Background: Various receptor proteins recruit Drp1 to drive fission of mitochondria and peroxisomes.
Results: MiD49 and MiD51 recruit Drp1 specifically to mitochondria independently of receptors Fis1 and Mff.
Conclusion: MiD49 and MiD51 appear to be specific to the mitochondrial fission apparatus of mammalian cells.
Significance: Mitochondrial and peroxisomal fission processes can be differentially regulated.
Keywords: Cell Biology, Confocal Microscopy, Mammal, Mitochondria, Peroxisomes, Drp1, MiD49, MiD51, Fission, Morphology
Abstract
Drp1 (dynamin-related protein 1) is recruited to both mitochondrial and peroxisomal membranes to execute fission. Fis1 and Mff are Drp1 receptor/effector proteins of mitochondria and peroxisomes. Recently, MiD49 and MiD51 were also shown to recruit Drp1 to the mitochondrial surface; however, different reports have ascribed opposing roles in fission and fusion. Here, we show that MiD49 or MiD51 overexpression blocked fission by acting in a dominant-negative manner by sequestering Drp1 specifically at mitochondria, causing unopposed fusion events at mitochondria along with elongation of peroxisomes. Mitochondrial elongation caused by MiD49/51 overexpression required the action of fusion mediators mitofusins 1 and 2. Furthermore, at low level overexpression when MiD49 and MiD51 form discrete foci at mitochondria, mitochondrial fission events still occurred. Unlike Fis1 and Mff, MiD49 and MiD51 were not targeted to the peroxisomal surface, suggesting that they specifically act to facilitate Drp1-directed fission at mitochondria. Moreover, when MiD49 or MiD51 was targeted to the surface of peroxisomes or lysosomes, Drp1 was specifically recruited to these organelles. Moreover, the Drp1 recruitment activity of MiD49/51 appeared stronger than that of Mff or Fis1. We conclude that MiD49 and MiD51 can act independently of Mff and Fis1 in Drp1 recruitment and suggest that they provide specificity to the division of mitochondria.
Introduction
Mitochondrial morphology is maintained through the opposing forces of fission and fusion, and the regulation of these processes governs the reticular nature of this organelle (1–3). Efficient control of the shape and distribution of mitochondria is important for a number of cellular processes (3). Regulation of mitochondrial dynamics has been shown to be crucial for neuronal cell function and transport of mitochondria along neuronal process (4–6) and for quality control and maintenance of a healthy mitochondrial network (7–11).
The key mediator of mitochondrial fission (division) is Drp1 (dynamin-related protein 1). Drp1 is a GTPase that is recruited to mitochondrial constriction sites, where it polymerizes around the organelle and, through the hydrolysis of GTP, changes conformation to constrict the outer and inner membranes to drive fission (12–14). The mitochondrial outer membrane proteins Fis1 and Mff have been proposed to act as receptors for Drp1 (15–21). However, deletion of Fis1 in cultured mammalian HCT116 cells results in no changes in mitochondrial morphology or Drp1 association with mitochondria, suggesting that Fis1 is dispensable for fission in mammalian cells (22). In contrast, a recent report found that Fis1-null mouse embryonic fibroblasts (MEFs)4 have reduced Drp1 puncta at mitochondria, which was compounded following the additional knock-out of Mff in these cells (23). Both Fis1 and Mff are also found on peroxisomal membranes and are involved in Drp1-mediated fission of that organelle (24). Recently, the mitochondrial outer membrane proteins MiD49 and MiD51 were found to also recruit Drp1 to the mitochondrial surface (25, 26). However, the proposed function of MiD49/51 in mitochondrial morphology has been under debate (1, 25–27). We proposed that MiD49 and MiD51 are mediators of mitochondrial fission (25), whereas Zhao et al. (26) reported that MiD51 (also termed MIEF1) promotes fusion rather than fission. This role in mitochondrial fusion was assigned based on the observation that the fused mitochondrial network seen following MiD51 overexpression was not blocked following knockdown of the fusion mediator mitofusin 2 (Mfn2) (26).
In this work, we set out to clarify the role of MiD49/51 in mitochondrial morphology. Herein, we provide additional evidence to validate the importance of the MiD proteins in Drp1 recruitment and mitochondrial fission. We found that mitochondrial fusion observed upon MiD49 or MiD51 overexpression is dependent on the presence of either known fusion mediator, mitofusin 1 (Mfn1) or Mfn2. We also established that the fused mitochondrial phenotype seen upon MiD49/51 overexpression is due to sequestration and inactivation of Drp1 on the mitochondrial surface, blocking fission and leading to unopposed fusion. Supporting this, we found that MiD49/51-mediated Drp1 sequestration at mitochondria results in peroxisome elongation. The Drp1 recruitment activity of MiD49/51 was stronger than that of Mff or Fis1. We suggest that unlike Fis1 and Mff, MiD49 and MiD51 specifically mediate the recruitment of Drp1 to mitochondria to execute fission.
EXPERIMENTAL PROCEDURES
Antibodies, Plasmids, and Chemicals
Antibodies specific for MiD49 were generated as described previously (25). Polyclonal antibodies against Mfn1 and Mfn2 were kindly provided by R. Youle (National Institutes of Health, Bethesda, MD), and anti-PEX14 antibody was a kind gift from David Crane (Griffith University, Queensland, Australia). Commercial antibodies used were anti-cytochrome c and anti-Drp1 (BD Biosciences), anti-β-actin (Sigma), and anti-Tom20 (Santa Cruz Biotechnology). Generation of GFP-Drp1, GFP-Drp1K38A, GFP-Fis1, mito-GFP, MiD49-GFP, and MiD51-GFP was described previously (17, 25). The mito-DsRed vector was purchased from BD Biosciences. The open reading frame of human Mff (GenBankTM accession number BC016597) was obtained from the Dana-Farber/Harvard Cancer Center. Mff was cloned downstream of GFP at BamHI and NotI. Plasmids encoding FKBP12/rapamycin-binding (FRB) domain-mito (FRB-Fis1TM), lyso-FRB (Lamp1-FRB), perox-FRB (PMP34-FRB), and GFP-FK506-binding protein (FKBP) (28) were kindly provided by R. Youle. MiD51 lacking the transmembrane domain (MiD51ΔTM) was cloned downstream of the FKBP domain, followed by GFP. Both Mff and Fis1 lacking the C-terminal transmembrane domains (MffΔTM and Fis1ΔTM) were cloned downstream of GFP, followed by the FKBP moiety. The plasmid encoding GFP-Sec61 (29) was a kind gift from Gia Voeltz (University of Colorado, Boulder, CO).
