Abstract
Left–right asymmetry in anatomical structures and functions of the nervous system is present throughout the animal kingdom. For example, language centers are localized in the left side of the human brain, while spatial recognition functions are found in the right hemisphere in the majority of the population. Disruption of asymmetry in the nervous system is correlated with neurological disorders. Although anatomical and functional asymmetries are observed in mammalian nervous systems, it has been a challenge to identify the molecular basis of these asymmetries. C. elegans has emerged as a prime model organism to investigate molecular asymmetries in the nervous system, as it has been shown to display functional asymmetries clearly correlated to asymmetric distribution and regulation of biologically relevant molecules. Small non-coding RNAs have been recently implicated in various aspects of neural development. Here, we review cases in which microRNAs are crucial for establishing left–right asymmetries in the C. elegans nervous system. These studies may provide insight into how molecular and functional asymmetries are established in the human brain.
Keywords: microRNA, neuronal asymmetry, sensory neurons, calcium signaling, C. elegans
INTRODUCTION
microRNAs (miRNAs) are endogenous 20–24 nt small non-coding RNAs that regulate gene expression through binding to complementary sequences in target messenger RNAs (mRNAs), leading to translational repression and/or cleavage of target mRNAs (Ambros, 2004; He and Hannon, 2004; Bartel, 2009; Chekulaeva and Filipowicz, 2009; Ghildiyal and Zamore, 2009). While most miRNAs downregulate gene expression, there are examples of miRNA-mediated upregulation of target gene expression during cell cycle arrest, suggesting that miRNA function is complex and context dependent (Vasudevan et al., 2007; Orom et al., 2008). miRNAs have been implicated in many aspects of development and disease including cell cycle, cell differentiation, apoptosis, life span, developmental timing, stress responses, neural development and regeneration, cancers, and neurodegenerative disorders (Boehm and Slack, 2005; Bushati and Cohen, 2007; Chang et al., 2009; Ambros, 2011; Sayed and Abdellatif, 2011; Zhang et al., 2011; Boulias and Horvitz, 2012; Cochella and Hobert, 2012a; Saito and Saito, 2012; Zou et al., 2012, 2013).
FUNCTIONS OF miRNAs IN NEURAL DEVELOPMENT
The importance of miRNAs in various aspects of neuronal development has been demonstrated in several animal models. In zebrafish, maternal–zygotic dicer mutants, that disrupt the processing of precursor miRNAs into mature miRNAs, display deleterious effects on the development of the brain, and injection of mature miR-430 rescues the early brain patterning defects (Giraldez et al., 2005). In mice, the neuron-specific miRNA miR-124 induces neuronal differentiation by directly targeting a global repressor of alternative pre-mRNA splicing and triggering a downstream switch to neuron-specific alternative splicing (Makeyev et al., 2007). In the Xenopus retina, a number of cell cycle related miRNAs target Xotx2 and Xvsx1 in early retinal progenitor cells to inhibit bipolar cell differentiation (Decembrini et al., 2009). In Drosophila, miR-9a targets senseless to inhibit neuronal fate in non-sensory organ precursors (Li et al., 2006). In C. elegans, the miRNA lin-4 targets the LIN-14 transcription factor to inhibit netrin-mediated axon attraction (Chang et al., 2004a; Zou et al., 2012), and the miRNA let-7 contributes to a developmental decline in neuronal regeneration (Zou et al., 2013). In addition, miRNAs lsy-6, mir-273, and mir-71 function in asymmetric differentiation of two pairs of C. elegans sensory neurons, which will be discussed later (Johnston and Hobert, 2003; Chang et al., 2004b; Hsieh et al., 2012). Thus, miRNAs are important factors that control neuronal development across the animal kingdom.
LEFT–RIGHT ASYMMETRY OF THE NERVOUS SYSTEM
Although the nervous systems of animals are largely symmetric across the left–right axis, there have been several observations of anatomical and functional brain lateralization throughout the animal kingdom. For example, zebrafish display asymmetry in the epithalamus (Snelson and Gamse, 2009; Taylor et al., 2010); mice have been shown to have paw preferences, indicating the presence of a dominant hemisphere in motor control (Signore et al., 1991; Biddle et al., 1993); and the majority of humans have language centers such as Wernicke’s and Broca’s area located in the left hemisphere of the brain (Sun and Walsh, 2006). This lateralization of the nervous system is thought to be beneficial, as it allows for an increase of functional capacity (Rogers et al., 2012).
