Abstract
The diverse morphology of orchid flowers and their complex, often deceptive strategies to become pollinated have fascinated researchers for a long time. However, it was not until the 20th century that the ontogeny of orchid flowers, the genetic basis of their morphology and the complex phylogeny of Orchidaceae were investigated. In parallel, the improvement of techniques for in vitro seed germination and tissue culture, together with studies on biochemistry, physiology, and cytology supported the progress of what is now a highly productive industry of orchid breeding and propagation. In the present century both basic research in orchid flower evo-devo and the interest for generating novel horticultural varieties have driven the characterization of many members of the MADS-box family encoding key regulators of flower development. This perspective summarizes the picture emerging from these studies and discusses the advantages and limitations of the comparative strategy employed so far. I address the growing role of natural and horticultural mutants in these studies and the emergence of several model species in orchid evo-devo and genomics. In this context, I make a plea for an increasingly integrative approach.
Keywords: Orchidaceae, evo-devo, MADS-box gene, peloric mutant, gene family, transcriptome, model species
The comparative approach to orchid evo-devo
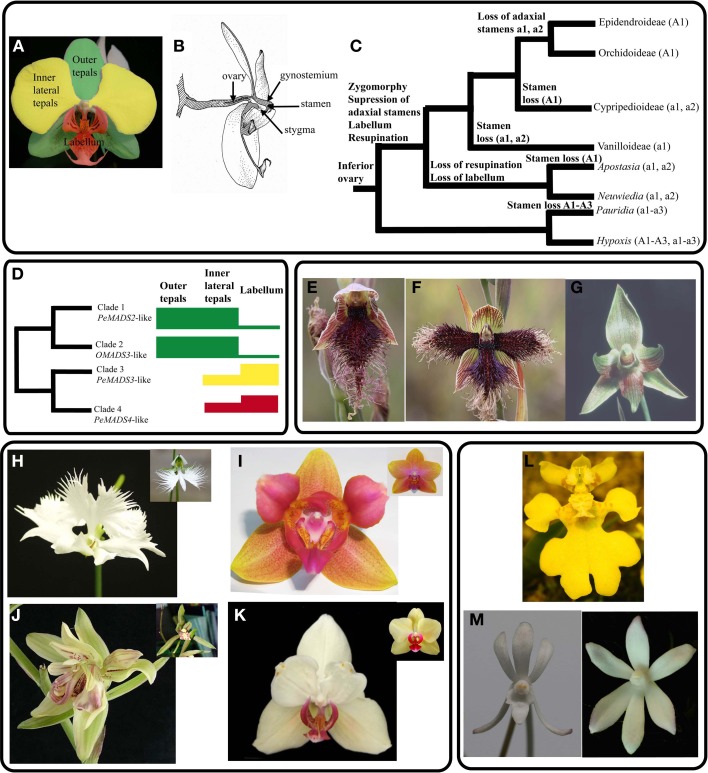
The unique diversification of flower morphology in Orchidaceae has taken place in the framework of a relatively conserved structure. Generally orchid flowers consist of three outer tepals similar to each other, two distinct inner lateral tepals and a highly differentiated inner median tepal or labellum (Figure 1A). Female and male reproductive organs are fused into a bilaterally symmetrical (zygomorphic) structure called gynostemium while the ovary is inferior with respect to the rest of the organs (Figure 1B). Along the diversification of this family there have been several major floral morphological transitions: from zygomorphy to actinomorphy, partial to complete suppression of three to five of the original six stamens and the differentiation of the inner median tepal into the distinct labellum (Figure 1C). These transitions and a 180° developmental rotation of the flower pedicel or ovary (resupination) yielded zygomorphic flowers where the abaxially oriented labellum serves pollinators as a landing platform and guide toward the pollinia (Bateman and Rudall, 2006).
Figure 1.
Wild type and peloric orchids in evo-devo and genomics. (A) Orchid perianth. The color coding indicates distinct organ identities. (B) Reproductive organs. The 180° turn of the pedicel (resupination) together with the opposing position of labellum and stamen enable pollinator attraction as well as precise pollinia removal and placement. (C) Perianth and pistils characters gained and lost along the evolution of Orchidaceae from an ancestor with actinomorphic perianth of identical organs and six stamens. Pauridia and Hypoxis are outgroups. Modified from (Rudall and Bateman, 2002). (D) The “orchid code” model associates the phylogenetic relationships of class B DEFICIENS-like genes with their differential expression in the perianth and their association to perianth organ identity specification. For example, a higher expression of clade 3 and clade 4 genes is associated to the development of the labellum (red-coded tepal in A). (E) Wild-type Calochilus robertsonii, (F) type A peloric Calochilus robertsonii, (G) type B peloric Calochilus robertsonii is recognized as species Chalochilus imberbis, (H) peloric and wild type (inset) Habenaria radiata, (I) Type A peloric and wild-type Phalaenopsis hyb. “Athens” (inset), (J) Multitepal mutant and wild-type Cymbidium ensifolum (inset), (K) Glyp mutant and wild-type Phalaenopsis “CD” (inset), (L) Flower of Erycina pusilla, (M) Flowers of Neofinetia falcata wild-type and Golden Star type B mutant.
Because of the key role of the gynostemium and labellum in orchid reproduction their origin has been a recurring question in botany and evolutionary biology since the 19th century. The finding that flower organ identity is specified by the genetic and physical interaction of MADS domain transcription factors (Bowman et al., 2012) served as a basis in the last 20 years for comparative studies on orchid flower evolutionary development (Table 1). So far the approach employed is essentially based on comparing the expression of orchid MADS-box genes with those of well-characterized model species. This method has generated informative associations between B- and C-like MADS-box genes from orchids with those of Arabidopsis thaliana and some non-model species like Tulipa gesneriana and Lilium regale.
Table 1.
Class A, B, C, D, and E MADS-box genes characterized in Orchidaceae.