Cell Culture and Treatments
MEFs and HeLa cells were grown as previously described (25). Mfn1−/−, Mfn2−/−, and Mfn1/Mfn2−/− (Mfn-DKO) MEFs were purchased from American Type Culture Collection (Manassas, VA). Generation, selection, and induction of stable MEF cell lines were performed as reported previously (30). Transfections were performed using Lipofectamine 2000 or Lipofectamine LTX (Invitrogen) according to the manufacturer's instructions. Cells were incubated with 50 nm MitoTracker Red CMXRos (Molecular Probes), 50 nm MitoTracker Deep Red (Molecular Probes), and 10 μg/ml Hoechst 33258 (Sigma). A/C Heterodimerizer (Clontech) was used at a final concentration of 250 nm for 3 h at 37 °C.
Immunofluorescence Assays
Immunofluorescence assays were performed as described previously (25). Briefly, cells were fixed in 4% (w/v) paraformaldehyde in PBS (pH 7.4) and incubated for 60 min at room temperature with primary antibody. Primary antibodies were labeled for 20 min at room temperature with Alexa Fluor 488-, Alexa Fluor 568-, or Alexa Fluor 647-conjugated anti-rabbit or anti-mouse (Molecular Probes) or FITC-conjugated anti-rabbit or anti-mouse (Sigma) secondary antibodies.
Electron Microscopy
Control, MiD49-induced, and MiD51-induced MEFs were treated with or without 100 nm 4-hydroxytamoxifen (4-OHT) for 72 h. Following protein induction, cytochemical staining for catalase was conducted as described previously (31). Briefly, cells were fixed in 4% paraformaldehyde, 0.05% glutaraldehyde, 2.5% sucrose, 3 mm CaCl2, and 100 mm Hepes (pH 7.4) for 1 h. The fixed cells were then washed and stored in PBS before incubation in 2 mg/ml 3, 3′-diaminobenzidine, 0.15% H2O2, and 100 mm glycine/NaOH (pH 10.5) at 37 °C for 1 h. Cells were dehydrated in successive washes of 70, 90, and 100% ethanol before embedding in resin and analysis by transmission electron microscopy (32).
PEG Cell Fusion Assay
The PEG cell fusion assay was conducted as reported previously (33, 34). Briefly, cells expressing either mito-EGFP or mito-DsRed were seeded onto coverslips at a 1:1 ratio. The following day, the cells were treated with cycloheximide (30 μg/ml) for 30 min prior to and at all times following PEG-mediated cell fusion. Cells were fused for 60 s with 50% PEG 1500 (Fluka) and washed thoroughly with PBS containing 30 μg/ml cycloheximide. Cells were then grown for 7 h in medium containing 30 μg/ml cycloheximide and subsequently fixed with 4% paraformaldehyde. Mfn-DKO MEFs expressing MiD49 were treated with 100 nm 4-OHT 24 h prior to PEG cell fusion.
Microscopy
Epifluorescence microscopy was conducted using an Olympus IX8I microscope equipped with an F-view2 camera and processing using Soft System SIS (Olympus). Confocal microscopy was performed with a Zeiss confocal microscope equipped with a ConfoCor 3 system containing an avalanche photodiode detector. Green fluorescence was detected using an argon laser, red fluorescence was detected using a DPSS laser, and deep red fluorescence was detected using a helium/neon laser. All images were processed using ImageJ (http://rsbweb.nih.gov/ij/index.html), Zeiss and/or MetaMorph software (Visitron Systems), ZEN lite 2011 (Blue edition, Zeiss), or Imaris imaging software (Bitplane AG). Average peroxisome length from confocal images was calculated by determining the volume of each individual peroxisome and the average peroxisome diameter using Imaris imaging software. The length of each peroxisome was then calculated from the volume according to volume = length × width × height. Analysis of peroxisome number was determined using MetaMorph software. Briefly, peroxisome images were converted into skeleton pictures, and the average length or number quantified. The method used to determine Drp1 fluorescence intensity at peroxisomes was adapted from Ref. 23. Briefly, images were binarized to select peroxisomes and subtracted from the Drp1 signal. These images were subsequently subtracted from the original Drp1 signal, producing a peroxisomal Drp1 image. The average fluorescence intensity per cell was then analyzed by MetaMorph software and normalized to the wild-type control. This method was also modified to determine cytosolic versus total Drp1 fluorescence. Briefly, images were thresholded and binarized for mitochondria and subtracted from the Drp1 signal, resulting in an image containing cytosolic Drp1. Measurements of fluorescence intensity of both the total and cytosolic Drp1 signals were obtained per cell. Analysis of colocalization following PEG fusion assay and A/C heterodimerization was conducted as described previously (25). Briefly, following a 3-h treatment with A/C Heterodimerizer or 24 h following PEG fusion, cells were fixed in 4% paraformaldehyde. Single plane images were obtained on a Zeiss 510 confocal microscope and analyzed by MetaMorph software. Colocalization statistics were obtained from 30–35-μm line scans of images and are presented as Pearson correlation units (r). Images in all experimental groups were obtained with the same settings, except for detector gain adjustments that were performed to normalize saturation levels.
Mitochondrial Treatments and Western Blotting
Tris/Tricine/SDS-PAGE was performed as described previously (35). ECL chemiluminescent substrate (Amersham Biosciences) was used to detect immunoreactive proteins in blots.
RESULTS
MiD49 and MiD51 Overexpression Induces Mfn1/2-dependent Mitochondrial Elongation
It has recently been proposed that MiD51 actively promotes mitochondrial fusion (26). However, these findings were largely based on the observation that mitochondria still appear fused in MiD51-overexpressing cells following knockdown of the fusion mediator Mfn2. Given that Mfn1 is also involved in mitochondrial fusion, we sought to determine how the complete absence of the mitochondrial fusion apparatus would influence MiD-induced mitochondrial morphology. To this end, we addressed whether MiD49 or MiD51 expression could promote mitochondrial fusion in MEFs lacking genes encoding Mfn1 and/or Mfn2. Stable cell lines were generated in which expression of MiD49 was induced by the addition of 4-OHT (25), and Western blot analysis confirmed the expression of MiD49 in these cells (Fig. 1A). Fluorescence microscopy revealed that up-regulation of MiD49 in wild-type MEFs induced the appearance of elongated mitochondria as seen by the presence of a largely interconnected network (Fig. 1, B and C) as reported previously (25). Interestingly, MiD49 expression in MEFs lacking Mfn1 or Mfn2 also resulted in the appearance of elongated mitochondria (Fig. 1, B and C). It was only in cells lacking both Mfn1 and Mfn2 (Mfn-DKO) that mitochondrial elongation was blocked following MiD49 up-regulation (Fig. 1, B and C), resulting in no change in mitochondrial phenotype from the uninduced fragmented morphology, as described by Chen et al. (33, 34). These results indicate that at least one mitofusin is required to drive the elongation phenotype seen upon MiD49 overexpression. Identical results were observed following transient overexpression of MiD51 in these Mfn1- and/or Mfn2-deficient cell lines (data not shown). Our results are consistent with previous reports showing that mitochondrial fusion requires a complement of mitofusins on opposing organelles so that membrane tethering can be facilitated (33, 34, 36).
