SUMMARY
Recent studies suggest a hierarchical model in which lineage-determining factors act in a collaborative manner to select and prime cell-specific enhancers, thereby enabling signal-dependent transcription factors to bind and function in a cell type-specific manner. Consistent with this model, TLR4 signaling primarily regulates macrophage gene expression through a pre-existing enhancer landscape. However, TLR4 signaling also induces priming of ~3000 enhancer-like regions de novo, enabling visualization of intermediates in enhancer selection and activation. Unexpectedly, we find that enhancer transcription precedes local mono- and di-methylation of histone H3 lysine 4 (H3K4me1/2). H3K4 methylation at de novo enhancers is primarily dependent on the histone methyltransferases Mll1, Mll2/4 and Mll3, and is significantly reduced by inhibition of RNA polymerase II elongation. Collectively, these findings suggest an essential role of enhancer transcription in H3K4me1/2 deposition at de novo enhancers that is independent of potential functions of the resulting eRNA transcripts.
INTRODUCTION
Molecular mechanisms enabling cell-specific transcriptional responses to intra- and extra-cellular signals remain poorly understood. Genome-wide location analysis of most signal-dependent transcription factors indicates that the vast majority of their binding sites are in distal intra- and intergenic locations that exhibit epigenomic features associated with enhancers (Barish et al., 2010; Carroll et al., 2006; Heinz et al., 2010; John et al., 2011; Lefterova et al., 2010; Nielsen et al., 2008). These findings are consistent with quantitatively important roles of enhancers in signal-dependent transcriptional responses, as well as evolutionary conservation of enhancer elements (Ghisletti et al., 2010; Pennacchio et al., 2006; Woolfe et al., 2005). An important insight enabling the identification of potential enhancer-like regions was provided by the definition of histone methylation signatures specific for enhancers, i.e. high enrichment histone H3 lysine 4 mono- and dimethylation (H3K4me1 and H3K4me2, respectively) and low enrichment of H3K4me3 compared to promoters (Heintzman et al., 2007). While genomic regions exhibiting these features are not necessarily functional enhancers, it appears that the vast majority of regions that do function as enhancers exhibit these characteristics (Heintzman et al., 2007; Heinz et al., 2010; Rada-Iglesias et al., 2011).
In cases in which cell signaling induces nuclear entry and DNA binding of regulated transcription factors, such as steroid hormone receptors and NF-κB, the majority of binding events take place at genomic locations that exhibit pre-existing enhancer-like features (Barish et al., 2010; John et al., 2011). Because the complement of active cis-regulatory elements changes across cell types, these findings introduced the notion that enhancers are largely responsible for cell type-specific gene expression (Heintzman et al., 2009; Thurman et al., 2012; Visel et al., 2009). Recent studies suggest a hierarchical model in which relatively simple combinations of the lineage determining factors required for differentiation of a particular cell type act in a collaborative manner to prime a large fraction of the enhancer-like regions in that cell type (Heinz et al., 2010). These regions provide cell-specific sites of open chromatin that enable access by signal-dependent factors. As a consequence, widely expressed signal-dependent transcription factors are localized to cell-specific enhancers, resulting in cell-specific transcriptional responses (Barish et al., 2010; Biddie et al., 2011; Heinz et al., 2010; Lefterova et al., 2010; Mullen et al., 2011; Nielsen et al., 2008; Trompouki et al., 2011). In macrophages, for example, collaborative interactions between the lineage-determining factors PU.1, C/EBPα/β, and AP-1 generate a large fraction of the cell-specific sites of open chromatin that are responsible for signal-dependent responses to liver X receptor (LXR) signaling (Heinz et al., 2010).
Recent studies of nuclear RNAs in neurons (Kim et al., 2010), cancer cells (Hah et al., 2011; Wang et al., 2011) and macrophages (De Santa et al., 2010) have led to the unexpected finding that many enhancers direct the expression of RNA transcripts, referred to as eRNAs, in a manner that is correlated with the expression of nearby genes. These observations raise a number of questions regarding the mechanism and purpose of enhancer-directed transcription. To investigate the link between enhancer transcription and signal-dependent gene activation we evaluated time-dependent changes in transcription factor binding, nascent RNA transcripts and genomic features of macrophage enhancers following ligation of TLR4 with Kdo2-lipid A (KLA), a highly specific TLR4 agonist (Raetz et al., 2006). TLR4 ligation results in activation of several latent, signal-dependent transcription factors, including NF-κB, interferon regulatory factors (IRFs), AP-1 factors, and STAT factors, that act in a combinatorial manner to regulate the expression of thousands of genes (Medzhitov and Horng, 2009; Smale, 2012). In addition to using pre-primed regulatory elements, we found that TLR4 signaling also alters macrophage gene expression through the selection and activation of a large number of new enhancers and inactivation of a subset of basally active enhancers. Unexpectedly, the evaluation of intermediates in the selection and activation of new enhancer-like elements suggests an essential role of enhancer transcription in the deposition of the H3K4me1 and H3K4me2 enhancer marks.
RESULTS
TLR4 signaling induces priming and activation of de novo enhancer-like regions
To investigate temporal effects of TLR4 activation on transcription, epigenetics and function of signal-dependent enhancers, we quantified nascent transcripts, enhancer-associated histone modifications, and transcription factor binding as a function of time following treatment of macrophages with the TLR4 agonist KLA (Raetz et al., 2006). Global nuclear run-on coupled to deep sequencing (GRO-Seq) analysis identified ~2200 nascent RNA transcripts induced by more than 2-fold (Figure S1A, Table S1) which were highly enriched for functional annotations related to inflammation and immunity (Figure S1B). Hierarchical clustering segregated these transcripts into early, late, and persistent subsets (Figures S1A and S1C), consistent with recent findings (Bhatt et al., 2012). Notably, 22% of the KLA-induced nascent transcripts were induced as early as 10 minutes, exemplified by Tnf and Irf1 (Figure S1D). Conversely, ~2800 genes were downregulated more than 2-fold upon TLR4 stimulation (Figure S1A, Table S1). Repressed genes, exemplified by Hhex (Figure S1D) were significantly enriched for annotations linked to lysosome function and metabolism (Figure S1B).
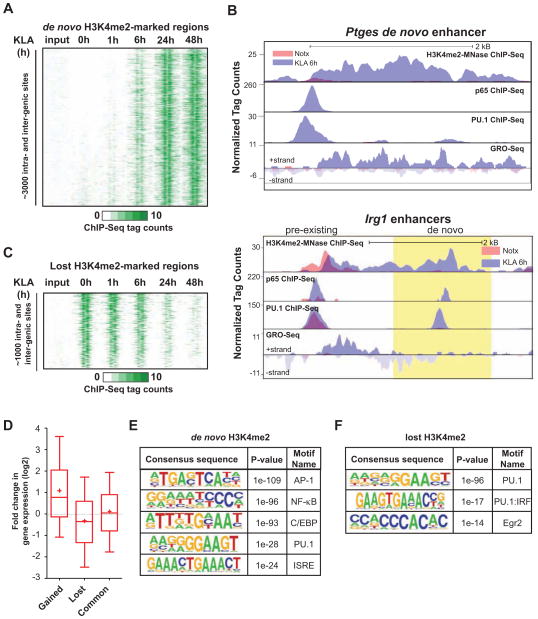
Potential enhancers were identified by performing H3K4me2 ChIP-Seq of MNase-treated chromatin obtained following 0, 1, 6, 24 and 48 hours of KLA treatment. This approach identified ~32,000 inter- and intra-genic marked locations containing a nucleosome-depleted region prior to KLA treatment, referred to as ‘pre-existing’ enhancers (Table S1). These regions were highly enriched for motifs recognized by PU.1, C/EBP and AP-1 factors, consistent with previous findings (Ghisletti et al., 2010; Heinz et al., 2010). Notably, ~3000 previously unmarked regions gained H3K4me2 upon KLA stimulation (Figure 1A, Table S1), exemplified by the new enhancer-like elements in the vicinity of Ptges and Irg1 (Figure 1B and S1E). We hereafter refer to these regions as ‘de novo’ enhancers. In contrast, ~1000 regions lost this mark following KLA treatment (Figure 1C, Table S1). Gain and loss of H3K4me2 at enhancer-like regions was highly correlated with expression of nearby genes (Figure 1D). Ten percent of KLA-induced genes, mostly belonging to the group of persistently up-regulated genes, were associated with the emergence of de novo enhancers in their vicinity.