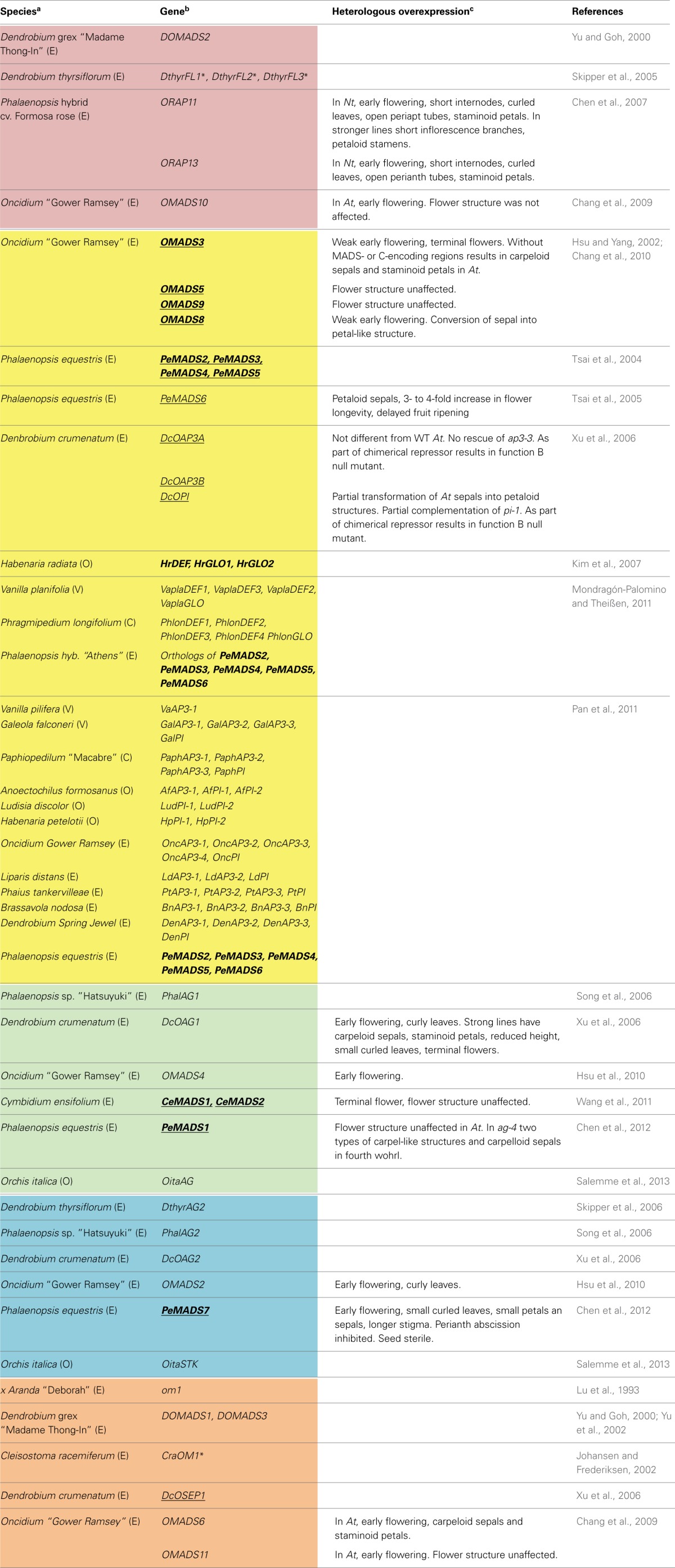
aE, Epidendroidea; O, Orchidoidea; V, Vanilloidea; C, Cypripedioidea.
bIn bold, Expression analyzed in wild-type and peloric orchids; Underlined, Protein interaction tested.
cAt, Arabidopsis thaliana; Nt, Nicotiana tabacum.
*Expression was measured in complete buds and inflorescences.
The description of the sequence and pattern of expression of the first MADS-box gene isolated from an orchid, om1 from x Aranda “Deborah” (Lu et al., 1993), illustrates the challenges and limitations of strongly relying on knowledge from model organisms. Back in 1993, based on sequence similarity om1 was considered homologous to FBP2 from Petunia hybrida. However, it was hard to explain the differences on their patterns of expression: while FBP2 was expressed in petals, stamens, carpels and at a very low level in sepals (Angenent et al., 1992), om1 was detected in the first and second perianth whorls of x Aranda “Deborah.” It was after 2000, when many more MADS-box genes from model species had been characterized and the phylogeny of this family was extensively investigated, that om1 was confirmed as a SEPALLATA3-like gene (Zahn et al., 2005) and expression of om1 orthologs DOMADS1 and DcOSEP1 from Dendrobium was confirmed in the perianth as well as in the gynostemium and ovary (Figures 1A,B) (Yu and Goh, 2000; Xu et al., 2006).
In the last 10 years isolation and characterization of individual MADS-box genes from orchids occurred at a faster pace (Table 1). However, because of their role in perianth and stamen specification, nearly all efforts focused on class B and C genes from species in Epidendroideae, the largest orchid subfamily containing most varieties of horticultural importance like Phalaenopsis, Dendrobium and Oncidium (Table 1). The picture emerging from the analysis of A-, C-, D- and E-like MADS-box genes is characterized by several instances of gene duplication in each of these groups, as well as a conserved pattern of expression of each duplicate gene. Specifically, FRUITFULL-like genes (class A) are expressed mostly in the gynostemium and in some instances also in the perianth (Yu and Goh, 2000; Skipper et al., 2005; Chen et al., 2007; Chang et al., 2009) while AGAMOUS- and SEEDSTICK-like genes (class C and D, respectively) are reproducibly expressed in the gynostemium and ovary (Song et al., 2006; Xu et al., 2006; Hsu et al., 2010; Wang et al., 2011; Chen et al., 2012; Salemme et al., 2013). Most of the SEPALLATA-like genes (E-like genes) isolated so far are expressed in all flower organs (Lu et al., 1993; Yu and Goh, 2000; Johansen and Frederiksen, 2002; Yu et al., 2002; Xu et al., 2006; Chang et al., 2009).