FIGURE 1.
Induction of MiD49 and MiD51 expression causes mitochondrial elongation by Drp1 inactivation and mitofusin-dependent elongation. A, MiD49-induced wild-type, Mfn1−/−, Mfn2−/−, and Mfn-DKO MEFs were treated with 4-OHT for 0–48 h to induce MiD49 expression. Following induction of MiD49, whole cell lysates were subjected to SDS-PAGE and analyzed by Western blotting with antibodies against Mfn1, Mfn2, human MiD49, and β-actin. B, following induction of MiD49 for either 0 or 24 h with 4-OHT (4HT), cells were stained with MitoTracker Red (red) and Hoechst (blue). The dashed boxes in the left panels are enlarged to enhance visibility in the right panels (Inset). Scale bars = 20 μm. C, Mfn1−/−, Mfn2−/−, and Mfn-DKO cells were treated with or without 4-OHT to induce MiD49 protein expression before being subjected to assessment of mitochondrial phenotypes (mean ± S.E. (n = 3), 100 cells counted per experiment). D, Mfn2−/− MEFs were transfected with plasmids encoding GFP-Drp1K38A, MiD49-GFP, MiD51-GFP, mito-GFP, or GFP-Drp1. Cells were stained with MitoTracker Red (red) and Hoechst (blue). Scale bars = 20 μm. E, cells from D were blind-counted for mitochondrial morphology (mean ± S.E. (n = 3), 100 cells counted per experiment).
From our previous data (25), we interpreted that mitochondrial elongation following MiD49/51 overexpression is due to sequestration of Drp1 at mitochondria, which blocks fission and leads to unopposed fusion. To look at this in a different way, we expressed the dominant-negative mutant Drp1K38A in Mfn2−/− cells. As shown in Fig. 1 (D and E), expression of GFP-tagged Drp1K38A caused mitochondria to elongate in a manner similar to that seen following overexpression of GFP-tagged MiD49 or MiD51 in these cells. Expression of mito-GFP or GFP-Drp1 did not result in the appearance of elongated mitochondria, as expected (Fig. 1, D and E). Similar results were also observed in Mfn1−/− cells expressing Drp1K38A (data not shown). Given this, it is likely that overexpression of MiD49 or MiD51 indeed impairs the organization of the mitochondrial fission machinery by blocking Drp1 function. Because the mitofusins remain active, the mitochondria elongate.
It remains possible that MiD49/51 could tether with a mitofusin on an opposing organelle to drive mitochondrial fusion, similar to the tethering described between Mfn1 and Mfn2 (Fig. 2A) (33, 34, 36). We therefore assessed this using PEG-mediated cell fusion analysis, in which one set of cells expressed mito-GFP and another mito-DsRed (33, 34). During cell fusion, cycloheximide was added to block further expression of the fluorescent protein. The addition of cycloheximide induces transient mitochondrial hyperfusion even in Mfn2−/− cells and, to a lesser extent, in Mfn1−/− cells, as reported previously (37). Fusion of mitochondria originating from distinct cells could be seen by the mixing of red and green mitochondrial contents to produce a yellow merged image (Fig. 2, A and B). Such an event was seen with combinations of wild-type MEFs (Fig. 2B) and single Mfn knock-out cells fused together (Fig. 2C). However, fusion of distinct mitochondrial populations was not observed when cells lacking both mitofusins were employed (Fig. 2, B and C), as described previously (33). Next, we performed cell fusion experiments to determine whether overexpressed MiD proteins could complement the lack of mitofusins (Fig. 2, A and B). When Mfn-DKO MEFs overexpressing MiD49 or MiD51 and mito-EGFP were fused to wild-type, Mfn1−/−, or Mfn2−/− MEFs expressing mito-DsRed, no mitochondrial fusion was observed (Fig. 2, B and C). From these results, we conclude that MiD49 or MiD51 cannot actively promote fusion with either Mfn partner and therefore cannot replace the function of either Mfn1 or Mfn2.
FIGURE 2.
MiD49- and MiD51-induced mitochondrial elongation is Mfn1/2-dependent. A, schematic depicting a potential model of MiD49/51 involvement in mitochondrial fusion. Mitochondrial mixing following fusion of a cell expressing mito-GFP with another cell expressing mito-DsRed can be detected by the presence of colocalization (upper panel, yellow). In the absence of both Mfn1 and Mfn2, fusion cannot occur in mitochondria due to the lack of tethering (middle panel). This method was used to test whether MiD49/51 overexpression could act in a manner similar to Mfn1 or Mfn2 in mitochondrial tethering (lower panel). B, PEG fusion assay of MEFs expressing mito-EGFP or mito-DsRed. Cell hybrids were formed from the fusion of wild-type to wild-type (WT × WT) cells, Mfn-DKO to Mfn1−/− or Mfn2−/− MEFs, and MiD49- or MiD51-induced Mfn-DKO cells to WT, Mfn1−/−, or Mfn2−/− MEFs. Cells were induced to express MiD49 or MiD51 for 24 h prior to cell fusion. The dashed boxes in the left panels are magnified in the right panels (Inset). Scale bars = 20 μm. g, green; r, red. C, colocalization of mito-GFP and mito-DsRed following PEG-mediated cell fusion was analyzed by line scans measuring relative fluorescence intensity. The degree of mitochondrial fusion is shown as Pearson correlation units (r; mean ± S.E., n = >10 cells/condition).
To support their role in fission, we performed time lapse imaging of a COS-7 cell cotransfected with MiD51-GFP and mito-DsRed. At very early stages of MiD51-GFP overexpression, mitochondrial morphology was normal, and fission events were indeed observed (see the supplemental movie). At longer time points when GFP expression became greater, the mitochondria became extensively elongated. Coupled with our previous RNAi results showing that knockdown of MiD49 and MiD51 causes a fission block (25) and recent findings from other groups (23, 38, 39), we conclude that the levels of the MiD proteins are precisely regulated to ensure that mitochondrial fission ensues.