Figure 1. TLR4-induced remodeling of the macrophage enhancer landscape.
(A) Heat map of normalized tag densities for the H3K4me2-MNase histone mark at inter- and intragenic de novo enhancers. Two kb regions are shown centered at the midpoints of the nucleosome free regions (NFR).
(B) UCSC genome browser images for Ptges and Irg1 enhancers ~10 kb upstream of the TSS of the coding genes. Normalized tag counts for the indicated features are shown under no treatment (Notx) and 6h KLA stimulation. The region of de novo enhancer formation upstream of Irg1 is highlighted in yellow.
See also Figure S1E.
(C) Heat map of normalized tag densities for the H3K4me2-MNase histone mark at inter- and intragenic enhancers lost upon KLA-stimulation. Two kb regions are shown centered at the midpoints of the NFRs.
(D) Box-and-whisker plots of the fold change in expression of genes located < 100 kB from the gained, lost or common enhancers. Boxes encompass the 25th to 75th % changes. Whiskers extend to 10th and 90th percentiles. The median fold change is indicated by the central horizontal bar and the mean fold change upon 1h KLA-treatment is indicated by +.
(E and F) Sequence motifs associated with (E) de novo and (F) lost H3K4me2-marked enhancers. See also Figure S1.
p65 collaborates with PU.1 and C/EBPs to select de novo enhancers
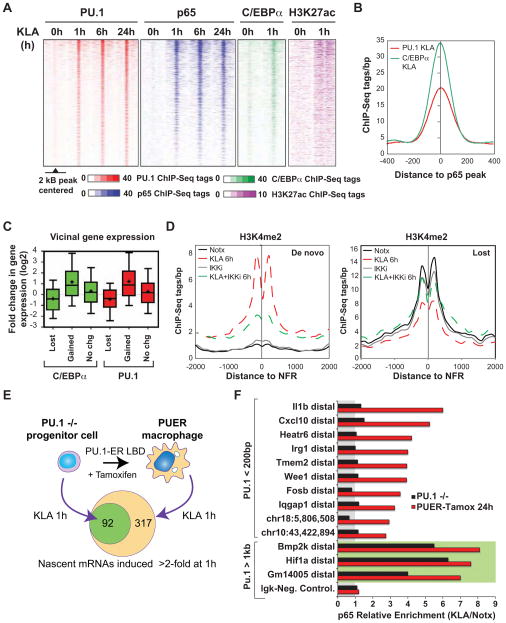
The appearance of de novo enhancers raised the question as to the mechanism of their selection. Genomic regions that gained H3K4me2 exhibited highly significant enrichment for motifs recognized by AP-1, NF-κB, C/EBP, IRF/STATs and PU.1 (Figure 1E), while regions exhibiting loss of the H3K4me2 mark were enriched for PU.1, a specific PU.1/ISRE composite, and Egr2-like motifs (Figure 1F). The marked enrichment of AP-1, NF-κB, C/EBP, PU.1, and ISRE motifs in de novo enhancer-like regions suggested that signal-dependent factors collaborate with macrophage lineage-determining factors to prime and activate these sites. To directly investigate these relationships, we performed ChIP-Seq for PU.1, C/EBPα, and the NF-κB subunit p65 following 1, 6 and 24 hours KLA treatment. In addition to binding at pre-existing enhancers, as expected, these factors also bound at de novo enhancers in a KLA-dependent manner (Figures 2A and 1B), with the majority of their binding sites located within 100 bp of each other (Figure 2B). This was also associated with a concomitant increase in H3K27Ac, a modification that marks active enhancers (Figure 2A) (Creyghton et al., 2010). Gain and loss of PU.1 and CEBPα binding at de novo and lost pre-existing enhancers was highly correlated with increased and decreased expression of nearby genes, respectively (Figure 2C), consistent with the pattern for H3K4me2 itself.
Figure 2. Mechanisms of de novo enhancer assembly.
(A) Heat maps of PU.1, p65, and C/EBPα binding and deposition of H3K27ac at de novo enhancers as a function of time following KLA treatment.
(B) Distance relationship between p65 and PU.1 or C/EBPα peaks at de novo enhancers exhibiting p65 binding upon 1h KLA treatment.
(C) Box-and-whisker plots of the fold change in expression of genes located < 100 kB from the C/EBPα and PU.1 peaks gained, lost or exhibiting no change upon 1h KLA stimulation. Data is plotted as shown in Figure 1D.
(D) Effect of 1 h pretreatment of macrophages with an IKK inhibitor (30 μM) on the profile of H3K4me2-MNase ChIP-Seq tags around de novo and lost enhancers. See also Figures S2A and S2B.
(E) Role of PU.1 in TLR4-dependent gene activation. PU.1−/− hematopoietic progenitors gain macrophage phenotype by the expression of tamoxifen- activatable PU.1 protein. Upon 1h KLA-stimulation 317 genes are induced in PUER cells whereas only 92 of these genes are induced in PU.1−/− cells (Fold change >2, RPKM> 0.5, FDR<0.01). See also Figure S2C.
(F) ChIP assay of p65 enrichment at PU1-dependent and PU1-independent (green background) enhancers quantified by ChIP-qPCR in PU.1−/− and in PUER cells treated with tamoxifen for 24 hours with or without KLA. Relative Enrichment for ratio of KLA/Notx is presented. The enhancer is associated to the nearest expressed gene if it is < 100 kb away from the amplicon location, otherwise the center position of the amplicon in the genome is indicated. Relative enrichments are representative of individual experiments performed in duplicate.
To investigate roles of NF-κB in directing formation of de novo enhancers, we evaluated gene expression and H3K4me2 deposition in control and KLA-treated macrophages in the presence or absence of an inhibitor of IκB kinase (IKK), which prevents signal-dependent degradation of IκBs (Kobori et al., 2004) required for nuclear entry of NF-κB. This inhibitor greatly reduced activation of NF-κB target genes, as expected (Figure S2A). Inhibition of IKK also greatly reduced the deposition of H3K4me2 at de novo enhancers and prevented the reduction of H3K4me2 at lost enhancer-like regions (Figure 2D), but did not affect H3K4me2 at pre-existing enhancers under basal conditions (Figure S2B).
These results suggested that NF-κB collaborates with PU.1 to establish a subset of de novo enhancers. To investigate this possibility, we performed ChIP-qPCR experiments for p65 in in PU.1-deficient myeloid progenitor cells (PU.1−/− cells) and PU.1−/− cells reconstituted with an inducible PU.1-ER fusion protein (PUER) (Walsh et al., 2002) following 1 h of KLA stimulation. These studies suggest that PU.1 is required for a substantial fraction of the KLA-induced gene expression observed in fully differentiated macrophages (Figure 2E, Table S1), as well as for the KLA-induced binding of p65 to enhancers where these factors bind within 200 bp of each other (Figure 2F). In contrast, p65 binding is intact at enhancer elements in PU.1−/− cells that were in the vicinity of genes that were induced by KLA in both cells types and where the nearest PU.1 binding sites are greater than 1 kb away (Figure 2F).