The orchid family has four ancient, highly conserved lineages of class B genes DEFICIENS-like genes (Tsai et al., 2004; Mondragón-Palomino and Theißen, 2008; Mondragón-Palomino et al., 2009). Genes in each of these clades follow a conserved combinatorial pattern of expression associated with the development of specific perianth organs (Figure 1D). Generally DEF-like genes in sister clades 1 and 2 are expressed in all flower organs, except the labellum (Chang et al., 2010; Mondragón-Palomino and Theißen, 2011), while genes in sister clades 3 and 4 are expressed in the inner perianth, gynostemium and ovary (Tsai et al., 2004; Xu et al., 2006; Kim et al., 2007; Chang et al., 2010; Mondragón-Palomino and Theißen, 2011; Pan et al., 2011). The specification of outer tepal identity is associated with the combinatorial expression of clade 1 and clade 2 genes, the identity of inner-lateral tepals with the lower expression of clade 3 and clade 4 genes (Figure 1D). Similarly labellum specification depends on the higher expression of clade 3 and clade 4 genes (Mondragón-Palomino and Theißen, 2011). The phylogenetic relationships and patterns of expression of these genes have been integrated in the “orchid code” a model to explain and test the evolution and regulatory relationships of these genes in wild-type and mutant flowers (Mondragón-Palomino and Theißen, 2008, Mondragón-Palomino and Theißen, 2011). The conserved, clade-specific pattern of expression of the four DEF-like genes in representatives from most Orchidaceae subfamilies suggests combinatorial regulation by these genes was established early on the evolution of the Orchidaceae and preserved for millions of years. Generally consistent with the revised “orchid code,” the “HOT model” describes the patterns of expression of class B genes on a temporal and spatial dimension starting from the floral primordial stage (Pan et al., 2011).
In contrast, GLOBOSA-like genes the second major lineage of class B genes, are expressed in all flower organs and have not duplicated on a family-wide scale but once in subfamily Orchidaceae (Tsai et al., 2005; Xu et al., 2006; Kim et al., 2007; Mondragón-Palomino et al., 2009; Chang et al., 2010; Mondragón-Palomino and Theißen, 2011; Pan et al., 2011) (Table 1).
ABCDE-class MADS domain transcription factors form dimers and higher order complexes (Egea-Cortines et al., 1999) that bind to CArG-box motifs in regulatory regions of a wide variety of target genes (Kaufmann et al., 2009). The dimeric and multimeric interactions of orchid MADS domain proteins have been described in Oncidium “Gower Ramsey,” Dendrobium crumenatum, Phalaenopsis equestris and Cymbidium ensifolium (Hsu and Yang, 2002; Xu et al., 2006; Tsai et al., 2008; Chang et al., 2010; Wang et al., 2011; Chen et al., 2012) (Table 1, underlined gene names).
Investigating the function of orchid MADS-box genes
Heterologous expression
Because of technical limitations to transform orchids, most functional analyses of MADS-box class A-, B-, C-, D-, and E-like genes have been performed by means of heterologous ectopic overexpression in Arabidopsis thaliana or Nicotiana tabacum (Table 1). These experiments employed the strong constitutive promoter CaMV35S and often resulted in early flowering of the species transformed, regardless of which gene was being overexpressed or whether the flower organs are affected or not (Table 1). Early flowering is not exclusively caused by orchid MADS-box genes, as suggested by similar experiments with Lilium longiflorum LMADS3 (Tzeng et al., 2003). Early flowering and a terminal flower-phenotype are in this context the outcome of activating many other genes, among them other class A, B, C MADS-box genes, which as described for SEP3, trigger the flower developmental program (Kaufmann et al., 2009).
Because of the widespread activation capabilities of MADS domain proteins, the effects of heterologous ectopic overexpression are often unpredictable or difficult to attribute to one or few specific genes. For example, in agreement with their role as GLO-like genes, the heterologous ectopic overexpression of DcOPI and OMADS8 results in transformation of sepals into petaloid structures and complementation of pi-1 from Arabidopsis thaliana (Xu et al., 2006; Chang et al., 2010). However, it is harder to interpret phenotypes such as a perianth formed by carpelloid sepals and staminoid petals, which result from overexpressing MADS-box genes from different classes like OMADS3 (DEF-like), DcOAG1 (AG-like) or OMADS6 (SEP3-like) (Hsu and Yang, 2002; Xu et al., 2006; Chang et al., 2009) (Table 1). Because this phenotype resembles an ap2 mutant, it has been proposed that these genes have diverged functionally, however, it is hard to distinguish these claims from the non-specific effects previously mentioned.
Additionally, heterologous overexpression experiments often do not yield phenotypic differences between wild-type and transgenic plants (Table 1). Alternatively, more direct methods for functional characterization are inducible or gene-specific promoters to limit expression to certain tissues and developmental stages.
Advances on the identification and experimental analysis orchid MADS-box promoters have already been made by employing orchid stable transformation to characterize the 5′ regions of DOMADS1 (Yu et al., 2002). More recently, the transcription factor OgMYB1 involved in anthocyanin biosynthesis was investigated by means of transient transformation of orchid perianth organs (Chiou and Yeh, 2007). A similar approach has been applied to identify flower-specific promoters in Oncidium Gower Ramsey (Hsu et al., 2011b).