MiD49/51 Overexpression Induces Peroxisome Elongation
Drp1 has a dual role in both mitochondrial and peroxisomal fission, with the formation of Drp1 puncta at regions of organelle division (40–43). Likewise, Fis1 and Mff are also found at peroxisomes, and in mammals, they are suggested to work with the peroxin PEX11 to facilitate Drp1-dependent fission (19, 20, 44). Because MiD49 and MiD51 recruit Drp1 to the mitochondrial surface, we investigated whether they are also involved in peroxisomal fission.
In contrast to Fis1 and Mff, MiD51 was not targeted to peroxisomes (Fig. 3A). MiD49 was similarly not found at peroxisomes (data not shown). This suggests that the MiD proteins have a primary role in mitochondrial dynamics. Following extended overexpression of MiD51, the length of peroxisomes markedly increased in comparison with wild-type cells (Fig. 3, B and C). This was also seen in cell sections analyzed by electron microscopy and stained for catalase to visualize peroxisomes (Fig. 3D). The non-continuous staining along the entire length of the peroxisome may be attributed to the thin sectioning required for electron microscopy. In both cases, the extended morphology of peroxisomes appeared even more striking than the mitochondrial elongation. There were no obvious morphological changes seen for the cis-Golgi, lysosomes, or endoplasmic reticulum following MiD51 overexpression (data not shown). With increased time of MiD51 induction, not only was there an increase in peroxisome length compared with wild-type MEFs (Fig. 3B), but also a concomitant reduction in the number of peroxisomes (Fig. 3E). The observed increase in peroxisome length and reduction in peroxisome number following MiD51 induction appeared similar to that observed following Drp1 knock-out or knockdown (31, 42, 45, 46), consistent with our findings that MiD49/51 overexpression renders Drp1 nonfunctional.
FIGURE 3.
Elongated peroxisomes are formed following induction of MiD51 expression. A, HeLa cells expressing GFP-Fis1, GFP-Mff, or MiD51-GFP were immunostained with antibodies against PEX14 to visualize peroxisomes (red). Cells were visualized by confocal microscopy. The dashed boxes in the left panels are enlarged to enhance visibility in the Inset panels. Arrows identify peroxisomes showing clear colocalization with GFP-Fis1 and GFP-Mff but not MiD51-GFP. Scale bars = 10 μm. B, wild-type and MiD51-induced MEFs were treated with 4-OHT for 0–72 h. Confocal images of cells immunostained with antibodies against PEX14 were analyzed, and average peroxisome length was determined (mean ± S.E. (n = 3), >15 images/experiment/condition) and normalized to uninduced control cells. C, MEFs before (−) and 72 h after (+) 4-OHT treatment to induce MiD51 expression were immunostained with antibodies against cytochrome c (mitochondria; red) and PEX14 (peroxisomes; green). Cells were analyzed by confocal microscopy. The dashed boxes in the merge panels are enlarged in the inset panels. Scale bars = 20 μm. D, peroxisomes in wild-type and MiD51-induced MEFs were analyzed by electron microscopy. Peroxisomes were visualized by staining for catalase. The dashed boxes in the upper panels are magnified in the lower panels (Inset). Asterisks indicate mitochondria, and arrows indicate peroxisomes. Scale bars = 2 μm. E, the average peroxisome number in wild-type and MiD51-induced MEFs before and after 4-OHT treatment was determined (mean ± S.E. (n = 3), >15 cells/experiment/condition).
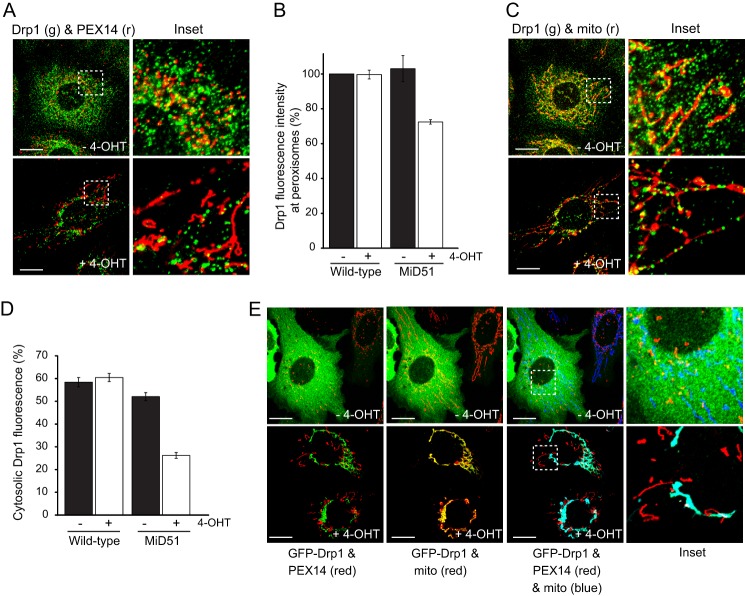
To confirm that peroxisome elongation is due to loss of Drp1 at peroxisomes, MiD51-uninduced (without 4-OHT) and MiD51-induced (with 4-OHT) MEFs were analyzed for Drp1 subcellular localization by immunofluorescence. Following induction of MiD51, endogenous Drp1 was reduced at peroxisomes compared with control cells, and peroxisomes adopted an elongated morphology (Fig. 4, A and B). In the same cells, Drp1 predominantly colocalized with mitochondria in discrete regions (Fig. 4C). The loss of cytosolic Drp1 could be observed upon MiD51 expression (Fig. 4, C and D). These results suggest that peroxisome elongation occurs following extended MiD51 induction due to the reduction of Drp1 association with peroxisomes. Even upon overexpression of Drp1 (fused to GFP), the signal for GFP-Drp1 was not observed at peroxisomes following MiD51 up-regulation (with 4-OHT) and instead was almost exclusively mitochondrial (Fig. 4E). Thus, ectopic expression of Drp1 is incapable of reversing the fused peroxisome morphology (Fig. 4E).
FIGURE 4.
