Abstract
The origin recognition complex (ORC) was first discovered in the baker’s yeast in 1992. Identification of ORC opened up a path for subsequent molecular level investigations on how eukaryotic cells initiate and control genome duplication each cell cycle. Twenty years after the first biochemical isolation, ORC is now taking on a three-dimensional shape, although a very blurry shape at the moment, thanks to the recent electron microscopy and image reconstruction efforts. In this chapter, we outline the current biochemical knowledge about ORC from several eukaryotic systems, with emphasis on the most recent structural and biochemical studies. Despite many species-specific properties, an emerging consensus is that ORC is a ATP-dependent machine that recruits other key proteins to form pre-Replicative Complexes (pre-RCs) at many origins of DNA replication, enabling the subsequent initiation of DNA replication in S phase.
Keywords: DNA replication, Replicative helicase loader, Eukaryotes, Replication initiators, ATPase, AAA+ protein, DNA binding, Origins of DNA replication, Origin Recognition Complex
Introduction
At the most fundamental level, the concept that at least one origin of DNA replication and multiple protein factors are required to initiate the physical process of genome duplication is conserved across the three domains of life (Kawakami and Katayama, 2010, Mendez and Stillman, 2003). In eukaryotes, an origin of DNA replication is a stretch of DNA sequence where the Origin Recognition Complex(Bell and Stillman, 1992) (ORC) binds and subsequently recruits other factors to establish a pre-replicative complex (pre-RC) (Bell and Dutta, 2002, Bielinsky and Gerbi, 2001, DePamphilis, 2003, Diffley and Labib, 2002, Kawakami and Katayama, 2010, Mendez and Stillman, 2003, Scholefield et al., 2011, Cocker et al., 1996). In budding yeast, the genetically defined replication origins are usually near and often overlap with the replication start sites (Marahrens and Stillman, 1992, Rao et al., 1994, Bielinsky and Gerbi, 1998, Brewer and Fangman, 1987, Theis and Newlon, 1994), but in mammalian species the start sites have not been genetically characterized in sufficient detail, but start sites have been reported new ORC binding sites (Abdurashidova et al., 2003, Bielinsky and Gerbi, 2001). In Drosophila, in which origins of DNA replication in a metazoan species been studied in most detail, ORC binding sites are located near actual sites of initiation of DNA replication and also correspond to start sites of transcription (Austin et al., 1999, Kim et al., 2011, Royzman et al., 1999, Xie and Orr-Weaver, 2008, Beall et al., 2002, Bielinsky et al., 2001, Chesnokov et al., 1999, Gossen et al., 1995, MacAlpine et al., 2010, Sher et al., 2012). Simple organisms such as virus and bacteria use a single origin of replication (Kawakami and Katayama, 2010). In eukaryotes, due to their vastly expanded genome size and the hierarchical structure of the chromosomes, there are hundreds to tens of thousands replication origins, depending on the organisms (Gilbert, 1998, Ryba et al., 2010, Cvetic and Walter, 2005, DePamphilis et al., 2006, Gilbert, 2010). The existence and utilization of the great number of origins is likely to ensure that the large genomes can be duplicated within a reasonable time frame and certainly within a single cell division cycle.
Aside from the fact that origins are located near to ORC binding sites and promote initiation of DNA replication, there is little consensus among eukaryotic species as to what constitutes an origin of replication. This is further complicated because ORC lacks sufficient DNA binding specificity to predict the location of origins of DNA replication, even in the budding yeasts where there is some sequence specificity to ORC binding (Chang et al., 2011). In this regard, the yeast S. cerevisiae is perhaps an exception rather than the normal. In S. cerevisiae, the replication origins are well defined and ScORC binds to the genetically defined origins, but even here the consensus sequence is rather variable (Chang et al., 2011, Marahrens and Stillman, 1992, Theis and Newlon, 2001, Bell and Stillman, 1992). The S. cerevisiae origins of replication are autonomously replicating sequences (ARS), 100 – 150 bp long, and constitute three of four elements termed A, B1, and B2, with an auxiliary element in some origins called B3 (Marahrens and Stillman, 1992). Element A contains the AT-rich 11-bp ARS consensus sequence (ACS) that is the most conserved and A and B1 contribute to ORC binding specificity (Bell and Stillman, 1992, Deshpande and Newlon, 1992, Van Houten and Newlon, 1990). The B2 element contains the double strand DNA unwinding element (DUE) where DNA replication starts and this element is required for loading the pre-RC and DNA helicase component MCM2-7 [mini-chromosome maintenance subunits 2-7] (Zou and Stillman, 2000, Wilmes and Bell, 2002). B3 is an accessary sequence 22-bp long and at the ARS1 origin binds the transcription factor Abf1 (Marahrens and Stillman, 1992). There are several hundred origins in S. cerevisiae and they all share the same ACS sequence and the general three-element architecture. In other eukaryotic organisms such as Schizosaccharomyces pombe, Drosophila melanogaster, Xenopus laevis, and Homo sapiens, the origin sequence pattern is not so well defined, except for the fact that they are generally contain AT rich sequences. It is clear now that, in these organisms, certain features outside ORC may be more important than ORC in defining replication origins. These additional determinants may include the local chromatin structure such as nucleosome positioning (Aggarwal and Calvi, 2004, Calvi et al., 2007, Chang et al., 2011, Eaton et al., 2010, Lipford and Bell, 2001, MacAlpine et al., 2010, Zou et al., 2006), chromatin modifications (Eaton et al., 2011, Liu et al., 2012, Weber et al., 2008, Hartl et al., 2007), transcription regulation (Karnani et al., 2010, MacAlpine et al., 2004), extra protein or RNA partners (Norseen et al., 2008, Thomae et al., 2008, Bartke et al., 2010, Shen et al., 2010), and potentially the physical properties, such as the rigidity or the malleability of the DNA fragment (Cao et al., 2008, Huang and Kowalski, 1993, Natale et al., 1993).
In contrast to the great divergence in the number and sequence of the eukaryotic replication origins, ORC, the ATP-dependent molecular machine that binds to those origins and helps to execute DNA replication, is well conserved throughout evolution, at least at the amino acid sequence level (Gavin et al., 1995, Tugal et al., 1998, Speck et al., 2005, Clarey et al., 2006). There are, however, considerable differences between species with regard to the stability and composition of ORC subunits during the cell division cycle, a topic that is addressed below with a discussion of selected species. Furthermore, ORC has functions and activities well beyond DNA replication (Sasaki and Gilbert, 2007). Here we limit our discussion to the principal role of ORC in replication initiation, although a summary of ORC activities and structures is summarized at the end of this review.
ORC is composed of six protein subunits Orc1-6, named initially in the yeast according to their molecular masses, with Orc1 being the largest subunit (120 kDa) and Orc6 the smallest (50 kDa) (Bell et al., 1995, Bell and Stillman, 1992). ORC subunits in other eukaryotes are named according to their function and amino acid sequence conservation with their yeast counterparts (Fig 1). ORC is an ATPase and its binding to origin DNA is usually ATP dependent (Klemm et al., 1997, Lee and Bell, 2000, Speck et al., 2005, Takenaka et al., 2004, Bell and Stillman, 1992). Despite the overall conservation, ORC has sufficiently evolved to warrant us to discuss ORC from different species individually. In the following sections, we will briefly survey four ORCs from S. cerevisiae, S. pombe, D. melanogaster, and H. sapiens, outlining some biochemical and structural studies haven been reported.
Figure 1.
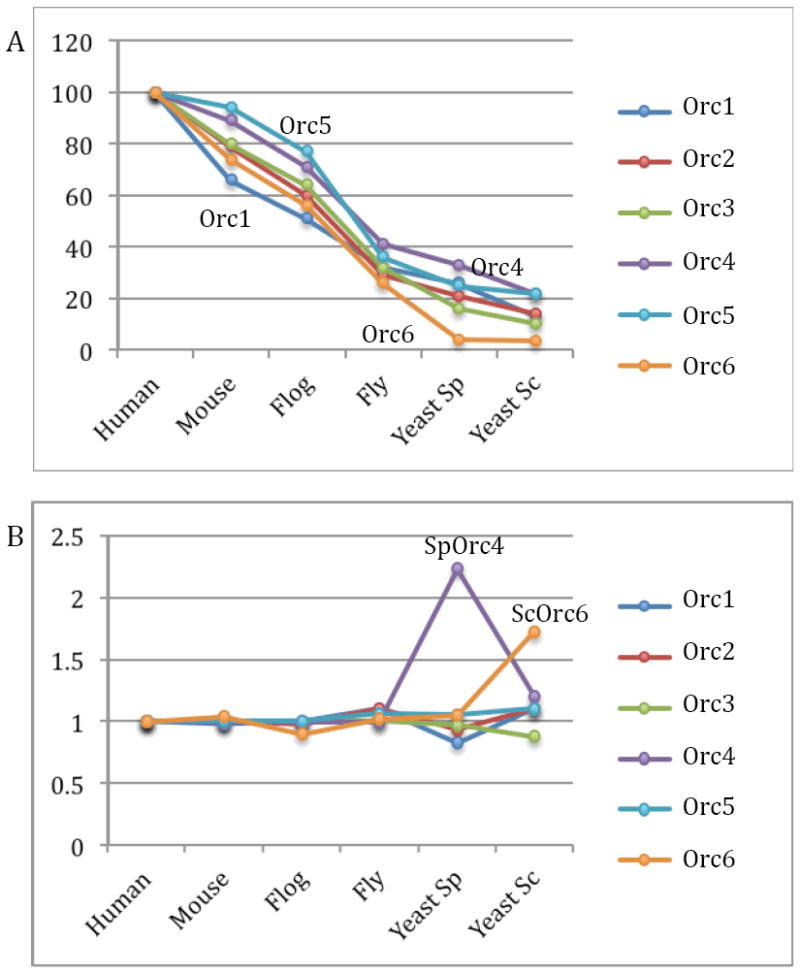
Conservation of the six ORC subunits among six selected eukaryotic species. (A) Sequence identity of Orc1 through Orc6 as compared to the corresponding human subunits. (B) The relative amino acid sequence length of Orc1 to Orc6 with that of the human subunit set to be 1.