Helpful monsters
Regardless of the underlying genetic or epigenetic causes teratological flowers are phenotypically different from their parental forms. Peloric terata are an special case involving a transition from zygomorphy (bilateral symmetry) to actinomorphy (radial symmetry) (Bateman and DiMichele, 2002; Rudall and Bateman, 2002). Peloric flowers have been reported in wild and cultivated orchids and are classified in type A when the two inner lateral tepals are replaced by labellum-like structures, type B when the labellum is replaced by a inner-lateral tepal (Figures 1E–G) and the more rare type C which involves the substitution of all inner perianth organs by structures resembling outer tepals (Rudall and Bateman, 2002).
Because of the difficulties involved in genetically manipulating orchids, peloric flowers are essential to investigate the developmental pathways specifying location, identity and patterning of each organ in the meristem. Orchid evo-devo has greatly profited from comparing flower ontogeny and patterns of gene expression between wild-type flowers and their peloric forms (Table 1; Figures 1F–M) (Tsai et al., 2004; Kim et al., 2007; Chang et al., 2010; Mondragón-Palomino and Theißen, 2011; Pan et al., 2011; Wang et al., 2011).
The first study comparing the expression of developmental genes in the flower organs of wild-type and type A peloric Phalaenopsis equestris suggested the differential expression of DEF-like genes PeMADS2, PeMADS3, PeMADS4, and PeMADS5 is associated with the development of specific flower organs (Tsai et al., 2004). The differential expression of PeMADS4 in the labellum and in the labellum-like inner lateral tepals of the peloric form suggested this gene is key to the development of this organ (Tsai et al., 2004). Later on, the finding that teratological flowers from Habenaria radiata ectopically expressed HrDEF (PeMADS3-like gene) in the outer perianth was associated to the transformation of outer tepals into organs resembling inner-lateral tepals and labellum, thus suggesting this gene is involved in the specification of the inner perianth (Kim et al., 2007).
A molecular phylogeny of DEF-like genes involving representatives from most orchid subfamilies showed that the paralogs first observed in Phalaenopsis equestris are actually part of four highly conserved, Orchidaceae-specific clades at least 70 million years old (Mondragón-Palomino and Theißen, 2008; Mondragón-Palomino et al., 2009). On this phylogenetic structure we mapped the expression information available for PeMADS2-PeMADS5, HrDEF, DcOAP3A, DcOAP3B and OMADS3 and realized the genes of each clade have a distinct and conserved pattern of expression in the perianth (Mondragón-Palomino and Theißen, 2008). These comparative analyses together with developmental principles derived from the analysis of peloric Phalaenopsis equestris (Tsai et al., 2004) and Habenaria radiata (Kim et al., 2007) suggested a combinatorial regulatory model for the determination of flower organ identity of orchids. Initially the “orchid code” proposed that specification of labellum identity depended on expression of the clade 4 gene. The “orchid code” was tested and modified accordingly to the results of qRT-PCR analyses on individual flower organs from wild-type and Type A peloric flowers of Phalaenopsis hyb. “Athens” (Figures 1H,I), as well as wild-type flowers from Vanilla planifolia, Phragmipedium longifolium. The comparison between wild-type and peloric flowers showed that in the inner lateral tepals there is a relatively lower amount of both clade 3 and clade 4 genes, while in the labellum and in the labellum-like inner lateral tepals of peloric flowers both genes are expressed at a higher level (Mondragón-Palomino and Theißen, 2011) (Figure 1D).
More recently, the study of floral terata from Cymbidium ensifolium and Phalaenopsis equestris advanced our understanding of AGAMOUS-like genes in orchid flower development. The phenotype of the multitepal mutant of Cymbidium ensifolium is analogous to agamous from A. thaliana in that the gynostemium is replaced by an ectopic flower which produces outer and inner tepal-like structures centripetally (Wang et al., 2011) (Figure 1J). Comparison of the patterns of expression of AGAMOUS-like genes CeMADS1 (DthryAG1-like) and CeMADS2 (DcOAG1-like) showed that while both genes are expressed in the gynostemium and buds of wild-type flowers, CeMADS1 is not expressed in developing buds of the multitepal mutant. This study thus suggests that CeMADS1 is a class C gene and both CeMADS1 and CeMADS2 are not functionally redundant in the specification of gynostemium identity.
In the glyp mutant of Phalaenopsis hyb. “CD1” the inner lateral tepals bear ectopic pollinia and their epidermal cells are morphologically intermediate between those of wild-type tepals and those of the gynostemium (Chen et al., 2012) (Figure 1K). The expression of PeMADS1 (DthyrAG1-like, class C) and PeMADS7 (DthyrAG2-like, class D gene) was detected exclusively in the column of the wild-type flowers, while only PeMADS1 was detected in the gynostemium-like inner lateral tepals of the glyp mutant. While this study suggests PeMADS1 (Chen et al., 2012) might be involved in the development of pollinia and epidermal cells, it is necessary to consider the pattern of expression of the ortholog of CeMADS2, the second non-redundant AGAMOUS-like gene previously described.
A major question underlying studies with peloric flowers is whether the mutant phenotypes actually result from changes in developmental regulators of flower symmetry determining the location of MADS-box gene expression. An elegant study on the loss of bilateral symmetry of peloric Linaria vulgaris flowers as well as analysis of orchid peloria suggest transcription factors from the TCP family could also be at play in the development of this kind of terata (Cubas et al., 1999; Rudall and Bateman, 2002, 2003; Mondragón-Palomino and Theißen, 2009).
A model for orchid flower evo-devo
The case of om1 from x Aranda “Deborah” discussed in the first section illustrates how the comparative approach to orchid evo-devo is limited by what is known about model species more amenable to genetic analysis and transformation. The recent growth of genomic and transcriptomic resources for Orchidaceae might soon eliminate these barriers.