Induction of MiD51 expression sequesters mitochondrial Drp1 and blocks peroxisomal fission. A, MiD51-induced MEFs were treated with or without 4-OHT for 72 h. Cells were then fixed and immunostained for peroxisomes and Drp1 (Drp1 (green, g) and PEX14 (red, r)). The dashed boxes in the left panels are magnified in the right panels (Inset). B, the intensity of Drp1 fluorescence at peroxisomes was quantified and normalized to the wild-type control (mean ± S.E. (n = 3), > 10 cells/experiment/condition). C, MiD51-induced MEFs were treated with or without 4-OHT for 72 h. Cells were then fixed and immunostained for Drp1 (green) and mitochondria (Tom20; red). D, the amount of cytosolic Drp1 relative to the total was quantified (mean ± S.E. (n = 3), > 9 cells/experiment/condition). E, MiD51-induced MEFs were transfected with plasmids encoding GFP-Drp1 (green) and then treated with or without 4-OHT. Cells were stained with MitoTracker Deep Red to visualize mitochondria and immunostained for peroxisomes (PEX14; red). Scale bars = 20 μm.
MiD49 and MiD51 Can Recruit Drp1 Independently of Fis1 and Mff
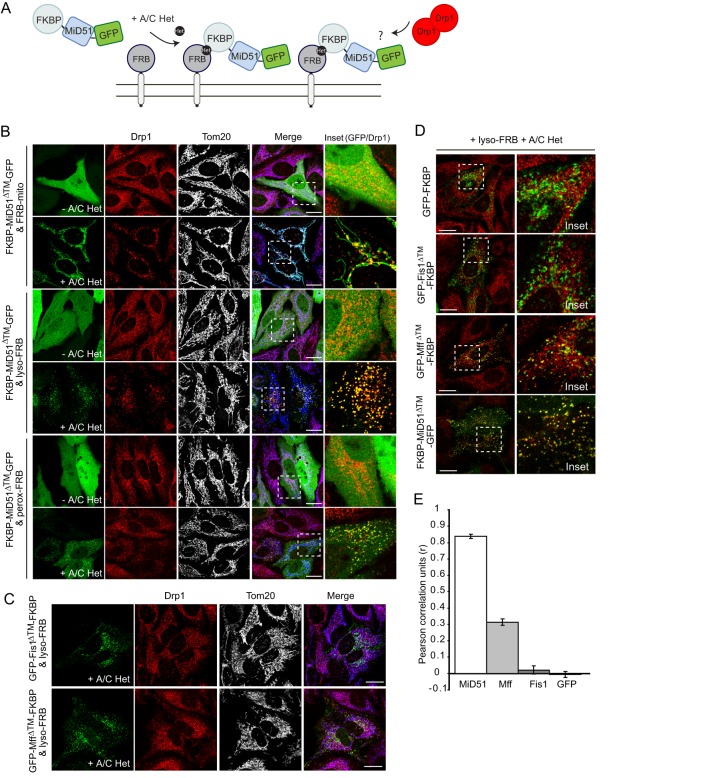
Because MiD49 and MiD51 are not found at peroxisomes with Mff and Fis1, it is possible that MiD49/51 may function independently of these proteins at mitochondria. To assess this, we asked whether MiD49/51 could recruit Drp1 to an independent organelle. We used a heterodimerization assay system based on the FKBP and FRB domains, which form a complex in the presence of the rapamycin analog A/C Heterodimerizer (28, 47). The cytosolic domain of MiD51 lacking its N-terminal transmembrane anchor (MiD51ΔTM) was fused to FKBP along with GFP, whereas the FRB domain was fused to membrane anchors of proteins targeted to the surface of lysosomes (lyso-FRB), peroxisomes (perox-FRB), and, as a control, mitochondria (FRB-mito) (Fig. 5A). The GFP moiety has previously been shown not to hinder MiD51 recruitment of Drp1 (25). Each FRB construct was cotransfected into HeLa cells with the GFP-tagged FKBP-MiD51ΔTM construct. In the absence of A/C Heterodimerizer, GFP-tagged FKBP-MiD51ΔTM was found in the cytosol and nucleus (Fig. 5B). When cells were treated with A/C Heterodimerizer for 3 h, the protein was redirected to the appropriate organelle based on the targeting of the FRB fusion construct as expected (complete targeting of FKBP fusion proteins to peroxisomes was not seen) (28). Immunostaining revealed that in all cases, Drp1 was redirected from the cytosol to the organelle decorated with FKBP-MiD51ΔTM-GFP (Fig. 5B). Although the time frame was short, some cells also showed the appearance of elongated mitochondria in A/C Heterodimerizer-treated cells, in which Drp1 became sequestered (Fig. 5B). As the MiD51 cytosolic domain can recruit Drp1 to lysosomes, we conclude that other mitochondrial outer membrane proteins such as Mff and Fis1 are not required for this activity. We next asked whether Fis1 and Mff have the ability to independently recruit Drp1 in a similar way as MiD51. The soluble domains of Fis1 or Mff (both lacking their individual C-terminal transmembrane anchors) were expressed as FKBP fusions along with GFP and targeted to lysosomes via expression of lyso-FRB and A/C Heterodimerizer treatment (Fig. 5C). Efficient targeting could be seen, and mitochondrial morphologies appeared normal. However, in neither case was strong Drp1 recruitment observed (Fig. 5C). Close inspection of Drp1 recruitment (Fig. 5D) and quantification of colocalization (Fig. 5E) revealed that there was no significant positive correlation of Drp1 recruitment when GFP or GFP-Fis1 was targeted to lysosomes. Some recruitment of Drp1 could be observed when Mff was directed to lysosomes, consistent with a previous report that plasma membrane-targeted Mff can recruit Drp1 (22). In contrast, the recruitment of Drp1 to lysosomes was highly effective when MiD51 was directed to the organelle (Fig. 5E). We conclude that MiD51 can recruit Drp1 to a membrane surface independently of other known components of the fission machinery.
FIGURE 5.