Disruption of asymmetry in the brain is seen in a number of neurodevelopmental diseases, including dyslexia, schizophrenia, autism, Alzheimer’s disease, and attention deficit/hyperactivity disorder (ADHD; Herbert et al., 2005; Derflinger et al., 2011; Oertel-Knochel and Linden, 2011; Renteria, 2012). In children diagnosed with ADHD, the prefrontal cortex was shown to have a loss in rightward asymmetric distribution of prefrontal cortex volume as compared to typically developing children (Shaw et al., 2009). In dyslexic patients, the planum temporale shows alteration in asymmetry, as the right temporale appears physically larger than the left counterpart (Galaburda et al., 1985).
There have been several reports on functional and anatomical nervous system asymmetries; however there have been comparatively fewer studies on identifying the molecular mechanisms that establish lateralization. Here we narrow our focus to the function and regulation of miRNAs in the development of neuronal asymmetry. The most interesting evidence of miRNAs in vertebrate neuronal asymmetry comes from the investigation of miRNA function in neocortex development. The simplest form of asymmetry is the division of a cell to give rise to two asymmetric fates. In the case of the mouse neocortex, a progenitor cell divides asymmetrically to give rise to a progenitor cell and a neuron. Asymmetric localization of the TRIM-NHL protein TRIM32 is observed in the daughter cell that becomes a neuron, while the cell lacking TRIM32 remains a progenitor (Schwamborn et al., 2009). TRIM32 increases the activity of specific miRNAs through binding of the RNAse argonaute-1 (Hammell et al., 2009; Schwamborn et al., 2009). It was further shown that the miRNA Let-7a, one of the TRIM32 targets, is required and sufficient for neuronal differentiation (Schwamborn et al., 2009).
A subset of zebrafish olfactory bulb output neurons called the mitral cells send axons asymmetrically to the right habenula, which is an asymmetric part of the brain where the higher olfactory processing center is located (Miyasaka et al., 2009; Taylor et al., 2010). Several molecules are asymmetrically expressed in adult-born olfactory neurons in the ventricular–subventricular zone (V–SVZ) of the zebrafish brain. The transcription factors Myt1 and Neurogenin1 are predominantly expressed in the left V–SVZ, while DeltaA and hairy/enhancer of split-related protein (a Notch effector) are mainly expressed on the right side (Kishimoto et al., 2013). In addition, members of the miR-200 family are involved in the proper differentiation of olfactory neurons of both mice and zebrafish (Choi et al., 2008). It would be interesting to see whether the miR-200 family or other miRNAs influences the asymmetric and lateral projection of olfactory axons to the higher olfactory centers, and whether Myt1, Neurogenin1, and Delta/Notch are potential targets of miR-200.
A study found that 27 genes are differentially expressed in the embryonic human cerebral cortex, and the Lim domain transcription factor LMO4 is more abundant in the right perisylvian cortex than the left, and may be involved in asymmetric development of the cortex (Sun et al., 2005). The hypothesis that differential expression of these genes between left and right sides of the cortex may be regulated by miRNAs is plausible and worth further investigation.
Although there are limited reports of the involvement of miRNAs in the development of neuronal asymmetry in vertebrates, C. elegans has proved to be a powerful model organism to study lateralization of the nervous system due to its genetic amenability, as well as evidence of functional asymmetry having clear molecular correlates. In this review, we highlight two cases of the roles that miRNAs play in establishing left/right asymmetry in the C. elegans nervous system. Both involve the specification of two types of chemosensory neurons: the pair of amphid neurons, single cilliated endings (ASE) taste neurons, in which lsy-6 and mir-273 miRNAs are involved, and the pair of amphid wing “C” (AWC) olfactory neurons, where mir-71 is crucial for establishment of asymmetry.
miRNAs IN TASTE NEURON ASYMMETRY
Like other animals, the C. elegans nervous system appears generally symmetric. However, the pair of taste neurons, called ASE left (ASEL) and ASE right (ASER) displays molecular and functional asymmetries. The ASE neurons are located in the nerve ring of the nematode, which is the brain equivalent in the worm. Although the neurons are derived from different cell lineages, they are anatomically symmetric in terms of cell position, morphologies, and axonal projections (White et al., 1986). The ASEL neuron, however, differentially expresses the putative chemoreceptor gcy-7, while the ASER neuron expresses another putative chemoreceptor gcy-5 (Yu et al., 1997). The pair of ASE neurons also senses different chemicals, as the ASEL neuron functions to detect sodium, while ASER senses chloride (Pierce-Shimomura et al., 2001).