The S. cerevisiae ORC
The most distinctive feature of ScORC is that it forms a hetero-hexamer consisting of the Orc1, Orc2, Orc3, Orc4, Orc5 and Orc6 subunits, forming a stable throughout the cell division cycle that constitutively binds all of the origins of replication (Gibson et al., 2006). ScORC binds in a DNA sequence specific manner, although compared to sequence specific transcription factors, ScORC has low binding affinity for its highly variable recognition site, which consists primarily of the A and B1 elements of the double strand DNA origins (Rao and Stillman, 1995, Rowley et al., 1995). ScORC exhibits a high affinity for single strand DNA (ssDNA) (Kd ≈10-8) in a sequence nonspecific manner and without the requirement for ATP, as long as the ssDNA is longer than 80 bases (Lee et al., 2000, Clarey et al., 2006, Lee and Bell, 1997, Speck et al., 2005)
Among the six subunits, the first five subunits, Orc1 through Orc5, all contain a predicted AAA+ domain, and are essential for DNA binding, although only Orc1, Orc2, Orc4, and Orc5 appear to have direct contact to the origin DNA (Clarey et al., 2006, Lee and Bell, 1997, Speck et al., 2005). This suggests that Orc3 may function to glue the DNA-contacting subunits together as a stable complex, but not bind DNA directly. Orc6 is the only subunit that does not contain a predicted AAA+ domain, and does not bind DNA. However, Orc6 is an essential subunit for DNA replication because Orc6 recruits multiple Cdt1 molecules during repeated loading of the replicative helicase core the Mcm2-7 hexamer (Asano et al., 2007, Chen and Bell, 2011, Chen et al., 2007, Takara and Bell, 2011).
The largest subunit Orc1 is unique among ORC subunits, because it has a N-terminal 235-residue bromo-adjacent homology (BAH) domain that interacts with the C-terminal region of silencing regulator Sir1 (Zhang et al., 2002, Bose et al., 2004, Fox et al., 1997, Gardner et al., 1999, Hou et al., 2005, Ozaydin and Rine, 2010, Triolo and Sternglanz, 1996). The BAH domain is not essential, but its presence in Orc1 can influence origin binding specificity, as it alos does in human cell ORC (Muller et al., 2010, Noguchi et al., 2006). This latter specificity may be related to the known nucleosome binding properties of the BAH domain present in the Orc1-related protein called Sir3 (Armache et al., 2011, Hickman and Rusche, 2010). Sir3 is a regulator of the silent mating type genes in yeast and maintains certain mating type gene loci transcriptionally silent. The BAH domain of Orc1 binds to the Sir1 protein, which is also required for efficient silencing of the silent mating type loci. In the absence of Sir1 or the Orc1 BAH domain, different epigenetic states of the mating type gene expression are established (Bell et al., 1993, Bell et al., 1995, Pillus and Rine, 1989, Pillus and Rine, 2004, Zhang et al., 2002). The crystal structure of the Sir3 AAA+ domain has recently been determined and although it does not have an ATPase activity (unlike the Orc1 AAA+ domain), the Sir3 AAA+ domain has evolved to bind to the partner Sir4 silent information regulator protein and to chromatin containing non-methylated H3K79 residues (Ehrentraut et al., 2011). Thus, after duplication of the Orc1 gene, the Sir3 allele evolved to acquire diverse biochemical functions for an AAA+ protein, even though it retained the same overall structural features of the AAA+ domain.
Both Orc1 and Orc5 can bind ATP (Klemm et al., 1997, Klemm and Bell, 2001, Takehara et al., 2008), but the ATPase activity of ScORC primarily resides in the Orc1 subunit, and relies on the presence of an arginine finger in Orc4 (Bowers et al., 2004, Randell et al., 2006). Orc1 has a predicted classic AAA+ ATPase domain, with functional Walker A and Walker B motifs and the ATPase activity controlled by the insertion of an arginine residue present in the Orc4 subunit into the active site of the Orc1 ATP binding site (Bowers et al., 2004). Orc1 ATPase activity is required for loading of multiple subunits of the MCM2-7 hexamer and is blocked by origin-specific double-stranded DNA (Klemm et al., 1997, Randell et al., 2006). Thus the ATPase activity of ORC is required for DNA replication. Many ORC subunits, including Orc1 have a predicted winged-helix (WH) domain that may contribute to DNA binding (Clarey et al., 2006, Speck et al., 2005), just like the DnaA protein uses both its AAA+ and its Helix-turn-helix domains for DNA sequence specific DNA binding (Kawakami and Katayama, 2010).
ScORC is subject to cyclin dependent kinase (CDK) activity regulation and is a substrate for the cell cycle regulatory kinase (Nguyen et al., 2001, Weinreich et al., 2001, Wilmes et al., 2004). ScOrc2 and ScOrc6 are phosphorylated by CDK only during S and G2 phases of the cell cycle. Phosphorylation on Orc2 enhances Orc5 to bind to ATP (Makise et al., 2009). Phosphorylation of Orc6 by CDK prevents it from interacting with Cdt1, thus regulating the Mcm2-7 helicase loading (Chen and Bell, 2011). CDK phosphorylation sites in ORC can be altered without much phenotypic consequence, unless additional sites in Cdc6 and MCM2-7 are simultaneously altered, leading to over-replication of DNA from certain origins of DNA replication in the genome (Nguyen et al., 2001).
The Orc1 subunit, in addition to having a primary amino acid sequence related to Sir3, is also highly related to Cdc6. In fact many archaea species only have a single Orc1/Cdc6 protein that binds to the origin in a DNA sequence-dependent manner (see Chapter 4) and has amino acid sequence similarity to both Orc1 and Cdc6 (Capaldi and Berger, 2004, Duncker et al., 2009, Gaudier et al., 2007, Tada et al., 2008). In S. cerevisiae, the Cdc6 protein is an AAA+ ATPase that is required for MCM2-7 loading onto the pre-RC, but then Cdc6 is degraded by ubiquitin-mediated proteolysis at the G1 to S phase transition, dependent on the activation of the S-phase cyclin-CDKs (Clb5-Cdc28 and Clb6-Cdc28) (Cocker et al., 1996, Liang et al., 1995, Santocanale and Diffley, 1996, Duncker et al., 1999, Perkins et al., 2001, Drury et al., 2000, Drury et al., 1997, Piatti et al., 1996). The destruction of Cdc6, while leaving ORC intact, is part of the mechanism that ensures that the pre-RC cannot be reassembled in S and G2 phases, thereby limiting the initiation of DNA replication to once per cell division.
Recent electron microscopic (EM) studies of the intact ScORC revealed a bipartite structure about 12 nm wide and 16 nm long (Fig 2) (Speck et al., 2005). Assuming that DNA binds along the length of the protein complex, the length of the ScORC structure is sufficient to interact with the observed 48-bp DNA fragment in the presence of ATP, as observed in the DNase I footprint assay (Bell and Stillman, 1992, Speck and Stillman, 2007). Systematic subunit mapping with a maltose binding protein fused at the N-terminus or the C-terminus of the individual ORC subunits showed that Orc1, 4, and 5 are located to one domain of the bi-globular structure, whereas Orc2 and Orc3 are located at the other end (Chen et al., 2008). Therefore, the most likely arrangement of the first five subunits is Orc1-Orc4-Orc5-Orc3-Orc2, with Orc1 and Orc2 at the two extremes of the pseudo-ringed structure. Such an architecture is consistent with in vivo and in vitro subunit interaction assays revealing direct contact between Orc2 and Orc3, and between Orc4 and Orc5, between Orc1 and Orc4, and between Orc4-5 and Orc2-3 (Chen et al., 2008, Matsuda et al., 2007). The physical position of Orc6 was mapped by comparing the EM structures of the intact ORC and Orc1-5 sub-complex missing Orc6 (Chen et al., 2008). In the 3D difference density map, two density peaks were identified: a larger one at the lower Orc2-3 lobe and a smaller one near the upper Orc1-4-5 lobe. This observation suggests that Orc6 binds mainly with Orc2-3 at the lower lobe but reaches up to Orc1-4-5 lobe. A weak interaction between Orc6 and Orc5 was indeed found in the yeast two-hybrid analysis (Matsuda et al., 2007).
Figure 2.
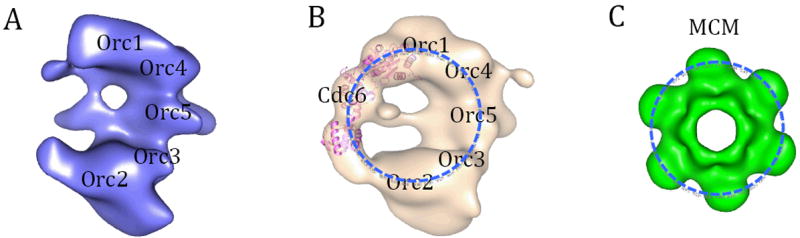
EM Structures of ScORC (A); ORC-Cdc6 (B). The approximate locations of the ORC subunits are marked based on the observed MBP location on ORC complex with MBP fused to N- or C-terminus of subunits, one subunit and one terminus at a time (Chen et al., 2008). Cdc6 binding to the left side of ORC, forming a ring-like feature as illustrated by a dashed blue circle. The crystal structure of an archaeal Cdc6 ortholog (PDB ID 1FNN) is docked into the EM density assigned to ScCdc6, and shown by pink ribbos. (C) The low-pass filtered crystal structure of the hexameric N-terminal domain of an archaeal MCM (PDB ID 1LTL). Figure modified from (Speck et al., 2005).
The S. cerevisiae Cdc6 is another important replication initiation factor beyond ORC. It is predicted to contain an AAA+ domain, including the DNA-binding initiator-specific motif (ISM), and the DNA-binding winged helix domain, features well conserved among known replication initiators (Dueber et al., 2011, Dueber et al., 2007, Gaudier et al., 2007). In the absence of Cdc6, ORC is merely an origin binder and cannot load the MCM2-7 complex. In order to establish a pre-RC at an origin, ORC has to be transformed from a passive origin binder or origin marker to the active replicative helicase loader. It appears that Cdc6 throws a molecular switch in ORC that enables such transformation (Lee and Bell, 2000, Speck et al., 2005). Cdc6 binding to ORC in vitro introduces an extended pre-RC-like footprint on several replication origins. Formation of the Pre-RC signature DNA footprint is dependent on specific origin sequence, and the intact ATPase activity of ORC as well as that of Cdc6 (Speck et al., 2005, Speck and Stillman, 2007). The ATP hydrolysis activity of ORC is inhibited upon binding to the specific origin DNAs, and the ATP hydrolysis activity of Cdc6 is suppressed upon binding to ORC that is bound on specific origin DNAs. Presumably, the ATP molecules in the ORC-Cdc6-DNA assembly are preserved for use in the subsequent recruitment and loading of MCM2-7 helicase that are likely to be a series of energy-requiring molecular events (Evrin et al., 2009, Remus et al., 2009, Lee and Bell, 2000, Randell et al., 2006). EM studies showed that Cdc6 binds to the side of the bipartite ORC, in close contact with Orc1, forming an apparently ring-like feature in the complex (Fig. 2). The ring-like feature may function as the landing pad for the ring-shaped MCM2-7 helicase core complex. Since MCM2-7 is first loaded on the dsDNA rather than ssDNA (Evrin et al., 2009, Remus et al., 2009), it is unlikely that ScORC will melt the dsDNA. Therefore, the function of initial dsDNA melting will then have to be executed by the replicative helicase.