MiD51ΔTM can recruit Drp1 to mitochondria, lysosomes, and peroxisomes. A, schematic of MiD51ΔTM-GFP translocation to subcellular organelles via heterodimerization. The FKBP domain at the N terminus of MiD51 is stimulated to interact with the membrane-localized FRB domain upon treatment with the rapamycin analog A/C Heterodimerizer (A/C Het). B, in the presence of A/C Heterodimerizer, cytosolic FKBP-MiD51ΔTM-GFP was directed to the mitochondrial (FRB-mito; upper panels), lysosomal (lyso-FRB; middle panels), and peroxisomal (perox-FRB; lower panels) outer membranes. Cells were immunostained for Drp1 (red) and mitochondria (Tom20; gray/blue). The dashed boxes in the Merge panels are magnified in the Inset panels, with GFP and Drp1 (red) shown. C, GFP-Fis1ΔTM-FKBP and GFP-MffΔTM-FKBP in the presence of lyso-FRB were directed to lysosomes following treatment with A/C Heterodimerizer. Cells were immunostained for Drp1 (red) and Tom20 (gray/blue). D, HeLa cells expressing GFP-FKBP, GFP-Fis1 ΔTM-FKBP, GFP-Mff ΔTM-FKBP, or FKBP-MiD51ΔTM-GFP together with lyso-FRB were treated with A/C Heterodimerizer and immunostained for Drp1 (red). The dashed boxes in the left panels are magnified in the right panels (Inset). Scale bars = 20 μm. E, line scans of images from D were analyzed for colocalization of Drp1 and GFP as described by Pearson correlation units (r; mean ± S.E. of three independent experiments, n > 55 measurements, two measurements/cell).
DISCUSSION
MiD49 and MiD51 Recruit Drp1 Independently of Additional Mitochondrial Proteins
In recent years, many studies have reported mechanisms by which Drp1 is regulated to exert its membrane constriction and fission activity. One of the major steps is the recruitment of Drp1 to the appropriate organelle and membrane region. In animals, this is achieved by a number of membrane receptor/effector proteins, including Fis1, Mff, MiD49, and MiD51. The involvement of Fis1 in Drp1 function may be indirect, as a number of reports have shown that it is not required for Drp1 recruitment in cultured mammalian cell lines (22, 48–50). In yeast, it is now established that Fis1 plays a part in scaffolding the soluble Drp1 effector protein Mdv1 at the mitochondrial outer membrane rather than engaging directly with Drp1 (51–54). Although Mdv1 appears to be confined to yeast, other Drp1 receptor/effector proteins are found in animals and plants. How they specifically function in mitochondrial fission is still not known.
Recently, Chan and co-workers (23) reported a study of mitochondrial dynamics in MEFs lacking Fis1 and/or Mff. The authors found some increase in mitochondrial elongation in contrast to a Fis1 knock-out human cell line (22, 23). This was more evident in the Mff knock-out cell line, whereas Fis1/Mff double knock-out MEFs were found to have strongly fused mitochondrial networks and reduction in Drp1 recruitment to mitochondria (23). In addition, the authors found that MiD49 and MiD51 overexpression in these cells alleviated the Drp1 recruitment defect (23). Here, we have confirmed this work and extended it by demonstrating that when MiD51 was mistargeted to other organelles, it could also directly recruit Drp1, suggesting that additional mitochondrial outer membrane proteins are not required for this activity. During the preparation of this manuscript, Shaw and co-workers (38) also demonstrated this by expressing these proteins in yeast. Interestingly, the degree by which lysosome-targeted MiD51 recruited Drp1 was greater compared with lysosome-targeted Fis1 or Mff.
Our analysis of MiD49 or MiD51 overexpression revealed that the appearance of an elongated mitochondrial network is due to sequestration and inactivation of Drp1 at the mitochondrial outer membrane, which causes a fission block, thereby leading to unopposed fusion events. In addition, it was recently found that MiD49/51 overexpression causes the accumulation of the inactive phosphorylated form of Drp1 (at Ser-637) (23). Whether this is a result of Drp1 sequestration or a direct effect caused by MiD49/51 action remains to be determined. We have also shown, that fusion events are strictly dependent on the presence of at least one mitofusin. The evidence that MiD49 and MiD51 are involved in mitochondrial fission rather than fusion is further supported by our previous study (25) and by recent publications from other groups (23, 38).
MiD49 and MiD51 May Specifically Regulate Mitochondrial Fission
The reduction in Drp1-mediated fission was most profoundly observed by analyzing peroxisome morphology in MiD49/51-overexpressing cells, with peroxisomal extensions representing a block in fission events due to reduced levels of Drp1 at the peroxisomal surface. Related to this, we found that unlike Mff and Fis1, MiD49 and MiD51 are not targeted to peroxisomes. The mechanism of organelle promiscuity for Fis1 and Mff most likely lies in the fact that they have C-terminal anchors. A number of reports have found that the degree of hydrophobicity, charge, and length of C-terminal anchors can alter targeting (55–58). In addition, the ability of Fis1 and Mff to interact with proteins of the PEX11 family may facilitate their localization to peroxisomes (59, 60). In contrast, MiD49 and MiD51 have an N-terminal membrane anchor, and although the mechanism of targeting is not clear, the sequence is highly conserved, suggesting that this is important for organelle selection. Moreover, we found that the MiD49/51 transmembrane anchors were not required for Drp1 recruitment activity.
Although Drp1 mediates both peroxisomal and mitochondrial fission, there is no evidence to suggest that the activities are linked, and hence, independently regulated mechanisms are required. The formation of mitochondrial constriction sites appears to be important for mitochondrial fission, with the wrapping of the endoplasmic reticulum and actin polymerization at the mitochondrial surface playing a role in this process (29, 61). Based on the results presented here, it is also likely that MiD49 and MiD51 confer a level of specificity in recruiting Drp1 to the mitochondrial surface independently of Fis1 and Mff.
Supplementary Material
Acknowledgments
We gratefully acknowledge Charles Ferguson for technical assistance. We acknowledge use of the facilities of the Australian Microscopy & Microanalysis Research Facility at the Centre for Microscopy and Microanalysis of The University of Queensland.
This work was supported in part by grants from the National Health and Medical Research Council (NHMRC) and the Australian Research Council (ARC).

This article contains a supplemental movie.
