Abstract
BACKGROUND
Family history is a significant risk factor for prostate cancer, although the molecular basis for this association is poorly understood. Linkage studies have implicated chromosome 17q21-22 as a possible location of a prostate-cancer susceptibility gene.
METHODS
We screened more than 200 genes in the 17q21-22 region by sequencing germline DNA from 94 unrelated patients with prostate cancer from families selected for linkage to the candidate region. We tested family members, additional case subjects, and control subjects to characterize the frequency of the identified mutations.
RESULTS
Probands from four families were discovered to have a rare but recurrent mutation (G84E) in HOXB13 (rs138213197), a homeobox transcription factor gene that is important in prostate development. All 18 men with prostate cancer and available DNA in these four families carried the mutation. The carrier rate of the G84E mutation was increased by a factor of approximately 20 in 5083 unrelated subjects of European descent who had prostate cancer, with the mutation found in 72 subjects (1.4%), as compared with 1 in 1401 control subjects (0.1%) (P = 8.5×10−7). The mutation was significantly more common in men with early-onset, familial prostate cancer (3.1%) than in those with late-onset, nonfamilial prostate cancer (0.6%) (P = 2.0×10−6).
CONCLUSIONS
The novel HOXB13 G84E variant is associated with a significantly increased risk of hereditary prostate cancer. Although the variant accounts for a small fraction of all prostate cancers, this finding has implications for prostate-cancer risk assessment and may provide new mechanistic insights into this common cancer. (Funded by the National Institutes of Health and others.)
Prostate cancer is the most common noncutaneous cancer diagnosed in men in the United States, with more than 240,000 new cases expected in 2011.1 Despite the demonstration of a strong familial component, identification of the genetic basis for hereditary prostate cancer has been challenging. Linkage studies of families with hereditary prostate cancer have provided inconsistent results.2 In contrast, genome-wide association studies have led to the identification of more than 30 single-nucleotide polymorphisms (SNPs) that are consistently associated with prostate cancer.3 However, the magnitude of risk elevation attributed to each individual SNP is low, with an increased elevation in risk by a factor of less than 1.3, and these SNPs in aggregate account for only an estimated one quarter of familial risk.4
One of the most intensely investigated regions of the genome for prostate cancer susceptibility loci is chromosome 17q21-22. Prostate-cancer linkage to this region was initially reported by the University of Michigan Prostate Cancer Genetics Project on the basis of analysis of 175 pedigrees of families with hereditary prostate cancer.5 Several collaborative studies that included families in the Prostate Cancer Genetics Project, including a large study by the International Consortium for Prostate Cancer Genetics, also showed evidence of linkage to 17q21-22.6,7 We performed a fine-mapping study of this region using 453 pedigrees from the Prostate Cancer Genetics Project and Johns Hopkins University. A subset of 147 families with 4 or more confirmed affected men and an average age at diagnosis of prostate cancer of 65 years or less provided strong evidence for linkage (logarithm of odds [LOD] near marker D17S1820, 5.49) and a narrow candidate interval for a putative susceptibility gene (1-LOD support interval, approximately 10 cM).8
Next-generation sequencing technologies have provided new opportunities to interrogate large genomic intervals that are implicated in human disease in a rapid and comprehensive manner. Given the consistent evidence of prostate-cancer linkage to 17q21-22 markers in our multiplex families with hereditary prostate cancer, we designed a targeted sequencing strategy to analyze 2009 exons of 202 genes contained in the most likely genomic interval defined by our fine-mapping studies.
METHODS
STUDY SUBJECTS
For data from the Prostate Cancer Genetics Project, subjects were restricted to men with prostate cancer who had at least one living first- or second-degree relative who also had prostate cancer or those in whom prostate cancer had been diagnosed at an age of 55 years or less, regardless of family history. We confirmed the diagnosis of prostate cancer by reviewing medical records whenever possible. Ancestry was self-reported. All subjects provided written informed consent to participate in the study. The protocol and consent documents were approved by the institutional review board at the University of Michigan Medical School.
For data from Johns Hopkins University, families with hereditary prostate cancer each had at least three first-degree relatives with prostate cancer. We verified the diagnosis of prostate cancer by reviewing medical records. Included in the study were men who had undergone radical prostatectomy for the treatment of clinically localized prostate cancer at Johns Hopkins Hospital. Advanced prostate cancer was defined as biochemical recurrence of prostate cancer or metastatic or castration-resistant disease that was identified at the Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins Hospital. The average age at diagnosis in this latter group was 60.9 years.