Several species have been put forward in the literature as candidates or de facto model species. Most notably Phalaenopsis species and hybrids are frequently employed for the study of orchid development (Table 1) because of their undisputable importance in horticultural breeding and trade (Tang and Chen, 2007) and the availability of horticultural peloric mutants. The genus Phalaenopsis is presently the best documented orchid group at the genomic level because the genomes of Phalaenopsis equestris (Hsu et al., 2011a) and Phalaenopsis aphrodite are being sequenced and genomic resources such as ESTs, transcriptomes and public databases (Su et al., 2013) (http://orchidstra.abrc.sinica.edu.tw/) have been generated for these species (Hsiao et al., 2006; Su et al., 2011; An and Chan, 2012).
Recently transcriptomic resources deposited in the OrchiBase 2.0 (Tsai et al., 2013) (http://orchidbase.itps.ncku.edu.tw) advance ten orchid species as candidate models from each of the five Orchidaceae subfamilies as well as the sister group Hypoxidaceae: Apostasia shenzhenica and Neuwiedia malipoensis (Apostasioideae); Vanilla shenzhenica and Galeola faberi (Vanilloideae); Paphiopedilum armeniacum and Cypripedium singchii (Cypripedioideae); Habenaria delavayi and Hemipilia forrestii (Orchidoideae); Cymbidium sinense as well as the previously mentioned Phalaenopsis equestris (Epidendroideae) and Sinocurculigo taishanica (Hypoxidaceae) (Tsai et al., 2013). Additionally transcriptomic information for Dendrobium nobile and Oncidium “Gower Ramsey” is deposited in the databases Orchidstra and Oncidium Orchid Genome Base, respectively (Chang et al., 2011) (predictor.nchu.edu.tw).
Erycina pusilla (Figure 1L) is an attractive candidate model species because of it can grow rapidly, produce flowers and fruits in vitro and has a small genome size (1C = 1.5 pg, P. equestris has 1.69 pg and A. thaliana has 0.30 pg). Recently, the chloroplast genome of E. pusilla and a transcriptome have been sequenced (Pan et al., 2012) (orchidstra.abrc.sinica.edu.tw).
Another promising candidate model species is the wind orchid Neofinetia falcata (Figure 1M) because there is a diverse collection of mutant flower morphologies that facilitate systematic analysis of the genes involved in perianth symmetry, spur development and flower organ specification. N. falcata grows in laboratory conditions and can be propagated by means of tissue culture and seed germination. Although transcriptomic resources still need to be generated, Agrobacterium-mediated transformation, selfing and outcrossing procedures might already enable genetic studies (Duttke et al., 2012).
While the availability of genomic information for a diverse group of species will be a major advance to efficiently isolate and investigate genes involved in orchid flower development, the ability to genetically manipulate orchids stably or transiently is key to directly associate specific genes with their functions. Although stable transformation mediated by Agrobacterium (Belarmino and Mii, 2000; Yu et al., 2001) and biolistic bombardment (Chia et al., 1994; Men et al., 2003) have been adapted to Phalaenopsis, Oncidium and Dendrobium, among others, their relatively low efficiency and long regeneration time to obtain flowering plants has limited their application in this area.
Alternatively, virus-induced gene silencing (VIGS) based on Cymbidium mosaic virus has been adapted to Phalaenopsis (Lu et al., 2006). Although in this study the levels of transcription of PeMADS6 and other MADS-box genes were significantly affected, flowers developed all their organs regularly, with the exception of greenish streaks on the back of inner and outer tepals (Lu et al., 2006).
Quo vadis orchid evo-devo?
At its beginnings orchid flower evo-devo greatly profited from knowledge on well- established model species like Arabidopsis thaliana and Antirrhinum majus as well as from research on other monocot species like Tulipa gesneriana and Lilium regale. On the other hand, this comparative approach and the technical limitations to genetically manipulate orchids have set important challenges to functionally approach the genetic basis of orchid flower development.
The systematic morphological and molecular characterization of flower terata offers a way around these limitations and has enabled the formulation of several testable models based on the large amount of information on class B MADS-box genes, the most studied developmental genes in this family.
The growing amount of transcriptomic information in a diverse group of orchid species calls for a second wave of integration and comparative analysis at an unprecedented scale. While the apparent number of “endless forms most beautiful,” the sinking prices of RNA-seq and the competitive nature of scientific endeavor might tempt us to sequence “yet another orchid transcriptome” the most significant advances on this subject will come from systematically integrating all available information and testing our findings experimentally by means of unifying developmental and evolutionary hypotheses and models. This process requires not only sharing and comparing information but also the agreement on common concepts for the developmental processes we are investigating. For example, because most studies describe orchid flowers buds based on their size it is not possible to make an objective comparison of transcriptomes or other patterns of gene expression within and between species. An alternative would be that for every species with a transcriptome a description of discrete stages of its development is generated and considered in the design of future expression studies as it is routinely done for Arabidopsis thaliana (Smyth et al., 1990; Niederhuth et al., 2013; Takeda et al., 2013).
Because of the prevalent occurrence of gene duplication in orchids the value of gene phylogenies and profiles of gene expression strongly depends on considering as many known duplicates as technically possible. By doing so it is possible to objectively compare studies and minimize the artifacts coming from simultaneously measuring the expression of highly similar paralogs.
Because orchid evo-devo is a relatively young area there are still many major challenges to overcome. In the near future the vitality of its research program depends on the consolidation of one or several model species amenable to genetic manipulation or with a rapid life cycle that enables the fruitful integration of genetic analysis and transcriptomic resources. In the long run, the scientific relevance and reach of orchid evo-devo will rely on its contribution to understanding orchid ecology and evolution in questions like the interaction between environmental variables, pollinators and the activity of developmental transcription factors.
Conflict of interest statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
I would like to thank the following colleagues for letting me use their valuable photographic material: James Wood (wild-type and type A peloric Calochilus robertsonii), Hans Wapstra (type B peloric Calochilus imberbis), Akira Kanno and So-Young Kim (Habenaria radiata), Wen-Chieh Tsai (wild-type and multitepal mutant from Cymbidium ensifolum; wild-type and glyp mutant from Phalaenopsis “CD1”) and to Minsung Kim (Neofinetia falcata). Mariana Mondragón-Palomino is funded by a fellowship for Habilitation from the Bayerisches Progamm zur Realisierung der Chancengleichheit für Frauen in Forschung.
