mm EGTA. NT, not tested. D, no changes in surface expression of the known MGL protein ligand CD45 after T cell stimulation. Expression of CD45 on unstimulated, PMA/ionomycin, and αCD3/CD28-activated T cells was determined by flow cytometry. Mean fluorescent intensities are depicted. One out of three donors is shown. E and F, only newly synthesized proteins carry MGL ligands. Biotinylated T cell proteins were captured on streptavidin-coated plates and probed with MGL-Fc (E) or anti-CD45 antibodies (F). One out of two independent experiments is shown.
The major MGL ligand on effector T cells is the cellular phosphatase CD45, although MGL on the T cell line Jurkat can also bind CD43 (Fig. 5C) (12). The enhanced MGL binding to activated T cells (∼5-fold, as shown in Fig. 1C) could not be explained by similar changes in expression levels of CD45 (Fig. 5D), suggesting that indeed the glycosylation of CD45 is changed on the activated T cells. In mice, CD45 was shown to have a slow turnover time, resulting in de novo TF moieties on newly synthesized CD45 following T cell activation (22). To validate that on human T cells MGL ligands also arise on de novo synthesized glycoproteins, we biotinylated T cells before or after T cell activation. Biotinylation following T cell stimulation will label all proteins, whereas labeling prior to T cell activation will not tag proteins that newly arrive on the cell surface. Clearly, MGL-Fc only bound biotinylated proteins that were labeled after T cell stimulation (Fig. 5E), even though abundant CD45 molecules were captured on the plate (Fig. 5F), suggesting that MGL ligands are indeed dependent on protein synthesis for their exposure on the T cell surface.
ERK-Calcineurin Axis Controls Expression of Tn Antigens
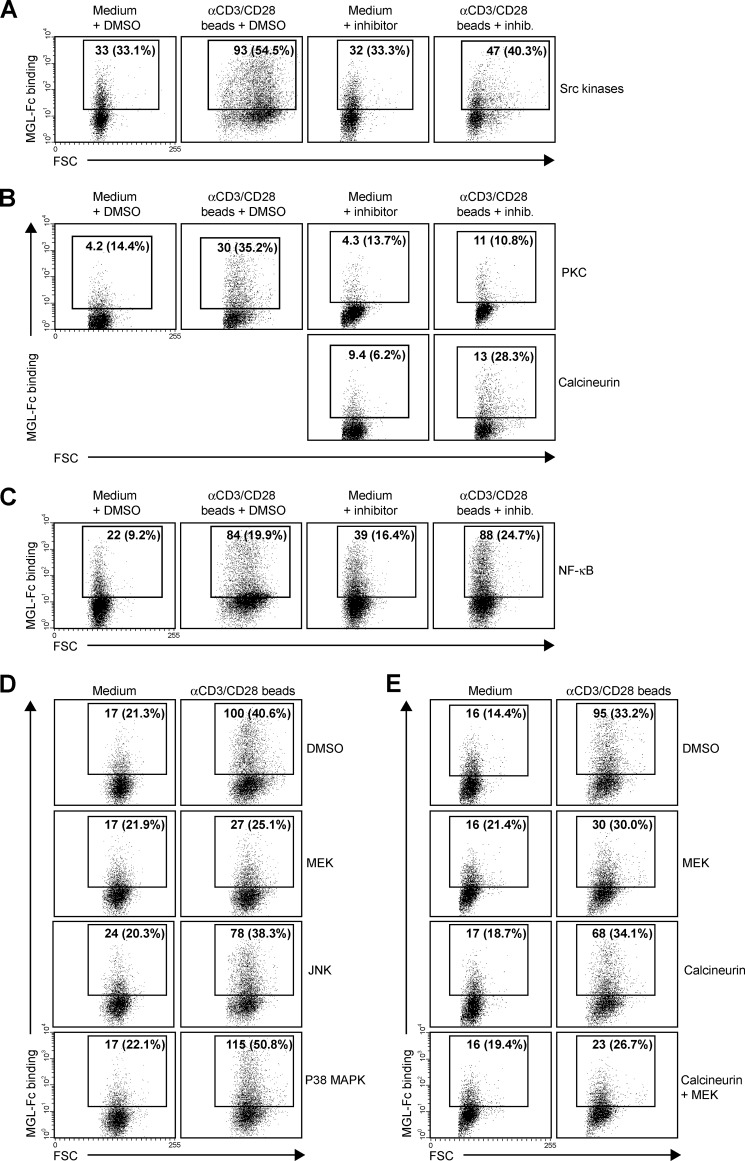
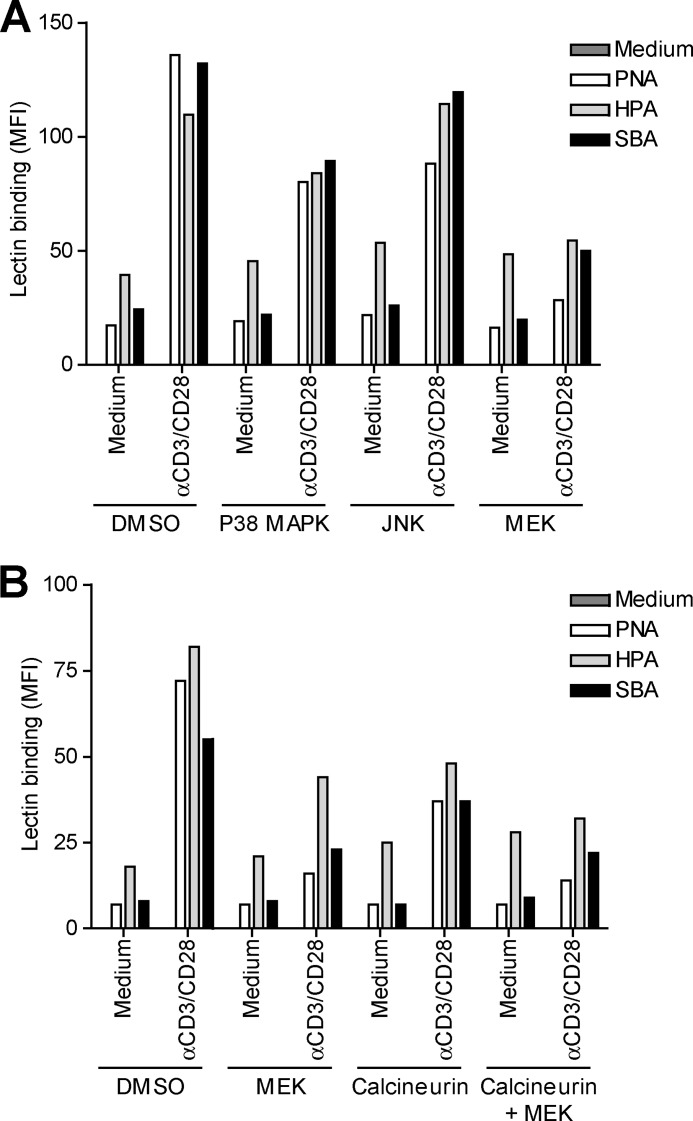
To gain more insight in the mechanisms that control expression of MGL ligands, we evaluated which cellular signaling pathways are involved in the exposure of Tn/GalNAc moieties on the T cell surface. T cell receptor stimulation was characterized by the activation of several intracellular cascades, which result in the mobilization of intracellular Ca2+ and the activation of NF-κB and the MAPK pathway. The initial step after T cell receptor triggering is the phosphorylation of protein-tyrosine kinases of the Src kinase family, of which Lck is the prototype member in T cells. Blocking Src kinases completely abrogated the up-regulation of MGL ligands (Fig. 6A), demonstrating the absolute necessity of T cell activation in this process. In general, αCD3/CD28 triggering and