Control subjects for this study included men who had undergone screening for prostate cancer. Such screening included measurement of serum prostate-specific antigen levels and digital rectal examination at Johns Hopkins Hospital, Johns Hopkins Bayview Medical Center, Johns Hopkins University Applied Physics Laboratory, and several other locations in the mid-Atlantic area. Inclusion criteria for control subjects included a knowledge of ancestry and no diagnosis of prostate cancer. For all studies at Johns Hopkins University, research proposals were reviewed and approved by the institutional review board.
TARGETED SEQUENCING OF GENES IN CANDIDATE REGION
We selected the youngest patient with prostate cancer who had available DNA from 94 families (54 families from the Prostate Cancer Genetics Project and 40 from Johns Hopkins University) on the basis of evidence of 17q21-22 linkage. Seven of the families were of African descent, 2 were of Asian descent, and the remaining 85 were of European descent. We identified 202 genes in our genetic region of interest (approximately 15.5 Mb).8 Details of sequence analysis are provided in the Supplementary Appendix, which is available with the full text of this article at NEJM.org.
GENOTYPING OF HOXB13 VARIANTS
We genotyped variants of HOXB13, a gene encoding transcription factor homeobox B13, which is within our candidate interval, using the MassARRAY system (Sequenom) and TaqMan assays (Applied Biosystems/Life Technologies). We confirmed all variants found on either of these platforms using Sanger sequencing.
STATISTICAL ANALYSIS
We performed association analyses for the HOXB13 G84E variant using Fisher’s exact tests and linear regression models implemented in the statistical program R (http://cran.r-project.org). We included genotype data for 5083 unrelated men in whom prostate cancer had been diagnosed and for 1401 unrelated men who were presumed to be free of prostate cancer. These subjects were not part of the discovery sequencing study and were of self-reported European descent. Additional case–control association analyses for G84E included the use of publicly available data for 1233 subjects of European descent from the Exome Sequencing Project,9 funded by the National Heart, Lung, and Blood Institute, and 28 unrelated genotyped samples from the Centre d’Etude du Polymorphisme Humain from Utah (CEU) HapMap.10 We tested the association between HOXB13 G84E and two quantitative clinical variables: the age at diagnosis and Gleason grade.
RESULTS
TARGETED SEQUENCING OF 202 CANDIDATE GENES
We reviewed all sequence data for the presence of nonsense or missense mutations in 202 genes in our genetic region of interest.8 Probands from four families were observed to have the same nonsynonymous mutation in HOXB13, a change of adenosine for guanine (transition, c.251G→A) in the second position of codon 84 (GGA→GAA), resulting in a nonconservative substitution of glutamic acid for glycine (G84E) (see Table 1 in the Supplementary Appendix for complete sequencing results). At the time of this analysis, this mutation, now identified as rs138213197, was not reported in dbSNP, the database of known DNA sequence variants of the National Center for Biotechnology Information (NCBI), nor in the May 2011 release of the NCBI 1000 Genomes sequencing project, which included 1094 subjects, including 381 of European descent.11
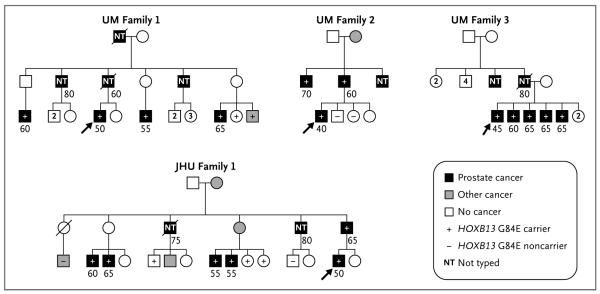
Given the importance and specificity of HOXB13 in prostate biology, we sought to further characterize this mutation. DNA samples from family members from each of these four pedigrees were sequenced to identify carriers of the HOXB13 G84E mutation. Cosegregation of the mutation with disease was observed in all 18 affected men with available DNA in the four families (Fig. 1).
Figure 1. Pedigrees of Four Subjects with the HOXB13 G84E Mutation on Initial Targeted Sequencing.
The proband who was selected for sequencing is indicated by the arrow in each pedigree. The remaining symbols are described in the key. Squares indicate male sex, and circles female sex. Ages of subjects, rounded to the nearest 5-year interval, are shown under the symbols. A slash through the symbol indicates that the subject is deceased. Two subjects in two families, Family 1 from the University of Michigan Prostate Cancer Genetics Project (UM) and Family 1 from Johns Hopkins University (JHU), who were inferred to be obligate carriers of the HOXB13 G84E mutation, died from prostate cancer. The unaffected G84E carrier in JHU Family 1 was 70 years of age at last contact.