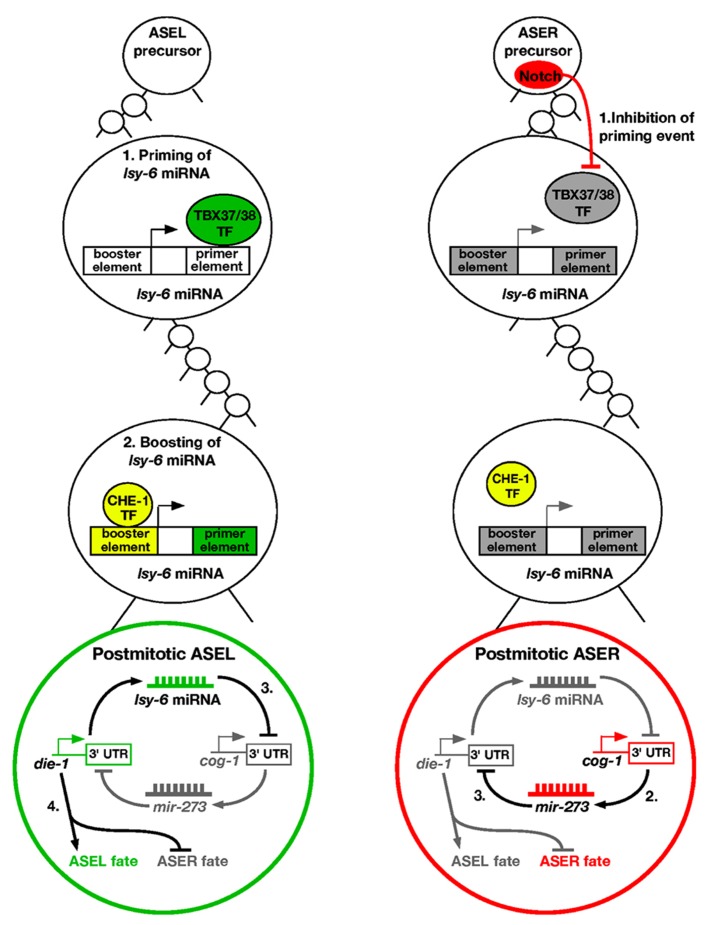
Over the past few years, there have been many studies investigating the molecular mechanism on how ASE asymmetry is established. Intriguingly, the first step of breaking symmetry of the taste neurons occurs several divisions before the ASEL and ASER neurons are born, during the early embryonic stage (Poole and Hobert, 2006). In the ASEL lineage, a pair of redundant T-box transcription factors TBX-37/38 are transiently expressed six cell divisions before the birth of ASEL (Good et al., 2004; Figure 1). These transcription factors work to “prime” a miRNA called lsy-6, which promotes the ASEL cell fate. This is achieved by binding of TBX-37/38 to a downstream primer element of lsy-6 and results in physical opening up of the lsy-6 chromatin (Cochella and Hobert, 2012b). The priming event is then “remembered” several cell divisions later in the ASEL mother cell. The open chromatin status of lsy-6 allows for the CHE-1 zinc finger transcription factor to bind to an upstream booster element of the lsy-6 locus (Cochella and Hobert, 2012b). This induces “boosting” of lsy-6 expression levels in the ASEL neuron. The “prime and boost” model is essential for establishing ASE asymmetry.
FIGURE 1.
Priming and boosting model of ASE asymmetry. ASEL: (1) The first step of promoting the ASEL cell fate occurs six cell divisions before the postmitotic ASEL is born. A transient pair of T-box transcription factors TBX-37/38 acts on the downstream primer element of lsy-6 miRNA to open up the chromatin. (2) The priming event allows boosting of lsy-6 miRNA levels by the CHE-1 transcription factor in the ASEL mother cell, leading to high levels of the miRNA in the postmitotic ASEL neuron. (3) The lsy-6 miRNA directly inhibits the ASER promoting transcription factor COG-1, and allows the ASEL promoting transcription factor DIE-1 to be expressed. (4) DIE-1 then activates ASEL effector genes and represses the ASER cell fate. ASER: (1) Notch is expressed in the ASER precursor and inhibits the lsy-6 priming event. Therefore, the lsy-6 miRNA expression level cannot be boosted as the chromatin remains compact. (2) Consequently, in the postmitotic ASER neuron, the COG-1 transcription factor is expressed, which activates another miRNA, mir-273. (3) mir-273 directly inhibits the ASEL promoting transcription factor DIE-1, and therefore promotes the ASER fate.
In the ASER lineage, a Delta/Notch signal in the ASER precursor cell causes the T-box transcription factors TBX-37/38 to be repressed (Good et al., 2004; Priess, 2005). Therefore, the lsy-6 “priming” event does not occur in the ASER lineage, allowing the lsy-6 chromatin to remain in a compact form (Cochella and Hobert, 2012b). This in turn leads to the inability of the CHE-1 transcription factor to physically bind the upstream element of lsy-6, and no boosting of the miRNA expression levels occurs in ASER. Overall, this causes asymmetric distribution of the lsy-6 miRNA in the ASEL neuron.