Following the determination of the structure of ORC and the ORC-Cdc6 structure using transmission electron microscopy, a recent study has shown a higher resolution structure of ORC-Cdc6 bound to an origin DNA [ARS1] (Sun et al., 2012). This structure shows that upon binding Cdc6 in a ATP-dependent manner, the BAH domain of Orc1 moves considerably to the back of the ring-like complex and new density, most likely the Orc6 subunit, protrudes to the front of the complex. Orc6 has been shown to bind to two copies of the Cdt1 protein and thus the major conformational change in Orc1 may facilitate the binding of two Ctd1-MCM2-7 heptamers to the origin DNA. Although the DNA was not visible in the cryo-EM structure of ORC-Cdc6-DNA complex, modeling of the archaeal Orc1/Cdc6 crystal structure into the individual ORC-Cdc6 subunits allows the prediction that the DNA forms a highly bent conformation within the ORC-Cdc6 complex, explaining the extended protection from nuclease in DNA footprinting experiments. The bending of the DNA is also consistent with other EM studies of ORC bound to DNA (Chastain et al., 2004).
The S. pombe ORC
It was thought that SpORC, like ScORC, remains bound to chromatin throughout the cell cycle (DePamphilis, 2005, Kong and DePamphilis, 2002). But a recent study showed that SpORC behaves much like the metazoan ORC rather than ScORC and binds replication origin periodically during the cell cycle, with the binding peaking at M to G1 transition stage (Wu and Nurse, 2009). Like in the budding yeast, SpORC, SpCdc18 and the SpCdt1 are required fro MCM loading and pre-RC assembly upon exit from mitosis (Kearsey et al., 2000, Kong and DePamphilis, 2002, Moon et al., 1999, Nishitani and Nurse, 1997, Ogawa et al., 1999, Takahashi et al., 2003).
SpOrc1, 2, and 5 subunits are highly conserved with their counterparts from S. cerevisiae (Fig. 1) (Moon et al., 1999). SpOrc4 is unique among ORC proteins in that it has an N-terminal extension containing nine AT-hook motifs that are not found in budding yeast or metazoan Orc4 homologs (Chuang and Kelly, 1999). The SpOrc4 AT hooks specifically bind the minor groove of AT-rich DNA tracts, and are necessary and sufficient for the DNA binding activity of SpORC (Chuang et al., 2002, Gaczynska et al., 2004, Lee et al., 2001). It appears that the SpOrc4 AT hooks are solely responsible for the DNA binding activity of the entire SpORC complex, as deletion of the Orc4 AT hooks not only abolish the DNA binding of Orc4, but that of the SpORC as well (Gaczynska et al., 2004). However, the AT-hook mediated initial binding of SpORC to origin DNA is salt sensitive, and this interaction is gradually converted to a salt-stable binding state in which the topology of origin DNA is changed into a negatively-supercoiled or under-wound state (Houchens et al., 2008). In agreement with this suggested DNA topology change, an atomic force microscopy measurement of SpORC bound to the autonomously replicating sequence 1 containing DNA fragments revealed shortening of the DNA length by 140 bp, a length sufficient for wrapping around SpORC by two turns (Gaczynska et al., 2004). SpCdc18, the homolog of ScCdc6, interacts with Cdt1 and together they further enhance the binding stability of SpORC on origin DNA, as if the SpCdc18-Cdt1 binary complex is an additional origin determinant in S. pombe (Kelly et al., 1993, Houchens et al., 2008). No structural characterization of the purified SpORC has been reported so far. Because of the unique AT hooks in SpOrc4, the mechanism of origin recognition of SpORC may be different from that of other eukaryotic systems.
The D. melanogaster ORC
DmORC can be isolated from a Drosophila embryo nuclear extract as a stable complex (Gossen et al., 1995). But the presence of DmORC is cell cycle dependent, and is regulated by the degradation of Orc1 via the ubiquitin proteasome pathway at the late M phase (Araki et al., 2003). DmOrc1 is resynthesized during late G1-phase. DmORC is also an ATPase, and like ScOrc1, DmOrc1 is essential for ATP hydrolysis and for ATP-dependent DNA binding (Chesnokov et al., 2001). Different from ScOrc6 that is not required for DNA binding, the DmOrc6 is required for the DNA binding of DmORC and it is an integral part of the complex (Chesnokov et al., 2001). The DmOrc6 alone has DNA binding activity, likely due to the predicted TFIIB-like DNA binding domain in the smallest subunit (Liu et al., 2011). Mutations to the predicted DNA binding region abolish its DNA binding activity (Liu et al., 2011). DmOrc6 contains a C-terminal domain that is important for cytokinesis and binds to the septin protein that mediates closure of the cytokinesis furrow at the end of cell division, and this feature seems to be conserved among metazoans (Chesnokov et al., 2003, Huijbregts et al., 2009, Prasanth et al., 2002).
Under EM, DmORC is an elongated structure with dimension of 170 Å by 115 Å, similar to ScORC (Clarey et al., 2006) (Fig. 3). The notable feature of DmORC is a spiral crescent that encompasses a 25-Å channel. Because there has been no biochemical or structural reports on the subunit arrangement, it is not clear if DmORC shares the same architecture with ScORC. But the overall dimension and basic shape of the EM reconstruction of the two protein complexes appear to be comparable, although not exactly the same. Similar features include the open-ring and the middle location of Orc5 that was mapped in ScORC by MBP-fusion approach, and in DmORC by a specific antibody (Fig. 3B).
Figure 3.
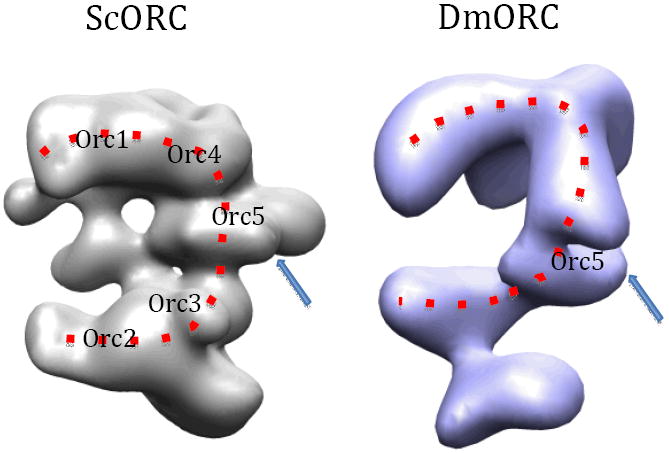
Comparison of EM structures of ScORC (EM Data Bank ID: EMD-1156) and DmORC (EMD-4820) reveals the similar basic overall architecture, including the opened ring feature outlined by a dashed curve, and the middle location of Orc5 in both ORC complexes. But the difference in the structural details is also obvious.
DmORC binds DNA with little sequence specificity. DmORC localizes to open chromatin regions that are depleted of nucleosomes (MacAlpine et al., 2004, MacAlpine et al., 2010). Interestingly, DmORC binds the negatively supercoiled DNA 30-fold better than a linear or relaxed DNA and therefore DmORC may target the topology rather than the sequence of the origin DNA (Remus et al., 2004). Because of the lack of sequence specificity, it has been unclear how DmORC binds origin DNA: which subunit does or does not directly contact DNA or what is the size of the DNA footprint of DmORC. An AFM study showed shortening of a linear DNA by ~130 bp upon DmORC binding. This length is similar to the 140-bp DNA shortening by the SpORC. Therefore, DmORC may also wrap around DNA, much like the SpORC (Clarey et al., 2006).
DmORC also is subject to CDK-mediated phosphorylation (Baldinger and Gossen, 2009). DmOrc1 and DmOrc2 each contain several phosphorylation sites. Hyper-phosphorylation at these sites does not affect the integrity of DmORC, but prevents DmORC from binding to DNA (Remus et al., 2005). This is different from the yeast ORC where phosphorylation does not appear to affect their DNA binding activity. At the single molecule EM level, the hyper-phosphorylated DmORC is structurally indistinguishable from the dephosphorylated version (Fig. 3) (Clarey et al., 2006). Therefore, it can be concluded that phosphorylation does not introduce substantial conformational changes in DmORC. To reconcile the phosphorylation-induced interference with the DNA binding, one could imagine that extensive phosphorylation will significantly alter the physicochemical property of the surface of DmORC, even in the absence of large structural changes. And this may interfere with DmORC interaction with the DNA.
The location of DmORC to the entire genome has been reported using a chromatin-immunoprecipitation assay and mapping the associated DNA fragments using microarrays or deep sequencing (Calvi et al., 2007, MacAlpine et al., 2004, MacAlpine et al., 2010, Spradling, 1999). This association occurs as cells exit mitosis (Baldinger and Gossen, 2009). DmORC is associated with regions of the genome enriched with the histone H3 variant H3.3 that is associated with transcribed regions and indeed DmORC is associated with transcription initiation sites (MacAlpine et al., 2010). The complex is also associated with origins of DNA replication that are amplified within a single cell division cycle during embryonic development, notably for the production of egg shell proteins in the follicle cells (Austin et al., 1999, Kim et al., 2011, Kim and Orr-Weaver, 2011, Xie and Orr-Weaver, 2008). Here, DmORC interactions with chromatin occur in regions enriched with the transcription factor E2F (Bosco et al., 2001, Royzman et al., 1999). The E2F transcription factor is associated with a larger complex called the Myb–MuvB (MMB)/dREAM complex that contains the Myb related protein that is also required for DNA replication and is also associated with developmentally regulated gene expression (Georlette et al., 2007, Beall et al., 2002). Thus ORC may associate with specific chromatin structures that differ in different cell types, although this aspect of ORC binding has not been well investigated.