HOXB13 G84E MUTATION
To further assess HOXB13 G84E as a prostate-cancer susceptibility allele, we studied several additional populations of European descent with a total of 5083 unrelated case subjects (see Table 2 in the Supplementary Appendix for clinical characteristics) and 1401 unrelated control subjects. The case subjects included a cohort of 1130 patients with early-onset or familial prostate cancer from the Prostate Cancer Genetics Project,5,12,13 161 patients with hereditary prostate cancer from Johns Hopkins University,14 a series of 3499 men with localized prostate cancer who were treated with radical prostatectomy at Johns Hopkins Hospital,15 and 293 men who were treated for advanced prostate cancer at Johns Hopkins Hospital; the control population of 1401 men were found to have no prostate cancer during screening.15 Among men in the latter screening group, the G84E mutation was found in only 1 man, resulting in a carrier-frequency estimate of 0.1%. We did not identify any homozygous carriers among either case or control subjects.
Overall, men with prostate cancer were significantly more likely to carry the HOXB13 G84E allele (carrier frequency, 1.4%) than were those without prostate cancer (carrier frequency, 0.1%) (P = 8.5×10−7; odds ratio, 20.1; 95% confidence interval [CI], 3.5 to 803.3) (Table 1). The carrier frequency varied as a function of age at diagnosis and family history, with the highest rates among men with both a positive family history and early diagnosis (≤55 years of age) (Table 2, and Table 3 in the Supplementary Appendix). The carrier frequency in this group (3.1%) was significantly higher than in men with early-on-set prostate cancer who did not have a family history of the disease (1.0%, P = 0.002) or in men with a family history in whom prostate cancer was diagnosed after the age of 55 years (1.2%, P = 0.004) (Table 2). The lowest carrier frequency was observed in men in whom prostate cancer was diagnosed after the age of 55 years and who did not have a family history, although this frequency was still higher than in control subjects (0.6%; odds ratio, 8.7; 95% CI, 1.2 to 381.3; P = 0.02) (Table 3 in the Supplementary Appendix). Carrier frequencies in men with early-onset prostate cancer or those who had a family history were similar in findings from both the Prostate Cancer Genetics Project and Johns Hopkins University (data not shown). Results were slightly more significant but odds ratios were attenuated for comparisons with data from an expanded control population, including subjects from the Exome Sequencing Project and HapMap (Table 1, and Table 3 in the Supplementary Appendix).
Table 1.
Summary of HOXB13 G84E Mutations in Prostate-Cancer Case Subjects and Control Subjects of European Descent.*
| Data Set |
G84E
Carriers |
G84E
Noncarriers |
Carrier
Frequency |
Comparison with 1401
Control Subjects † |
Comparison with Expanded
Pool of Control Subjects ‡ |
||
|---|---|---|---|---|---|---|---|
| Odds Ratio | P Value | Odds Ratio | P Value | ||||
| no. | % | ||||||
| UM-PCGP and Johns Hopkins University | |||||||
| All subjects§ | 72 | 5011 | 1.4 | 20.1 | 8.5×10−7 | 9.5 | 2.4×10−9 |
| Subjects from 85 sequenced families with hereditary prostate cancer |
4 | 81 | 4.7 | 68.4 | 4.8×10−5 | 32.6 | 5.4×10−5 |
| UM-PCGP | |||||||
| Subjects with early-onset or hereditary prostate cancer§ |
26 | 1104 | 2.3 | 33.0 | 1.0×10−8 | 15.6 | 1.3×10−10 |
| Johns Hopkins University | |||||||
| Subjects with hereditary prostate cancer§ and those undergoing prostatectomy or other oncologic therapy |
46 | 3907 | 1.2 | 16.5 | 1.6×10−5 | 7.8 | 6.0×10−7 |
| Control subjects | |||||||
| Johns Hopkins University | 1 | 1400 | 0.1 | NA | NA | NA | NA |
| Exome Sequencing Project plus CEU HapMap | 3 | 1258 | 0.2 | NA | NA | NA | NA |
The HOXB13 G84E mutation was detected by means of both direct sequencing and high-throughput genotyping approaches. CEU denotes Centre d’Etude du Polymorphisme Humain from Utah, NA not applicable, and UM-PCGP University of Michigan Prostate Cancer Genetics Project.
Case subjects were compared with 1401 control subjects at Johns Hopkins University.