PMA/ionomycin treatment share activation of PKC activation and Ca2+ mobilization. Indeed, inhibition at the level of PKC resulted in a complete loss of MGL binding after stimulation, whereas a consistent although never full block was observed using inhibitors of calcineurin, the effector molecule downstream of Ca2+ release (Fig. 6B). Commonly, the transcription factor NF-κB is the major pathway downstream of PKC; however, after NF-κB inhibition, we did not detect a block in MGL binding, although blast formation is clearly abrogated (Fig. 6C, notice the decease in forward scatter in combination with the NF-κB inhibitor). As NF-κB seemed not to be involved in the regulation of MGL ligands on stimulated T cells, we focused our attention to the MAPK route, also known to operate downstream of PKC. Impeding JNK or p38 MAPK function had little or no effect on the expression of MGL-binding epitopes on the T cell surface. Yet, when blocking ERK activation through inhibition of MEK, a consistent decrease in MGL ligands was observed (Fig. 6D). Strikingly, a complete block was observed when MEK and calcineurin inhibitors were combined (Fig. 6E), confirming that MGL binding is regulated through the ERK-calcineurin axis. The binding of MGL-Fc displayed a strong correlation with the expression of GalNAc moieties on the T cells, which was also dependent on the MEK-ERK and calcineurin pathways (Fig. 7). The MAPK and Calcineurin pathways are known to converge at the formation of the AP-1 transcriptional complex; however, AP-1 inhibitors or blocking peptides do not prevent the up-regulation of MGL ligands after T cell stimulation (data not shown). Except for the inhibition of Src kinases, which acts upstream of PMA/ionomycin, similar results were obtained in PMA/ionomycin-activated T cells (data not shown).
FIGURE 6.
MGL ligand expression is controlled by an ERK- and calcineurin-dependent pathway. A–E, CD4+ T cells were left untreated or stimulated with αCD3/CD28 beads in the presence or absence of PP2 (Src kinases, 12.5 μm), GO6983 (PKC, 10 μm), FK506 (calcineurin, 10 nm), Bay11-7082 (IκB-α, 1 μm), U0126 (MEK/ERK, 25 μm), SB203580 (P38 MAPK, 12.5 μm), and SP600125 (JNK, 25 μm). DMSO was added as a vehicle control. All inhibitors were titrated and used at concentrations that allowed for at least 90% cell survival. 7-AAD was included to exclude dead cells. Mean fluorescent intensities and percentage of positive cells are indicated in the plots. All inhibitors were tested in at least three independent donors, yielding similar results. One representative donor is shown. FSC, forward scatter.
FIGURE 7.