References
- An F. M., Chan M. T. (2012). Transcriptome-wide characterization of miRNA-directed and non-miRNA-directed endonucleolytic cleavage using degradome analysis under low ambient temperature in Phalaenopsis aphrodite subsp. formosana. Plant Cell Physiol. 53, 1737–1750 10.1093/pcp/pcs118 [DOI] [PubMed] [Google Scholar]
- Angenent G. C., Busscher M., Franken J., Mol J. N., Van Tunen A. J. (1992). Differential expression of two MADS box genes in wild-type and mutant petunia flowers. Plant Cell 4, 983–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman R. M., DiMichele W. A. (2002). Generating and filtering major phenotypic novelties: neoGoldschmidtian saltation revisited, in Developmental Genetics and Plant Evolution, eds Cronk Q. C. B., Bateman R. M., Hawkins J. A. (London: Taylor and Francis; ), 109–159 10.1201/9781420024982.ch7 [DOI] [Google Scholar]
- Bateman R. M., Rudall P. J. (2006). The good, the bad and the ugly: using naturally occurring terata to distinguish the possible from the impossible in orchid floral evolution, in Monocots: Comparative Biology and Evolution. Excluding Poales, eds Columbus J. T., Friar E. A., Porter J. M., Prince L. M., Simpson M. G. (Claremont, CA: Rancho Santa Ana Botanical Garden; ), 481–496 [Google Scholar]
- Belarmino M. M., Mii M. (2000). Agrobacterium-mediated genetic transformation of a Phalaenopsis orchid. Plant Cell Rep. 19, 435–442 10.1007/s002990050752 [DOI] [PubMed] [Google Scholar]
- Bowman J. L., Smyth D. R., Meyerowitz E. M. (2012). The ABC model of flower development: then and now. Development 139, 4095–4098 10.1242/dev.083972 [DOI] [PubMed] [Google Scholar]
- Chang Y. Y., Chiu Y. F., Wu J. W., Yang C. H. (2009). Four orchid (Oncidium ‘Gower Ramsey’) AP1/AGL9-like MADS box genes show novel expression patterns and cause different effects on floral transition and formation in Arabidopsis thaliana. Plant Cell Physiol. 50, 1425–1438 10.1093/pcp/pcp087 [DOI] [PubMed] [Google Scholar]
- Chang Y. Y., Chu Y. W., Chen C. W., Leu W. M., Hsu H. F., Yang C. H. (2011). Characterization of Oncidium ‘Gower Ramsey’ transcriptomes using 454 GS-FLX pyrosequencing and their application to the identification of genes associated with flowering time. Plant Cell Physiol. 52, 1532–1545 10.1093/pcp/pcr101 [DOI] [PubMed] [Google Scholar]
- Chang Y. Y., Kao N. H., Li J. Y., Hsu W. H., Liang Y. L., Wu J. W., et al. (2010). Characterization of the possible roles for B class MADS box genes in regulation of perianth formation in orchid. Plant Physiol. 152, 837–853 10.1104/pp.109.147116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Guo B., Hexige S., Zhang T., Shen D., Ming F. (2007). SQUA-like genes in the orchid Phalaenopsis are expressed in both vegetative and reproductive tissues. Planta 226, 369–380 10.1007/s00425-007-0488-0 [DOI] [PubMed] [Google Scholar]
- Chen Y. Y., Lee P. F., Hsiao Y. Y., Wu W. L., Pan Z. J., Lee Y. I., et al. (2012). C- and D-class MADS-box genes from Phalaenopsis equestris (Orchidaceae) display functions in gynostemium and ovule development. Plant Cell Physiol. 53, 1053–1067 10.1093/pcp/pcs048 [DOI] [PubMed] [Google Scholar]
- Chia T. F., Chan Y. S., Chua N. H. (1994). The firefly luciferase gene as a non-invasive reporter for Dendrobium transformation. Plant J. 6, 441–446 10.1046/j.1365-313X.1994.06030441.x [DOI] [Google Scholar]
- Chiou C.-Y., Yeh K.-W. (2007). Differential expression of MYB gene (OgMYB1) determines color patterning in floral tissue of Oncidium ‘Gower Ramsey’. Plant Mol. Biol. 66, 379–388 10.1007/s11103-007-9275-3 [DOI] [PubMed] [Google Scholar]
- Cubas P., Vincent C., Coen E. (1999). An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401, 157–161 10.1038/43657 [DOI] [PubMed] [Google Scholar]
- Duttke S., Zoulias N., Kim M. (2012). Mutant flower morphologies in the wind orchid, a novel orchid model species. Plant Physiol. 158, 1542–1547 10.1104/pp.111.191643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea-Cortines M., Saedler H., Sommer H. (1999). Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 18, 5370–5379 10.1093/emboj/18.19.5370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao Y.-Y., Tsai W.-C., Kuoh C.-S., Huang T.-H., Wang H.-C., Wu T.-S., et al. (2006). Comparison of transcripts in Phalaenopsis bellina and Phalaenopsis equestris (Orchidaceae) flowers to deduce monoterpene biosynthesis pathway. BMC Plant Biol. 6:14 10.1186/1471-2229-6-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C.-C., Chung Y.-L., Chen T.-C., Lee Y.-L., Kuo Y.-T., Tsai W.-C., et al. (2011a). An overview of the Phalaenopsis orchid genome through BAC end sequence analysis. BMC Plant Biol. 11:3 10.1186/1471-2229-11-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C.-T., Liao D.-C., Wu F.-H., Liu N.-T., Shen S.-C., Chou S.-J., et al. (2011b). Integration of molecular biology tools for identifying promoters and genes abundantly expressed in flowers of Oncidium ‘Gower Ramsey’. BMC Plant Biol. 11:60 10.1186/1471-2229-11-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H. F., Hsieh W. P., Chen M. K., Chang Y. Y., Yang C. H. (2010). C/D class MADS box genes from two monocots, orchid (Oncidium ‘Gower Ramsey’) and lily (Lilium longiflorum), exhibit different effects on floral transition and formation in Arabidopsis thaliana. Plant Cell Physiol. 51, 1029–1045 10.1093/pcp/pcq052 [DOI] [PubMed] [Google Scholar]