Case subjects were compared with 1401 control subjects at Johns Hopkins University plus 1233 subjects in the Exome Sequencing Project and 28 unrelated genotyped subjects from the CEU HapMap.
This category includes unrelated subjects with prostate cancer, excluding the 85 families of European descent used for discovery. The youngest available subject with prostate cancer was selected from families that had more than one subject with the disease.
Table 2.
Frequency of HOXB13 G84E Carriers in Subjects of European Descent with Prostate Cancer, According to Family History and Age at Diagnosis.*
| Variable |
G84E
Carriers |
G84E
Noncarriers |
Carrier
Frequency |
Comparison Group † |
Odds Ratio (95% CI) |
P Value |
|---|---|---|---|---|---|---|
| no. | % | |||||
| Family history of prostate cancer | ||||||
| Positive | 45 | 2019 | 2.2 | Negative family history | 2.8 (1.6–5.1) | 1.2×10−4 |
| Negative | 19 | 2391 | 0.8 | NA | NA | NA |
| Age at prostate-cancer diagnosis | ||||||
| ≤55 yr | 46 | 2084 | 2.2 | Age at diagnosis, >55 yr | 2.7 (1.6–4.7) | 1.1×10−4 |
| >55 yr | 22 | 2681 | 0.8 | NA | NA | NA |
| Positive family history | ||||||
| Age at diagnosis, ≤55 yr | 33 | 1040 | 3.1 | Negative family history; age at diagnosis, >55 yr |
5.1 (2.4–12.2) | 2.0×10−6 |
| Age at diagnosis, >55 yr | 12 | 993 | 1.2 | Negative family history; age at diagnosis, >55 yr |
1.9 (0.75–5.2) | 0.18 |
| Negative family history | ||||||
| Age at diagnosis, ≤55 yr | 10 | 943 | 1.0 | Negative family history; age at diagnosis, >55 yr |
1.7 (0.62–4.8) | 0.25 |
| Age at diagnosis, >55 yr | 9 | 1447 | 0.6 | NA | NA | NA |
Included are 4474 subjects with data on family history and 4833 subjects with data on the age at diagnosis from the University of Michigan Prostate Cancer Genetics Project and Johns Hopkins University.
Additional comparisons are as follows: Among subjects with a positive family history, the odds ratio for the presence of the HOXB13 G84E mutation among those with an early-onset diagnosis (≤55 years), as compared with a later-onset diagnosis (>55 years), was 2.6 (95% CI, 1.3 to 5.6; P = 0.004). Among subjects with early-onset disease, the odds ratio among those with a positive family history, as compared with a negative family history, was 3.0 (95% CI, 1.4 to 6.8; P = 0.002). NA denotes not applicable.
G84E carriers were significantly younger than noncarriers (52.9 vs. 57.1 years, P = 7.4×10−7) (Table 4 in the Supplementary Appendix). On the basis of clinical data collected for patients who had undergone radical prostatectomy at the Prostate Cancer Genetics Project and Johns Hopkins University, we found no evidence supporting significant differences in Gleason grade between G84E carriers and noncarriers before or after accounting for the age at diagnosis (Table 4 in the Supplementary Appendix). The G84E mutation was found in 6 of 293 men (2.0%) who were being treated for metastatic disease. Finally, we did not identify any additional G84E carriers among 84 unrelated subjects of African descent with prostate cancer (i.e., unrelated to the 7 subjects of African descent who were included in the initial sequencing data set) (see the Supplementary Appendix).
ADDITIONAL NOVEL NONSYNONYMOUS HOXB13 MUTATIONS
In the initial targeted sequencing study of 94 families with hereditary prostate cancer, 1 proband from an African-American family was observed to have a novel HOXB13 missense mutation (transversion c.685C→G), resulting in the substitution of glycine for arginine at position 229 (R229G). The same mutation was detected in the patient’s 2 brothers with prostate cancer. To search for additional HOXB13 variants that were not observed in the original sequence analysis, we sequenced both exons of HOXB13 in additional men of European and African descent from the Prostate Cancer Genetics Project and Johns Hopkins University. A novel substitution of cysteine for glycine at codon 216 (transversion c.646G→T, p.G216C) was found in an African-Caribbean family. This mutation was present in both subjects with prostate cancer (2 half-brothers) for whom DNA was available. Neither the R229G nor the G216C mutation was observed in approximately 1100 African-American subjects in the Exome Sequencing Project.