Mutations in DmORC subunits cause defects in DNA replication as expected, but cells are also observed to arrest in mitosis, although this has been attributed to DNA damage as a result of incomplete DNA replication (Chesnokov et al., 2001, Loupart et al., 2000, Pflumm and Botchan, 2001). DmORC subunits, however, localize to centromeric heterochromatin and also bind the HP1 protein that is associated with heterochromatin (Badugu et al., 2005, Huang et al., 1998, Pak et al., 1997, Shareef et al., 2003, Shareef et al., 2001).
The H. sapiens ORC
Human cells contain origins of DNA replication that are perhaps as many as, or even more than the number of genes in the genome; although their usage is not well understood (Falaschi et al., 2007). Only a little over a dozen origins have been studied with any depth and even these have not been well characterized. One of the first specific origins identified was the lamin B2 origin (Abdurashidova et al., 2000). The recombinant human Orc4 alone was shown to bind the lamin B2 origin of DNA in vitro and in the ATP-independent manner (Stefanovic et al., 2003), perhaps reminiscent of the SpOrc4 in the origin DNA binding capacity as an individual ORC subunit, although the human protein contains no discernible AT hooks. However it is unlikely that this biochemical interaction is functionally significant for DNA replication. Human ORC binds to the latent replication origin of Epstein-Barr virus in B cells where it is required for the maintenance of the EBV plasmid (Chaudhuri et al., 2001, Dhar et al., 2001b, Julien et al., 2004). ORC also has been reported to bind in HeLa S3 cells to intergenic AT-rich regions (Ladenburger et al., 2002) and to the DBF4 promoter locus that is an efficient replication origin (Romero and Lee, 2008). This origin of replication contains two initiation zones and two ORC binding sites that are approximately 400 bp apart. The two-zone origin appears to promote a novel mode of asymmetric bidirectional replication at the DBF4 origin. The recombinant and purified human ORC has intrinsic DNA binding activity with preference for AT-rich sequences (Vashee et al., 2003). The purified human ORC seems to be capable of promoting the initiation of DNA replication from any DNA sequence in vitro, with no preference for human origin sequences, when DNA is added to ORC-depleted Xenopus egg extracts (Vashee et al., 2003). This specificity of initiation of DNA replication reflects the lack of DNA sequence specificity for DNA injected into activated Xenopus eggs (Harland and Laskey, 1980) and may say more about the nature of the egg system then origin specificity in human cells.
The six subunits of human ORC were first identified by sequence similarity to their yeast counterparts ((Dhar et al., 2001a, Dhar and Dutta, 2000, Gavin et al., 1995, Siddiqui and Stillman, 2007, Tugal et al., 1998, Vashee et al., 2001); See also fig. 1). The assembly of ORC and the stability of the complex are both ATP dependent. In the absence of ATP, the full complex doesn’t assemble, and in the absence of ATP the assembled complex is so fragile that it can’t survive the glycerol gradient fractionation (Ranjan and Gossen, 2006). ATP can be replaced by ATPγS for the purpose of assembly or maintenance of structural integrity, suggesting ATP is a structural cofactor for ORC assembly. The assembly of human ORC is a stepwise process in vitro: First, Orc2 and Orc3 form a binary complex, then the binary complex recruits Orc5. The newly formed ternary complex subsequently recruits Orc4, forming the quaternary complex. Incorporation of Orc4 into the growing complexes is ATP dependent (Ranjan and Gossen, 2006, Siddiqui and Stillman, 2007). The Orc2-5 quaternary complex in turn recruits the Orc1. ATP binding by Orc1 is not essential for this step, but ATP binding of Orc4 is essential for assembly of both Orc1-5 and Orc2-5 (Siddiqui and Stillman, 2007). It is possible that Orc4 is physically located between Orc1 and Orc5, and ATP binding converts Orc4 into an assembly competent configuration with which Orc1 can interact from one side, and Orc5 interacts from the other. Mutations in ATP binding sites of Orc4 and Orc5 impair complex assembly. Thus human ORC is unique in that ATP is not only required for the function in replication initiation, but also in the assembly and stability of the complex. Although the ATP binding motif in Orc1, Orc4, and Orc5 subunits are important for replication activity (Giordano-Coltart et al., 2005), the ATPase activity of human ORC is largely contained in Orc1.
The HsORC is a very dynamic complex in vivo and indeed the period of the cell division cycle during which ORC exists as a complete complex may be temporally restricted to G1 phase (Kreitz et al., 2001, Mendez et al., 2002, Tatsumi et al., 2003). The HsOrc1 subunit is degraded at the G1 to S phase transition in a Skp2-ubiquitin ligase-dependent manner, only to be re-appear as cells enter mitosis (Mendez et al., 2002, Tatsumi et al., 2003). The Orc2, Orc3, Orc4, Orc5 and Orc6 subunits are displaced form chromatin as cells progress through S phase, but a heterodimer of Orc2 and Orc3 remain bound to the centromere and function during mitosis (Craig et al., 2003, Prasanth et al., 2004, Siddiqui and Stillman, 2007). Thus the HsORC is a very dynamic complex with respect to the cell division cycle. It has also been reported to be associated with centrosomes and there it controls the cyclin E-CDK2-dependent duplication of centrioles (Hemerly et al., 2009). Orc6 and Orc1 each contain their own NLS, and are targeted to nucleus independently of Orc2-5. Such a mechanism allows for the formation of different sub-complex for the different functions (Ghosh et al., 2011). Additional proteins other than ORC subunits, such as the WD-repeat protein ORCA (LRWD1) may enhance the human ORC binding to origins and facilitate pre-RC assembly (Bartke et al., 2010, Chakraborty et al., 2011, Shen et al., 2010, Vermeulen et al., 2010). The LRWD1 protein is interesting since it is associated with repressive marks on histone H3, such as H3K9 and H3K27 methylation, suggesting that the ORC may recognize specialized chromatin structures via histone modifications.
Overexpressing all subunits yields only Orc1-5 sub-complex, with Orc6 only loosely associated with the complex. The human Orc6 joins Orc1-5 only at the G1/M phase, and dissociates from Orc1-5 in the S phase. Orc6 contains a middle domain with a structure similar to the helical domain of the TFIIB transcription factor (Liu et al., 2011). Interestingly, this middle domain of Orc6 binds to dsDNA. Orc6 is shown to interact directly with Orc3, as result that is different to the interaction between S. cerevisiae Orc2 and Orc6 (Siddiqui and Stillman, 2007). Other than that, the subunit interaction pattern of the human ORC appears to be similar to that of the ScORC, suggesting a conserved ORC architecture across evolution.
A prominent feature of human ORC is the transient association of both Orc1 and Orc6 with the Orc2-5 quaternary core complex (Dhar et al., 2001a, Siddiqui and Stillman, 2007, Vashee et al., 2003, Vashee et al., 2001). In addition to sites that correspond to origins of DNA replication, ORC also binds to heterochromatin and interacts with the heterochromatin protein HP1 (Chakraborty et al., 2011, Duncker et al., 2009, Lidonnici et al., 2004, Prasanth et al., 2010, Wallace and Orr-Weaver, 2005). Human Orc1-5, together with HP1, localizes to heterochromatin, and may be involved in organizing higher order chromatin structure (Prasanth et al., 2010). Interestingly, the association of the Orc2, Orc3, Orc4 and Orc5 subunits at sites of heterochromatin are very dynamic, with a half-life of association in vivo of approximately 4-5 seconds. This dynamic association of ORC subunits is similar to the dynamic association of HP1 to heterochromatin. In stark contrast, the Orc1 subunit, once bound, is stably bound to heterochromatin, suggesting that this subunit has a distinct interaction with either DNA or HP1. Both the Orc1 and Orc3 subunits bind directly to HP1, perhaps explaining the different chromatin binding kinetics, but Orc1 must interact with other components of the chromatin other than HP1.
Future perspective
Crystal structures of archaeal ORC proteins in complex with DNA have provided tantalizing clues about how the eukaryotic ORC may interact with DNA (Berquist and DasSarma, 2003, Dueber et al., 2007, Duncker et al., 2009, Gaudier et al., 2007, Grainge et al., 2003, Wigley, 2009). But clearly, the crystal structure of ORC is a critical missing piece of information that when available will greatly advance the field. Many eukaryotic ORC have been expressed in various heterologous systems and then purified. It is hoped that the next several years may see a series of crystal structures of the eukaryotic replication initiators, likely in the form of individual ORC subunits or as the stable sub-complexes. However, given the large size, the dynamic nature, and interaction with extended stretch of DNA sequences, the structure of the entire ORC, and its further assembly with other replication factors, such as the replicative helicases, could be exceedingly difficult for such crystallographic approaches. We anticipate that single particle cryo-EM will continue to play an important role in elucidating the molecular mechanism of eukaryotic replication initiation. Indeed, the anticipated crystal structures of the individual subunits or sub-complexes will facilitate the interpretation of cryo-EM studies of the various replication initiation complexes, including ORC, Cdc6, Cdt1 bound to MCM2-7 and the MCM2-7 double hexamer. And without a doubt, EM and crystallographic observations will raise deeper mechanistic questions that will prompt more specific biochemical experiments.
Acknowledgments
Research in the authors’ labs is supported by grants from the National Institutes of Health, particularly CA13106 and GM45436.
Contributor Information
Huilin Li, Department of Biochemistry and Cell Biology, Stony Brook University, Stony Brook, NY 11794, USA, And, Biology Department, Brookhaven National Laboratory, Upton, NY 11973, USA, hli@bnl.gov, Tel: 631-344-2931, Fax: 631-344-3407.
Bruce Stillman, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY 11724, USA, bstillman@cshl.edu, Tel: 516-367-8383.