GalNAc exposure is associated with ERK activation. CD4+ T cells were left untreated or stimulated overnight with αCD3/CD28 beads in the presence or absence of the inhibitor FK506 (calcineurin, 10 nm) and/or the MAPK kinase inhibitors U0126 (MEK/ERK, 25 mm), SB203580 (p38 MAPK, 12.5 mm), SP600125 (JNK, 25 mm), and subsequently characterized for their surface glycosylation by lectin binding. Glycan specificities of the lectins used HPA, α-GalNAc/Tn, PNA, Galβ1–3GalNAc, and SBA, α/β-GalNAc. DMSO was added as a vehicle control. All inhibitors were titrated and used at concentrations that allowed for at least 90% cell survival. 7-AAD was included to exclude dead cells. All inhibitors were tested in at least three independent donors, yielding similar results. One representative donor is shown.
ERK-Calcineurin Axis Regulates Expression of the Glycosyltransferase Core 1 β3GalT and Its Chaperone Cosmc
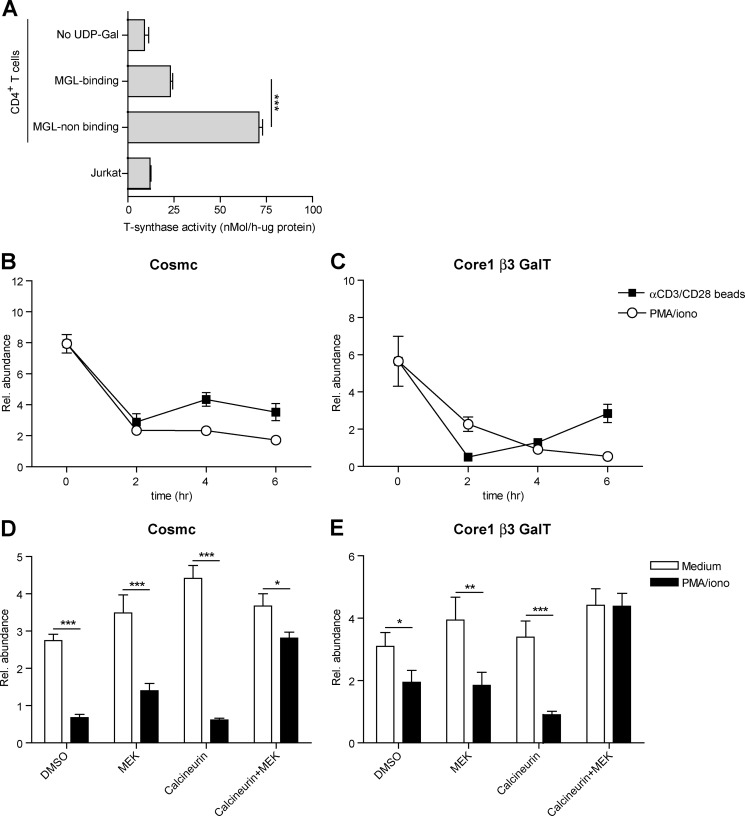
In lymphocytes, O-glycosylation is initiated through the addition of a single GalNAc residue to serine or threonine, thereby forming the Tn antigen, one the major ligands of MGL (15). Normally, this Tn antigen is elongated to the core 1 glycan (Galβ1–3GalNac) through the action of the glycosyltransferase core 1 β3GalT and its chaperone Cosmc (23). Strikingly, after T cell activation, the activity of the core 1 β3GalT-Cosmc complex was strongly reduced in the MGL-binding population compared with the cells that did not bind MGL after T cell stimulation (Fig. 8A). Moreover, the activity of this complex in the MGL-binding cells was only slightly higher than the negative control (without the sugar donor UDP-Gal) or the Cosmc-deficient Jurkat cells (Fig. 8A). These changes in enzymatic activity of the core 1 β3GalT-Cosmc complex likely result from the rapid and time-dependent decline in mRNA levels of Cosmc and core 1 β3GalT observed after both αCD3/CD28 as well as PMA/ionomycin-induced T cell activation (Fig. 8, B and C, respectively). In line with the binding of MGL (Fig. 6E), the mRNA levels of both Cosmc and core 1 β3GalT were completely dependent on the activation the MEK-ERK and calcineurin axis, as inhibitors of these pathways could prevent the down-regulation in stimulated T cells (Fig. 8, D and E).
FIGURE 8.
Expression levels and activity of Cosmc and core 1 β3GalT are down-regulated after T cell activation. A, MGL-binding CD4+ T cells have a diminished activity of the core 1 β3GalT-Cosmc complex. CD4+ T cells were stimulated overnight with PMA/ionomycin and subsequently sorted into a MGL-binding and MGL nonbinding fraction. Both fractions were analyzed for their glycosyltransferase activity. Activities were normalized to the protein content of the samples. B and C, mRNA levels of Cosmc and core 1 β3GalT were analyzed in time by RT-PCR using GAPDH as an endogenous reference gene. One out of three donors is shown. D and E, CD4+ T cells were left untreated or stimulated with αCD3/CD28 beads in the presence or absence of FK506 (calcineurin, 10 nm) and/or U0126 (MEK/ERK, 25 μm). DMSO was added as a vehicle control. mRNA levels of Cosmc and core 1 β3GalT were analyzed by RT-PCR using GAPDH as an endogenous reference gene.