- Hsu H. F., Yang C. H. (2002). An Orchid (Oncidium Gower Ramsey) AP3-like MADS gene regulates floral formation and initiation. Plant Cell Physiol. 43, 1198–1209 10.1093/pcp/pcf143 [DOI] [PubMed] [Google Scholar]
- Johansen B., Frederiksen S. (2002). Orchid flowers: evolution and molecular development, in Developmental Genetics and Plant Evolution, eds Cronk Q. C. B., Bateman R. M., Hawkins J. A. (London: Taylor and Francis; ), 206–219 10.1201/9781420024982.ch10 [DOI] [Google Scholar]
- Kaufmann K., Muiño J. M., Jauregui R., Airoldi C. A., Smaczniak C., Krajewski P., et al. (2009). Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol. 7:e1000090 10.1371/journal.pbio.1000090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-Y., Yun P.-Y., Fukuda T., Ochiai T., Yokoyama J., Kameya T., et al. (2007). Expression of a DEFICIENS-like gene correlates with the differentiation between sepal and petal in the orchid, Habenaria radiata (Orchidaceae). Plant Sci. 172, 319–326 10.1016/j.plantsci.2006.09.009 [DOI] [Google Scholar]
- Lu H. C., Chen H. H., Tsai W. C., Chen W. H., Su H. J., Chang D. C. N., et al. (2006). Strategies for functional validation of genes involved in reproductive stages of orchids. Plant Physiol. 143, 558–569 10.1104/pp.106.092742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z. X., Wu M., Loh C. S., Yeong C. Y., Goh C. J. (1993). Nucleotide sequence of a flower-specific MADS box cDNA clone from orchid. Plant Mol. Biol. 23, 901–904 10.1007/BF00021545 [DOI] [PubMed] [Google Scholar]
- Men S., Ming X., Wang Y., Liu R., Wei C., Li Y. (2003). Genetic transformation of two species of orchid by biolistic bombardment. Plant Cell Rep. 21, 592–598 10.1007/s00299-002-0559-4 [DOI] [PubMed] [Google Scholar]
- Mondragón-Palomino M., Hiese L., Härter A., Koch M. A., Theissen G. (2009). Positive selection and ancient duplications in the evolution of class B floral homeotic genes of orchids and grasses. BMC Evol. Biol. 9:81 10.1186/1471-2148-9-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondragón-Palomino M., Theißen G. (2008). MADS about the evolution of orchid flowers. Trends Plant Sci. 13, 51–59 10.1016/j.tplants.2007.11.007 [DOI] [PubMed] [Google Scholar]
- Mondragón-Palomino M., Theißen G. (2009). Why are orchid flowers so diverse? Reduction of evolutionary constraints by paralogues of class B floral homeotic genes. Ann. Bot. 104, 583 10.1093/aob/mcn258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondragón-Palomino M., Theißen G. (2011). Conserved differential expression of paralogous DEFICIENS- and GLOBOSA-like MADS-box genes in the flowers of Orchidaceae: refining the ‘orchid code’ Plant J. 66, 1008–1019 10.1111/j.1365-313X.2011.04560.x [DOI] [PubMed] [Google Scholar]
- Niederhuth C. E., Patharkar O. R., Walker J. C. (2013). Transcriptional profiling of the Arabidopsis abscission mutant hae hsl2 by RNA-Seq. BMC Genomics 14:37 10.1186/1471-2164-14-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan I.-C., Liao D.-C., Wu F.-H., Daniell H., Singh N. D., Chang C., et al. (2012). Complete chloroplast genome sequence of an orchid model plant candidate: Erycina pusilla apply in tropical Oncidium breeding. PLoS ONE 7:e34738 10.1371/journal.pone.0034738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z. J., Cheng C. C., Tsai W. C., Chung M. C., Chen W. H., Hu J. M., et al. (2011). The duplicated B-class MADS-box genes display dualistic characters in orchid floral organ identity and growth. Plant Cell Physiol. 52, 1515–1531 10.1093/pcp/pcr092 [DOI] [PubMed] [Google Scholar]
- Rudall P. J., Bateman R. M. (2002). Roles of synorganisation, zygomorphy and heterotopy in floral evolution: the gynostemium and labellum of orchids and other lilioid monocots. Biol. Rev. 77, 403–441 10.1017/S1464793102005936 [DOI] [PubMed] [Google Scholar]
- Rudall P. J., Bateman R. M. (2003). Evolutionary change in flowers and inflorescences: evidence from naturally occurring terata. Trends Plant. Sci. 8, 76–82 10.1016/S1360-1385(02)00026-2 [DOI] [PubMed] [Google Scholar]
- Salemme M., Sica M., Gaudio L., Aceto S. (2013). The OitaAG and OitaSTK genes of the orchid Orchis italica: a comparative analysis with other C- and D-class MADS-box genes. Mol. Biol. Rep. 40, 3523–3535 10.1007/s11033-012-2426-x [DOI] [PubMed] [Google Scholar]