HOXB13 was also sequenced in eight available prostate-cancer cell lines (LNCaP, PC3, DU145, CRW22Rv1, E006AA, VCaP, MDAPCa2b, and LAPC4).16 LNCaP and LAPC4, both androgen-sensitive human prostate adenocarcinoma cell lines, were found to have nonsynonymous mutations: substitution of proline for leucine at codon 144 (transition c.431T→C, L144P) in LNCaP and aspartic acid for tyrosine at codon 88 (transversion c.262T→G, p.Y88D) in LAPC4. Neither missense mutation was observed in sequencing of the 94 probands or the database of the Exome Sequencing Project, although a lack of available germline DNA for these cell lines precluded the determination of a definitive origin for these changes as somatic or germline.
The locations of the HOXB13 G84E mutation and the four additional rare HOXB13 mutations are shown in Figure 2. All the changes are in highly conserved functional domains of HOXB13 and are predicted to be damaging to protein function on the Sorting Intolerant from Tolerant (SIFT)17 or PolyPhen18 algorithms. The G84E and the Y88D mutations are located in the same non-homeobox domain that was previously shown to mediate the binding of HOX13 paralogues (including HOXB13) to the MEIS family of HOX cofactor proteins.19 The L144P change is in the second of two MEIS-binding domains. Both mutations that were found in subjects of African descent, R229G and G216C, reside in the N-terminal portion of the homeobox domain, and both changes affect highly conserved amino acid residues.
Figure 2. Structure of HOXB13.
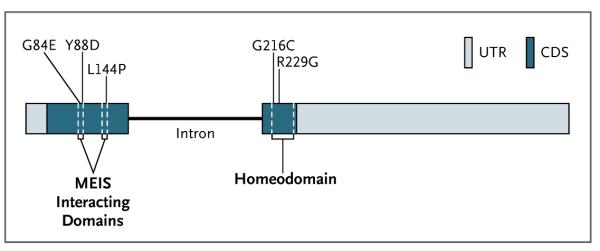
The HOXB13 gene is the most 5′ member of the HOXB gene cluster on chromosome 17q21-22. The locations of the five missense mutations are indicated in the two exons of HOXB13. The homeodomain region and MEIS interacting domains are indicated. CDS denotes coding sequences, and UTR untranslated regions.
DISCUSSION
Targeted sequencing of exons in 202 genes on chromosome 17q21-22 resulted in the identification of a recurrent mutation, G84E in HOXB13, in four probands of families with a history of prostate cancer. Analysis of additional case and control subjects revealed that this variant was associated with prostate-cancer risk and, in particular, early-onset and hereditary prostate cancer. The identification of several additional rare missense HOXB13 variants (Y88D, L144P, G216C, and R229G) further implicates HOXB13 in prostate carcinogenesis. These rare HOXB13 variants appear to be independent of chromosome 17q risk alleles that have previously been identified in genomewide association studies (see the Supplementary Appendix).20,21
The HOX genes are a subfamily of the homeobox superfamily of transcription factors characterized by a highly conserved DNA-binding domain, or homeodomain. In humans, there are four HOX clusters, with each spanning approximately 200 kb on chromosomes 7 (HOXA), 17 (HOXB), 12 (HOXC), and 2 (HOXD). The combination of coordinated HOX expression provides a so-called HOX code that is essential for the pattern formation of the animal body.22 The genes within each HOX cluster are expressed temporally during development; 3′ genes are expressed early in anterior and proximal regions, whereas 5′ genes are expressed late in posterior and distal regions.23 HOX genes in paralogue group 13 are members of the abdominal B subfamily of such genes, which have posterior domains of expression, including in the developing urogenital system in vertebrates. Whereas multiple HOX13 paralogues are expressed during embryonic development of the prostate, HOXB13 maintains a high expression level into adulthood in normal prostate and, to a lesser level, in distal colon.
In a study by Economides et al.,24 mice that had been generated from embryonic stem cells with targeted disruption in HOXB13 had overgrowth of structures arising from the tail bud, including the spinal cord and tail vertebrae, with decreased apoptosis proposed as a possible mechanism. Further characterization of these animals showed subtle but definitive, lobe-specific abnormalities of the prostate gland but without evidence of preneoplastic lesions.25
The mechanisms by which the HOXB13 G84E mutation might act to promote prostate carcinogenesis are unknown. One clue may be provided by the location of this change in a conserved domain of the HOXB13 protein that has been shown to mediate binding to members of the MEIS protein family. This binding is thought to modulate the interaction of HOX proteins with specific DNA or other proteins and thus finetune HOX function. MEIS expression has been implicated in collaboration with HOX genes in the development of leukemia.26 Understanding how G84E affects the interaction between HOXB13 and the MEIS proteins and what effect this has on HOXB13–MEIS function is an obvious area for future research.