Another ligand of MGL, the LacdiNAc epitope (GalNAcβ1–4Gal), is dependent on the action of the enzymes β4GAlNAcT4 and β4GAlNAcT3; however, no mRNA could be detected in activated T cells coding for these enzymes (data not shown), again confirming that the MGL ligands on activated T cells are decorated with Tn antigens.
Together, our results indicate that T cell activation is accompanied by a dramatic change in cell surface glycosylation, thereby exposing newly synthesized Tn epitopes for binding the C-type lectin MGL. Expression of these Tn antigens is independent of NF-κB activation and instead is dependent on the MEK-ERK and calcineurin pathways (Fig. 9).
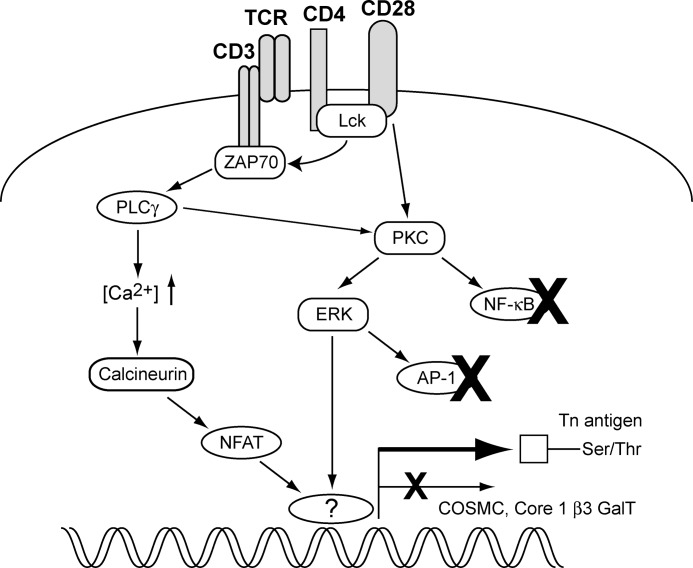
FIGURE 9.
Schematic representation of the T cell signaling pathways that control the exposure of MGL-binding ligands on activated CD4+ T cells. Triggering of the ERK-MAPK and calcineurin-NFAT cascades results in a diminished expression and activity of the core1 β3GalT-Cosmc complex via an as yet unidentified factor, which is neither AP-1 nor NF-κB.
DISCUSSION
Here, we describe how activation of human CD4+ and CD8+ T cells enhances expression of terminal GalNAc moieties, which can subsequently be recognized by the DC-expressed C-type lectin MGL. T cell receptor triggering of human T cells led to an up-regulation of Tn antigen on the cell surface, as visualized by the increased staining with the Tn-specific lectin HPA and the elevated binding of a Tn-specific antibody. This process was dependent on signaling via the MEK-ERK and calcineurin pathways. T cell activation also coincided with a decreased sialylation and an increase in the expression of the PNA epitope Galβ1–3GalNAc. Our data expand the observations made by Antonopoulos et al. (24) that show the remodeling of the N- and O-glycosylation profile of recently activated human CD8+ T cells, leading to more multiantennary structures and longer N-acetyllactosamine units, thereby increasing the binding of galectin-3 in the tumor microenvironment. Similar to humans, a reduction in sialylated biantennary N-glycans and an increased PNA binding specifically marks activated murine lymphocytes (22). However, activation of murine T cells is also accompanied by the appearance of glycans carrying the Galα1–3Gal sequence (25), an epitope not found on human cells due to an inactivation of the corresponding enzyme, UDP-Gal:β-galactosyl α1–3-galactosyltransferase (26). It is currently unclear whether antigen-stimulated murine T cells also carry the GalNAc epitope.
Post-translational modification by glycosylation is a dynamic, but tightly controlled process that is closely coupled to the physiology of the cell. Although our understanding on the repertoire of glycosylated structures on mouse immune cells has greatly increased over the last decades, the exact cellular pathways that regulate glycosylation remain to be elucidated to a great extent. Factors known to influence glycosylation include the expression and location of glycosyltransferases, as well as the availability of sugar donors and scaffold (glyco)proteins and (glyco)lipids. O-Glycosylation is initiated by the addition of a single GalNAc residue to a threonine or serine (αGalNAc-Thr/Ser or Tn antigen) by a polypeptide-GalNAc transferase (ppGalNAcT). Strikingly, the human genome encodes for a large family of 20 ppGalNAcTs (27), whereas the elongation of the Tn antigen with galactose is facilitated by only one enzyme, the core1 β3GalT, which requires the unique but crucial chaperone Cosmc (28). Cosmc appears to play a central role in Tn expression as mutations or reduced levels of Cosmc lead to loss of core1 β3GalT activity and increased exposure of the Tn antigen on the cell surface (23, 29, 30). Indeed, we observed a reduction in core1 β3GalT and Cosmc mRNA levels, likely leading to a decreased activity of this glycosyltransferase complex. Of course, we cannot rule out that a changed repertoire of ppGalNAcT enzymes during T cell activation also contributes to the expression of Tn antigen on the T cell surface, and this will be the focus of future studies.