The lsy-6 miRNA functions in a double negative feedback loop to control the asymmetry of ASEL/R neurons (Johnston et al., 2005). In the ASEL neuron, lsy-6 directly represses an ASER promoting transcription factor COG-1, through physical binding of complementary bases in the cog-1 3′UTR (Johnston and Hobert, 2003). Repression of COG-1 allows for yet another transcription factor DIE-1 to be expressed, which is the output of the feedback loop. DIE-1 then activates ASEL effector genes and suppresses ASER effector genes (Johnston et al., 2005; Figure 1).
In the ASER neuron, lsy-6 expression is relatively low, and cannot effectively repress the COG-1 transcription factor. This allows activation of another miRNA, mir-273, which displays complementarity to the 3′UTR of the previously described die-1 output (Chang et al., 2004b). mir-273 therefore represses DIE-1, resulting in the de-repression of the ASER fate (Figure 1). Mutating any of the factors involved results in a loss of asymmetry, with both cells adopting either the ASEL or ASER cell fate.
miRNAs IN OLFACTORY NEURON ASYMMETRY
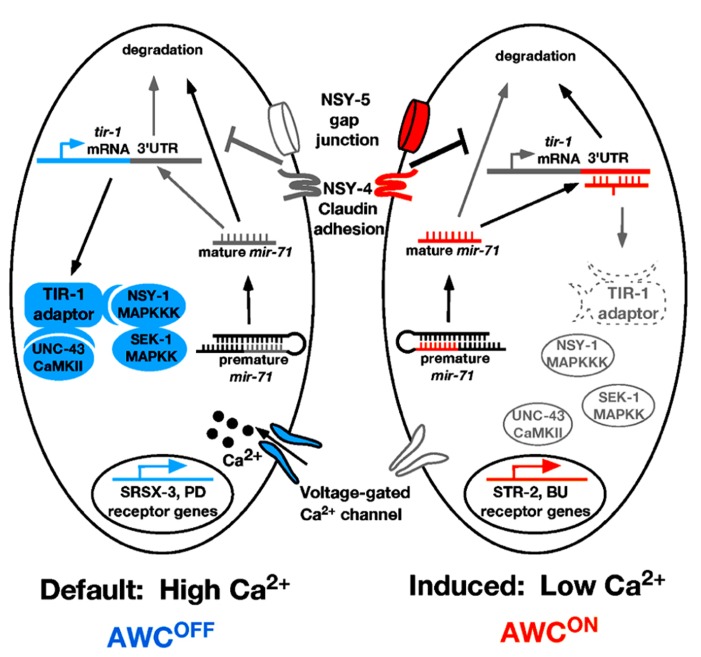
Like the ASE neurons, the C. elegans AWC olfactory neurons are bilaterally symmetrical at the morphological level. However, AWC left (AWCL) and AWC right (AWCR) neurons express different odorant receptors and sense different odors (Figure 2; Troemel et al., 1999). The AWCON neuron expresses the odorant receptor gene str-2 and specifically senses the odor butanone; while the contralateral AWCOFF neuron expresses the odorant receptor gene srsx-3 and specifically senses the odor pentanedione (Figure 2; Troemel et al., 1999; Wes and Bargmann, 2001; Bauer Huang et al., 2007). In wild-type animals, only one of the AWC neurons expresses str-2. AWC asymmetry is stochastic and coordinated, so that 50% of the worms in a population express str-2 in AWCL, while the other 50% express str-2 in AWCR (Troemel et al., 1999; Taylor et al., 2010). The default state of the AWC neurons is AWCOFF, which is specified by a calcium-regulated and microtubule-dependent MAP kinase pathway including UNC-43/CaMKII, TIR-1/Sarm1, NSY-1/MAPKKK, and SEK-1/MAPKK (Figure 2; Sagasti et al., 2001; Tanaka-Hino et al., 2002; Chuang and Bargmann, 2005; Chang et al., 2011). NSY-4 claudin-like protein and NSY-5 gap junction protein act in parallel to inhibit the calcium signaling pathway in the induced AWCON cell (Vanhoven et al., 2006; Chuang et al., 2007). In addition, intercellular calcium signaling between AWCs and non-AWC neurons via a NSY-5 gap junction-dependent neural network coordinates precise AWC asymmetry (Schumacher et al., 2012).
FIGURE 2.