Enhancer acetylation and transcription precede enhancer H3K4 methylation
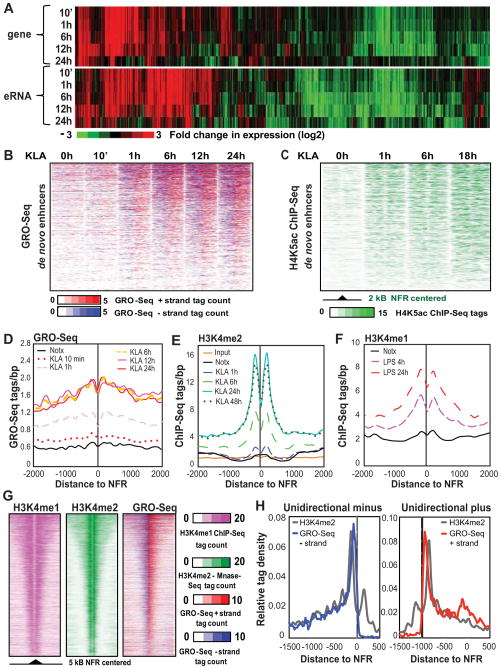
We next compared the kinetics of eRNA production to signal-dependent changes in mRNA transcription and histone modifications associated with de novo enhancers. This analysis was limited to the ~16,000 pre-existing and ~1550 de novo enhancers residing in intergenic regions to enable discrimination of enhancer-derived RNA transcripts from mRNAs (Table S1). GRO-Seq analysis revealed widespread and inducible transcription from enhancers, in agreement with previous studies (De Santa et al., 2010; Hah et al., 2011; Kim et al., 2010; Wang et al., 2011). Hierarchical clustering of genes that were associated with vicinal intergenic eRNAs within 100 kb revealed that neighboring eRNAs and mRNAs are significantly co-regulated over the entire time course (Figures 3A–B and S3A–D).
Figure 3. Relationship between eRNA expression and deposition of H4K5ac and H3K4me2.
(A) Hierarchical clustering and heat map of the fold change in eRNA and coding gene expression (eRNAs: RPKM>1, genes: FDR 0.01, RPKM>0.5). See also Figure S3A–S3D.
(B) Heat maps of normalized tag densities for GRO-Seq around 2 kb of de novo enhancers centered to nucleosome free regions as a function of time following KLA treatment.
(C) Temporal profile of H4K5ac normalized tag densities tag counts around 2 kb of de novo enhancers as a function of time following KLA treatment. See also Figure S3E–S3G.
(D) Distribution of GRO-Seq tags at around NFRs at de novo H3K4me2-associated enhancers as a function of time following KLA treatment.
(E) Temporal profile of change in H3K4me2-MNase ChIP-Seq tags at de novo enhancers following KLA treatment. See also Figure S3H.
(F) Coverage of H3K4me1 ChIP-Seq tags at de novo enhancers following LPS treatment (Ostuni et al., 2013).
(G) Heat map comparing the distribution of intergenic H3K4me1 and H3K4me2 regions and GRO-Seq signal. Pre-existing H3K4me2 regions centered to NFR are presented. See also Figure S3I.
(H) Comparison of GRO-Seq and H3K4me2 cumulative tag densities at enhancers characterized by exclusive unidirectional eRNA initiation from minus (left) or plus (strands). GRO-Seq signal is multiplied by ten compared to H3K4me2 signal.
We next investigated temporal relationships between histone acetylation and eRNA production. Acetylation of histone H3 at K9, K14 and K27, and of histone H4 at K5, K8 and K12 is correlated with active enhancers and promoters and is often rapidly altered in a signal-dependent manner. Many of these marks result from recruitment of p300 histone acetyltransferase (HAT) and play an important role in subsequent binding of transcriptional effectors (Schiltz et al., 1999; Szerlong et al., 2010). In particular, H4K5/8ac is rapidly induced at TLR4-responsive promoters following treatment with TLR4 agonists (Escoubet-Lozach et al., 2011; Hargreaves et al., 2009). Time course analysis of H4K5 acetylation indicated that the kinetics of KLA-induced eRNA expression and deposition of H4K5 acetylation were highly concordant at de novo enhancers (Figure 3B and 3C). Similarly, changes in eRNA expression and H4K8ac at pre-existing enhancers were highly correlated (Figure S3E). H4K8ac and H3K27ac showed very strong correlation (r=0.9) in line with the common mechanism of deposition (Figure S3F).
Acetylation of histone H3 and H4 has been shown to modulate the association of bromodomain-containing 4 protein (Brd4) with chromatin and the subsequent recruitment of initiation cofactor Mediator and positive transcription elongation factor b (P-TEFb) (Jang et al., 2005; Yang et al., 2005). Brd4 converts P-TEFb into an active form which then phosphorylates the negative-acting elongation factor complex, NELF, the DRB-sensitivity inducing complex, DSIF (Spt4/5) and the C-terminal domain Serine 2 (Ser2P) of RNA polymerase II (RNAPII) to convert the pre-initiation complex to the elongation complex (Marshall et al., 1996; Renner et al., 2001). Consistent with this, acquisition of acetylation at induced enhancers and eRNA elongation was associated with the presence of increased Ser2P RNAPII at 1 h of KLA treatment (Figure S3G).
We next compared the kinetics of eRNA transcription at enhancers to the timing of appearance of H3K4-modified histones. In striking contrast to eRNA production, which was clearly induced at 1 h, reached near maximal levels of expression by 6 h and persisted for 24 h (Figure 3D), increases in H3K4me2 were barely detectable at 1 h, and accumulated progressively from 6 to 24 h, (Figure 1A and 3E). Analysis of recently reported genome wide data for H3K4me1 upon LPS treatment of macrophages (Ostuni et al., 2013) revealed a similar time-dependent increase at de novo enhancers (Figure 3F). This temporal pattern was independently confirmed by locus-specific qPCR, (Figure S3H), indicating that H3K4 methylation is initially detected concomitantly with or after initial enhancer transcription, and that H3K4 methylation increases progressively in the face of persistent eRNA expression (Figure 3D and 3E). Of note, comparison of the length of intergenic GRO-Seq signal at all enhancer-like regions showed a strong correlation with spread of H3K4me1 and H3K4me2 (Figure 3G, S3I). In line with this, H3K4me2-marked intergenic regions had on average 70% coverage by GRO-Seq signal (Figure S3I). Although most enhancers direct some degree of bi-directional transcription, a subset was identified that selectively direct uni-directional transcription. Comparison of averaged GRO-Seq and H3K4me2 tag densities at these enhancers yielded a close correlation with asymmetrically distributed H3K4me2 (Figure 3H).