- MEF
- mouse embryonic fibroblast
- Mfn2
- mitofusin 2
- Mfn1
- mitofusin 1
- FRB
- FKBP12/rapamycin-binding
- FKBP
- FK506-binding protein
- Mfn-DKO
- Mfn1/Mfn2−/− double knock-out
- 4-OHT
- 4-hydroxytamoxifen
- EGFP
- enhanced GFP
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Elgass K., Pakay J., Ryan M. T., Palmer C. S. (2013) Recent advances into the understanding of mitochondrial fission. Biochim. Biophys. Acta 1833, 150–161 [DOI] [PubMed] [Google Scholar]
- 2. Westermann B. (2010) Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 11, 872–884 [DOI] [PubMed] [Google Scholar]
- 3. Chan D. C. (2012) Fusion and fission: interlinked processes critical for mitochondrial health. Annu. Rev. Genet. 46, 265–287 [DOI] [PubMed] [Google Scholar]
- 4. Baloh R. H., Schmidt R. E., Pestronk A., Milbrandt J. (2007) Altered axonal mitochondrial transport in the pathogenesis of Charcot-Marie-Tooth disease from mitofusin 2 mutations. J. Neurosci. 27, 422–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li Z., Okamoto K., Hayashi Y., Sheng M. (2004) The importance of dendritic mitochondria in the morphogenesis and plasticity of spines and synapses. Cell 119, 873–887 [DOI] [PubMed] [Google Scholar]
- 6. Shirendeb U., Reddy A. P., Manczak M., Calkins M. J., Mao P., Tagle D. A., Reddy P. H. (2011) Abnormal mitochondrial dynamics, mitochondrial loss and mutant huntingtin oligomers in Huntington's disease: implications for selective neuronal damage. Hum. Mol. Genet. 20, 1438–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Osellame L. D., Blacker T. S., Duchen M. R. (2012) Cellular and molecular mechanisms of mitochondrial function. Best Pract. Res. Clin. Endocrinol. Metab. 26, 711–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gomes L. C., Di Benedetto G., Scorrano L. (2011) During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 13, 589–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Twig G., Elorza A., Molina A. J., Mohamed H., Wikstrom J. D., Walzer G., Stiles L., Haigh S. E., Katz S., Las G., Alroy J., Wu M., Py B. F., Yuan J., Deeney J. T., Corkey B. E., Shirihai O. S. (2008) Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 27, 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rambold A. S., Lippincott-Schwartz J. (2011) Mechanisms of mitochondria and autophagy crosstalk. Cell Cycle 10, 4032–4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Okamoto K., Kondo-Okamoto N. (2012) Mitochondria and autophagy: critical interplay between the two homeostats. Biochim. Biophys. Acta 1820, 595–600 [DOI] [PubMed] [Google Scholar]
- 12. Mears J. A., Lackner L. L., Fang S., Ingerman E., Nunnari J., Hinshaw J. E. (2011) Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat. Struct. Mol. Biol. 18, 20–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ingerman E., Perkins E. M., Marino M., Mears J. A., McCaffery J. M., Hinshaw J. E., Nunnari J. (2005) Dnm1 forms spirals that are structurally tailored to fit mitochondria. J. Cell Biol. 170, 1021–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yoon Y., Pitts K. R., McNiven M. A. (2001) Mammalian dynamin-like protein DLP1 tubulates membranes. Mol. Biol. Cell 12, 2894–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yoon Y., Krueger E. W., Oswald B. J., McNiven M. A. (2003) The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol. Cell. Biol. 23, 5409–5420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. James D. I., Parone P. A., Mattenberger Y., Martinou J. C. (2003) hFis1, a novel component of the mammalian mitochondrial fission machinery. J. Biol. Chem. 278, 36373–36379 [DOI] [PubMed] [Google Scholar]
- 17. Stojanovski D., Koutsopoulos O. S., Okamoto K., Ryan M. T. (2004) Levels of human Fis1 at the mitochondrial outer membrane regulate mitochondrial morphology. J. Cell Sci. 117, 1201–1210 [DOI] [PubMed] [Google Scholar]
- 18. Zhang X. C., Hu J. P. (2008) FISSION1A and FISSION1B proteins mediate the fission of peroxisomes and mitochondria in Arabidopsis. Mol. Plant 1, 1036–1047 [DOI] [PubMed] [Google Scholar]
- 19. Kobayashi S., Tanaka A., Fujiki Y. (2007) Fis1, DLP1, and Pex11p coordinately regulate peroxisome morphogenesis. Exp. Cell Res. 313, 1675–1686 [DOI] [PubMed] [Google Scholar]
- 20. Koch A., Yoon Y., Bonekamp N. A., McNiven M. A., Schrader M. (2005) A role for Fis1 in both mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell 16, 5077–5086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Scott I., Tobin A. K., Logan D. C. (2006) BIGYIN, an orthologue of human and yeast FIS1 genes functions in the control of mitochondrial size and number in Arabidopsis thaliana. J. Exp. Bot. 57, 1275–1280 [DOI] [PubMed] [Google Scholar]
- 22. Otera H., Wang C., Cleland M. M., Setoguchi K., Yokota S., Youle R. J., Mihara K. (2010) Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J. Cell Biol. 191, 1141–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Losón O. C., Song Z., Chen H., Chan D. C. (2013) Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol. Biol. Cell 24, 659–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schrader M., Bonekamp N. A., Islinger M. (2012) Fission and proliferation of peroxisomes. Biochim. Biophys. Acta 1822, 1343–1357 [DOI] [PubMed] [Google Scholar]
- 25. Palmer C. S., Osellame L. D., Laine D., Koutsopoulos O. S., Frazier A. E., Ryan M. T. (2011) MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep. 12, 565–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhao J., Liu T., Jin S., Wang X., Qu M., Uhlén P., Tomilin N., Shupliakov O., Lendahl U., Nistér M. (2011) Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. EMBO J. 30, 2762–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhao J., Lendahl U., Nistér M. (2013) Regulation of mitochondrial dynamics: convergences and divergences between yeast and vertebrates. Cell. Mol. Life Sci. 70, 951–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lazarou M., Jin S. M., Kane L. A., Youle R. J. (2012) Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Dev. Cell 22, 320–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Friedman J. R., Lackner L. L., West M., DiBenedetto J. R., Nunnari J., Voeltz G. K. (2011) ER tubules mark sites of mitochondrial division. Science 334, 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dunning C. J. R., McKenzie M., Sugiana C., Lazarou M., Silke J., Connelly A., Fletcher J. M., Kirby D. M., Thorburn D. R., Ryan M. T. (2007) Human CIA30 is involved in the early assembly of mitochondrial complex I and mutations in its gene cause disease. EMBO J. 26, 3227–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wakabayashi J., Zhang Z., Wakabayashi N., Tamura Y., Fukaya M., Kensler T. W., Iijima M., Sesaki H. (2009) The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J. Cell Biol. 186, 805–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Howes M. T., Kirkham M., Riches J., Cortese K., Walser P. J., Simpson F., Hill M. M., Jones A., Lundmark R., Lindsay M. R., Hernandez-Deviez D. J., Hadzic G., McCluskey A., Bashir R., Liu L., Pilch P., McMahon H., Robinson P. J., Hancock J. F., Mayor S., Parton R. G. (2010) Clathrin-independent carriers form a high capacity endocytic sorting system at the leading edge of migrating cells. J. Cell Biol. 190, 675–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen H., Chomyn A., Chan D. C. (2005) Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J. Biol. Chem. 280, 26185–26192 [DOI] [PubMed] [Google Scholar]