References
- Abdurashidova G, Deganuto M, Klima R, Riva S, Biamonti G, Giacca M, Falaschi A. Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science. 2000;287:2023–6. doi: 10.1126/science.287.5460.2023. [DOI] [PubMed] [Google Scholar]
- Abdurashidova G, Danailov MB, Ochem A, Triolo G, Djeliova V, Radulescu S, Vindigni A, Riva S, Falaschi A. Localization of proteins bound to a replication origin of human DNA along the cell cycle. The EMBO journal. 2003;22:4294–303. doi: 10.1093/emboj/cdg404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal BD, Calvi BR. Chromatin regulates origin activity in Drosophila follicle cells. Nature. 2004;430:372–6. doi: 10.1038/nature02694. [DOI] [PubMed] [Google Scholar]
- Araki M, Wharton RP, Tang Z, Yu H, Asano M. Degradation of origin recognition complex large subunit by the anaphase-promoting complex in Drosophila. The EMBO journal. 2003;22:6115–26. doi: 10.1093/emboj/cdg573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armache KJ, Garlick JD, Canzio D, Narlikar GJ, Kingston RE. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 A resolution. Science. 2011;334:977–82. doi: 10.1126/science.1210915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano T, Makise M, Takehara M, Mizushima T. Interaction between ORC and Cdt1p of Saccharomyces cerevisiae. FEMS yeast research. 2007;7:1256–62. doi: 10.1111/j.1567-1364.2007.00299.x. [DOI] [PubMed] [Google Scholar]
- Austin RJ, Orr-Weaver TL, Bell SP. Drosophila ORC specifically binds to ACE3, an origin of DNA replication control element. Genes & development. 1999;13:2639–49. doi: 10.1101/gad.13.20.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badugu R, Yoo Y, Singh PB, Kellum R. Mutations in the heterochromatin protein 1 (HP1) hinge domain affect HP1 protein interactions and chromosomal distribution. Chromosoma. 2005;113:370–84. doi: 10.1007/s00412-004-0324-2. [DOI] [PubMed] [Google Scholar]
- Baldinger T, Gossen M. Binding of Drosophila ORC proteins to anaphase chromosomes requires cessation of mitotic cyclin-dependent kinase activity. Molecular and cellular biology. 2009;29:140–9. doi: 10.1128/MCB.00981-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143:470–84. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall EL, Manak JR, Zhou S, Bell M, Lipsick JS, Botchan MR. Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature. 2002;420:833–7. doi: 10.1038/nature01228. [DOI] [PubMed] [Google Scholar]
- Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–34. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annual review of biochemistry. 2002;71:333–74. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Bell SP, Kobayashi R, Stillman B. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science. 1993;262:1844–9. doi: 10.1126/science.8266072. [DOI] [PubMed] [Google Scholar]
- Bell SP, Mitchell J, Leber J, Kobayashi R, Stillman B. The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell. 1995;83:563–8. doi: 10.1016/0092-8674(95)90096-9. [DOI] [PubMed] [Google Scholar]
- Berquist BR, DasSarma S. An archaeal chromosomal autonomously replicating sequence element from an extreme halophile, Halobacterium sp. strain NRC-1. Journal of bacteriology. 2003;185:5959–66. doi: 10.1128/JB.185.20.5959-5966.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielinsky AK, Gerbi SA. Discrete start sites for DNA synthesis in the yeast ARS1 origin. Science. 1998;279:95–8. doi: 10.1126/science.279.5347.95. [DOI] [PubMed] [Google Scholar]
- Bielinsky AK, Gerbi SA. Where it all starts: eukaryotic origins of DNA replication. Journal of cell science. 2001;114:643–51. doi: 10.1242/jcs.114.4.643. [DOI] [PubMed] [Google Scholar]
- Bielinsky AK, Blitzblau H, Beall EL, Ezrokhi M, Smith HS, Botchan MR, Gerbi SA. Origin recognition complex binding to a metazoan replication origin. Current biology : CB. 2001;11:1427–31. doi: 10.1016/s0960-9822(01)00444-4. [DOI] [PubMed] [Google Scholar]
- Bosco G, Du W, Orr-Weaver TL. DNA replication control through interaction of E2F-RB and the origin recognition complex. Nature cell biology. 2001;3:289–95. doi: 10.1038/35060086. [DOI] [PubMed] [Google Scholar]
- Bose ME, McConnell KH, Gardner-Aukema KA, Muller U, Weinreich M, Keck JL, Fox CA. The origin recognition complex and Sir4 protein recruit Sir1p to yeast silent chromatin through independent interactions requiring a common Sir1p domain. Molecular and cellular biology. 2004;24:774–86. doi: 10.1128/MCB.24.2.774-786.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JL, Randell JC, Chen S, Bell SP. ATP hydrolysis by ORC catalyzes reiterative Mcm2-7 assembly at a defined origin of replication. Molecular cell. 2004;16:967–78. doi: 10.1016/j.molcel.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–71. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- Calvi BR, Byrnes BA, Kolpakas AJ. Conservation of epigenetic regulation, ORC binding and developmental timing of DNA replication origins in the genus Drosophila. Genetics. 2007;177:1291–301. doi: 10.1534/genetics.107.070862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao XQ, Zeng J, Yan H. Structural properties of replication origins in yeast DNA sequences. Physical biology. 2008;5:036012. doi: 10.1088/1478-3975/5/3/036012. [DOI] [PubMed] [Google Scholar]
- Capaldi SA, Berger JM. Biochemical characterization of Cdc6/Orc1 binding to the replication origin of the euryarchaeon Methanothermobacter thermoautotrophicus. Nucleic acids research. 2004;32:4821–32. doi: 10.1093/nar/gkh819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty A, Shen Z, Prasanth SG. “ORCanization” on heterochromatin: linking DNA replication initiation to chromatin organization. Epigenetics : official journal of the DNA Methylation Society. 2011;6:665–70. doi: 10.4161/epi.6.6.16179. [DOI] [PubMed] [Google Scholar]
- Chang F, May CD, Hoggard T, Miller J, Fox CA, Weinreich M. High-resolution analysis of four efficient yeast replication origins reveals new insights into the ORC and putative MCM binding elements. Nucleic acids research. 2011;39:6523–35. doi: 10.1093/nar/gkr301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastain PD, 2nd, Bowers JL, Lee DG, Bell SP, Griffith JD. Mapping subunit location on the Saccharomyces cerevisiae origin recognition complex free and bound to DNA using a novel nanoscale biopointer. The Journal of biological chemistry. 2004;279:36354–62. doi: 10.1074/jbc.M403501200. [DOI] [PubMed] [Google Scholar]
- Chaudhuri B, Xu H, Todorov I, Dutta A, Yates JL. Human DNA replication initiation factors, ORC and MCM, associate with oriP of Epstein-Barr virus. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10085–9. doi: 10.1073/pnas.181347998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Bell SP. CDK prevents Mcm2-7 helicase loading by inhibiting Cdt1 interaction with Orc6. Genes & development. 2011;25:363–72. doi: 10.1101/gad.2011511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, de Vries MA, Bell SP. Orc6 is required for dynamic recruitment of Cdt1 during repeated Mcm2-7 loading. Genes & development. 2007;21:2897–907. doi: 10.1101/gad.1596807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Speck C, Wendel P, Tang C, Stillman B, Li H. The architecture of the DNA replication origin recognition complex in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:10326–31. doi: 10.1073/pnas.0803829105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesnokov I, Remus D, Botchan M. Functional analysis of mutant and wild-type Drosophila origin recognition complex. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:11997–2002. doi: 10.1073/pnas.211342798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesnokov I, Gossen M, Remus D, Botchan M. Assembly of functionally active Drosophila origin recognition complex from recombinant proteins. Genes & development. 1999;13:1289–96. doi: 10.1101/gad.13.10.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesnokov IN, Chesnokova ON, Botchan M. A cytokinetic function of Drosophila ORC6 protein resides in a domain distinct from its replication activity. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9150–5. doi: 10.1073/pnas.1633580100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang RY, Kelly TJ. The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AT-hooks. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:2656–61. doi: 10.1073/pnas.96.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang RY, Chretien L, Dai J, Kelly TJ. Purification and characterization of the Schizosaccharomyces pombe origin recognition complex: interaction with origin DNA and Cdc18 protein. The Journal of biological chemistry. 2002;277:16920–7. doi: 10.1074/jbc.M107710200. [DOI] [PubMed] [Google Scholar]
- Clarey MG, Erzberger JP, Grob P, Leschziner AE, Berger JM, Nogales E, Botchan M. Nucleotide-dependent conformational changes in the DnaA-like core of the origin recognition complex. Nature structural & molecular biology. 2006;13:684–90. doi: 10.1038/nsmb1121. [DOI] [PubMed] [Google Scholar]
- Cocker JH, Piatti S, Santocanale C, Nasmyth K, Diffley JF. An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature. 1996;379:180–2. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- Craig JM, Earle E, Canham P, Wong LH, Anderson M, Choo KH. Analysis of mammalian proteins involved in chromatin modification reveals new metaphase centromeric proteins and distinct chromosomal distribution patterns. Human molecular genetics. 2003;12:3109–21. doi: 10.1093/hmg/ddg330. [DOI] [PubMed] [Google Scholar]
- Cvetic C, Walter JC. Eukaryotic origins of DNA replication: could you please be more specific? Seminars in cell & developmental biology. 2005;16:343–53. doi: 10.1016/j.semcdb.2005.02.009. [DOI] [PubMed] [Google Scholar]
- DePamphilis ML. The ‘ORC cycle’: a novel pathway for regulating eukaryotic DNA replication. Gene. 2003;310:1–15. doi: 10.1016/s0378-1119(03)00546-8. [DOI] [PubMed] [Google Scholar]
- DePamphilis ML. Cell cycle dependent regulation of the origin recognition complex. Cell cycle. 2005;4:70–9. doi: 10.4161/cc.4.1.1333. [DOI] [PubMed] [Google Scholar]
- DePamphilis ML, Blow JJ, Ghosh S, Saha T, Noguchi K, Vassilev A. Regulating the licensing of DNA replication origins in metazoa. Current opinion in cell biology. 2006;18:231–9. doi: 10.1016/j.ceb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Deshpande AM, Newlon CS. The ARS consensus sequence is required for chromosomal origin function in Saccharomyces cerevisiae. Molecular and cellular biology. 1992;12:4305–13. doi: 10.1128/mcb.12.10.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar SK, Dutta A. Identification and characterization of the human ORC6 homolog. The Journal of biological chemistry. 2000;275:34983–8. doi: 10.1074/jbc.M006069200. [DOI] [PubMed] [Google Scholar]