Although differences in surface glycosylation can sometimes be directly linked to enhanced or decreased levels of glycosyltransferases (31) or availability of sugar donors, little is known about the cellular signaling cascades or transcription factors that control the expression of glycosylation-related enzymes and thus particular carbohydrate epitopes. Here, we report that activation through the ERK-MAPK kinase pathway initiated the appearance of Tn epitopes on the T cell surface, whereby the calcineurin-NFAT axis also played a minor but significant role in this process. Strikingly, NF-κB activation and IκB-α degradation did not seem to be involved, although this transcription factor regulates many pro-inflammatory genes induced after T cell receptor triggering (32). Signaling via the ERK axis is well known for its function in cell fate determination. Not only is thymic positive selection of T cells coupled to ERK activation (33), the ERK cascade is required for proper IL-4 receptor function and the differentiation of CD4+ T helper 2 cells in vivo as well (34). Triggering of ERK and the subsequent exposure of Tn epitopes has important parallels to thymic decisions in T cell life and death, as Tn antigen expression on the CD45 scaffold induces MGL binding and apoptosis of effector T cells (12). NFAT is considered to be a master transcriptional regulator in effector T cells; however, it is now also implicated in the induction of T cell anergy (35), again linking Tn exposure to a silencing of T cell immune responses. As ERK and NFAT converge at the formation of the AP-1 complex (36), our results suggest an involvement of other transcriptional partners in controlling Tn exposure; however, this needs to be addressed in future experiments.
Noticeably, expression of the sialyltransferase ST6Gal-I and the UDP-N-acetylglucosamine:α-6-d-mannoside β1,6 N-acetylglucosaminyltransferases II and V (GnT-II and GnT-V are also controlled by the ERK pathway in both tumor cells and immune cells (37, 38). GnT-V, which facilitates β1,6-branching on N-glycan intermediates, is overexpressed during malignant transformation and is associated with enhanced onset of mammary tumor formation (39). GnT-V gene transcription is mediated by the transcription factor Ets-1, a direct downstream target of ERK (40, 41). Whether the appearance of Tn antigen on activated T cells can also be attributed to Ets-1 remains to be determined. Activation of ERK, however, does not appear to be a general regulatory mechanism that applies to all glycosyltransferases as the enzyme β4GalT-1 in endothelial cells is regulated through mRNA stability rather than mRNA expression levels (42). In resting endothelial cells, the β4GalT-1 mRNA is bound by a complex of tristetraprolin and 14-3-3β targeting it for degradation. After TNFα activation, the β4GalT-1 mRNA is released from this complex due to the phosphorylation of 14-3-3β by Iκβ kinase and protein kinase Cδ (43). Furthermore, even tissue-specific transcriptional regulation through alternative splicing and promoter utilization should be considered, as has been observed for the different β-galactoside α2,3-sialyltransferase genes (44). The fact that GnT-V is cleaved by γ-secretase, suggests that glycosyltransferases might even be post-transcriptionally modified themselves (45).
In addition to transcriptional regulation, the correct localization of glycosyltransferases is imperative for proper glycosylation. The multisubunit conserved oligomeric Golgi complex is crucial in the positioning of glycosyltransferases in the Golgi, and conserved oligomeric Golgi defects have been shown to disrupt both the N- and O-glycosylation machinery (46). In addition, the correct compartmentalization of the Golgi apparatus itself is imperative for proper glycosylation, and changes in Golgi morphology directly affect the synthesis of both N- and O-glycans (47). Signaling via all four MAPK pathways modulates the structural organization of the Golgi network, yet in our experiments only ERK-mediated signals appeared to regulate the expression of MGL ligands, whereas inhibition of JNK and p38 function had little to no effect.
Recently, Gill et al. (48) described the retrograde trafficking of multiple ppGalNAcT from the Golgi to the endoplasmic reticulum, allowing more time for O-glycosylation initiation due to a lower degree of competition with other glycosyltransferases. This re-location of ppGalNAcT in HeLa cells was dependent on activation of Src kinases, a signaling molecule also involved in regulating Tn expression on stimulated T cells (Fig. 6A). Furthermore, ERK controls polarization and remodeling of the Golgi in migrating cells (49), suggesting that the ERK-dependent exposure of Tn epitopes might also implicate a redistribution of ppGalNAcT in activated T cells. However, initial experiments showed an exclusive localization of ppGalNacT to the Golgi in both resting as well as activated CD4+ T cells implicating that the localization of glycosyltransferases may not be altered during the synthesis of Tn epitopes on activated T cells (data not shown). Further research will be needed to evaluate if the Golgi network organization is influenced by T cell activation and whether changes in Golgi structure can explain the differential glycosylation in activated T cells.
In conclusion, expression of GalNAc moieties is controlled by ERK- and calcineurin-dependent pathways reflecting the activation status of human effector T cells. Thus, only highly stimulated T cells are predisposed to the suppressive action of MGL. Understanding the mechanisms that control GalNAc exposure may aid the development of novel strategies to intervene in inappropriate T cell activation as observed in chronic inflammatory and autoimmune disorders.
Acknowledgments
We thank T. O'Toole for help with the cell sorting and Dr. R. Cummings for the specific Tn antibody.
This work was supported by Dutch Organization for Health and Disease (NWO) VENI Grant 863.10.017, VU Medical Center Institute for Cancer and Immunology Grant CCA2011-5-03, Dutch Technology Foundation (STW) Grant 10622, and Mizutani Foundation for Glycoscience Research Grant 090032.