eRNA elongation and H3K4 methylation
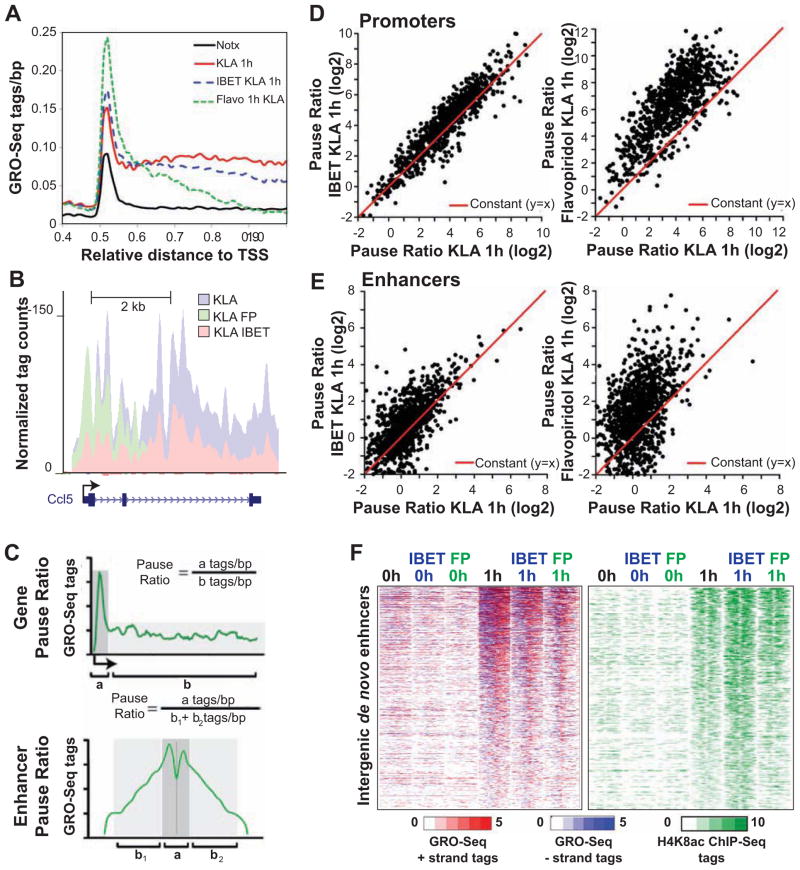
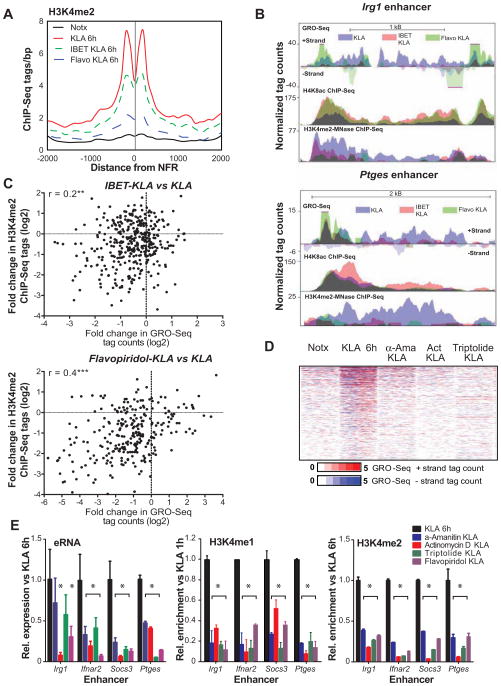
Taken together, these findings suggested that H3K4 methylation could be a consequence of transcriptional elongation at enhancers. To investigate this possibility, we assessed the effects of inhibiting RNA PolII elongation on the H3K4me2 status after KLA treatment. We initially tested two different elongation inhibitors: the cyclin-dependent kinase (cdk) inhibitor flavopiridol, which at low concentrations preferentially inhibits the Cdk9 activity of P-TEFb (Chao and Price, 2001); and IBET151, a selective inhibitor of BET (bromodomain and extra terminal domain) protein binding to acetylated histones, which disrupts the assembly of histone acetylation-dependent chromatin complexes that regulate inflammatory gene expression (Dawson et al., 2011; Nicodeme et al., 2010). At 1 h, the induction of 47% of KLA-up-regulated genes was inhibited by more than 1.5-fold by at least one of the two drugs (Table S1). Of these, 56% were repressed by both drugs. In line with previous data, both drugs suppressed the transcription of multiple inflammatory mediators (Table S1).
Both drugs affected the elongation of KLA-induced nascent transcripts as evidenced by a decrease in the cumulative GRO-Seq tags beyond the TSS (Figure 4A), exemplified by Ccl5 (Figure 4B), with the effect of flavopiridol being more pronounced. In addition, flavopiridol, and to a lesser extent the IBET151 inhibitor, increased GRO-Seq tag densities just downstream of the TSS (Figure 4A), consistent with accumulation of paused RNAPII. We quantified the effect on elongation efficiency by dividing promoter-proximal tag counts by gene body tag counts (Gene Pause Ratio; Figure 4C). This analysis indicated that 44% and 73% of all KLA-induced genes exhibited an over 2-fold increase in Pause Ratio after pretreatment with IBET151 and flavopiridol, respectively (Figure 4D), indicating the expected inhibitory effects on transcriptional elongation. In addition, transcription of 46% and 60% of all KLA-induced eRNAs exhibited an over 2-fold increase in Enhancer Pause Ratio after pretreatment with IBET151 and flavopiridol, respectively (Figure 4E). Inhibition of elongation was also evident at pre-existing induced inflammatory enhancers (Figure S4). Inhibition of eRNA elongation had minimal effects on the overall deposition of H4K8ac at KLA-induced novel enhancers, as flavopiridol and IBET151 pretreatment, respectively, left acetylation of 70–90 % of these sites largely unchanged (Figure 4F).
Figure 4. IBET151 and flavopiridol decrease mRNA and eRNA elongation without affecting H4K8ac levels.
(A) Distribution of average GRO-Seq tag densities on the + strand around the TSS of KLA- induced genes subject to inhibition by IBET151 or flavopiridol (Flavo) pretreatment (1 μM).
(B) UCSC Genome browser image depicting normalized GRO-Seq tag counts at Ccl5 coding gene in KLA-stimulated cells pretreated or not with Flavopiridol or IBET151 inhibitor for 1h.
(C) The Gene Pause Ratio (upper figure) was defined as the ratio of strand-specific GRO-Seq tag density found within the proximal promoter (a = −25 bp to +175 bp) to the GRO-Seq tag density found at the gene body (b = +175 to end of the gene). The Enhancer Pause Ratio (lower figure) was calculated as the ratio of GRO-Seq tag density found within the proximal enhancer region (a = +/− 250 bp) to the GRO-Seq tag density found at the distal enhancer region (b1/2= +/− 250 bp to +/− 1250 bp).
(D) Scatter plot of mRNA Pause Ratios comparing KLA-stimulated control to IBET151-pretreated (left) or Flavopiridol pretreated samples. KLA-induced genes exhibiting promoter RPKM > 2 were included in the analysis.
(E) Scatter plot of the change in enhancer Pause Ratios comparing KLA-stimulated control to IBET151-pretreated (left) or Flavopiridol pretreated samples. De novo enhancers exhibiting >1.5-induction in eRNA level and RPKM > 0.5 were included in the analysis. See also Figure S4.
(F) Heat maps of normalized tag densities for H4K8ac ChIP-Seq and GRO-Seq around 2 kb of de novo enhancers in KLA-stimulated control and IBET151 or Flavopiridol (FP) pretreated samples
To investigate the relationship between eRNA transcription and deposition of H3K4me2 at de novo enhancers, we performed MNase ChIP-Seq for H3K4me2, with or without drug pretreatment, six hours after KLA stimulation. The time point was chosen to minimize any side effects due to drug treatment while capturing changes in H3K4me2 evident from this point onwards (Figure 1A). We found that inhibition of eRNA elongation by IBET151 and flavopiridol was correlated with a decrease in the deposition of H3K4me2 by at least 1.5-fold at ~40 % and ~70 % of these de novo enhancers (Figure 5A), respectively, exemplified by Irg1, Ptges and Socs3 enhancers (Figures 5B and S5A). The effectiveness of drug treatment on reducing eRNA expression at individual enhancers was significantly correlated with a corresponding local reduction of H3K4me2 (Figure 5C). A similar effect was seen at pre-existing induced enhancers. Although some genes exhibited decreased H3K4me2 without an apparent decrease in total GRO-Seq signal, this was primarily due to offsetting effects of the drugs on increasing short transcripts due to stalled RNA Pol II near the enhancer transcriptional start sites, while decreasing elongating transcripts, particularly in the case of flavopiridol (e.g. Figure 5B). In contrast to the highly significant correlation between effects of drugs on enhancer transcription and local H3K4me2, there was no significant correlation between effects of drugs on transcription at gene promoters and H3K4me2 at the nearest de novo enhancer (Figure S5B).