- Dhar SK, Delmolino L, Dutta A. Architecture of the human origin recognition complex. The Journal of biological chemistry. 2001a;276:29067–71. doi: 10.1074/jbc.M103078200. [DOI] [PubMed] [Google Scholar]
- Dhar SK, Yoshida K, Machida Y, Khaira P, Chaudhuri B, Wohlschlegel JA, Leffak M, Yates J, Dutta A. Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell. 2001b;106:287–96. doi: 10.1016/s0092-8674(01)00458-5. [DOI] [PubMed] [Google Scholar]
- Diffley JF, Labib K. The chromosome replication cycle. Journal of cell science. 2002;115:869–72. doi: 10.1242/jcs.115.5.869. [DOI] [PubMed] [Google Scholar]
- Drury LS, Perkins G, Diffley JF. The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. The EMBO journal. 1997;16:5966–76. doi: 10.1093/emboj/16.19.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury LS, Perkins G, Diffley JF. The cyclin-dependent kinase Cdc28p regulates distinct modes of Cdc6p proteolysis during the budding yeast cell cycle. Current biology : CB. 2000;10:231–40. doi: 10.1016/s0960-9822(00)00355-9. [DOI] [PubMed] [Google Scholar]
- Dueber EC, Costa A, Corn JE, Bell SD, Berger JM. Molecular determinants of origin discrimination by Orc1 initiators in archaea. Nucleic acids research. 2011;39:3621–31. doi: 10.1093/nar/gkq1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueber EL, Corn JE, Bell SD, Berger JM. Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science. 2007;317:1210–3. doi: 10.1126/science.1143690. [DOI] [PubMed] [Google Scholar]
- Duncker BP, Chesnokov IN, McConkey BJ. The origin recognition complex protein family. Genome biology. 2009;10:214. doi: 10.1186/gb-2009-10-3-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncker BP, Pasero P, Braguglia D, Heun P, Weinreich M, Gasser SM. Cyclin B-cdk1 kinase stimulates ORC-and Cdc6-independent steps of semiconservative plasmid replication in yeast nuclear extracts. Molecular and cellular biology. 1999;19:1226–41. doi: 10.1128/mcb.19.2.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton ML, Galani K, Kang S, Bell SP, MacAlpine DM. Conserved nucleosome positioning defines replication origins. Genes & development. 2010;24:748–53. doi: 10.1101/gad.1913210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton ML, Prinz JA, MacAlpine HK, Tretyakov G, Kharchenko PV, MacAlpine DM. Chromatin signatures of the Drosophila replication program. Genome research. 2011;21:164–74. doi: 10.1101/gr.116038.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrentraut S, Hassler M, Oppikofer M, Kueng S, Weber JM, Mueller JW, Gasser SM, Ladurner AG, Ehrenhofer-Murray AE. Structural basis for the role of the Sir3 AAA+ domain in silencing: interaction with Sir4 and unmethylated histone H3K79. Genes & development. 2011;25:1835–46. doi: 10.1101/gad.17175111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20240–5. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falaschi A, Abdurashidova G, Sandoval O, Radulescu S, Biamonti G, Riva S. Molecular and structural transactions at human DNA replication origins. Cell cycle. 2007;6:1705–12. doi: 10.4161/cc.6.14.4495. [DOI] [PubMed] [Google Scholar]
- Fox CA, Ehrenhofer-Murray AE, Loo S, Rine J. The origin recognition complex, SIR1, and the S phase requirement for silencing. Science. 1997;276:1547–51. doi: 10.1126/science.276.5318.1547. [DOI] [PubMed] [Google Scholar]
- Gaczynska M, Osmulski PA, Jiang Y, Lee JK, Bermudez V, Hurwitz J. Atomic force microscopic analysis of the binding of the Schizosaccharomyces pombe origin recognition complex and the spOrc4 protein with origin DNA. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:17952–7. doi: 10.1073/pnas.0408369102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner KA, Rine J, Fox CA. A region of the Sir1 protein dedicated to recognition of a silencer and required for interaction with the Orc1 protein in saccharomyces cerevisiae. Genetics. 1999;151:31–44. doi: 10.1093/genetics/151.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudier M, Schuwirth BS, Westcott SL, Wigley DB. Structural basis of DNA replication origin recognition by an ORC protein. Science. 2007;317:1213–6. doi: 10.1126/science.1143664. [DOI] [PubMed] [Google Scholar]
- Gavin KA, Hidaka M, Stillman B. Conserved initiator proteins in eukaryotes. Science. 1995;270:1667–71. doi: 10.1126/science.270.5242.1667. [DOI] [PubMed] [Google Scholar]
- Georlette D, Ahn S, MacAlpine DM, Cheung E, Lewis PW, Beall EL, Bell SP, Speed T, Manak JR, Botchan MR. Genomic profiling and expression studies reveal both positive and negative activities for the Drosophila Myb MuvB/dREAM complex in proliferating cells. Genes & development. 2007;21:2880–96. doi: 10.1101/gad.1600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Vassilev AP, Zhang J, Zhao Y, DePamphilis ML. Assembly of the human origin recognition complex occurs through independent nuclear localization of its components. The Journal of biological chemistry. 2011;286:23831–41. doi: 10.1074/jbc.M110.215988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Bell SP, Aparicio OM. Cell cycle execution point analysis of ORC function and characterization of the checkpoint response to ORC inactivation in Saccharomyces cerevisiae. Genes to cells : devoted to molecular & cellular mechanisms. 2006;11:557–73. doi: 10.1111/j.1365-2443.2006.00967.x. [DOI] [PubMed] [Google Scholar]
- Gilbert DM. Replication origins in yeast versus metazoa: separation of the haves and the have nots. Current opinion in genetics & development. 1998;8:194–9. doi: 10.1016/s0959-437x(98)80141-x. [DOI] [PubMed] [Google Scholar]
- Gilbert DM. Evaluating genome-scale approaches to eukaryotic DNA replication. Nature reviews. Genetics. 2010;11:673–84. doi: 10.1038/nrg2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano-Coltart J, Ying CY, Gautier J, Hurwitz J. Studies of the properties of human origin recognition complex and its Walker A motif mutants. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:69–74. doi: 10.1073/pnas.0408690102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Pak DT, Hansen SK, Acharya JK, Botchan MR. A Drosophila homolog of the yeast origin recognition complex. Science. 1995;270:1674–7. doi: 10.1126/science.270.5242.1674. [DOI] [PubMed] [Google Scholar]
- Grainge I, Scaife S, Wigley DB. Biochemical analysis of components of the pre-replication complex of Archaeoglobus fulgidus. Nucleic acids research. 2003;31:4888–98. doi: 10.1093/nar/gkg662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland RM, Laskey RA. Regulated replication of DNA microinjected into eggs of Xenopus laevis. Cell. 1980;21:761–71. doi: 10.1016/0092-8674(80)90439-0. [DOI] [PubMed] [Google Scholar]
- Hartl T, Boswell C, Orr-Weaver TL, Bosco G. Developmentally regulated histone modifications in Drosophila follicle cells: initiation of gene amplification is associated with histone H3 and H4 hyperacetylation and H1 phosphorylation. Chromosoma. 2007;116:197–214. doi: 10.1007/s00412-006-0092-2. [DOI] [PubMed] [Google Scholar]
- Hemerly AS, Prasanth SG, Siddiqui K, Stillman B. Orc1 controls centriole and centrosome copy number in human cells. Science. 2009;323:789–93. doi: 10.1126/science.1166745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman MA, Rusche LN. Transcriptional silencing functions of the yeast protein Orc1/Sir3 subfunctionalized after gene duplication. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:19384–9. doi: 10.1073/pnas.1006436107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z, Bernstein DA, Fox CA, Keck JL. Structural basis of the Sir1-origin recognition complex interaction in transcriptional silencing. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:8489–94. doi: 10.1073/pnas.0503525102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houchens CR, Lu W, Chuang RY, Frattini MG, Fuller A, Simancek P, Kelly TJ. Multiple mechanisms contribute to Schizosaccharomyces pombe origin recognition complex-DNA interactions. The Journal of biological chemistry. 2008;283:30216–24. doi: 10.1074/jbc.M802649200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Fanti L, Pak DT, Botchan MR, Pimpinelli S, Kellum R. Distinct cytoplasmic and nuclear fractions of Drosophila heterochromatin protein 1: their phosphorylation levels and associations with origin recognition complex proteins. The Journal of cell biology. 1998;142:307–18. doi: 10.1083/jcb.142.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RY, Kowalski D. A DNA unwinding element and an ARS consensus comprise a replication origin within a yeast chromosome. The EMBO journal. 1993;12:4521–31. doi: 10.1002/j.1460-2075.1993.tb06141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijbregts RP, Svitin A, Stinnett MW, Renfrow MB, Chesnokov I. Drosophila Orc6 facilitates GTPase activity and filament formation of the septin complex. Molecular biology of the cell. 2009;20:270–81. doi: 10.1091/mbc.E08-07-0754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien MD, Polonskaya Z, Hearing J. Protein and sequence requirements for the recruitment of the human origin recognition complex to the latent cycle origin of DNA replication of Epstein-Barr virus oriP. Virology. 2004;326:317–28. doi: 10.1016/j.virol.2004.05.026. [DOI] [PubMed] [Google Scholar]
- Karnani N, Taylor CM, Malhotra A, Dutta A. Genomic study of replication initiation in human chromosomes reveals the influence of transcription regulation and chromatin structure on origin selection. Molecular biology of the cell. 2010;21:393–404. doi: 10.1091/mbc.E09-08-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami H, Katayama T. DnaA, ORC, and Cdc6: similarity beyond the domains of life and diversity. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2010;88:49–62. doi: 10.1139/o09-154. [DOI] [PubMed] [Google Scholar]
- Kearsey SE, Montgomery S, Labib K, Lindner K. Chromatin binding of the fission yeast replication factor mcm4 occurs during anaphase and requires ORC and cdc18. The EMBO journal. 2000;19:1681–90. doi: 10.1093/emboj/19.7.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly TJ, Martin GS, Forsburg SL, Stephen RJ, Russo A, Nurse P. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell. 1993;74:371–82. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- Kim JC, Orr-Weaver TL. Analysis of a Drosophila amplicon in follicle cells highlights the diversity of metazoan replication origins. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16681–6. doi: 10.1073/pnas.1114209108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JC, Nordman J, Xie F, Kashevsky H, Eng T, Li S, MacAlpine DM, Orr-Weaver TL. Integrative analysis of gene amplification in Drosophila follicle cells: parameters of origin activation and repression. Genes & development. 2011;25:1384–98. doi: 10.1101/gad.2043111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm RD, Bell SP. ATP bound to the origin recognition complex is important for preRC formation. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:8361–7. doi: 10.1073/pnas.131006898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm RD, Austin RJ, Bell SP. Coordinate binding of ATP and origin DNA regulates the ATPase activity of the origin recognition complex. Cell. 1997;88:493–502. doi: 10.1016/s0092-8674(00)81889-9. [DOI] [PubMed] [Google Scholar]