Several studies have examined the role of HOXB13 in normal and cancerous prostate biology, although substantially different conclusions have been reached, with HOXB13 being implicated as both a tumor suppressor and an oncogene in prostate and other cancers. For example, the growth of the prostate-cancer cell line LNCaP has been shown to be suppressed by both experimental overexpression of HOXB13 by transfection and by reduction of endogenous HOXB13 expression by RNA interference.27 It is clear that HOXB13 physically interacts with the androgen receptor, one of the most important growth and differentiation regulators in normal and cancerous prostate biology.27,28 Direct experimentation with the G84E allele will be necessary to determine its effect on cell development and differentiation in prostate cancer.
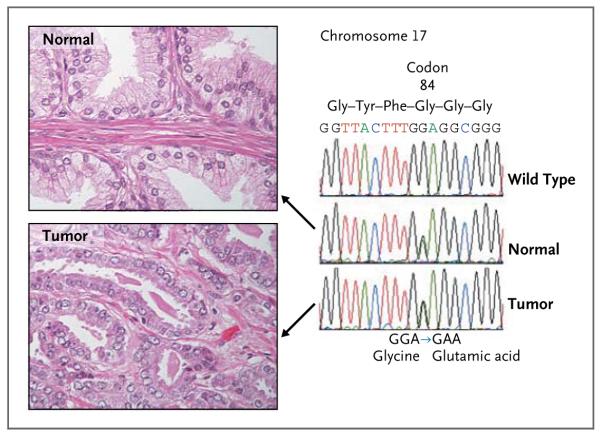
The recurrent nature of the G84E change and a reported lack of any truncating mutations in HOXB13 in patients with prostate cancer suggest a carcinogenic mechanism that is more consistent with a gain of function (oncogene) than with a loss of function (tumor-suppressor gene). Both deletions29 and amplifications30 of 17q21 have been described in prostate-cancer specimens. However, such analyses need to be performed in tumors from known HOXB13 G84E carriers. Multiple studies have shown the prostate specificity of high-level HOXB13 expression, and this expression appears to be maintained in the formation and progression of prostate cancer.31 Our preliminary analyses indicate that tumors in G84E carriers continue to express HOXB13 and maintain the mutant allele (Fig. 3, and Fig. 1 in the Supplementary Appendix).
Figure 3. DNA Sequence Chromatograms Obtained from Normal Prostate and Prostate-Cancer Tissue from a Heterozygous Carrier of the HOXB13 G84E Variant, with Associated Histologic Findings.
Wild-type and mutant DNA are present in both normal prostate tissue and prostate-cancer tissue from HOXB13 G84E carriers. For this experiment, DNA was extracted from sections of paraffin-embedded blocks of tissue obtained during a radical prostatectomy performed in a patient who was heterozygous for the HOXB13 G84E variant. The blocks were selected and trimmed to contain either normal or tumor tissue, as shown on hematoxylin and eosin staining (at left), and were subjected to Sanger sequencing. The chromatograms (at right) show the presence of both wild-type (GGA) and mutant (GAA) alleles at codon 84 in normal prostate tissue (middle) and the maintenance of both alleles in the matched sample of prostate tumor tissue (bottom). The top chromatogram is a homozygous wild-type sequence from a subject without the G84E mutation. The genome position that is shown (44,160,704) is based on the National Center for Biotechnology Information database, build 36 (hg18).
In summary, we have used linkage analysis in combination with targeted massively parallel sequencing to identify a recurrent mutation in HOXB13 that is associated with early-onset and hereditary prostate cancer. From a clinical perspective, testing for germline mutations in BRCA1/2 is recommended in some families, since mutations in these breast-cancer–susceptibility genes are associated with elevations in the risk of prostate cancer, particularly for BRCA2.32,33 However, neither of these genes has been shown to contribute to hereditary prostate cancer.34,35 HOXB13 G84E is associated with a significantly increased risk of hereditary prostate cancer. This work suggests that future DNA sequencing studies using next-generation technology and study populations enriched for genetic influence (as evidenced by an early age at onset and positive family history) may identify additional rare variants that will contribute to familial clustering of prostate cancer. Although HOXB13 mutations will be identified in a minority of men with prostate cancer, rare genetic lesions can identify pathways that are found to be abnormal in more common, sporadic cases.