Model of mir-71 function in AWC asymmetry. In the AWCOFF cell, high calcium level activates the calcium-regulated UNC-43/TIR-1/MAPK cascade, leading to srsx-3 expression. In the AWCON cell, nsy-4 and nsy-5 promote the stability of mature mir-71 through an unknown mechanism. The mature mir-71 miRNA targets the 3′UTR of tir-1 mRNA for degradation, leading to the inhibition of the calcium signaling pathway and the subsequent induction of str-2 expression. Orange color represents factors that promote AWCON. Blue color represents factors that promote AWCOFF. Gray/white color represents inactive or less active factors. Dotted line represents factors being downregulated. PD, pentanedione; BU, butanone.
One of the important questions on AWC asymmetry is how the calcium signaling pathway is downregulated by nsy-4 and nsy-5 in the AWCON cell. A recent study showed that the miRNA mir-71 acts downstream of nsy-4 and nsy-5 to promote AWCON in a cell autonomous manner through inhibiting the expression of the calcium signaling adaptor protein gene tir-1 (Figure 2; Hsieh et al., 2012). The TIR-1/Sarm1 adaptor protein assembles a calcium-signaling complex to cell-autonomously specify the default AWCOFF identity (Chuang and Bargmann, 2005). Thus downregulation of tir-1 expression by mir-71 is an efficient mechanism to inhibit calcium signaling in the cell becoming AWCON.
mir-71 is regulated at both transcriptional and post-transcriptional levels in AWC (Hsieh et al., 2012). At the transcriptional level, the expression level of mir-71 is higher in the AWCON cell than in the AWCOFF cell. This transcriptional bias of mir-71 is not dependent on nsy-4 or nsy-5; thus, the mechanisms that regulate differential expression of mir-71 in the two AWC cells are yet to be elucidated. At the post-transcriptional level, the stability of mature mir-71 is dependent on nsy-4 and nsy-5. The C. elegans 5′ → 3′ exoribonuclease XRN-2 has been shown to be involved in degradation of mature miRNAs (Chatterjee and Grosshans, 2009). It is possible that nsy-4 and nsy-5 may antagonize the xrn-2-mediated miRNA turnover pathway to increase the level of mature mir-71. However, RNA interference (RNAi) knockdown of xrn-2 did not cause a defect in AWC asymmetry, suggesting that the stability of mature mir-71 may be independent of xrn-2. In support of this idea, not all miRNAs accumulate in xrn-2 RNAi worms (Chatterjee and Grosshans, 2009), suggesting the existence of alternative miRNA turnover pathways that may be inhibited by nsy-4 and nsy-5.
PERSPECTIVES
Understanding the molecular mechanisms involved in establishing left–right asymmetry in the C. elegans nervous system can lay the groundwork for identifying the processes used in higher organisms, as the methods used may be evolutionarily conserved. Because of the highly conserved nature of miRNAs, insights into how they are involved to control asymmetric fates will help facilitate our understanding in vertebrate neuronal asymmetry. The involvement of miRNAs in asymmetry may also be reflective of the principles these small non-coding RNAs use in directing other neurodevelopmental processes.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Grethel Millington and Chieh Chang for comments on the manuscript. Amel Alqadah is supported by a Choose Ohio First Scholarship, Yi-Wen Hsieh by a NIH Organogenesis Training Grant, and Chiou-Fen Chuang by a Whitehall Foundation Research Award, an Alfred P. Sloan Research Fellowship, and a NIH R01GM098026 grant.