- Kong D, DePamphilis ML. Site-specific ORC binding, pre-replication complex assembly and DNA synthesis at Schizosaccharomyces pombe replication origins. The EMBO journal. 2002;21:5567–76. doi: 10.1093/emboj/cdf546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitz S, Ritzi M, Baack M, Knippers R. The human origin recognition complex protein 1 dissociates from chromatin during S phase in HeLa cells. The Journal of biological chemistry. 2001;276:6337–42. doi: 10.1074/jbc.M009473200. [DOI] [PubMed] [Google Scholar]
- Ladenburger EM, Keller C, Knippers R. Identification of a binding region for human origin recognition complex proteins 1 and 2 that coincides with an origin of DNA replication. Molecular and cellular biology. 2002;22:1036–48. doi: 10.1128/MCB.22.4.1036-1048.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DG, Bell SP. Architecture of the yeast origin recognition complex bound to origins of DNA replication. Molecular and cellular biology. 1997;17:7159–68. doi: 10.1128/mcb.17.12.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DG, Bell SP. ATPase switches controlling DNA replication initiation. Current opinion in cell biology. 2000;12:280–5. doi: 10.1016/s0955-0674(00)00089-2. [DOI] [PubMed] [Google Scholar]
- Lee DG, Makhov AM, Klemm RD, Griffith JD, Bell SP. Regulation of origin recognition complex conformation and ATPase activity: differential effects of single-stranded and double-stranded DNA binding. The EMBO journal. 2000;19:4774–82. doi: 10.1093/emboj/19.17.4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JK, Moon KY, Jiang Y, Hurwitz J. The Schizosaccharomyces pombe origin recognition complex interacts with multiple AT-rich regions of the replication origin DNA by means of the AT-hook domains of the spOrc4 protein. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:13589–94. doi: 10.1073/pnas.251530398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Weinreich M, Stillman B. ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell. 1995;81:667–76. doi: 10.1016/0092-8674(95)90528-6. [DOI] [PubMed] [Google Scholar]
- Lidonnici MR, Rossi R, Paixao S, Mendoza-Maldonado R, Paolinelli R, Arcangeli C, Giacca M, Biamonti G, Montecucco A. Subnuclear distribution of the largest subunit of the human origin recognition complex during the cell cycle. Journal of cell science. 2004;117:5221–31. doi: 10.1242/jcs.01405. [DOI] [PubMed] [Google Scholar]
- Lipford JR, Bell SP. Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Molecular cell. 2001;7:21–30. doi: 10.1016/s1097-2765(01)00151-4. [DOI] [PubMed] [Google Scholar]
- Liu J, McConnell K, Dixon M, Calvi BR. Analysis of model replication origins in Drosophila reveals new aspects of the chromatin landscape and its relationship to origin activity and the prereplicative complex. Molecular biology of the cell. 2012;23:200–12. doi: 10.1091/mbc.E11-05-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Balasov M, Wang H, Wu L, Chesnokov IN, Liu Y. Structural analysis of human Orc6 protein reveals a homology with transcription factor TFIIB. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7373–8. doi: 10.1073/pnas.1013676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loupart ML, Krause SA, Heck MS. Aberrant replication timing induces defective chromosome condensation in Drosophila ORC2 mutants. Current biology : CB. 2000;10:1547–56. doi: 10.1016/s0960-9822(00)00844-7. [DOI] [PubMed] [Google Scholar]
- MacAlpine DM, Rodriguez HK, Bell SP. Coordination of replication and transcription along a Drosophila chromosome. Genes & development. 2004;18:3094–105. doi: 10.1101/gad.1246404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine HK, Gordan R, Powell SK, Hartemink AJ, MacAlpine DM. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome research. 2010;20:201–11. doi: 10.1101/gr.097873.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makise M, Takehara M, Kuniyasu A, Matsui N, Nakayama H, Mizushima T. Linkage between phosphorylation of the origin recognition complex and its ATP binding activity in Saccharomyces cerevisiae. The Journal of biological chemistry. 2009;284:3396–407. doi: 10.1074/jbc.M804293200. [DOI] [PubMed] [Google Scholar]
- Marahrens Y, Stillman B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science. 1992;255:817–23. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- Matsuda K, Makise M, Sueyasu Y, Takehara M, Asano T, Mizushima T. Yeast two-hybrid analysis of the origin recognition complex of Saccharomyces cerevisiae: interaction between subunits and identification of binding proteins. FEMS yeast research. 2007;7:1263–9. doi: 10.1111/j.1567-1364.2007.00298.x. [DOI] [PubMed] [Google Scholar]
- Mendez J, Stillman B. Perpetuating the double helix: molecular machines at eukaryotic DNA replication origins. BioEssays : news and reviews in molecular, cellular and developmental biology. 2003;25:1158–67. doi: 10.1002/bies.10370. [DOI] [PubMed] [Google Scholar]
- Mendez J, Zou-Yang XH, Kim SY, Hidaka M, Tansey WP, Stillman B. Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Molecular cell. 2002;9:481–91. doi: 10.1016/s1097-2765(02)00467-7. [DOI] [PubMed] [Google Scholar]
- Moon KY, Kong D, Lee JK, Raychaudhuri S, Hurwitz J. Identification and reconstitution of the origin recognition complex from Schizosaccharomyces pombe. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:12367–72. doi: 10.1073/pnas.96.22.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P, Park S, Shor E, Huebert DJ, Warren CL, Ansari AZ, Weinreich M, Eaton ML, MacAlpine DM, Fox CA. The conserved bromo-adjacent homology domain of yeast Orc1 functions in the selection of DNA replication origins within chromatin. Genes & development. 2010;24:1418–33. doi: 10.1101/gad.1906410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natale DA, Umek RM, Kowalski D. Ease of DNA unwinding is a conserved property of yeast replication origins. Nucleic acids research. 1993;21:555–60. doi: 10.1093/nar/21.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen VQ, Co C, Li JJ. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature. 2001;411:1068–73. doi: 10.1038/35082600. [DOI] [PubMed] [Google Scholar]
- Nishitani H, Nurse P. The cdc18 protein initiates DNA replication in fission yeast. Progress in cell cycle research. 1997;3:135–42. doi: 10.1007/978-1-4615-5371-7_11. [DOI] [PubMed] [Google Scholar]
- Noguchi K, Vassilev A, Ghosh S, Yates JL, DePamphilis ML. The BAH domain facilitates the ability of human Orc1 protein to activate replication origins in vivo. The EMBO journal. 2006;25:5372–82. doi: 10.1038/sj.emboj.7601396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norseen J, Thomae A, Sridharan V, Aiyar A, Schepers A, Lieberman PM. RNA-dependent recruitment of the origin recognition complex. The EMBO journal. 2008;27:3024–35. doi: 10.1038/emboj.2008.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y, Takahashi T, Masukata H. Association of fission yeast Orp1 and Mcm6 proteins with chromosomal replication origins. Molecular and cellular biology. 1999;19:7228–36. doi: 10.1128/mcb.19.10.7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaydin B, Rine J. Expanded roles of the origin recognition complex in the architecture and function of silenced chromatin in Saccharomyces cerevisiae. Molecular and cellular biology. 2010;30:626–39. doi: 10.1128/MCB.00614-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak DT, Pflumm M, Chesnokov I, Huang DW, Kellum R, Marr J, Romanowski P, Botchan MR. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell. 1997;91:311–23. doi: 10.1016/s0092-8674(00)80415-8. [DOI] [PubMed] [Google Scholar]
- Perkins G, Drury LS, Diffley JF. Separate SCF(CDC4) recognition elements target Cdc6 for proteolysis in S phase and mitosis. The EMBO journal. 2001;20:4836–45. doi: 10.1093/emboj/20.17.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflumm MF, Botchan MR. Orc mutants arrest in metaphase with abnormally condensed chromosomes. Development. 2001;128:1697–707. doi: 10.1242/dev.128.9.1697. [DOI] [PubMed] [Google Scholar]
- Piatti S, Bohm T, Cocker JH, Diffley JF, Nasmyth K. Activation of S-phase-promoting CDKs in late G1 defines a “point of no return” after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes & development. 1996;10:1516–31. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- Pillus L, Rine J. Epigenetic inheritance of transcriptional states in S. cerevisiae. Cell. 1989;59:637–47. doi: 10.1016/0092-8674(89)90009-3. [DOI] [PubMed] [Google Scholar]
- Pillus L, Rine J. SIR1 and the origin of epigenetic states in Saccharomyces cerevisiae. Cold Spring Harbor symposia on quantitative biology. 2004;69:259–65. doi: 10.1101/sqb.2004.69.259. [DOI] [PubMed] [Google Scholar]
- Prasanth SG, Prasanth KV, Stillman B. Orc6 involved in DNA replication, chromosome segregation, and cytokinesis. Science. 2002;297:1026–31. doi: 10.1126/science.1072802. [DOI] [PubMed] [Google Scholar]
- Prasanth SG, Shen Z, Prasanth KV, Stillman B. Human origin recognition complex is essential for HP1 binding to chromatin and heterochromatin organization. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15093–8. doi: 10.1073/pnas.1009945107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth SG, Prasanth KV, Siddiqui K, Spector DL, Stillman B. Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. The EMBO journal. 2004;23:2651–63. doi: 10.1038/sj.emboj.7600255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randell JC, Bowers JL, Rodriguez HK, Bell SP. Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2-7 helicase. Molecular cell. 2006;21:29–39. doi: 10.1016/j.molcel.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Ranjan A, Gossen M. A structural role for ATP in the formation and stability of the human origin recognition complex. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4864–9. doi: 10.1073/pnas.0510305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao H, Stillman B. The origin recognition complex interacts with a bipartite DNA binding site within yeast replicators. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:2224–8. doi: 10.1073/pnas.92.6.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao H, Marahrens Y, Stillman B. Functional conservation of multiple elements in yeast chromosomal replicators. Molecular and cellular biology. 1994;14:7643–51. doi: 10.1128/mcb.14.11.7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D, Beall EL, Botchan MR. DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. The EMBO journal. 2004;23:897–907. doi: 10.1038/sj.emboj.7600077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D, Blanchette M, Rio DC, Botchan MR. CDK phosphorylation inhibits the DNA-binding and ATP-hydrolysis activities of the Drosophila origin recognition complex. The Journal of biological chemistry. 2005;280:39740–51. doi: 10.1074/jbc.M508515200. [DOI] [PubMed] [Google Scholar]
- Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JF. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–30. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero J, Lee H. Asymmetric bidirectional replication at the human DBF4 origin. Nature structural & molecular biology. 2008;15:722–9. doi: 10.1038/nsmb.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley A, Cocker JH, Harwood J, Diffley JF. Initiation complex assembly at budding yeast replication origins begins with the recognition of a bipartite sequence by limiting amounts of the initiator, ORC. The EMBO journal. 1995;14:2631–41. doi: 10.1002/j.1460-2075.1995.tb07261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royzman I, Austin RJ, Bosco G, Bell SP, Orr-Weaver TL. ORC localization in Drosophila follicle cells and the effects of mutations in dE2F and dDP. Genes & development. 1999;13:827–40. doi: 10.1101/gad.13.7.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, Gilbert DM. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome research. 2010;20:761–70. doi: 10.1101/gr.099655.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santocanale C, Diffley JF. ORC- and Cdc6-dependent complexes at active and inactive chromosomal replication origins in Saccharomyces cerevisiae. The EMBO journal. 1996;15:6671–9. [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Gilbert DM. The many faces of the origin recognition complex. Current opinion in cell biology. 2007;19:337–43. doi: 10.1016/j.ceb.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Scholefield G, Veening JW, Murray H. DnaA and ORC: more than DNA replication initiators. Trends in cell biology. 2011;21:188–94. doi: 10.1016/j.tcb.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Shareef MM, Badugu R, Kellum R. HP1/ORC complex and heterochromatin assembly. Genetica. 2003;117:127–34. doi: 10.1023/a:1022963223220. [DOI] [PubMed] [Google Scholar]
- Shareef MM, King C, Damaj M, Badagu R, Huang DW, Kellum R. Drosophila heterochromatin protein 1 (HP1)/origin recognition complex (ORC) protein is associated with HP1 and ORC and functions in heterochromatin-induced silencing. Molecular biology of the cell. 2001;12:1671–85. doi: 10.1091/mbc.12.6.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z, Sathyan KM, Geng Y, Zheng R, Chakraborty A, Freeman B, Wang F, Prasanth KV, Prasanth SG. A WD-repeat protein stabilizes ORC binding to chromatin. Molecular cell. 2010;40:99–111. doi: 10.1016/j.molcel.2010.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher N, Bell GW, Li S, Nordman J, Eng T, Eaton ML, Macalpine DM, Orr-Weaver TL. Developmental control of gene copy number by repression of replication initiation and fork progression. Genome research. 2012;22:64–75. doi: 10.1101/gr.126003.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui K, Stillman B. ATP-dependent assembly of the human origin recognition complex. The Journal of biological chemistry. 2007;282:32370–83. doi: 10.1074/jbc.M705905200. [DOI] [PubMed] [Google Scholar]
- Speck C, Stillman B. Cdc6 ATPase activity regulates ORC × Cdc6 stability and the selection of specific DNA sequences as origins of DNA replication. The Journal of biological chemistry. 2007;282:11705–14. doi: 10.1074/jbc.M700399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck C, Chen Z, Li H, Stillman B. ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA. Nature structural & molecular biology. 2005;12:965–71. doi: 10.1038/nsmb1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC. ORC binding, gene amplification, and the nature of metazoan replication origins. Genes & development. 1999;13:2619–23. doi: 10.1101/gad.13.20.2619. [DOI] [PubMed] [Google Scholar]
- Stefanovic D, Stanojcic S, Vindigni A, Ochem A, Falaschi A. In vitro protein-DNA interactions at the human lamin B2 replication origin. The Journal of biological chemistry. 2003;278:42737–43. doi: 10.1074/jbc.M307058200. [DOI] [PubMed] [Google Scholar]
- Sun J, Kawakami H, Zech J, Speck C, Stillman B, Li H. Cdc6-induced Conformational Changes in ORC Bound To Origin DNA Revealed by Cryo-Electron Microscopy. Structure. 2012 doi: 10.1016/j.str.2012.01.011. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada S, Kundu LR, Enomoto T. Insight into initiator-DNA interactions: a lesson from the archaeal ORC. BioEssays : news and reviews in molecular, cellular and developmental biology. 2008;30:208–11. doi: 10.1002/bies.20726. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Ohara E, Nishitani H, Masukata H. Multiple ORC-binding sites are required for efficient MCM loading and origin firing in fission yeast. The EMBO journal. 2003;22:964–74. doi: 10.1093/emboj/cdg079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takara TJ, Bell SP. Multiple Cdt1 molecules act at each origin to load replication-competent Mcm2-7 helicases. The EMBO journal. 2011;30:4885–96. doi: 10.1038/emboj.2011.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehara M, Makise M, Takenaka H, Asano T, Mizushima T. Analysis of mutant origin recognition complex with reduced ATPase activity in vivo and in vitro. The Biochemical journal. 2008;413:535–43. doi: 10.1042/BJ20070484. [DOI] [PubMed] [Google Scholar]
- Takenaka H, Makise M, Kuwae W, Takahashi N, Tsuchiya T, Mizushima T. ADP-binding to origin recognition complex of Saccharomyces cerevisiae. Journal of molecular biology. 2004;340:29–37. doi: 10.1016/j.jmb.2004.04.045. [DOI] [PubMed] [Google Scholar]
- Tatsumi Y, Ohta S, Kimura H, Tsurimoto T, Obuse C. The ORC1 cycle in human cells: I. cell cycle-regulated oscillation of human ORC1. The Journal of biological chemistry. 2003;278:41528–34. doi: 10.1074/jbc.M307534200. [DOI] [PubMed] [Google Scholar]
- Theis JF, Newlon CS. Domain B of ARS307 contains two functional elements and contributes to chromosomal replication origin function. Molecular and cellular biology. 1994;14:7652–9. doi: 10.1128/mcb.14.11.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis JF, Newlon CS. Two compound replication origins in Saccharomyces cerevisiae contain redundant origin recognition complex binding sites. Molecular and cellular biology. 2001;21:2790–801. doi: 10.1128/MCB.21.8.2790-2801.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomae AW, Pich D, Brocher J, Spindler MP, Berens C, Hock R, Hammerschmidt W, Schepers A. Interaction between HMGA1a and the origin recognition complex creates site-specific replication origins. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:1692–7. doi: 10.1073/pnas.0707260105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triolo T, Sternglanz R. Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature. 1996;381:251–3. doi: 10.1038/381251a0. [DOI] [PubMed] [Google Scholar]
- Tugal T, Zou-Yang XH, Gavin K, Pappin D, Canas B, Kobayashi R, Hunt T, Stillman B. The Orc4p and Orc5p subunits of the Xenopus and human origin recognition complex are related to Orc1p and Cdc6p. The Journal of biological chemistry. 1998;273:32421–9. doi: 10.1074/jbc.273.49.32421. [DOI] [PubMed] [Google Scholar]
- Van Houten JV, Newlon CS. Mutational analysis of the consensus sequence of a replication origin from yeast chromosome III. Molecular and cellular biology. 1990;10:3917–25. doi: 10.1128/mcb.10.8.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashee S, Simancek P, Challberg MD, Kelly TJ. Assembly of the human origin recognition complex. The Journal of biological chemistry. 2001;276:26666–73. doi: 10.1074/jbc.M102493200. [DOI] [PubMed] [Google Scholar]
- Vashee S, Cvetic C, Lu W, Simancek P, Kelly TJ, Walter JC. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes & development. 2003;17:1894–908. doi: 10.1101/gad.1084203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, Mann M. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142:967–80. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- Wallace JA, Orr-Weaver TL. Replication of heterochromatin: insights into mechanisms of epigenetic inheritance. Chromosoma. 2005;114:389–402. doi: 10.1007/s00412-005-0024-6. [DOI] [PubMed] [Google Scholar]
- Weber JM, Irlbacher H, Ehrenhofer-Murray AE. Control of replication initiation by the Sum1/Rfm1/Hst1 histone deacetylase. BMC molecular biology. 2008;9:100. doi: 10.1186/1471-2199-9-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich M, Liang C, Chen HH, Stillman B. Binding of cyclin-dependent kinases to ORC and Cdc6p regulates the chromosome replication cycle. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:11211–7. doi: 10.1073/pnas.201387198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigley DB. ORC proteins: marking the start. Current opinion in structural biology. 2009;19:72–8. doi: 10.1016/j.sbi.2008.12.010. [DOI] [PubMed] [Google Scholar]
- Wilmes GM, Bell SP. The B2 element of the Saccharomyces cerevisiae ARS1 origin of replication requires specific sequences to facilitate pre-RC formation. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:101–6. doi: 10.1073/pnas.012578499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes GM, Archambault V, Austin RJ, Jacobson MD, Bell SP, Cross FR. Interaction of the S-phase cyclin Clb5 with an “RXL” docking sequence in the initiator protein Orc6 provides an origin-localized replication control switch. Genes & development. 2004;18:981–91. doi: 10.1101/gad.1202304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu PY, Nurse P. Establishing the program of origin firing during S phase in fission Yeast. Cell. 2009;136:852–64. doi: 10.1016/j.cell.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Orr-Weaver TL. Isolation of a Drosophila amplification origin developmentally activated by transcription. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9651–6. doi: 10.1073/pnas.0804146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Hayashi MK, Merkel O, Stillman B, Xu RM. Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing. The EMBO journal. 2002;21:4600–11. doi: 10.1093/emboj/cdf468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Stillman B. Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase. Molecular and cellular biology. 2000;20:3086–96. doi: 10.1128/mcb.20.9.3086-3096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Yu Q, Bi X. Asymmetric positioning of nucleosomes and directional establishment of transcriptionally silent chromatin by Saccharomyces cerevisiae silencers. Molecular and cellular biology. 2006;26:7806–19. doi: 10.1128/MCB.01197-06. [DOI] [PMC free article] [PubMed] [Google Scholar]