Figure 5. Inhibition of eRNA elongation prevents deposition of H3K4me2.
(A) Effect of IBET151 and flavopiridol pretreatment on the profile of H3K4me2-MNase ChIP-Seq tags around de novo enhancers which exhibit > 1.5-fold drug-induced decrease in eRNA induction.
(B) UCSC genome browser images for Irg1 and Ptges enhancers. Normalized tag counts for GRO-Seq (1h) H4K8ac ChIP-Seq (1h) and H3K4me2 MNase ChIP-Seq (6h) in KLA-stimulated macrophages pretreated with IBET151 or Flavopiridol. See also Figure S5A.
(C) Scatter plots depicting the relationship between the fold change in GRO-Seq tag count and H3K4me2-MNAse ChIP-Seq tag count at de novo enhancers. Enhancers with GRO-Seq RPKM levels above 1 within de novo enhancer regions were included in the analysis. Pearson correlation values (r) are also shown: *** p<0.0001, **p=0.0012. See also S5B.
(D) Heat maps of normalized tag densities for GRO-Seq around 2 kb of de novo enhancers in KLA-stimulated control and α-amanitin (a-Ama), actinomycin D (ActD) and triptolide pretreated samples. Data are presented as mean ± SD. See also S5D and S5E.
(E) Fold change in eRNA expression and H3K4me1/2 deposition at Irg1, Ifnar2, Socs3 and Ptges de novo enhancers quantified by GRO-Seq (1h) or ChIP-qPCR (6h) in KLA-stimulated macrophages pretreated with transcriptional inhibitors. Data are presented as mean ± SD. *P<0.05 versus KLA treated. See also S5F.
To investigate whether the drug effects on H3K4me2 might be due to changes in transcription factor recruitment, we assessed the influence of flavopiridol and IBET151 on the binding of p65 and PU.1. Some locus-specific effects on the binding of p65 were observed following flavopiridol treatment, whereas some locus-specific effects on PU.1 recruitment were observed following treatment with both drugs. However, the degree to which drugs inhibited H3K4me2 deposition was similar at enhancers that exhibited no decrease in p65/PU.1 binding compared to those at which binding was affected (Figure S5C), suggesting that the changes in H3K4me2 are not explained by the altered TF binding. This interpretation is consistent with the general lack of effect of the drugs on deposition of H4K8ac (Figure 4F), which is dependent on recruitment of PU.1 and p65.
To further exclude potential off-target effects of inhibitors of Pol II on enhancer H3K4 methylation, we analyzed effects of three additional drugs known to affect different stages of transcription: α-amanitin, which traps the conformation of RNAPII to prevent nucleotide incorporation and translocation of the transcript (Brueckner and Cramer, 2008), actinomycin D, which intercalates to DNA (Perry and Kelley, 1970) and triptolide, which blocks the formation of the “transcription bubble” and hence inhibits initiation of transcription (Titov et al., 2011). At 1 h, α-amanitin, actinomycin D and triptolide reduced the induction of 40%, 80% and 90% of KLA-up-regulated genes, respectively, by more than 1.5-fold (Table S1). In each case, inhibition of eRNA transcription was associated with a corresponding decrease in the deposition of local H3K4me2 and H3K4me1 at de novo enhancers (Figure 5D and S5D). α-amanitin and triptolide had a limited effect on H4K8ac, but actinomycin D also abolished this mark (Figure S5D and S5E).
Finally, we sought to determine whether H3K4me2 deposition requires the presence of the eRNA transcript itself. We used locked nucleic acid antisense oligos (ASOs), which mediate nuclear RNA degradation via an RNase H pathway (Bennett and Swayze, 2010), to knock down the eRNAs for Irg1 and Ifnar2 enhancers (Figure S5F). Importantly, reduction in eRNA expression was not associated with a decrease in H3K4me2 deposition (Figure S5F). Altogether these results suggest that H3K4me2 deposition requires enhancer transcription but not the eRNA transcript itself.
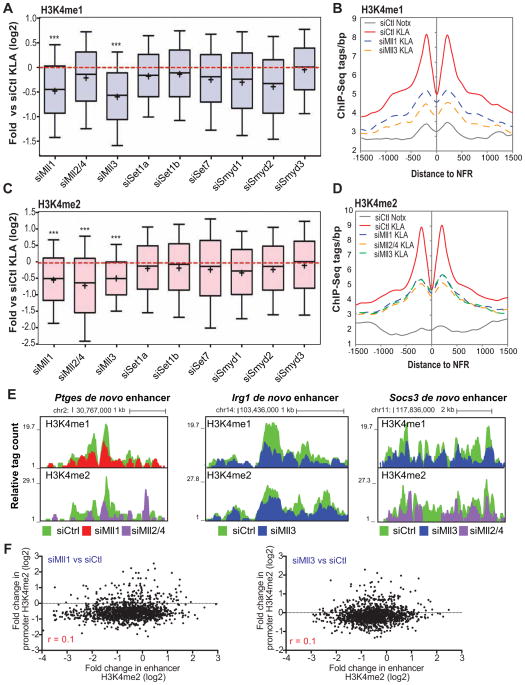
Roles of Mll family members in H3K4 methylation at de novo enhancers
The requirement for active enhancer transcription for H3K4 mono- and dimethylation of de novo enhancers raised the question how methylation is deposited to the enhancers. To investigate this question, we knocked down all known HMTs capable of H3K4 methylation in primary macrophages using specific siRNAs (Figure S6A) and performed ChIP-sequencing for H3K4me1 and H3K4me2. The knockdown of several of the HMTs resulted in significant decreases in the deposition of H3K4me1 at de novo enhancers, with the most pronounced effects (P<1E-100) seen with siRNAs against Mll1 and especially Mll3 (Figure 6A and 6B). In addition to Mll1and Mll3, knockdown of Mll2/4 led to a significant decrease in H3K4me2 at de novo enhancers (Figure 6C, 6D and 6E). A similar effect was seen at pre-existing enhancers exhibiting over 2-fold increase in H3K4me1 (Figure S6A) or H3K4me2 (Figure S6B) upon 6 h KLA-treatment. Importantly, the change in enhancer H3K4me2 caused by the Mll knockdowns did not correlate with nearest promoter methylation, consistent with local effects (Figure 6F).
Figure 6. Mll-family of histone methylatransferases are responsible for the deposition of H3K4me1/2.
(A) Box-and-whisker plots of the fold change in H3K4me1 ChIP-Seq tags at de novo enhancers compared to control siRNA treated sample upon 6h KLA treatment. Data is plotted as shown in Figure 1D. Student’s paired 2-tailed t-test, only P<1E-100 = *** are shown. See also S6A.
(B) Distribution of average H3K4me1 tag densities on the around the NFR-centered de novo enhancers with indicated siRNA treatments.