This article was selected as a Paper of the Week.
- MGL
- macrophage galactose-type lectin
- DC
- dendritic cell
- HPA
- H. pomatia agglutinin
- MAA II
- M. amurensis agglutinin II
- MU
- methylumbelliferone
- PNA
- peanut agglutinin
- pNp
- p-nitrophenyl
- SBA
- soybean agglutinin
- SNA
- S. nigra agglutinin
- PMA
- phorbol 12-myristate 13-acetate
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- ppGalNAcT
- polypeptide-GalNAc transferase
- NFAT
- nuclear factor of activated T cell
- β3GalT
- β3-galactosyltransferase
- 7-AAD
- 7-aminoactinomycin.
REFERENCES
- 1. Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R., Hart G. W., Etzler M. E. (2009) Essentials of Glycobiology, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 2. Freeze H. H., Aebi M. (2005) Altered glycan structures: the molecular basis of congenital disorders of glycosylation. Curr. Opin. Struct. Biol. 15, 490–498 [DOI] [PubMed] [Google Scholar]
- 3. Green R. S., Stone E. L., Tenno M., Lehtonen E., Farquhar M. G., Marth J. D. (2007) Mammalian N-glycan branching protects against innate immune self-recognition and inflammation in autoimmune disease pathogenesis. Immunity 27, 308–320 [DOI] [PubMed] [Google Scholar]
- 4. Morgan R., Gao G., Pawling J., Dennis J. W., Demetriou M., Li B. (2004) N-Acetylglucosaminyltransferase V (Mgat5)-mediated N-glycosylation negatively regulates Th1 cytokine production by T cells. J. Immunol. 173, 7200–7208 [DOI] [PubMed] [Google Scholar]
- 5. Hakomori S. (2002) Glycosylation defining cancer malignancy: new wine in an old bottle. Proc. Natl. Acad. Sci. U.S.A. 99, 10231–10233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Garcia G. G., Berger S. B., Sadighi Akha A. A., Miller R. A. (2005) Age-associated changes in glycosylation of CD43 and CD45 on mouse CD4 T cells. Eur. J. Immunol. 35, 622–631 [DOI] [PubMed] [Google Scholar]
- 7. Moody A. M., Chui D., Reche P. A., Priatel J. J., Marth J. D., Reinherz E. L. (2001) Developmentally regulated glycosylation of the CD8αβ coreceptor stalk modulates ligand binding. Cell 107, 501–512 [DOI] [PubMed] [Google Scholar]
- 8. Hernandez J. D., Klein J., Van Dyken S. J., Marth J. D., Baum L. G. (2007) T-cell activation results in microheterogeneous changes in glycosylation of CD45. Int. Immunol. 19, 847–856 [DOI] [PubMed] [Google Scholar]
- 9. Toscano M. A., Bianco G. A., Ilarregui J. M., Croci D. O., Correale J., Hernandez J. D., Zwirner N. W., Poirier F., Riley E. M., Baum L. G., Rabinovich G. A. (2007) Differential glycosylation of TH1, TH2, and TH-17 effector cells selectively regulates susceptibility to cell death, Nat. Immunol. 8, 825–834 [DOI] [PubMed] [Google Scholar]
- 10. Earl L. A., Bi S., Baum L. G. (2010) N- and O-glycans modulate galectin-1 binding, CD45 signaling, and T cell death. J. Biol. Chem. 285, 2232–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grigorian A., Lee S. U., Tian W., Chen I. J., Gao G., Mendelsohn R., Dennis J. W., Demetriou M. (2007) Control of T cell-mediated autoimmunity by metabolite flux to N-glycan biosynthesis. J. Biol. Chem. 282, 20027–20035 [DOI] [PubMed] [Google Scholar]
- 12. van Vliet S. J., Gringhuis S. I., Geijtenbeek T. B., van Kooyk Y. (2006) Regulation of effector T cells by antigen-presenting cells via interaction of the C-type lectin MGL with CD45. Nat. Immunol. 7, 1200–1208 [DOI] [PubMed] [Google Scholar]
- 13. Holzberg D., Knight C. G., Dittrich-Breiholz O., Schneider H., Dörrie A., Hoffmann E., Resch K., Kracht M. (2003) Disruption of the c-JUN-JNK complex by a cell-permeable peptide containing the c-JUN δ domain induces apoptosis and affects a distinct set of interleukin-1-induced inflammatory genes. J. Biol. Chem. 278, 40213–40223 [DOI] [PubMed] [Google Scholar]
- 14. Lanzavecchia A., Abrignani S., Scheidegger D., Obrist R., Dörken B., Moldenhauer G. (1988) Antibodies as antigens. The use of mouse monoclonal antibodies to focus human T cells against selected targets. J. Exp. Med. 167, 345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van Vliet S. J., van Liempt E., Saeland E., Aarnoudse C. A., Appelmelk B., Irimura T., Geijtenbeek T. B., Blixt O., Alvarez R., van Die I., van Kooyk Y. (2005) Carbohydrate profiling reveals a distinctive role for the C-type lectin MGL in the recognition of helminth parasites and tumor antigens by dendritic cells. Int. Immunol. 17, 661–669 [DOI] [PubMed] [Google Scholar]