Supplementary Material
Acknowledgments
Supported by grants (R01-CA79596, R01-CA079596-10-S1, R01-CA136621, P50-CA69568, U01-CA89600, and U01-CA86323) from the National Institutes of Health; by William Gerrard, Mario Duhon, John and Jennifer Chalsty, and P. Kevin Jaffe; and by the Patrick C. Walsh Cancer Research Fund (to Dr. Isaacs).
We thank the men with prostate cancer and their family members for their participation in this research; Drs. M. Carducci, S. Denmeade, M. Eisenberger, R. Pili, J. Nelson, D. Wood, M. Hussain, D. Smith, and K. Pienta for their willingness to refer many patients to our studies; Dr. B. Trock for contributing of a portion of the control DNA samples; H. Fedor and Dr. A. DeMarzo for assistance with tissue resources; Dr. E. Fearon for critical review of the manuscript; the Network and Computing Systems department of TGen for facilitating the use of supercomputing resources; and the Exome Sequencing Project for providing exome variant calls for comparison.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
REFERENCES
- 1.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–36. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Langeberg WJ, Isaacs WB, Stanford JL. Genetic etiology of hereditary prostate cancer. Front Biosci. 2007;12:4101–10. doi: 10.2741/2374. [DOI] [PubMed] [Google Scholar]
- 3.Kim ST, Cheng Y, Hsu FC, et al. Prostate cancer risk-associated variants reported from genome-wide association studies: meta-analysis and their contribution to genetic variation. Prostate. 2010;70:1729–38. doi: 10.1002/pros.21208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kote-Jarai Z, Olama AA, Giles GG, et al. Seven prostate cancer susceptibility loci identified by a multi-stage genome-wide association study. Nat Genet. 2011;43:785–91. doi: 10.1038/ng.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lange EM, Gillanders EM, Davis CC, et al. Genome-wide scan for prostate cancer susceptibility genes using families from the University of Michigan Prostate Cancer Genetics Project finds evidence for linkage on chromosome 17 near BRCA1. Prostate. 2003;57:326–34. doi: 10.1002/pros.10307. [DOI] [PubMed] [Google Scholar]
- 6.Gillanders EM, Xu J, Chang BL, et al. Combined genome-wide scan for prostate cancer susceptibility genes. J Natl Cancer Inst. 2004;96:1240–7. doi: 10.1093/jnci/djh228. [DOI] [PubMed] [Google Scholar]
- 7.Xu J, Dimitrov L, Chang BL, et al. A combined genomewide linkage scan of 1,233 families for prostate cancer-susceptibility genes conducted by the International Consortium for Prostate Cancer Genetics. Am J Hum Genet. 2005;77:219–29. doi: 10.1086/432377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lange EM, Robbins CM, Gillanders EM, et al. Fine-mapping the putative chromosome 17q21-22 prostate cancer susceptibility gene to a 10 cM region based on linkage analysis. Hum Genet. 2007;121:49–55. doi: 10.1007/s00439-006-0274-2. [DOI] [PubMed] [Google Scholar]
- 9.Exome variant server. NHLBI Exome Sequencing Project; Seattle: http://snp.gs.washington.edu/EVS. [Google Scholar]
- 10.The International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.1000 Genomes: a deep catalog of human variation. http://www.1000genomes.org/home.