- Skipper M., Johansen L. B., Pedersen K. B., Frederiksen S., Johansen B. (2006). Cloning and transcription analysisi of an AGAMOUS- and SEEDSTICK orthologo in the orchid Dendrobium thyrsiflorum (Reichb. f.). Gene 366, 266–274 10.1016/j.gene.2005.08.014 [DOI] [PubMed] [Google Scholar]
- Skipper M., Pedersen K. B., Johansen L. B., Frederiksen S., Irish V. F., Johansen B. (2005). Identification and quantification of expression levels of three FRUITFULL-like MADS-box genes from the orchid Dendrobium thyrsiflorum (Reich. f.). Plant Sci. 169, 579–586 10.1016/j.plantsci.2005.04.011 [DOI] [Google Scholar]
- Smyth D. R., Bowman J. L., Meyerowitz E. M. (1990). Early flower development in Arabidopsis. Plant cell 2, 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song I.-J., Nakamura T., Fukuda T., Yokoyama J., Ito T., Ichikawa H., et al. (2006). Spatiotemporal expression of duplicate AGAMOUS orthologues during floral development in Phalaenopsis. Dev. Genes. Evol. 216, 301–313 10.1007/s00427-005-0057-0 [DOI] [PubMed] [Google Scholar]
- Su C. L., Chao Y. T., Alex Chang Y. C., Chen W. C., Chen C. Y., Lee A. Y., et al. (2011). De novo assembly of expressed transcripts and global analysis of the Phalaenopsis aphrodite transcriptome. Plant Cell Physiol. 52, 1501–1514 10.1093/pcp/pcr097 [DOI] [PubMed] [Google Scholar]
- Su C.-L., Chao Y.-T., Yen S.-H., Chen C.-Y., Chen W.-C., Chang Y.-C. A., et al. (2013). Orchidstra: an integrated orchid functional genomics database. Plant Cell Physiol. 54, e11 10.1093/pcp/pct004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S., Iwasaki A., Matsumoto N., Uemura T., Tatematsu K., Okada K. (2013). Physical interaction of floral organs controls petal morphogenesis in arabidopsis. Plant Physiol. 161, 1242–1250 10.1104/pp.112.212084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C.-Y., Chen W.-H. (2007). Breeding and development of new varieties in Phalaenopsis, in Orchid Biotechnology, eds Chen W.-H., Chen H.-H. (Singapore: World Scientific Publishing; ), 1–22 10.1142/9789812775900_0001 [DOI] [Google Scholar]
- Tsai W. C., Fu C. H., Hsiao Y. Y., Huang Y. M. (2013). OrchidBase 2.0: comprehensive collection of Orchidaceae floral transcriptomes. Plant Cell Physiol. 54, e7 10.1093/pcp/pcs187 [DOI] [PubMed] [Google Scholar]
- Tsai W. C., Kuoh C. S., Chuang M. H., Chen W. H., Chen H. H. (2004). Four DEF-like MADS box genes displayed distinct floral morphogenetic roles in Phalaenopsis orchid. Plant Cell Physiol. 46, 831–844 10.1093/pcp/pch095 [DOI] [PubMed] [Google Scholar]
- Tsai W. C., Lee P. F., Chen H. I., Hsiao Y. Y., Wei W. J., Pan Z. J., et al. (2005). PeMADS6, a GLOBOSA/PISTILLATA-like gene in Phalaenopsis equestris involved in petaloid formation, and correlated with flower longevity and ovary development. Plant Cell Physiol. 46, 1125–1139 10.1093/pcp/pci125 [DOI] [PubMed] [Google Scholar]
- Tsai W. C., Pan Z. J., Hsiao Y. Y., Jeng M. F., Wu T. F., Chen W. H., et al. (2008). Interactions of B-class complex proteins involved in tepal development in Phalaenopsis orchid. Plant Cell Physiol. 2008, 814–824 10.1093/pcp/pcn059 [DOI] [PubMed] [Google Scholar]
- Tzeng T. Y., Hsiao C. C., Chi P. J., Yang C. H. (2003). Two lily SEPALLATA-like genes cause different effects on floral formation and floral transition in Arabidopsis. Plant Physiol. 133, 1091–1101 10.1104/pp.103.026997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. Y., Lee P. F., Lee Y. I., Hsiao Y. Y., Chen Y. Y., Pan Z. J., et al. (2011). Duplicated C-class MADS-box genes reveal distinct roles in gynostemium development in cymbidium ensifolium (Orchidaceae). Plant Cell Physiol. 52, 563–577 10.1093/pcp/pcr015 [DOI] [PubMed] [Google Scholar]
- Xu Y., Teo L. L., Zhou J., Kumar P. P., Yu H. (2006). Floral organ identity genes in the orchid Dendrobium crumenatum. Plant J. 46, 54–68 10.1111/j.1365-313X.2006.02669.x [DOI] [PubMed] [Google Scholar]
- Yu H., Goh C. J. (2000). Identification and characterization of three orchid MADS-box genes of the AP1/AGL9 subfamily during floral transition. Plant Physiol. 123, 1325–1336 10.1104/pp.123.4.1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Hua S., Goh C. J. (2002). Spatial and temporal expression of the orchid floral homeotic gene DOMADS1 is mediated by its upstram regulatory regions. Plant Mol. Biol. 49, 225–237 10.1023/A:1014958118852 [DOI] [PubMed] [Google Scholar]
- Yu H., Yang S. H., Goh C. J. (2001). Agrobacterium-mediated transformation of a Dendrobium orchid with the class 1 knox gene DOH1. Plant Cell Rep. 20, 301–305 10.1007/s002990100334 [DOI] [Google Scholar]
- Zahn L. M., Kong H., Leebens-Mack J. H., Kim S., Soltis P. S., Landherr L. L., et al. (2005). The evolution of the SEPALLATA subfamily of MADS-box genes: a pre-angiosperm origin with multiple duplications throughout angiosperm history. Genetics 169, 2209–2223 10.1534/genetics.104.037770 [DOI] [PMC free article] [PubMed] [Google Scholar]