REFERENCES
- Ambros V. (2004). The functions of animal microRNAs. Nature 431 350–355 10.1038/nature02871 [DOI] [PubMed] [Google Scholar]
- Ambros V. (2011). MicroRNAs and developmental timing. Curr. Opin. Genet. Dev. 21 511–517 10.1016/j.gde.2011.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136 215–233 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer Huang S. L., Saheki Y., VanHoven M. K., Torayama I., Ishihara T., Katsura I., et al. (2007). Left–right olfactory asymmetry results from antagonistic functions of voltage-activated calcium channels and the Raw repeat protein OLRN-1 in C. elegans. Neural Dev. 2 24 10.1186/1749-8104-2-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddle F. G., Coffaro C. M., Ziehr J. E., Eales B. A. (1993). Genetic variation in paw preference (handedness) in the mouse. Genome 36 935–943 10.1139/g93-123 [DOI] [PubMed] [Google Scholar]
- Boehm M., Slack F. (2005). A developmental timing microRNA and its target regulate life span in C. elegans. Science 310 1954–1957 10.1126/science.1115596 [DOI] [PubMed] [Google Scholar]
- Boulias K., Horvitz H. R. (2012). The C. elegans microRNA mir-71 acts in neurons to promote germline-mediated longevity through regulation of DAF-16/FOXO. Cell Metab. 15 439–450 10.1016/j.cmet.2012.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N., Cohen S. M. (2007). microRNA functions. Annu. Rev. Cell Dev. Biol. 23 175–205 10.1146/annurev.cellbio.23.090506.123406 [DOI] [PubMed] [Google Scholar]
- Chang C., Hsieh Y. W., Lesch B. J., Bargmann C. I., Chuang C. F. (2011). Microtubule-based localization of a synaptic calcium-signaling complex is required for left–right neuronal asymmetry in C. elegans. Development 138 3509–3518 10.1242/dev.069740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C., Yu T. W., Bargmann C. I., Tessier-Lavigne M. (2004a). Inhibition of netrin-mediated axon attraction by a receptor protein tyrosine phosphatase. Science 305 103–106 10.1126/science.1096983 [DOI] [PubMed] [Google Scholar]
- Chang S., Johnston R. J., Jr., Frokjaer-Jensen C., Lockery S., Hobert O. (2004b). MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature 430 785–789 10.1038/nature02752 [DOI] [PubMed] [Google Scholar]
- Chang S., Wen S., Chen D., Jin P. (2009). Small regulatory RNAs in neurodevelopmental disorders. Hum. Mol. Genet. 18 R18–R26 10.1093/hmg/ddp072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S., Grosshans H. (2009). Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature 461 546–549 10.1038/nature08349 [DOI] [PubMed] [Google Scholar]
- Chekulaeva M., Filipowicz W. (2009). Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr. Opin. Cell Biol. 21 452–460 10.1016/j.ceb.2009.04.009 [DOI] [PubMed] [Google Scholar]
- Choi P. S., Zakhary L., Choi W. Y., Caron S., Alvarez-Saavedra E., Miska E. A., et al. (2008). Members of the miRNA-200 family regulate olfactory neurogenesis. Neuron 57 41–55 10.1016/j.neuron.2007.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang C. F., Bargmann C. I. (2005). A toll-interleukin 1 repeat protein at the synapse specifies asymmetric odorant receptor expression via ASK1 MAPKKK signaling. Genes Dev. 19 270–281 10.1101/gad.1276505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang C. F., Vanhoven M. K., Fetter R. D., Verselis V. K., Bargmann C. I. (2007). An innexin-dependent cell network establishes left–right neuronal asymmetry in C. elegans. Cell 129 787–799 10.1016/j.cell.2007.02.052 [DOI] [PubMed] [Google Scholar]
- Cochella L., Hobert O. (2012a). Diverse functions of microRNAs in nervous system development. Curr. Top. Dev. Biol. 99 115–143 10.1016/B978-0-12-387038-4.00005-7 [DOI] [PubMed] [Google Scholar]
- Cochella L., Hobert O. (2012b). Embryonic priming of a miRNA locus predetermines postmitotic neuronal left/right asymmetry in C. elegans. Cell 151 1229–1242 10.1016/j.cell.2012.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decembrini S., Bressan D., Vignali R., Pitto L., Mariotti S., Rainaldi G., et al. (2009). MicroRNAs couple cell fate and developmental timing in retina. Proc. Natl. Acad. Sci. U.S.A. 106 21179–21184 10.1073/pnas.0909167106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derflinger S., Sorg C., Gaser C., Myers N., Arsic M., Kurz A., et al. (2011). Grey-matter atrophy in Alzheimer’s disease is asymmetric but not lateralized. J. Alzheimers Dis. 25 347–357 [DOI] [PubMed] [Google Scholar]
- Galaburda A. M., Sherman G. F., Rosen G. D., Aboitiz F., Geschwind N. (1985). Developmental dyslexia: four consecutive patients with cortical anomalies. Ann. Neurol. 18 222–233 10.1002/ana.410180210 [DOI] [PubMed] [Google Scholar]