(C) Box-and-whisker plots of the fold change in H3K4me2 ChIP-Seq tags at de novo enhancers compared to control siRNA treated sample upon 6h KLA treatment. Data is plotted as shown in Figure 1D. Student’s paired 2-tailed t-test, P<1E-100 = *** are shown. See also S6B and S6C.
(D) Profile of H3K4me2-MNase ChIP-Seq tags around de novo after the indicated siRNA treatments.
(E) UCSC Genome browser image depicting normalized H3K4me1 and H3K4me2 ChIP-Seq tag counts upstream of Irg1 gene in KLA-stimulated cells pretreated or not with siRNAs against the indicated histone methyltransferases. De novo enhancers are highlighted in yellow. See also S6D.
(F) Scatter plot of fold change in H3K4me2 ChIP-Seq tags comparing KLA-stimulated control siRNA (siCtl) and siRNAs against Mll1 and Mll3. Pearson correlation values (r) are shown in red.
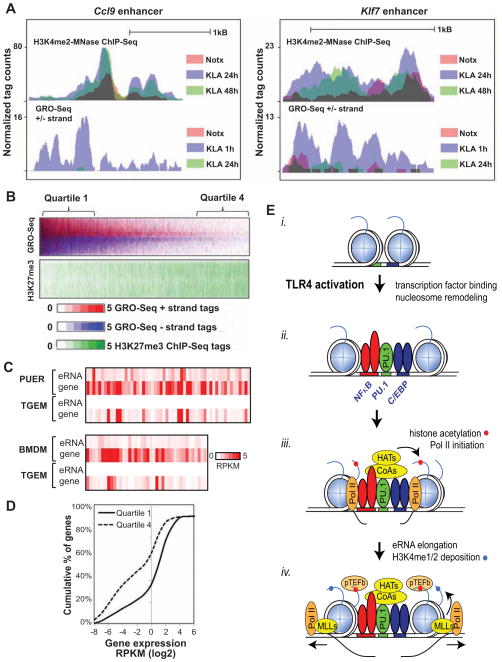
Relationship of H3K4me2 to eRNA expression and H3K27me3 deposition
While the molecular functions of H3K4me2 remain to be established, this mark has previously been suggested to represent a molecular memory of prior transcription of mRNA coding regions (Ng et al., 2003). To investigate this possibility at enhancers, we studied enhancers associated with genes that are induced early in inflammatory response but had returned to basal levels by 24 hours. Such enhancers, exemplified by Ccl9, Klf7, Notch2 and Kcnn4, exhibited a persistent H3K4me2 signal up to 24–48 h after treatment in the absence of continued eRNA transcription at late time points (Figures 7A and S7).
Figure 7. eRNA transcription reflects functionality of enhancers.
(A) UCSC Genome browser image showing the normalized H3K4me2-MNase ChIP-Seq and GRO-Seq tag densities at Ccl9 and Klf7 enhancer. See also ure S7.
(B) Heat map of normalized tag densities for GRO-Seq and H3K27me3 ChIP-Seq 2 kB around the nucleosome-free regions of basal intergenic enhancers.
(C) Heat-map of normalized GRO-seq reads (RPKM) at enhancers exhibiting no eRNA expression in thioglycollate elicited macrophages (TGEM) and at the associated genes compared to myeloid progenitor cells (PUER) and bone marrow derived macrophages (BMDM).
(D) The enhancers with highest (quartile 1) and lowest (quartile 4) eRNA expression in B were associated with expression of the nearest genes. The plot illustrates a cumulative percentage histogram of genes based on their normalized expression level (RPKM).
(E) Model for de novo enhancer assembly. See discussion for details.
Importantly, 10% of the total set of H3K4me2-marked enhancer-like regions in macrophages showed no GRO-Seq tags at any time point, and 30% exhibited fewer than 10 tags in basal conditions (Figure 7B). We therefore investigated whether these regions might have been previously transcribed during the differentiation of macrophages from progenitor cells by examining GRO-Seq signal in the PUER macrophage progenitor cell line and in bone marrow-derived macrophages. Notably, ~70% of the regions defined by the presence of H3K4me2 and absence of detectable GRO-Seq signal in thioglycollate-elicited macrophages exhibited detectable eRNA expression in one or both of these macrophage progenitors (Figure 7C). To investigate what separates enhancer-like regions (defined by H3K4me2 and a nucleosome-free region) associated with low eRNA expression from those of high expression, we divided these regions from high (quartile 1) to low (quartile 4) quartiles based on eRNA expression (Figure 7B) and correlated with H3K27me3, a mark of transcriptional repression (Cao et al., 2002). Notably, eRNA expression was inversely correlated with H3K27me3 deposition, consistent with previous observations of genomic regions that exhibit enhancer-like features being actively repressed (Dunham et al., 2012)(Figure 7B). Moreover, 55% of the mRNA encoding genes vicinal to quartile 4 enhancers exhibited very low expression, whereas only 25% of quartile 1 genes shared similar characteristics (Figure 7D), indicating that the repression of enhancer transcription is correlated with nearby genic silencing.
DISCUSSION
Here, we investigated the impact of TLR4 activation on macrophage gene expression by analyzing time-dependent changes in genomic features that are associated with functional enhancers. While transcriptional responses to TLR4 ligation appeared to primarily use pre-existing enhancers, we also observed the selection of thousands of de novo enhancers at genomic locations exhibiting very low or absent occupancy of transcription factors under basal conditions, in agreement with a recent study (Ostuni et al., 2013). Consistent with this, p65 bound primarily to pre-existing enhancers, but also localized to genomic regions destined to acquire enhancer-like chromatin and transcription signatures in association with newly acquired binding of PU.1 and C/EBPα. Based on the observation that PU.1 binding at de novo enhancers was dependent on the activation of NF-κB, and binding of p65 at these sites was conversely dependent on nuclear PU.1, these factors appear to function in a collaborative manner to direct selection of many de novo enhancer elements. Furthermore, these findings suggest that p65 functions as both a signal-dependent and lineage-determining factor. Based on similar co-induced binding of p65 and C/EBPα and the enrichment of motifs for NF-κB, IRFs, PU.1, C/EBPs and AP-1 factors, the assembly of de novo enhancers appears to proceed in a cell-specific manner that is analogous to that of pre-existing enhancers in resting macrophages, with the additional requirement for the collaborative actions of a broadly expressed signal-dependent factor. We speculate that similar strategies will be used to establish the distinct phenotypes of other macrophage subsets, such as Kupffer cells in liver, microglia in the CNS, etc.
The signal-dependent selection of de novo enhancers provided an opportunity to evaluate intermediates in the pathway of enhancer priming and activation. By combining temporal measurement of transcription factor binding, histone acetylation, histone methylation and nascent RNA transcription, we observed a sequence of events at de novo enhancers proceeding from i) unmarked chromatin to ii), transcription factor binding to iii), histone H4K5/8 acetylation coupled to eRNA transcription, followed by iv) progressive mono- and di-methylation of H3K4 (Figure 7E). Several lines of evidence suggest that enhancer transcription is mechanistically linked to H3K4 methylation at de novo enhancers. First, within the limits of sensitivity of GRO-Seq measurements, virtually all de novo enhancers defined by acquisition of H3K4me1/2 also exhibit evidence of enhancer transcription. Second, the initial detection of H3K4me1/2 deposition at de novo enhancers coincides with or occurs after the appearance of eRNAs. Third, H3K4 methylation increases progressively in the setting of continuing enhancer transcription. Fourth, the distribution of H3K4me1/2 is highly correlated with length of eRNA transcripts. Fifth, deposition of H3K4me2 is asymmetrically deposited at asymmetrically transcribed enhancers. Sixth, inhibition of RNAPII transcription with five different inhibitors that act through different molecular targets reduces local H3K4 methylation at TLR4-induced enhancers. Similar relationships between transcription and augmented H3K4 methylation are observed at pre-existing enhancers and promoters that are activated by TLR4 ligation, suggesting that H3K4 methylation may be broadly coupled to transcription.