- 12.Lange EM, Salinas CA, Zuhlke KA, et al. Early onset prostate cancer has a significant genetic component. Prostate. 2011 May 2; doi: 10.1002/pros.21414. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lange EM, Beebe-Dimmer JL, Ray AM, et al. Genome-wide linkage scan for prostate cancer susceptibility from the University of Michigan Prostate Cancer Genetics Project: suggestive evidence for linkage at 16q23. Prostate. 2009;69:385–91. doi: 10.1002/pros.20891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu J, Zheng SL, Komiya A, et al. Germline mutations and sequence variants of the macrophage scavenger receptor 1 gene are associated with prostate cancer risk. Nat Genet. 2002;32:321–5. doi: 10.1038/ng994. [DOI] [PubMed] [Google Scholar]
- 15.Zheng SL, Sun J, Cheng Y, et al. Association between two unlinked loci at 8q24 and prostate cancer risk among European Americans. J Natl Cancer Inst. 2007;99:1525–33. doi: 10.1093/jnci/djm169. [DOI] [PubMed] [Google Scholar]
- 16.Liu W, Xie CC, Zhu Y, et al. Homozygous deletions and recurrent amplifications implicate new genes involved in prostate cancer. Neoplasia. 2008;10:897–907. doi: 10.1593/neo.08428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu Rev Genomics Hum Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- 18.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams TM, Williams ME, Innis JW. Range of HOX/TALE superclass associations and protein domain requirements for HOXA13:MEIS interaction. Dev Biol. 2005;277:457–71. doi: 10.1016/j.ydbio.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 20.Haiman CA, Chen GK, Blot WJ, et al. Genome-wide association study of prostate cancer in men of African ancestry identifies a susceptibility locus at 17q21. Nat Genet. 2011;43:570–3. doi: 10.1038/ng.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gudmundsson J, Sulem P, Steinthorsdottir V, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39:977–83. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 22.Graham A. Developmental patterning: the Hox code out on a limb. Curr Biol. 1994;4:1135–7. doi: 10.1016/s0960-9822(00)00256-6. [DOI] [PubMed] [Google Scholar]
- 23.Goodman FR, Scambler PJ. Human HOX gene mutations. Clin Genet. 2001;59:1–11. doi: 10.1034/j.1399-0004.2001.590101.x. [DOI] [PubMed] [Google Scholar]
- 24.Economides KD, Zeltser L, Capecchi MR. Hoxb13 mutations cause overgrowth of caudal spinal cord and tail vertebrae. Dev Biol. 2003;256:317–30. doi: 10.1016/s0012-1606(02)00137-9. [DOI] [PubMed] [Google Scholar]
- 25.Economides KD, Capecchi MR. Hoxb13 is required for normal differentiation and secretory function of the ventral prostate. Development. 2003;130:2061–9. doi: 10.1242/dev.00432. [DOI] [PubMed] [Google Scholar]
- 26.Thorsteinsdottir U, Kroon E, Jerome L, Blasi F, Sauvageau G. Defining roles for HOX and MEIS1 genes in induction of acute myeloid leukemia. Mol Cell Biol. 2001;21:224–34. doi: 10.1128/MCB.21.1.224-234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung C, Kim RS, Zhang HJ, Lee SJ, Jeng MH. HOXB13 induces growth suppression of prostate cancer cells as a repressor of hormone-activated androgen receptor signaling. Cancer Res. 2004;64:9185–92. doi: 10.1158/0008-5472.CAN-04-1330. [DOI] [PubMed] [Google Scholar]
- 28.Norris JD, Chang CY, Wittmann BM, et al. The homeodomain protein HOXB13 regulates the cellular response to androgens. Mol Cell. 2009;36:405–16. doi: 10.1016/j.molcel.2009.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim JH, Dhanasekaran SM, Mehra R, et al. Integrative analysis of genomic aberrations associated with prostate cancer progression. Cancer Res. 2007;67:8229–39. doi: 10.1158/0008-5472.CAN-07-1297. [DOI] [PubMed] [Google Scholar]
- 30.Fukasawa S, Kino M, Kobayashi M, et al. Genetic changes in pT2 and pT3 prostate cancer detected by comparative genomic hybridization. Prostate Cancer Prostatic Dis. 2008;11:303–10. doi: 10.1038/sj.pcan.4501017. [DOI] [PubMed] [Google Scholar]
- 31.Edwards S, Campbell C, Flohr P, et al. Expression analysis onto microarrays of randomly selected cDNA clones high-lights HOXB13 as a marker of human prostate cancer. Br J Cancer. 2005;92:376–81. doi: 10.1038/sj.bjc.6602261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson D, Easton DF. Cancer incidence in BRCA1 mutation carriers. J Natl Cancer Inst. 2002;94:1358–65. doi: 10.1093/jnci/94.18.1358. [DOI] [PubMed] [Google Scholar]
- 33.Ostrander EA, Udler MS. The role of the BRCA2 gene in susceptibility to prostate cancer revisited. Cancer Epidemiol Biomarkers Prev. 2008;17:1843–8. doi: 10.1158/1055-9965.EPI-08-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schehl-Sinclair C, Berry R, Schaid D, Thibodeau SN, Couch FJ. BRCA1 and BRCA2 have a limited role in familial prostate cancer. Cancer Res. 2000;60:1371–5. [PubMed] [Google Scholar]
- 35.Zuhlke KA, Madeoy JJ, Beebe-Dimmer J, et al. Truncating BRCA1 mutations are uncommon in a cohort of hereditary prostate cancer families with evidence of linkage to 17q markers. Clin Cancer Res. 2004;10:5975–80. doi: 10.1158/1078-0432.CCR-04-0554. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.