- Ghildiyal M., Zamore P. D. (2009). Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 10 94–108 10.1038/nrg2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez A. J., Cinalli R. M., Glasner M. E., Enright A. J., Thomson J. M., Baskerville S., et al. (2005). MicroRNAs regulate brain morphogenesis in zebrafish. Science 308 833–838 10.1126/science.1109020 [DOI] [PubMed] [Google Scholar]
- Good K., Ciosk R., Nance J., Neves A., Hill R. J., Priess J. R. (2004). The T-box transcription factors TBX-37 and TBX-38 link GLP-1/Notch signaling to mesoderm induction in C. elegans embryos. Development 131 1967–1978 10.1242/dev.01088 [DOI] [PubMed] [Google Scholar]
- Hammell C. M., Lubin I., Boag P. R., Blackwell T. K., Ambros V. (2009). nhl-2 modulates microRNA activity in Caenorhabditis elegans. Cell 136 926–938 10.1016/j.cell.2009.01.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Hannon G. J. (2004). MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5 522–531 10.1038/nrg1379 [DOI] [PubMed] [Google Scholar]
- Herbert M. R., Ziegler D. A., Deutsch C. K., O’Brien L. M., Kennedy D. N., Filipek P. A., et al. (2005). Brain asymmetries in autism and developmental language disorder: a nested whole-brain analysis. Brain 128 213–226 10.1093/brain/awh330 [DOI] [PubMed] [Google Scholar]
- Hsieh Y. W., Chang C., Chuang C. F. (2012). The microRNA mir-71 inhibits calcium signaling by targeting the TIR-1/Sarm1 adaptor protein to control stochastic L/R neuronal asymmetry in C. elegans. PLoS Genet. 8:e1002864 10.1371/journal.pgen.1002864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R. J., Jr., Chang S., Etchberger J. F., Ortiz C. O., Hobert O. (2005). MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc. Natl. Acad. Sci. U.S.A. 102 12449–12454 10.1073/pnas.0505530102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R. J., Hobert O. (2003). A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426 845–849 10.1038/nature02255 [DOI] [PubMed] [Google Scholar]
- Kishimoto N., Asakawa K., Madelaine R., Blader P., Kawakami K., Sawamoto K. (2013). Interhemispheric asymmetry of olfactory input-dependent neuronal specification in the adult brain. Nat. Neurosci. 16 884–888 10.1038/nn.3409 [DOI] [PubMed] [Google Scholar]
- Li Y., Wang F., Lee J. A., Gao F. B. (2006). MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 20 2793–2805 10.1101/gad.1466306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev E. V., Zhang J., Carrasco M. A., Maniatis T. (2007). The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell 27 435–448 10.1016/j.molcel.2007.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyasaka N., Morimoto K., Tsubokawa T., Higashijima S., Okamoto H., Yoshihara Y. (2009). From the olfactory bulb to higher brain centers: genetic visualization of secondary olfactory pathways in zebrafish. J. Neurosci. 29 4756–4767 10.1523/JNEUROSCI.0118-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oertel-Knochel V., Linden D. E. (2011). Cerebral asymmetry in schizophrenia. Neuroscientist 17 456–467 10.1177/1073858410386493 [DOI] [PubMed] [Google Scholar]
- Orom U. A., Nielsen F. C., Lund A. H. (2008). MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell 30 460–471 10.1016/j.molcel.2008.05.001 [DOI] [PubMed] [Google Scholar]
- Pierce-Shimomura J. T., Faumont S., Gaston M. R., Pearson B. J., Lockery S. R. (2001). The homeobox gene lim-6 is required for distinct chemosensory representations in C. elegans. Nature 410 694–698 10.1038/35070575 [DOI] [PubMed] [Google Scholar]
- Poole R. J., Hobert O. (2006). Early embryonic programming of neuronal left/right asymmetry in C. elegans. Curr. Biol. 16 2279–2292 10.1016/j.cub.2006.09.041 [DOI] [PubMed] [Google Scholar]
- Priess J. R. (2005). “Notch signaling in the C. elegans embryo,” in WormBook ed. The C. elegans Research Community (Pasadena, CA: WormBook; ) 1–16 Available at: http://www.wormbook.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renteria M. E. (2012). Cerebral asymmetry: a quantitative, multifactorial, and plastic brain phenotype. Twin Res. Hum. Genet. 15 401–413 10.1017/thg.2012.13 [DOI] [PubMed] [Google Scholar]
- Rogers L. J., Vallortigara G., Andrew R. J. (2012). Divided Brains: the Biology and Behaviour of Brain Asymmetries. Cambridge: Cambridge University Press [Google Scholar]
- Sagasti A., Hisamoto N., Hyodo J., Tanaka-Hino M., Matsumoto K., Bargmann C. I. (2001). The CaMKII UNC-43 activates the MAPKKK NSY-1 to execute a lateral signaling decision required for asymmetric olfactory neuron fates. Cell 105 221–232 10.1016/S0092-8674(01)00313-0 [DOI] [PubMed] [Google Scholar]