A small but significant subset of pre-existing enhancers lack detectable evidence of transcription. This set of enhancers is enriched for H3K27me3, suggesting that they are actively repressed. As we find that the majority of these sites in thioglycollate-elicited macrophages do have evidence of transcription in macrophage progenitor cells, and that the H3K4me2 mark persists at de novo enhancers following cessation of active enhancer transcription, it is also possible that the initial deposition of H3K4 methylation at pre-existing enhancers was transcription-dependent.
We further identify Mll1, Mll2/4 and Mll3 as the major HMTs responsible for induced methylation at de novo enhancers. These results are consistent with the recent finding that Drosophila Trr is linked to the deposition of H3K4me1 at enhancers in various tissues of the fruitfly and the mammalian homologue MLL3/MLL4 contributes to H3K4me1 in mouse embryonic fibroblasts (Herz et al., 2012). The partial effects observed following selective knockdown of Mll1, Mll2/4 and Mll3 in mouse macrophages suggest that they function in an overlapping and redundant manner. Previous studies have shown that members of the Mll family of HMTs are able to associate with the C terminal domain (CTD) of RNA PolII, in some cases dependent on phosphorylation of CTD serine 2 (Hughes et al., 2004; Krogan et al., 2003; MacConaill et al., 2006; Milne et al., 2005; Ng et al., 2003; Rana et al., 2011; Wood et al., 2003). The reduced H3K4 methylation observed at de novo enhancers following treatment of macrophages with flavopiridol is consistent with the possibility that Mll complexes are recruited to enhancers in response to CTD phosphorylation by the Cdk9 component of pTEFb. This would provide a potential explanation for the relationship between length of eRNA transcript and spread of H3K4 methylation from the enhancer center.
While the molecular functions of H3K4me1 and H3K4me2 at enhancers remain to be elucidated, several lines of evidence indicate that these marks contribute to enhancer activity. Genes associated with newly selected enhancers marked by H3K4me1 in macrophages were recently shown to exhibit faster and stronger responses upon secondary stimulation (Ostuni et al., 2013). These observations are consistent with the idea that H3K4 methylation in some way ‘licenses’ an enhancer for function. Additional evidence is provided by studies of the histone lysine demethylase 1 (LSD1). Studies indicate that LSD1 directly binds to and is required for the ‘de-commissioning’ of embyonic stem cell-specific enhancers during ES cell differentiation by removing the H3K4me1/2 marks (Whyte et al., 2012). It remains to be determined whether the loss of H3K4me2 observed at a subset of pre-existing enhancers following KLA stimulation follows a similar mechanism. Our findings are consistent with the idea that H3K4me1/2 provides a molecular memory of prior transcriptional activity that can persist for extended times after enhancer activation (Bonn et al., 2012; Ostuni et al., 2013).
In concert, these findings provide evidence that enhancer transcription is mechanistically linked to H3K4 methylation at signal-inducible enhancers. Recent studies suggest functional roles of eRNAs themselves in contributing to enhancer activity, at least in part by promoting enhancer-promoter looping and regulating mediator recruitment (Lai et al., 2013; Lam et al., 2013; Li et al., 2013; Melo et al., 2013). In the present studies, transcription-coupled H3K4 methylation of de novo enhancers did not require the eRNA products. This observation is most consistent with the possibility that H3K4 methylation and eRNAs serve distinct molecular functions and suggests that enhancer transcription contributes to multiple facets of enhancer activation.
EXPERIMENTAL PROCEDURES
Cells
Primary cells were isolated from 6–8 week-old C57Bl/6 (Harlan Laboratories) male mice. All studies were conducted in accordance with the GSK Policy on the Care, Welfare and Treatment of Laboratory Animals and were reviewed by the UCSD Institutional Animal Care and Use Committee. Thioglycollate-elicited macrophages were isolated by peritoneal lavage 3–4 days following peritoneal injection of 2.5 ml thioglycollate. BMDM were generated as described (Valledor et al., 2004). PU.1−/− and PUER cells were propagated and the PU.1-ER fusion protein was activated with 100 nM 4-hydroxy-tamoxifen as described (Walsh et al., 2002).
Stimulations
After overnight serum starvation the cells were treated with 100 ng/ml of Kdo2-Lipid A for 10 min – 48 hours (Raetz et al., 2006). The inhibitors were added one hour prior to KLA, except α-amanitin pretreatment was 4h due to its slow uptake. Final concentrations of the drugs were 30 μM of Inhibitor of IKK II (Calbiochem La Jolla, CA, USA), 10 μg/ml of alpha-amanitin (Sigma), 5 μg/ml of actinomycin D (Sigma), 1μM of IBET151 (Dawson et al., 2011), flavopiridol (Sigma, St. Louis, MO, USA) and triptolide (Sigma),
Preparation of GRO-Seq and RNA-Seq Libraries
GRO-Seq libraries were prepared from 1–7 biological replicates per condition. Global run-on (Core et al., 2008) and library preparation for sequencing (Ingolia et al., 2009) were done as described. The protocol was performed as described in (Wang et al., 2011) with minor modifications(see the Supplemental Experimental Procedures).
Preparation of ChIP-Seq Libraries
The ChIP-Seq libraries for PU.1, H4K5/8ac, C/EBPα and H3K27me3 were prepared as previously described (Escoubet-Lozach et al., 2011). Antibodies against PU.1 (sc-352) and p65 (sc-372) were purchased from Santa Cruz Biotechnology (CA, USA). Antibodies recognizing H4K5/8ac (07-327, 07-328), H3K4me2 (07-030) and H3K27me3 (07-449) were from Millipore (Billerica, MA, USA) and those against H3K4me1 (ab8895) and H3K27ac (ab4729) were from Abcam. Detailed procedures are described in the Supplemental Experimental Procedures.
High Throughput Sequencing
Libraries were sequenced for 36 or 50 cycles on an Illumina Genome Analyzer II or HiSeq 2000, respectively, according to the manufacturer’s instructions. Each sequence tag returned by the Illumina Pipeline was aligned to the mm9 assembly.
Data Analysis
Data analysis was performed using HOMER and the detailed instructions for analysis can be found at http://biowhat.ucsd.edu/homer/ (Heinz et al., 2010) and in the Supplemental Experimental Procedures.
ACCESION NUMBERS
Sequencing data supporting these studies can be found at the Gene Expression Omnibus database under accession numbers GSE48759. Data from previously published ChIP-Seq experiments can be found under accession numbers GSE21512 and GSE23622.
Supplementary Material
HIGHLIGHTS.