- 16. Borgert A., Heimburg-Molinaro J., Song X., Lasanajak Y., Ju T., Liu M., Thompson P., Ragupathi G., Barany G., Smith D. F., Cummings R. D., Live D. (2012) Deciphering structural elements of mucin glycoprotein recognition. ACS Chem. Biol. 7, 1031–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tefsen B., Lagendijk E. L., Park J., Akeroyd M., Schachtschabel D., Winkler R., van Die I., Ram A. F. (2012) Fungal α-arabinofuranosidases of glycosyl hydrolase families 51 and 54 show a dual arabinofuranosyl- and galactofuranosyl-hydrolyzing activity. Biol. Chem. 393, 767–775 [DOI] [PubMed] [Google Scholar]
- 18. Ju T., Xia B., Aryal R. P., Wang W., Wang Y., Ding X., Mi R., He M., Cummings R. D. (2011) A novel fluorescent assay for T-synthase activity. Glycobiology 21, 352–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. García-Vallejo J. J., van het Hof B., Robben J., van Wijk J. A., van Die I., Joziasse D. H., van Dijk W. (2004) Approach for defining endogenous reference genes in gene expression experiments. Anal. Biochem. 329, 293–299 [DOI] [PubMed] [Google Scholar]
- 20. García-Vallejo J. J., Gringhuis S. I., van Dijk W., van Die I. (2006) Gene expression analysis of glycosylation-related genes by real-time polymerase chain reaction. Methods Mol. Biol. 347, 187–209 [DOI] [PubMed] [Google Scholar]
- 21. Wu A. M., Lisowska E., Duk M., Yang Z. (2009) Lectins as tools in glycoconjugate research. Glycoconj. J. 26, 899–913 [DOI] [PubMed] [Google Scholar]
- 22. Amado M., Yan Q., Comelli E. M., Collins B. E., Paulson J. C. (2004) Peanut agglutinin high phenotype of activated CD8+ T cells results from de novo synthesis of CD45 glycans. J. Biol. Chem. 279, 36689–36697 [DOI] [PubMed] [Google Scholar]
- 23. Ju T., Cummings R. D. (2002) A unique molecular chaperone Cosmc required for activity of the mammalian core 1 β3-galactosyltransferase. Proc. Natl. Acad. Sci. U.S.A. 99, 16613–16618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Antonopoulos A., Demotte N., Stroobant V., Haslam S. M., van der Bruggen P., Dell A. (2012) Loss of effector function of human cytolytic T lymphocytes is accompanied by major alterations in N- and O-glycosylation. J. Biol. Chem. 287, 11240–11251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Comelli E. M., Sutton-Smith M., Yan Q., Amado M., Panico M., Gilmartin T., Whisenant T., Lanigan C. M., Head S. R., Goldberg D., Morris H. R., Dell A., Paulson J. C. (2006) Activation of murine CD4+ and CD8+ T lymphocytes leads to dramatic remodeling of N-linked glycans. J. Immunol. 177, 2431–2440 [DOI] [PubMed] [Google Scholar]
- 26. Galili U., Shohet S. B., Kobrin E., Stults C. L., Macher B. A. (1988) Man, apes, and Old World monkeys differ from other mammals in the expression of α-galactosyl epitopes on nucleated cells. J. Biol. Chem. 263, 17755–17762 [PubMed] [Google Scholar]
- 27. Bennett E. P., Mandel U., Clausen H., Gerken T. A., Fritz T. A., Tabak L. A. (2012) Control of mucin-type O-glycosylation-A classification of the polypeptide GalNAc transferase gene family. Glycobiology 22, 736–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang Y., Ju T., Ding X., Xia B., Wang W., Xia L., He M., Cummings R. D. (2010) Cosmc is an essential chaperone for correct protein O-glycosylation. Proc. Natl. Acad. Sci. U.S.A. 107, 9228–9233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yamada K., Kobayashi N., Ikeda T., Suzuki Y., Tsuge T., Horikoshi S., Emancipator S. N., Tomino Y. (2010) Down-regulation of core 1 β1,3-galactosyltransferase and Cosmc by Th2 cytokine alters O-glycosylation of IgA1. Nephrol. Dial. Transplant. 25, 3890–3897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schietinger A., Philip M., Yoshida B. A., Azadi P., Liu H., Meredith S. C., Schreiber H. (2006) A mutant chaperone converts a wild-type protein into a tumor-specific antigen. Science 314, 304–308 [DOI] [PubMed] [Google Scholar]
- 31. Marcos N. T., Bennett E. P., Gomes J., Magalhaes A., Gomes C., David L., Dar I., Jeanneau C., DeFrees S., Krustrup D., Vogel L. K., Kure E. H., Burchell J., Taylor-Papadimitriou J., Clausen H., Mandel U., Reis C. A. (2011) ST6GalNAc-I controls expression of sialyl-Tn antigen in gastrointestinal tissues. Front. Biosci. 3, 1443–1455 [DOI] [PubMed] [Google Scholar]
- 32. Cheng J., Montecalvo A., Kane L. P. (2011) Regulation of NF-κB induction by TCR/CD28. Immunol. Res. 50, 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Delgado P., Fernández E., Dave V., Kappes D., Alarcón B. (2000) CD3δ couples T-cell receptor signalling to ERK activation and thymocyte positive selection. Nature 406, 426–430 [DOI] [PubMed] [Google Scholar]