- Saito Y., Saito H. (2012). MicroRNAs in cancers and neurodegenerative disorders. Front. Genet. 3:194. 10.3389/fgene.2012.00194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed D., Abdellatif M. (2011). MicroRNAs in development and disease. Physiol. Rev. 91 827–887 10.1152/physrev.00006.2010 [DOI] [PubMed] [Google Scholar]
- Schumacher J. A., Hsieh Y. W., Chen S., Pirri J. K., Alkema M. J., Li W. H., et al. (2012). Intercellular calcium signaling in a gap junction-coupled cell network establishes asymmetric neuronal fates in C. elegans. Development 139 4191–4201 10.1242/dev.083428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwamborn J. C., Berezikov E., Knoblich J. A. (2009). The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell 136 913–925 10.1016/j.cell.2008.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P., Lalonde F., Lepage C., Rabin C., Eckstrand K., Sharp W., et al. (2009). Development of cortical asymmetry in typically developing children and its disruption in attention-deficit/hyperactivity disorder. Arch. Gen. Psychiatry 66 888–896 10.1001/archgenpsychiatry.2009.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signore P., Nosten-Bertrand M., Chaoui M., Roubertoux P. L., Marchaland C., Perez-Diaz F. (1991). An assessment of handedness in mice. Physiol. Behav. 49 701–704 10.1016/0031-9384(91)90305-8 [DOI] [PubMed] [Google Scholar]
- Snelson C. D., Gamse J. T. (2009). Building an asymmetric brain: development of the zebrafish epithalamus. Semin. Cell Dev. Biol. 20 491–497 10.1016/j.semcdb.2008.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T., Patoine C., Abu-Khalil A., Visvader J., Sum E., Cherry T. J., et al. (2005). Early asymmetry of gene transcription in embryonic human left and right cerebral cortex. Science 308 1794–1798 10.1126/science.1110324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T., Walsh C. A. (2006). Molecular approaches to brain asymmetry and handedness. Nat. Rev. Neurosci. 7 655–662 10.1038/nrn1930 [DOI] [PubMed] [Google Scholar]
- Tanaka-Hino M., Sagasti A., Hisamoto N., Kawasaki M., Nakano S., Ninomiya-Tsuji J., et al. (2002). SEK-1 MAPKK mediates Ca2+ signaling to determine neuronal asymmetric development in Caenorhabditis elegans. EMBO Rep. 3 56–62 10.1093/embo-reports/kvf001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R. W., Hsieh Y. W., Gamse J. T., Chuang C. F. (2010). Making a difference together: reciprocal interactions in C. elegans and zebrafish asymmetric neural development. Development 137 681–691 10.1242/dev.038695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel E. R., Sagasti A., Bargmann C. I. (1999). Lateral signaling mediated by axon contact and calcium entry regulates asymmetric odorant receptor expression in C. elegans. Cell 99 387–398 10.1016/S0092-8674(00)81525-1 [DOI] [PubMed] [Google Scholar]
- Vanhoven M. K., Bauer Huang S. L., Albin S. D., Bargmann C. I. (2006). The claudin superfamily protein nsy-4 biases lateral signaling to generate left–right asymmetry in C. elegans olfactory neurons. Neuron 51 291–302 10.1016/j.neuron.2006.06.029 [DOI] [PubMed] [Google Scholar]
- Vasudevan S., Tong Y., Steitz J. A. (2007). Switching from repression to activation: microRNAs can up-regulate translation. Science 318 1931–1934 10.1126/science.1149460 [DOI] [PubMed] [Google Scholar]
- Wes P. D., Bargmann C. I. (2001). C. elegans odour discrimination requires asymmetric diversity in olfactory neurons. Nature 410 698–701 10.1038/35070581 [DOI] [PubMed] [Google Scholar]
- White J. G., Southgate E., Thomson J. N., Brenner S. (1986). The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 314 1–340 10.1098/rstb.1986.0056 [DOI] [PubMed] [Google Scholar]
- Yu S., Avery L., Baude E., Garbers D. L. (1997). Guanylyl cyclase expression in specific sensory neurons: a new family of chemosensory receptors. Proc. Natl. Acad. Sci. U.S.A. 94 3384–3387 10.1073/pnas.94.7.3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zabinsky R., Teng Y., Cui M., Han M. (2011). microRNAs play critical roles in the survival and recovery of Caenorhabditis elegans from starvation-induced L1 diapause. Proc. Natl. Acad. Sci. U.S.A. 108 17997–18002 10.1073/pnas.1105982108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y., Chiu H., Domenger D., Chuang C. F., Chang C. (2012). The lin-4 microRNA targets the LIN-14 transcription factor to inhibit netrin-mediated axon attraction. Sci. Signal. 5 ra43 10.1126/scisignal.2002437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y., Chiu H., Zinovyeva A., Ambros V., Chuang C. F., Chang C. (2013). Developmental decline in neuronal regeneration by the progressive change of two intrinsic timers. Science 340 372–376 10.1126/science.1231321 [DOI] [PMC free article] [PubMed] [Google Scholar]