- 34. Chen H., Detmer S. A., Ewald A. J., Griffin E. E., Fraser S. E., Chan D. C. (2003) Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 160, 189–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schägger H., von Jagow G. (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379 [DOI] [PubMed] [Google Scholar]
- 36. Hoppins S., Edlich F., Cleland M. M., Banerjee S., McCaffery J. M., Youle R. J., Nunnari J. (2011) The soluble form of Bax regulates mitochondrial fusion via MFN2 homotypic complexes. Mol. Cell 41, 150–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tondera D., Grandemange S., Jourdain A., Karbowski M., Mattenberger Y., Herzig S., Da Cruz S., Clerc P., Raschke I., Merkwirth C., Ehses S., Krause F., Chan D. C., Alexander C., Bauer C., Youle R., Langer T., Martinou J. C. (2009) SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J. 28, 1589–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Koirala S., Guo Q., Kalia R., Bui H. T., Eckert D. M., Frost A., Shaw J. M. (2013) Interchangeable adaptors regulate mitochondrial dynamin assembly for membrane scission. Proc. Natl. Acad. Sci. U.S.A. 110, E1342–E1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Otera H., Ishihara N., Mihara K. (2013) New insights into the function and regulation of mitochondrial fission. Biochim. Biophys. Acta 1833, 1256–1268 [DOI] [PubMed] [Google Scholar]
- 40. Smirnova E., Griparic L., Shurland D. L., van der Bliek A. M. (2001) Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell 12, 2245–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Smirnova E., Shurland D. L., Ryazantsev S. N., van der Bliek A. M. (1998) A human dynamin-related protein controls the distribution of mitochondria. J. Cell Biol. 143, 351–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li X., Gould S. J. (2003) The dynamin-like GTPase DLP1 is essential for peroxisome division and is recruited to peroxisomes in part by PEX11. J. Biol. Chem. 278, 17012–17020 [DOI] [PubMed] [Google Scholar]
- 43. Koch A., Thiemann M., Grabenbauer M., Yoon Y., McNiven M. A., Schrader M. (2003) Dynamin-like protein 1 is involved in peroxisomal fission. J. Biol. Chem. 278, 8597–8605 [DOI] [PubMed] [Google Scholar]
- 44. Gandre-Babbe S., van der Bliek A. M. (2008) The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell 19, 2402–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Waterham H. R., Koster J., van Roermund C. W., Mooyer P. A., Wanders R. J., Leonard J. V. (2007) A lethal defect of mitochondrial and peroxisomal fission. N. Engl. J. Med. 356, 1736–1741 [DOI] [PubMed] [Google Scholar]
- 46. Koch A., Schneider G., Lüers G. H., Schrader M. (2004) Peroxisome elongation and constriction but not fission can occur independently of dynamin-like protein 1. J. Cell Sci. 117, 3995–4006 [DOI] [PubMed] [Google Scholar]
- 47. Narendra D. P., Jin S. M., Tanaka A., Suen D. F., Gautier C. A., Shen J., Cookson M. R., Youle R. J. (2010) PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8, e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Onoue K., Jofuku A., Ban-Ishihara R., Ishihara T., Maeda M., Koshiba T., Itoh T., Fukuda M., Otera H., Oka T., Takano H., Mizushima N., Mihara K., Ishihara N. (2013) Fis1 acts as a mitochondrial recruitment factor for TBC1D15 that is involved in regulation of mitochondrial morphology. J. Cell Sci. 126, 176–185 [DOI] [PubMed] [Google Scholar]
- 49. Lee Y. J., Jeong S. Y., Karbowski M., Smith C. L., Youle R. J. (2004) Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Mol. Biol. Cell 15, 5001–5011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wasiak S., Zunino R., McBride H. M. (2007) Bax/Bak promote sumoylation of DRP1 and its stable association with mitochondria during apoptotic cell death. J. Cell Biol. 177, 439–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Karren M. A., Coonrod E. M., Anderson T. K., Shaw J. M. (2005) The role of Fis1p-Mdv1p interactions in mitochondrial fission complex assembly. J. Cell Biol. 171, 291–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Koirala S., Bui H. T., Schubert H. L., Eckert D. M., Hill C. P., Kay M. S., Shaw J. M. (2010) Molecular architecture of a dynamin adaptor: implications for assembly of mitochondrial fission complexes. J. Cell Biol. 191, 1127–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Naylor K., Ingerman E., Okreglak V., Marino M., Hinshaw J. E., Nunnari J. (2006) Mdv1 interacts with assembled Dnm1 to promote mitochondrial division. J. Biol. Chem. 281, 2177–2183 [DOI] [PubMed] [Google Scholar]
- 54. Zhang Y., Chan D. C. (2007) Structural basis for recruitment of mitochondrial fission complexes by Fis1. Proc. Natl. Acad. Sci. U.S.A. 104, 18526–18530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Borgese N., Brambillasca S., Colombo S. (2007) How tails guide tail-anchored proteins to their destinations. Curr. Opin. Cell Biol. 19, 368–375 [DOI] [PubMed] [Google Scholar]
- 56. Kaufmann T., Schlipf S., Sanz J., Neubert K., Stein R., Borner C. (2003) Characterization of the signal that directs Bcl-xL, but not Bcl-2, to the mitochondrial outer membrane. J. Cell Biol. 160, 53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kriechbaumer V., Shaw R., Mukherjee J., Bowsher C. G., Harrison A. M., Abell B. M. (2009) Subcellular distribution of tail-anchored proteins in Arabidopsis. Traffic 10, 1753–1764 [DOI] [PubMed] [Google Scholar]
- 58. Lan L., Isenmann S., Wattenberg B. W. (2000) Targeting and insertion of C-terminally anchored proteins to the mitochondrial outer membrane is specific and saturable but does not strictly require ATP or molecular chaperones. Biochem. J. 349, 611–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Koch J., Brocard C. (2012) PEX11 proteins attract Mff and human Fis1 to coordinate peroxisomal fission. J. Cell Sci. 125, 3813–3826 [DOI] [PubMed] [Google Scholar]
- 60. Koch J., Pranjic K., Huber A., Ellinger A., Hartig A., Kragler F., Brocard C. (2010) PEX11 family members are membrane elongation factors that coordinate peroxisome proliferation and maintenance. J. Cell Sci. 123, 3389–3400 [DOI] [PubMed] [Google Scholar]
- 61. Korobova F., Ramabhadran V., Higgs H. N. (2013) An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science 339, 464–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.