- 34. Yamashita M., Kimura M., Kubo M., Shimizu C., Tada T., Perlmutter R. M., Nakayama T. (1999) T cell antigen receptor-mediated activation of the Ras/mitogen-activated protein kinase pathway controls interleukin 4 receptor function and type-2 helper T cell differentiation. Proc. Natl. Acad. Sci. U.S.A. 96, 1024–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Macián F., García-Cózar F., Im S. H., Horton H. F., Byrne M. C., Rao A. (2002) Transcriptional mechanisms underlying lymphocyte tolerance. Cell 109, 719–731 [DOI] [PubMed] [Google Scholar]
- 36. Genot E., Cleverley S., Henning S., Cantrell D. (1996) Multiple p21ras effector pathways regulate nuclear factor of activated T cells. EMBO J. 15, 3923–3933 [PMC free article] [PubMed] [Google Scholar]
- 37. Chen H. L., Li C. F., Grigorian A., Tian W., Demetriou M. (2009) T cell receptor signaling co-regulates multiple Golgi genes to enhance N-glycan branching. J. Biol. Chem. 284, 32454–32461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Seales E. C., Shaikh F. M., Woodard-Grice A. V., Aggarwal P., McBrayer A. C., Hennessy K. M., Bellis S. L. (2005) A protein kinase C/Ras/ERK signaling pathway activates myeloid fibronectin receptors by altering β1 integrin sialylation. J. Biol. Chem. 280, 37610–37615 [DOI] [PubMed] [Google Scholar]
- 39. Guo H. B., Johnson H., Randolph M., Nagy T., Blalock R., Pierce M. (2010) Specific posttranslational modification regulates early events in mammary carcinoma formation. Proc. Natl. Acad. Sci. U.S.A. 107, 21116–21121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ko J. H., Miyoshi E., Noda K., Ekuni A., Kang R., Ikeda Y., Taniguchi N. (1999) Regulation of the GnT-V promoter by transcription factor Ets-1 in various cancer cell lines. J. Biol. Chem. 274, 22941–22948 [DOI] [PubMed] [Google Scholar]
- 41. Kang R., Saito H., Ihara Y., Miyoshi E., Koyama N., Sheng Y., Taniguchi N. (1996) Transcriptional regulation of the N-acetylglucosaminyltransferase V gene in human bile duct carcinoma cells (HuCC-T1) is mediated by Ets-1. J. Biol. Chem. 271, 26706–26712 [DOI] [PubMed] [Google Scholar]
- 42. García-Vallejo J. J., van Dijk W., van Die I., Gringhuis S. I. (2005) Tumor necrosis factor-α up-regulates the expression of β1,4-galactosyltransferase I in primary human endothelial cells by mRNA stabilization. J. Biol. Chem. 280, 12676–12682 [DOI] [PubMed] [Google Scholar]
- 43. Gringhuis S. I., García-Vallejo J. J., van het Hof B., van Dijk W. (2005) Convergent actions of IκB kinase β and protein kinase Cδ modulate mRNA stability through phosphorylation of 14-3-3β complexed with tristetraprolin. Mol. Cell. Biol. 25, 6454–6463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Taniguchi A. (2008) Promoter structure and transcriptional regulation of human β-galactoside α2,3-sialyltransferase genes. Curr. Drug Targets 9, 310–316 [DOI] [PubMed] [Google Scholar]
- 45. Nakahara S., Saito T., Kondo N., Moriwaki K., Noda K., Ihara S., Takahashi M., Ide Y., Gu J., Inohara H., Katayama T., Tohyama M., Kubo T., Taniguchi N., Miyoshi E. (2006) A secreted type of β1,6 N-acetylglucosaminyltransferase V (GnT-V), a novel angiogenesis inducer, is regulated by γ-secretase. FASEB J. 20, 2451–2459 [DOI] [PubMed] [Google Scholar]
- 46. Wu X., Steet R. A., Bohorov O., Bakker J., Newell J., Krieger M., Spaapen L., Kornfeld S., Freeze H. H. (2004) Mutation of the COG complex subunit gene COG7 causes a lethal congenital disorder. Nat. Med. 10, 518–523 [DOI] [PubMed] [Google Scholar]
- 47. Chia J., Goh G., Racine V., Ng S., Kumar P., Bard F. (2012) RNAi screening reveals a large signaling network controlling the Golgi apparatus in human cells. Mol. Syst. Biol. 8, 629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gill D. J., Chia J., Senewiratne J., Bard F. (2010) Regulation of O-glycosylation through Golgi-to-endoplasmic reticulum relocation of initiation enzymes. J. Cell Biol. 189, 843–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bisel B., Wang Y., Wei J. H., Xiang Y., Tang D., Miron-Mendoza M., Yoshimura S., Nakamura N., Seemann J. (2008) ERK regulates Golgi and centrosome orientation toward the leading edge through GRASP65. J. Cell Biol. 182, 837–843 [DOI] [PMC free article] [PubMed] [Google Scholar]