TLR4 signaling substantially remodels the macrophage enhancer landscape
Intermediates in enhancer selection and activation can be visualized
Signal-dependent enhancer transcription is linked to H3K4 methylation
Enhancer H3K4 methylation is primarily dependent on Mll1, Mll2/4 and Mll3
Acknowledgments
We thank Dr. Gary Hardiman, James Sprague, Colleen Ludka and Michael Harabaglia for assistance with Illumina sequencing, Jana Collier and Jesse N. Fox for technical assistance, and Lynn Bautista for assistance with manuscript preparation. We acknowledge Dr. Michael T. Y. Lam, Dr. David Gosselin and Dr. Evan Muse for experimental assistance and comments. We thank Dr. Leighton Core for advice with the GRO-Seq protocol. M.U.K. was supported by Fondation Leducq Career Development award, Sigrid Jusélius fellowship and grants from Academy of Finland, ASLA-Fulbright, Finnish Foundation for Cardiovascular Research, Finnish Cultural Foundation (North Savo Regional Fund) and Orion-Farmos Research Foundation. Studies were primarily supported by NIH grants DK091183, CA17390, and DK063491.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barish GD, Yu RT, Karunasiri M, Ocampo CB, Dixon J, Benner C, Dent AL, Tangirala RK, Evans RM. Bcl-6 and NF-kappaB cistromes mediate opposing regulation of the innate immune response. Genes & development. 2010;24:2760–2765. doi: 10.1101/gad.1998010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annual review of pharmacology and toxicology. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- Bhatt DM, Pandya-Jones A, Tong AJ, Barozzi I, Lissner MM, Natoli G, Black DL, Smale ST. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell. 2012;150:279–290. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddie SC, John S, Sabo PJ, Thurman RE, Johnson TA, Schiltz RL, Miranda TB, Sung MH, Trump S, Lightman SL, et al. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol Cell. 2011;43:145–155. doi: 10.1016/j.molcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, Ghavi-Helm Y, Wilczynski B, Riddell A, Furlong EE. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nature Genetics. 2012;44:148–156. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- Brueckner F, Cramer P. Structural basis of transcription inhibition by alpha-amanitin and implications for RNA polymerase II translocation. Nature structural & molecular biology. 2008;15:811–818. doi: 10.1038/nsmb.1458. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- Chao SH, Price DH. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. The Journal of biological chemistry. 2001;276:31793–31799. doi: 10.1074/jbc.M102306200. [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escoubet-Lozach L, Benner C, Kaikkonen MU, Lozach J, Heinz S, Spann NJ, Crotti A, Stender J, Ghisletti S, Reichart D, et al. Mechanisms establishing TLR4-responsive activation states of inflammatory response genes. PLoS Genet. 2011;7:e1002401. doi: 10.1371/journal.pgen.1002401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, Kraus WL. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145:622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz HM, Mohan M, Garruss AS, Liang K, Takahashi YH, Mickey K, Voets O, Verrijzer CP, Shilatifard A. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes & development. 2012;26:2604–2620. doi: 10.1101/gad.201327.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CM, Rozenblatt-Rosen O, Milne TA, Copeland TD, Levine SS, Lee JC, Hayes DN, Shanmugam KS, Bhattacharjee A, Biondi CA, et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Molecular Cell. 2004;13:587–597. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobori M, Yang Z, Gong D, Heissmeyer V, Zhu H, Jung YK, Gakidis MA, Rao A, Sekine T, Ikegami F, et al. Wedelolactone suppresses LPS-induced caspase-11 expression by directly inhibiting the IKK complex. Cell Death Differ. 2004;11:123–130. doi: 10.1038/sj.cdd.4401325. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Molecular and Cellular Biology. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013 doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam MT, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013 doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefterova MI, Steger DJ, Zhuo D, Qatanani M, Mullican SE, Tuteja G, Manduchi E, Grant GR, Lazar MA. Cell-specific determinants of peroxisome proliferator-activated receptor gamma function in adipocytes and macrophages. Mol Cell Biol. 2010;30:2078–2089. doi: 10.1128/MCB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013 doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacConaill LE, Hughes CM, Rozenblatt-Rosen O, Nannepaga S, Meyerson M. Phosphorylation of the menin tumor suppressor protein on serine 543 and serine 583. Molecular cancer research: MCR. 2006;4:793–801. doi: 10.1158/1541-7786.MCR-06-0123. [DOI] [PubMed] [Google Scholar]
- Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol. 2009;9:692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit E, Oude Vrielink JA, Elkon R, Melo SA, Leveille N, Kalluri R, et al. eRNAs Are Required for p53-Dependent Enhancer Activity and Gene Transcription. Molecular Cell. 2013;49:524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- Milne TA, Dou Y, Martin ME, Brock HW, Roeder RG, Hess JL. MLL associates specifically with a subset of transcriptionally active target genes. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:14765–14770. doi: 10.1073/pnas.0503630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AC, Orlando DA, Newman JJ, Loven J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, Young RA. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Molecular Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, Denissov S, Borgesen M, Francoijs KJ, Mandrup S, et al. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 2008;22:2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Perry RP, Kelley DE. Inhibition of RNA synthesis by actinomycin D: characteristic dose-response of different RNA species. Journal of cellular physiology. 1970;76:127–139. doi: 10.1002/jcp.1040760202. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz CR, Garrett TA, Reynolds CM, Shaw WA, Moore JD, Smith DC, Jr, Ribeiro AA, Murphy RC, Ulevitch RJ, Fearns C, et al. Purification and properties of Escherichia coli Kdo2-lipid A, a defined endotoxin that activates macrophages via TLR-4. J Lipid Res. 2006 doi: 10.1194/jlr.M600027-JLR200. [DOI] [PubMed] [Google Scholar]
- Rana R, Surapureddi S, Kam W, Ferguson S, Goldstein JA. Med25 is required for RNA polymerase II recruitment to specific promoters, thus regulating xenobiotic and lipid metabolism in human liver. Molecular and Cellular Biology. 2011;31:466–481. doi: 10.1128/MCB.00847-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner DB, Yamaguchi Y, Wada T, Handa H, Price DH. A highly purified RNA polymerase II elongation control system. J Biol Chem. 2001;276:42601–42609. doi: 10.1074/jbc.M104967200. [DOI] [PubMed] [Google Scholar]
- Schiltz RL, Mizzen CA, Vassilev A, Cook RG, Allis CD, Nakatani Y. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. The Journal of biological chemistry. 1999;274:1189–1192. doi: 10.1074/jbc.274.3.1189. [DOI] [PubMed] [Google Scholar]
- Smale ST. Transcriptional regulation in the innate immune system. Curr Opin Immunol. 2012;24:51–57. doi: 10.1016/j.coi.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szerlong HJ, Prenni JE, Nyborg JK, Hansen JC. Activator-dependent p300 acetylation of chromatin in vitro: enhancement of transcription by disruption of repressive nucleosome-nucleosome interactions. The Journal of biological chemistry. 2010;285:31954–31964. doi: 10.1074/jbc.M110.148718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titov DV, Gilman B, He QL, Bhat S, Low WK, Dang Y, Smeaton M, Demain AL, Miller PS, Kugel JF, et al. XPB, a subunit of TFIIH, is a target of the natural product triptolide. Nature chemical biology. 2011;7:182–188. doi: 10.1038/nchembio.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompouki E, Bowman TV, Lawton LN, Fan ZP, Wu DC, DiBiase A, Martin CS, Cech JN, Sessa AK, Leblanc JL, et al. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell. 2011;147:577–589. doi: 10.1016/j.cell.2011.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valledor AF, Hsu LC, Ogawa S, Sawka-Verhelle D, Karin M, Glass CK. Activation of liver X receptors and retinoid X receptors prevents bacterial-induced macrophage apoptosis. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:17813–17818. doi: 10.1073/pnas.0407749101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh JC, DeKoter RP, Lee HJ, Smith ED, Lancki DW, Gurish MF, Friend DS, Stevens RL, Anastasi J, Singh H. Cooperative and antagonistic interplay between PU.1 and GATA-2 in the specification of myeloid cell fates. Immunity. 2002;17:665–676. doi: 10.1016/s1074-7613(02)00452-1. [DOI] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. The Journal of biological chemistry. 2003;278:34739–34742. doi: 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS biology. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.